DOI:
10.1039/D2CC06556H
(Feature Article)
Chem. Commun., 2023,
59, 2056-2071
Excited-state intramolecular proton transfer (ESIPT)-based fluorescent probes for biomarker detection: design, mechanism, and application
Received
1st December 2022
, Accepted 16th January 2023
First published on 18th January 2023
Abstract
Biomarkers are essential in biology, physiology, and pharmacology; thus, their detection is of extensive importance. Fluorescent probes provide effective tools for detecting biomarkers exactly. Excited state intramolecular proton transfer (ESIPT), one of the significant photophysical processes that possesses specific photoisomerization between Keto and Enol forms, can effectively avoid annoying interference from the background with a large Stokes shift. Hence, ESIPT is an excellent choice for biomarker monitoring. Based on the ESIPT process, abundant probes were designed and synthesized using three major design methods. In this review, we conclude probes for 14 kinds of biomarkers based on ESIPT explored in the past five years, summarize these general design methods, and highlight their application for biomarker detection in vitro or in vivo.

Hao Gu
| Hao Gu graduated from Shenyang University of Chemical Technology (China) with BS degree in 2017. He is pursuing his doctorate degree under the guidance of Prof. Chen's group, College of Chemical Engineering at Nanjing Tech University (China). He has been mainly involved in the research of fluorescent dyes, fluorescent chemosensors and photochromic dyes. |

Wenjing Wang
| Wenjing Wang graduated from Taizhou University (China) with BS degree in 2020 and is a master's student under the guidance of Prof. Chen, College of Chemical Engineering at Nanjing Tech University (China). She has been mainly involved in the research of fluorescent chemosensors and photochromic dyes. |

Wenyan Wu
| Wenyan Wu graduated from Huainan Normal University (China) with BS degree in 2020 and is a master‘s student under the guidance of Prof. Chen, College of Chemical Engineering at Nanjing Tech University (China). She has been mainly involved in the research of polymer self-assembly and chiral induction for chiral and achiral molecules. |

Maolin Wang
| Maolin Wang graduated from Xi’an University of Science and Technology (China) with BS degree in 2020 and is a master's student under the guidance of Prof. Chen, College of Chemical Engineering at Nanjing Tech University (China). She has been mainly involved in the research of fluorescent dyes, fluorescent chemosensors and photo-controlled release of active substance. |

Yongrong Liu
| Yongrong Liu graduated from Yancheng Normal University (China) with BS degree in 2020 and is a master's student under the guidance of Prof. Chen, College of Chemical Engineering at Nanjing Tech University (China). She has been mainly involved in the research of fluorescent chemosensors and chemiluminescent molecular probes. |

Yanjun Jiao
| Yanjun Jiao graduated from Henan University of Science and Technology (China) with BS degree in 2020 and is a master's student under the guidance of Prof. Chen, College of Chemical Engineering at Nanjing Tech University (China). He has been mainly involved in the research of fluorescent dyes, fluorescent chemosensors and defaming agent. |

Fan Wang
| Fan Wang graduated from Anhui Science and Technology University (China) with his BS degree in 2019 and is a master's student under the guidance of Prof. Chen's group. He has been mainly involved in the research of developing chemiluminescent sensors. |

Fang Wang
| Fang Wang received her PhD from Ewha Womans University under the direction of Prof. Yoon (Korea) in 2012. She is currently a professor at Nanjing Tech University (China) and has joined Prof. Chen's group. Her research focuses on the design and synthesis of fluorescent chemosensors. |

Xiaoqiang Chen
| Xiaoqiang Chen received his PhD in Applied Chemistry from Dalian University of Technology (China) in 2007. He joined Prof. Yoon's group at Ewha Womans University (Korea) as a postdoctoral fellow in 2008. In March 2010, he moved to Nanjing Tech University, where he is currently a professor of College of Chemical Engineering. He visited Ewha Womans University as a visiting professor in 2015. His current research interests mainly focus on fluorescent dyes, fluorescent probes, and light-controlled release of active substance. |
1. Introduction
Biomarkers, which can objectively measure and evaluate physiological or pathological processes, have aroused widespread attention and research interest.1–7 As shown in Table 1, we list some important biomarkers, their participating physiological processes, and related diseases. Evidently, these biomarkers are closely related to many diseases; therefore, there is an urgent need to monitor the indicators of these biomarkers to accurately reflect changes or even real-time changes in biomarkers.8–12 Fluorescent probes are routinely utilized to monitor biomarkers because of their high sensitivity,13–18 fast response19–23 and structural adjustability.24–28 Four mechanisms of fluorescent probes have been identified: PET (Photo-induced Electron Transfer),29,30 FRET (Fluorescence Resonance Energy Transfer),31–33 ICT (Intramolecular Charge Transfer),34–36 and Excited-state Intramolecular Proton Transfer (ESIPT).37–39 In this review, ESIPT is emphasized, and the relative combination of mechanisms, such as FRET-ESITP, PET-ESIPT, and ICT-ESIPT, are also involved.
Table 1 Important biomarkers, their participating physiological processes, and related diseases
| Name of biomarkers |
Abbreviation |
Physiological processes |
Related diseases |
| Acid and base |
pH |
Cell proliferation and apoptosis, ion transport, enzyme activity, and protein degradation55,56 |
Cancer,57–59 sepsis,60 hyperkalemia,61 and chronic kidney disease62 |
| Carbon monoxide |
CO |
Cytoprotection, anti-inflammation, improvement of organ transplantation survival, and inhibition of bacterial growth63,64 |
Neurodegenerations, hypertension, heart failure, and inflammation63,64 |
| Sulfites |
SO32−/HSO3− |
Regulation of vascular smooth muscle tone and lowering blood pressure65 |
Lung cancer, cardiovascular diseases, and many neuro-logical disorders66–69 |
| Cysteine |
Cys |
Redox homeostasis, metabolism, protein synthesis, and signal transduction70–72 |
Cardiovascular complications, neurotoxicity, Parkinson's disease, Alzheimer's disease, edema, lethargy, and liver damage70–73 |
| Glutathione |
GSH |
Redox homeostasis, growth and metabolism, oxidative stress and immune responses, and apoptosis74–77 |
Parkinson's disease, liver injury, and cancer78–81 |
| Peroxynitrite |
ONOO− |
Host defense, cell proliferation, and cell growth82–84 |
Inflammation, cancer, atherosclerosis and neurodegenerative diseases85–88 |
| Hydrogen peroxide |
H2O2 |
Immune responses, cellular signal transduction, regulation of oxidative stress, and stimulation of antioxidant defenses89,90 |
Cancer,91 diabetes,92 cardiovascular diseases,93,94 and neurodegenerative disorders95 |
| Hypochlorous acid |
ClO− |
Neuronal aging, pathogen response, and anti-inflammatory regulation96–101 |
Cardiovascular and kidney diseases, inflammatory disease, neuron degeneration, and cancer57,58,102 |
| Formaldehyde |
HCHO |
The carbon cycle metabolism process103,104 |
Alzheimer's disease, cognitive impairment, neurodegenerative diseases, and atherosclerosis105–108 |
| β-Glucuronidase |
GUS |
Tumor invasion, and metastasis109 |
/ |
| β-Galactosidase |
β-Gal |
Gene expression and cellular senescence110,111 |
Primary ovarian cancer110,111 |
| Phosphatase |
ALP |
Regulating phosphate metabolism112,113 |
Diabetes, bone disease, liver dysfunction, and prostate cancer114,115 |
| Esterase |
— |
Gene expression, ester metabolism, material transport, and detoxification116–118 |
Wolman disease, obesity, atherosclerosis, cancer, hyperlipidemia, and hepatic steatosis119–124 |
| Acetylcholinesterase |
AChE |
Metabolic processes125 |
Salivation, convulsions, tremors, and even death126,127 |
| Leucine aminopeptidase |
LAP |
Leucine regulation128,129 |
Hepatic dysfunction or liver cancer130,131 |
| Serum albumin |
HSA |
Maintaining colloid osmotic pressure, regulating free radicals, and transporting nutrients, fatty acids, and metabolites132 |
Cardiovascular disease, kidney disease, hypertension, liver cirrhosis, and cancers133 |
ESIPT, as one of the most important photo-physical processes, has attracted substantial attention since its discovery by Weller in the 1950s.40 The extraordinary structural feature of ESIPT emitters is that they always have an H-bond between the hydrogen bond donors (–OH, –NH2, and –SH)41–44 and hydrogen bond acceptors (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N,
N, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S, and –C
S, and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).45–47 Upon irradiation, ESIPT emitters absorb the energy to achieve E* (excited state of Enol form). Then, ESIPT, a photoisomerization process, appears to form K* (excited state of Keto form) in less than 1 ps.48,49 The decay of K* emits a much longer fluorescence than E* owing to the inner ICT effect arising from the electron-withdrawing property of ketone. Finally, the ground state of the Enol form (E) changes to its original state (ground state of the Keto form, K) called GSIPT (Ground-state Intramolecular Proton Transfer) (Fig. 1).49,50 Benefitting from the transfer from the Enol form to the Keto form, the ESIPT emitters with two emissions could assist in obtaining ratiometric sensing, which could reduce the interference from the background.48,50,51 Simultaneously, compared with traditional probes, such as fluorescein and rhodamine, annoying self-reabsorption and inner-filter effects are prevented in ESIPT emitters.52–54 Consequently, ESIPT emitters are extensively used in biomarker detection.
O).45–47 Upon irradiation, ESIPT emitters absorb the energy to achieve E* (excited state of Enol form). Then, ESIPT, a photoisomerization process, appears to form K* (excited state of Keto form) in less than 1 ps.48,49 The decay of K* emits a much longer fluorescence than E* owing to the inner ICT effect arising from the electron-withdrawing property of ketone. Finally, the ground state of the Enol form (E) changes to its original state (ground state of the Keto form, K) called GSIPT (Ground-state Intramolecular Proton Transfer) (Fig. 1).49,50 Benefitting from the transfer from the Enol form to the Keto form, the ESIPT emitters with two emissions could assist in obtaining ratiometric sensing, which could reduce the interference from the background.48,50,51 Simultaneously, compared with traditional probes, such as fluorescein and rhodamine, annoying self-reabsorption and inner-filter effects are prevented in ESIPT emitters.52–54 Consequently, ESIPT emitters are extensively used in biomarker detection.
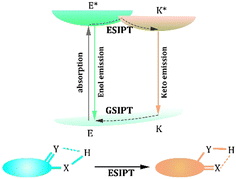 |
| | Fig. 1 Process of excited-state intramolecular proton transfer (ESIPT). | |
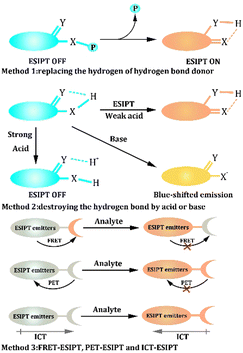
Based on the feature of the ESIPT process, the general strategy for designing ESIPT-based probes is to influence the ESIPT process to achieve the alteration of fluorescence. The first and most common method involves replacing the hydrogen of the hydrogen bond donor to restrict the ESIPT process. Thus, the probes emit a short wavelength from the Enol form. However, the hydrogen bond is reconstructed by adding a specific biomarker that could activate the ESIPT process and emit long-wavelength fluorescence.134–136 Another method of influencing the ESIPT process is destroying the hydrogen bond.137–139 Strong acids are much more acidic than hydroxyl groups and can combine with hydrogen bond acceptors, preventing the ESIPT process. Considering the acidity of the hydroxyl group or amine group, it could react with the base, and the emission of the ESIPT-based probe would be blue shifted. Moreover, FRET-ESIPT, PET-ESIPT, and ICT-ESIPT processes are effective procedures to achieve the modification of fluorescence.53,140 These combinatorial processes are instrumental in obtaining diversified fluorescence signals, so they are promising for designing multi-functional probes.
In this review, we highlight the design features of ESIPT emitters that have shown potential for detecting biomarkers. Moreover, the mechanism of the response to the biomarker is described. We focus on a method to optimize the optical properties and characteristics involved in biomarker detection. For example, some pesky problems, such as short wavelengths, inhibit the use of ESIPT-based fluorescent probes.141–143 Thus, the Near-Infrared Red (NIR) emission is useful in biomarker detection. Finally, this review outlines the application of these probes in biomarker detection, disease treatment, clinical diagnosis, and high-accuracy imaging fields.
2. Inorganic biomarkers and detection methods
2.1 Acid and base
Acids and bases play essential roles in many physiological processes, such as cell proliferation and apoptosis, ion transport, enzyme activity, and protein degradation (Table 1). As shown in Fig. 2, ESIPT relies on photoisomerization, which can be influenced effectively by acids and bases. Feng et al. synthesized a ratiometric fluorescent sensor, Probe 1, for pH sensing based on the amino-type ESIPT process.144 It is a promising sensor that responds rapidly and reversibly to pH changes after the introduction of a strong electron-withdrawing tosyl group. Probe 1 had a large Stokes shift in an acidic medium exhibiting yellow emission. In contrast, blue-shifted emission was observed in an alkaline medium owing to the deprotonation of the sulfonamide fraction, which suppressed the ESIPT process. A series of dual-factor-activated ESIPT probes are displayed in Fig. 3b, and we used Probe 2 as a typical example.145Probe 2 displayed three distinct fluorescent signals, indicating that it could sort acidic, neutrophils, and basic granulocytes in the presence of naphthol-AS-D-chloroacetate esterase. Tang et al. reported a pH jump probe with a midpoint of 7.5, which is closely related to physiological pH.146 The alterations in Probe 3 originated from the changes in Keto and Enol forms. Moreover, some applications, including cell imaging and pH fluctuation monitoring, are mentioned in these studies. For example, as the pH increased from 6.86 to 8.07, the fluorescence of Probe 3 increased in channel I (450–520 nm), accompanied by a decrease in channel II (520–600 nm), which demonstrated that the pH fluctuations could be monitored excellently by Probe 3 as a candidate molecule in live cells.
 |
| | Fig. 2 The design strategy of ESIPT-based probes. | |
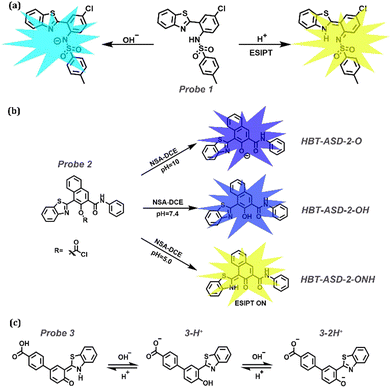 |
| | Fig. 3 (a) Proposed mechanisms for Probe 1 changes in acidic or basic media. (b) The mechanism of Probe 2 with NSA-DCE and different pH values. (c) The mechanism of Probe 3 to sense pH. | |
2.2 Carbon monoxide (CO)
CO is considered a signal molecule that regulates various physiological processes, including cytoprotection, anti-inflammation, organ transplantation survival, and bacterial growth inhibition. Furthermore, many diseases, including neurodegeneration, hypertension, heart failure, and inflammation, are associated with abnormalities in CO function (Table 1). The Tsuji-Trost reaction is a nucleophilic substitution reaction of allylation between allyl ether and CO, making it a promising choice for CO detection (Fig. 4a).147,148 Furthermore, Probes 4 and 5 are fluorophores comprising ESIPT and ICT along with the Aggregation-Induced emission (AIE) phenomenon.149Probe 4 encompasses three functional components (Fig. 4a): (a) AIE that originated from the rotators belonging to tetraarylimidazole; (b) two methoxyl groups introduced as electron donors and steric hindrance units; and (c) 2-(2′-hydroxyphenyl)-benzothiazole (HBT), a traditional fluorophore providing ESIPT property. In this review, AIE is not discussed because this phenomenon is widely observed in ESIPT materials.150–152 Liu et al. replaced the hydroxyl groups with allyl formate and allyl ether to construct two ratiometric probes, Probes 4 and 5, respectively (Fig. 4b). In the presence of CO, Pd2+ is reduced to Pd0, which further catalyzed the cleavage of the acceptor fragment to release free hydroxyl groups. Both probes could be loaded onto a test strip to visualize CO in the air. It is noteworthy that they also exhibit great potential for observing the fluctuation of exogenous and endogenous CO induced by heme, lipopolysaccharides (LPS) and hypoxia.
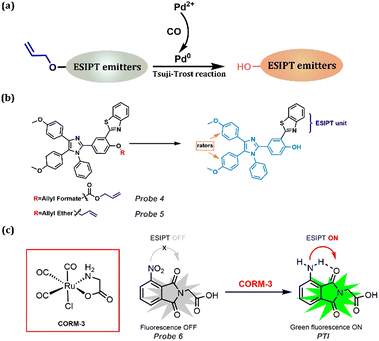 |
| | Fig. 4 (a) General procedure for CO detection using the Tsuji-Trost reaction. (b) Design strategy for Probes 4 and 5 with multiple phenomena. (c) CO detection method using its reducing property. | |
Another method for CO detection is based on its reducing property. As shown in Fig. 4c, by reducing NO2 to NH2, the ESIPT is released, and this process could be seen as a kind of H-bond donor replacement method.153Probe 6 was non-fluorescent; CORM-3 impelled the reduction of the nitro group, generating a highly fluorescent phthalimide (PTI), which emitted bright green fluorescence due to the ESIPT process. More importantly, this probe is promising for CO detection in living cells, zebrafish, and other animals.
2.3 Sulfites (SO32−/HSO3−)
As a key role in regulating vascular smooth muscle tone and lowering blood pressure, Sulfur dioxide (SO2) has attracted substantial research interest and has proven to induce many respiratory responses. It is related to serious diseases, including lung cancer, cardiovascular diseases, and neurological disorders (Table 1). Because of the high reactivity between SO2 and the double bond, FRET-ESIPT and ICT-ESIPT are widely considered in detecting SO2 (Fig. 5a and b). Peng's group constructed a new ratiometric Probe 7 based on the ESIPT-FRET platform.154 3-Hydroxyflavone (an ESIPT dye) was used as an energy donor, and a 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF) fluorophore was used as a “two-in-one” ptor, which is an energy acceptor and SO2 derivatives acceptor (Fig. 5c). More importantly, high resolution of the two emission peaks could be achieved because of the effective FRET before reacting with SO2 and quenching FRET after the reaction. A similar procedure could be performed using probes with an ICT-ESIPT mechanism. Probes 8 and 9 with NIR emission at 716 nm and 686 nm, respectively, displayed good water-solubility, attributed to the hydrophilicity and electron-withdrawing property of thiazole salt moiety.155 Then, with the addition of SO2, the double bond between HBT or HBO and thiazole salt moiety disappeared, leading to the fluorescent changes in Probe 9. These probes were also successfully applied to visualize SO2 levels in cells under oxidative stress.
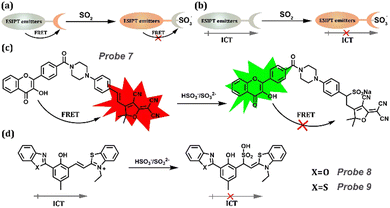 |
| | Fig. 5 (a) General procedure for SO2 detection based on FRET-ESIPT. (b) General procedure for SO2 detection based on ICT-ESIPT. (c) Probe 7 and its detection procedure based on FRET-ESIPT. (d) Detection procedure for Probes 8 and 9 based on FRET-ESIPT. | |
Another method for monitoring SO2 is introducing the 4-Oxopentanoic ester to replace the hydroxyl group. Lin's group developed a novel ratiometric probe, Probe 10, to monitor SO2 using a unique fluorescent signal in mammalian cells, mouse embryonic fibroblasts, and zebrafish via ESIPT and AIE.156 This probe was based on the novel AIE + ESIPT platform Probe 10-O with the sensing site levulinate moiety as the receptor and fluorescence quencher. When the hydroxyl group is substituted by levulinate, the inefficient ESIPT process forbids the long wavelength of Probe 10-O. Thus, Probe 10 would show blue fluorescence arising from the enol form. After treating with SO2, the ESIPT was released with the deprotection of levulinate in Probe 10 exhibiting a yellow aggregation fluorescence signal. Therefore, the AIE + ESIPT dye in Probe 10 is promising for monitoring SO2 (Fig. 6).
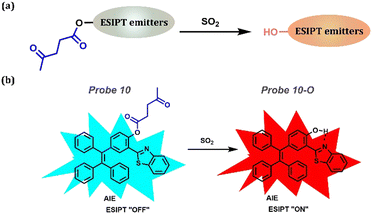 |
| | Fig. 6 (a) General procedure for SO2 detection: introducing the 4-Oxopentanoic ester to replace the hydroxyl group. (b) Probe 10 and its detection procedure. | |
2.4 Peroxynitrite (ONOO−)
ONOO−, which is closely related to various significant physiological processes, such as host defense, cell proliferation, and cell growth, is a significant biomarker for detecting Alzheimer's disease, one of the neurodegenerative diseases (Table 1). Because peroxide has strong oxidability, borate esters are widely selected as substituents owing to their excellent reducibility. Simultaneously, the borate ester substance could block the ESIPT process; thus, the probes emitted weak fluorescence corresponding to the Keto form. Upon reacting with ONOO−, the benzyl borate group is shed, activating the ESIPT process and releasing long-wavelength fluorescence (Fig. 7a). Fig. 7b shows a series of probes for ONOO− detection containing benzyl borate, which are reported by James group.135,136,157–159 They reported a ratiometric fluorescent, Probe 11.157 The detection limit for ONOO− was 21.4 nm, resulting in excellent detection efficiency at extremely low concentrations in few seconds. Additionally, Probe 11 could perform proportional imaging of endogenous and exogenous ONOO− in HeLa cells. Prominently, the probe had excellent selectivity for the endoplasmic reticulum. Comparing the co-staining in HeLa cells with Probe 11 and commercial dyes, such as ER-, Lyso-, and Mito-Tracker Red, Pearson's correlation coefficient with ER-Tracker Red was 0.93. Then, they synthesized a boronate-based ESIPT fluorescent (Probe 12) that can bind amyloid-β aggregates to construct a ratiometric probe for ONOO−.135 The probe blocked the ESIPT process by protecting the group with benzylborate, which could be particularly evacuated by ONOO−, leading to an “On” fluorescent probe suitable for detecting ONOO−. Furthermore, the probe exhibited excellent selectivity for ONOO− compared with other reactive oxygen species (ROS). They performed fluorescence imaging of AD-transgenic mouse brains to investigate the potential of the probes for practical applications. They observed that brain slices of Aβ42 aggregates exhibit N-state fluorescence with Probe 12. After ONOO− treatment, probes bound to the peptide aggregates in the brain slice and changed the fluorescence emission to the T* (the photo tautomeric form) state. Fluorescent Probe 13 with two-photon excitation was developed to detect ONOO−.158Probe 13 was non-fluorescent. However, the phenol fraction was released when ONOO− was added. Furthermore, Probe 13 exhibited high photo-stability under two-photon illumination at 750 nm, enabling the observation of endogenous ONOO− in HeLa cells and exogenous ONOO− in rat hippocampal slices at a depth of 110 μm. Moreover, two-photon ONOO−ex vivo and in vitro imaging was performed using an ESIPT-based benzimidazole fluorescence platform.136 Each platform enabled the rapid detection of micromolar concentrations of ONOO−. Importantly, Probe 14 was used for the ratio detection of endogenous ONOO− in RAW264.7 macrophages and rat hippocampus tissue.
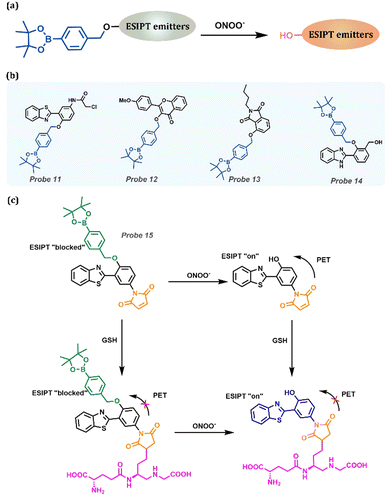 |
| | Fig. 7 (a) General procedure for detecting ONOO− based on ESIPT-based probes. (b) Probes for ONOO− detection. (c) Multifunctional Probe 15 and its “AND” logic detecting processes for ONOO− and GSH. | |
Probe 15 was highlighted as the special double function with “AND” logic (Fig. 7c).159 The ESIPT process of Probe 15 with benzyl borate was blocked, which acted as a reaction unit of ONOO−. Probe 15 was functionalized as an amino group to recognize thioalcohol, which assisted in fluorescence quenching through the PET process. The fluorescence intensity rapidly recovered in the presence of biothiols. Therefore, the probe yielded an efficient PET + ESIPT with “AND” logic. Additionally, the probe exhibited high sensitivity and selectivity for biological thiols and ONOO−, and it visualized exogenous ONOO− and GSH in living cells.
2.5 Hydrogen peroxide (H2O2)
Hydrogen peroxide (H2O2), which is a typical member of reactive oxygen species (ROS), plays an essential role in immune responses, cellular signal transduction, oxidative stress regulation, and antioxidant stimulation in high-oxygen-stressed tissues. Similar to other ROS, abnormal levels of H2O2 in the body can lead to cancer and neurodegenerative diseases. In addition, H2O2 is closely related to other diseases, such as diabetes and cardiovascular diseases (Table 1). Considering that H2O2 has similar oxidative properties with ONOO−, benzyl borate substituent is used as the recognition group for H2O2 owing to the high oxidative activity of H2O2 (Fig. 8a). Zeng et al. designed and synthesized a HBT-based fluorescent Probe 16.160 Boronesterification of phenol could interrupt the intramolecular hydrogen bond, leading to the blockage of the ESIPT process and inactivating the AIE effect (Fig. 8b). H2O2 and Probe 16 underwent phenylboronic oxidation and 1,6-elimination to regenerate p-BHBT and recover ESIPT-AIE fluorescence. Probe 16 could selectively and sensitively monitor H2O2 in an aqueous solution or living cells with the assistance of AIE + ESIPT, so it is a promising contender for H2O2 detection accurately and highly efficient. Liu et al. explored an H2O2 fluorescent probe based on the ESIPT dye, 1,3-bis(bispyridin-2-irimo) isoindole-4-ol, by choosing an aromatic boronic ester group as the responsive moiety (Fig. 8b).161 The introduced functional unit masked the hydroxyl group to block the proton transfer process and quench its fluorescence. In the occurrence of H2O2, the responsive moiety is cleaved in Probe 17, followed by fluorescence recovery. Significantly, Probe 17 exhibited outstanding performance in detecting intracellular H2O2 in living cells. Tang et al. reported a new ratiometric H2O2 fluorescent probe, Probe 18 (Fig. 8b), by utilizing phenylboronate as the recognizing group and an ESIPT blocking moiety.162 In the occurrence of H2O2, oxidation and 1,6-elimination reactions were conducted to release the phenolic hydroxyl group and then regain the long-wavelength emission belonging to the Enol form. It is noteworthy that Probe 18 had been used to image the exogenous and endogenous H2O2 in A549 cells. Chen et al. proposed a water-soluble ESIPT ratiometric fluorescent probe, Probe 19, with NIR emission, which specifically reacts with H2O2 through oxidation and 1,6-elimination with the release of Probe 19 appearing as NIR emission (Fig. 8b).163 Moreover, Probe 19 was applied to detect intracellular H2O2 in live cells and zebrafish. When the zebrafish were treated with Probe 19 alone, green fluorescence corresponding to the Enol form was observed. Exogenous H2O2 injection into zebrafish would provoke the crack of phenylboronate and a visual red fluorescence could be clearly witnessed. Compared with Probe 19 alone, the red channel fluorescence signal was significantly increased after stimulation with PMA (phorbol ester), indicating an increase in endogenous H2O2 (Fig. 8c). Subsequently, by treating with the inhibitor NAC (acetylcysteine), the red fluorescence in zebrafish was reduced, along with the enhancement of green fluorescence, demonstrating that NAC could effectively eliminate the increase in endogenous H2O2 arisen from PMA. These studies confirmed that Probe 19 is a promising tool for assessing and inspecting exo-/endogenous H2O2 in living zebrafish.
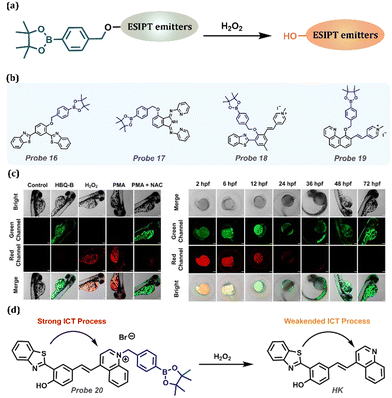 |
| | Fig. 8 (a) General procedure for detecting H2O2 with ESIPT-based probes. (b) Probes for H2O2 detection. (c) Confocal fluorescence images for H2O2 in zebrafish. Control group: untreated zebrafish; Probe 19 (HBQ-B) group: zebrafish treated with Probe 19 (10 μM); H2O2 group: zebrafish treated with H2O2 (40 μM) and Probe 19 (10 μM); PMA group: treated with PMA (3 μg mL−1) and Probe 19 (10 μM); PMA + NAC group: zebrafish treated with PMA (3 μg mL−1), NAC (1 mM), and Probe 19 (10 μM). (left) Confocal fluorescence images of H2O2 in various developmental stages of zebrafish. Zebrafish at 2, 6, 12, 24, 36, 48, and 72 hpf. (right) (d) The proposed reaction mechanism of Probe 20 with the ICT-ESIPT process. (c) Reprinted (adapted) from ref. 163 with permission from Elsevier, copyright 2022. | |
He et al. developed the ESIPT-based Probe 20, a novel mitochondria-targeting fluorescent probe, with distinctive ICT changes in contact with H2O2.164 Phenylboronate substituted the quinoline moiety, forming a quinoline salt with considerable electron-withdrawing property, which prolonged the emission to achieve the NIR/red fluorescence of Probe 20. In the presence of H2O2, double step reactions occurred containing borate oxidation and 1,6-elimination reactions, resulting in the appearance of HK with fluorescence at 594 nm (Fig. 8d). Remarkably, laser scanning confocal microscopy was employed to observe exogenous and endogenous H2O2 in live HeLa cells after treating Probe 20. The excellent results demonstrate that Probe 20 possesses outstanding prospects in biological applications.
2.6 Hypochlorous acid (HClO)
Hypochlorous acid (HClO) is a vital member of ROS, which is linked to the risk of cardiovascular, inflammatory, kidney diseases, and neuron degeneration (Table 1). N,N-Dimethylamino-thiomethyl ester is an excellent substrate for recognizing HClO, with the assistance of a hydrolyzing reaction (Fig. 9a). Wu et al. constructed an ESIPT-based Probe 21 with excellent HClO/ClO− responsiveness.165 On adding HClO, the protecting group was removed, and a strong emission could be observed. With great sensitivity and selectivity, the probe was applied to the endogenous and exogenous detection of HClO/ClO− in HeLa cells. Moreover, considering that HClO has relative acidity, a novel on–off Probe 22 was reported by Young et al., which could selectively monitor HClO with high solubility in 100% aqueous solution (Fig. 9c).166 With the appearance of HClO, the ESIPT process was forbidden, exhibiting shorter wavelength emission than the Keto form. Probe 22 possessed a low limit level of 8.2 × 10−9 M over a wide pH range of 5–11. Moreover, Probe 22 was successfully applied to the physiological system.
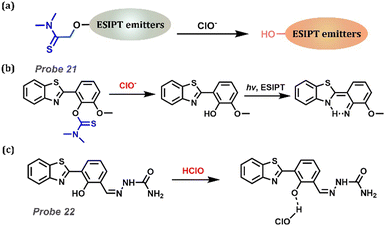 |
| | Fig. 9 (a) General procedure for detecting HClO with ESIPT-based probes. (b) The proposed reaction mechanism of Probe 21. (c) The proposed reaction mechanism of Probe 22. | |
2.7 Formaldehyde (HCHO)
Excess HCHO in the human body can cause many diseases, including Alzheimer's disease, cognitive impairment, neurodegenerative diseases, and atherosclerosis (Table 1). HCHO with exceptional reactivity can be monitored using the ICT-ESIPT and PET-ESIPT methods. As shown in Fig. 10a, Zhou et al. designed a new ratio of fluorescence Probe 23 for detecting HCHO in air and water based on the aza-Cope reaction.167 Before reacting with HCHO, the weak ICT endowed probes with short-wavelength emission at 462 nm owing to the electron-donating moiety weakening ICT. After adding HCHO, the aldehyde group as an electron-withdrawing group strengthened the ICT, and emission at 525 nm appeared. Additionally, the probe could be applied for the visual detection of gas FA and its fluorescence ratio in calf serum. Moreover, Probe 24 with PET-ESIPT was designed by Zeng et al. (Fig. 10b).168 Secondary ammonia has a strong nucleophilic attacking ability, which promoted the formation of β-hydroxy tertiary ammonia. Next, the formative hydroxyl group attacked the five-membered ring, linked to nitrogen (an electronic donor in the PET process), forming a six-membered azalactone. After the rearrangement of 2-aza-Cope in an aqueous solution, the C-N bond was broken owing to chemical structural instability. This structural change was followed by PET removal, leading to turn-on emission. Then, Probe 24 was used in imaging for FA in live AD mouse brain tissues.
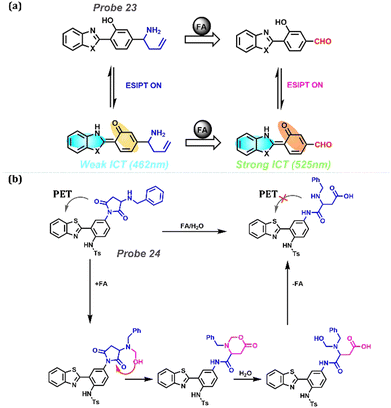 |
| | Fig. 10 (a) Reaction between probe Probe 23 and FA (formaldehyde) with weakened ICT-ESIPT and strengthened ICT-ESIPT. (b) The proposed reaction mechanism of Probe 24 with the PET-ESIPT process. | |
3. Organic biomarkers and detection methods
3.1 Cysteine (Cys)
Cys is an amino acid that participates in physiological processes, such as redox homeostasis, metabolism, protein synthesis, and signal transduction. Abnormal levels of Cys can trigger many diseases, including cardiovascular disease, neurotoxicity, Parkinson's disease, Alzheimer's disease, edema, lethargy, and liver damage (Table 1). As shown in Fig. 11a, β-aminothiol can react with acrylic esters in two steps containing Michael addition and nucleophilic addition reactions under ambient conditions. With the sloughing of the acrylic ester moiety, the exposed hydroxyl group ensures the ESIPT process, and the probe emits strong fluorescence.169,170 Based on the method, many probes have been synthesized in the past few years. In Fig. 11b, we list some of them that could release red or NIR emissions after reacting with Cys.171–176 NIR probes are outstanding in expanding tissue penetration depth, and one of the promising routes to obtaining NIR probes is introducing an electron-withdrawing moiety, such as pyranonitrile and pyridine salt. As shown in Fig. 11, this method was successfully utilized in in vitro and in vivo experiments, as discussed below. Probe 25 is an HBT-based (benzothiazole-based) fluorescent probe with a large Stokes shift (263 nm) emitting NIR fluorescence at 673 nm.171 It was successfully applied in Cys detection with high selectivity and sensitivity, and its detection limit was as low as 6.51 × 10−6 M. By introducing a strong electronic acceptor group dicyanoisophorone to the HBT fluorophore, Probe 26 emitted NIR fluorescence and was sensitive to Cys with a detection limit of 0.20 μM at pH 8.0 and 0.98 μM at pH 7.4.172 When Probe 26 reacted with Cys, a kinetically advantageous seven-membered ring was formed rapidly. In contrast, an eight-membered ring was formed slowly when in contact with homocysteine (Hcy), and the reaction with glutathione (GSH) or H2S could not be observed, so ESIPT-based Probe 26 was an excellent choice for the selective detection of Cys. Chen et al. designed a novel biflavone derivative, Probe 27, that could be located in the mitochondria without targeted groups, which exhibited ESIPT and showed visible monitoring of Cys with a large Stokes shift (approximately 260 nm).173Probe 28 was synthesized and grafted HBT in SCM-based ICT scaffolds by converting electron-withdrawing members from DCM to SCM and detected Cys with short-wave infrared (SWIR, 900–1700 nm).174 In collaboration with the ICT and ESIPT processes, SH-OH exhibited a NIR maximum absorption, SWIR emission, large Stokes shift, and excellent photostability. Long et al. designed an NIR two-photon fluorescence Probe 29 using an HBT derivative as fluorophore and implanting a butadienyl-N-methylpyridinium moiety.175 The selective detection of the photoluminescence mechanism of Cys by Probe 29 was considered ESIPT and ICT. It had a superior NIR fluorescence emission enhancement (713 nm), a significant Stokes shift (302 nm) and a two-photon absorption cross-section (213.5 GM at 820 nm). Successful application in Cys imaging was conducted by one/two photon imaging in live cells, zebrafish, live mouse brains or abdominal cavities. Zhao et al. reported a novel ESIPT-ICT-based NIR fluorescent Probe 30, with acrylate as the recognition group for the detection of Cys.176 The positively charged methylquinoline group of Probe 30 provided ICT and increased the emission wavelength. When Probe 30 reacted with Cys, the product further underwent intramolecular cyclisation, thus releasing Probe 30 with the advantages of long-wavelength emission at 740 nm and a large Stokes shift of 310 nm. Probe 30 could recognize Cys with high selectivity and sensitivity at pH = 5.5–8.0, and it has been successfully used to specifically recognize Cys in A549 cells. Moreover, we chose Probe 28 as a typical example for cell and living mouse imaging, owing to the near 950 nm emission (SWIR region) of Probe 28 (Fig. 11c–e). As shown in Fig. 11c, the SWIR signal was obviously obtained when treated with Probe 28. Moreover, they evaluated the in vivo performance of Probe 28 as a cis-activated SWIR probe using PerkinElmer IVIS spectroscopy. Then, real-time in vivo monitoring of Cys in mouse tumours was implemented. The SWIR emission was immediately displayed after injecting with Probe 28 in nude mice, indicating that Probe 28 is a sensitive probe for endogenous Cys.
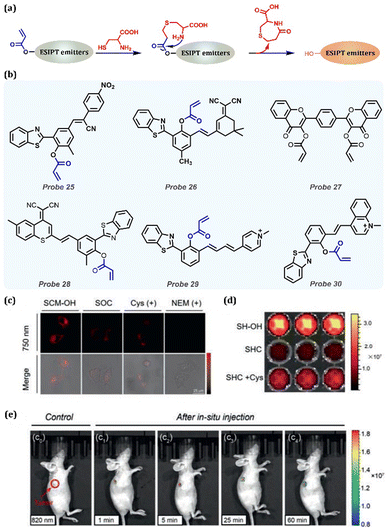 |
| | Fig. 11 (a) General procedure for Cys detection by replacing the hydrogen of the hydrogen donor with ESIPT-based probes. (b) Probes for Cys detection. (c) Confocal laser scanning microscope (CLSM) images of HeLa cells incubated with different compounds (100 μmol L−1), such as SCM-OH, probe SOC, exogenous Cys and SOC, NEM, and SOC, for 1 h. Note: the NIR fluorescence intensity monitored channel was obtained in the range of 740 nm to 760 nm, λex = 605 nm. (d) Plate fluorescence imaging of SH-OH (10 μmol L−1), Probe 28 (SHC) (10 μmol L−1) and Probe 28 (10 μmol L−1) incubated with Cys (100 μmol L−1), yellow-red scale. (e) In vivo SWIR fluorescence imaging (rainbow scale) of mice with HeLa xenograft after in situ injections of Probe 28 at various times (1 min, e1; 5 min, e2; 25 min, e3; 60 min, e4). Note: λex = 600 nm, SWIR fluorescence signal monitored at 950 nm. (c–e) Reprinted (adapted) from ref. 173 with permission from Elsevier, copyright 2022. | |
Despite the method mentioned above, PET is a tool to lock the emission before reacting with Cys and release the fluorescence after reacting with Cys. Maleimide is a useful recognition functional group that provides lone-pair electrons to occupy the empty orbit after excitation, and it can react with Cys, which is attributed to the reducibility of Cys (Fig. 12a). Feng et al. reported a fluorescent Probe 31 that could successfully distinguish Cys from other biothiols (GSH and Hcy) having a similar structure and reactivity to Cys, with high sensitivity (the ultralow detection limit at 9.4 nM) (Fig. 12b).177 Strong fluorescence emission at 503 nm occurred by yielding amino-adducts when Probe 31 reacted with Cys. In the case of GSH, the thermodynamic hindrance of a 10-membered cyclic transition state was created, and the adjacent amino group was interrupted by the glutamyl group, preventing visual observation. Although the cyclic transition state of Hcy was thermodynamically allowed, the increased carbon chain led to a slower cascade reaction, reducing the number of amino adducts produced at the same reaction time with only weak fluorescence. Moreover, they successfully applied Probe 31 to detect Cys in test papers and to the imaging of Cys in A549 live cells. This method is significant in Cys detection, especially for multi-functional applications, owing to the rich functionalizing possibilities of free hydroxyl groups. Thus, detailed examples are mentioned again in the next section.
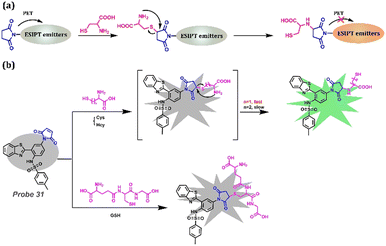 |
| | Fig. 12 (a) General procedure for Cys detection based on PET-ESIPT-based probes. (b) Probe 31 and its mechanism for distinguishing Cys, GSH, and Hcy. | |
3.2 Glutathione (GSH)
When discussing intracellular redox homeostasis, growth and metabolism, oxidative stress, immune responses, and apoptosis, GSH is always considered to play a significant role in these biological processes. Simultaneously, abnormal concentrations of GSH are important transmission signals for many diseases, including Parkinson's disease, liver injury, and cancer (Table 1). GSH can be monitored by introducing an acrylic ester moiety, which is similar to the detection method of Cys (Fig. 13a). Zhang et al. reported an HBT-based fluorescent, Probe 32, that has specific double functions, which could emit infrared fluorescence (684 nm) reacting with GSH and blue fluorescence (469 nm) reacting with Arg (Arginine).178 The detection of GSH, which depends on the Michael addition reaction and ester lysis, exhibited NIR emission owing to the ESIPT process. However, ESIPT was inhibited in the presence of Arg attributed to the suitable basicity and acidity of benzothiazole and Arg, respectively. Hence, this strategy provides an outstanding method for preparing multi-functional probes using various procedures, including chemical reactions and weak intermolecular interactions.
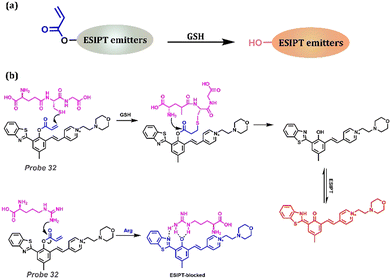 |
| | Fig. 13 (a) General procedure for GSH detection based on the ESIPT process. (b) Proposed mechanisms of Probe 32 for GSH and Arg detection. | |
Considering the acidity of GSH, it could influence the ESIPT process with a dominant blue shift (Fig. 2, Method 2). Ren et al. reported a series of fluorescent Probes 33, 34, and 35 based on HBT derivatives for detecting GSH (Fig. 14).179Probe 33 could respond to GSH reversibly and rapidly compared with Probes 34 and 35. The proton transfer from OH to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N in the excited state contributed to Probe 33 emitting double emission (Enol and Keto forms). Conversely, the presence of GSH prevented the formation of the hydrogen bond between “C
N in the excited state contributed to Probe 33 emitting double emission (Enol and Keto forms). Conversely, the presence of GSH prevented the formation of the hydrogen bond between “C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N” and “COOH”, which blocked ESIPT and stabilized the conformation of the probe-GSH adduct. Consequently, this process assisted in emitting the blue-shifted emission arising from the ICT process.
N” and “COOH”, which blocked ESIPT and stabilized the conformation of the probe-GSH adduct. Consequently, this process assisted in emitting the blue-shifted emission arising from the ICT process.
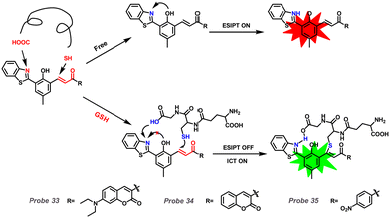 |
| | Fig. 14 Proposed mechanisms of Probes 34, 33, and 35 with GSH. | |
3.3 β-Glucuronidase (GUS)/β-galactosidase (β-gal)
GUS, a matrix degradation enzyme, is involved in tumor invasion and metastasis, and it acts as an important biomarker for Escherichia coli (E. coli) (Table 1). β-Gal is generally considered a reporter for monitoring gene expression and a significant biomarker for primary ovarian cancer and cellular senescence (Table 1). Galactoside is an efficient substrate for the recognition of β-glucuronidase and β-galactosidase because of its enzyme specificity (Fig. 15a). Wu and his colleagues introduced a GUS activation unit into a water-insoluble fluorophore HBT with ESIPT and AIE effects (Fig. 15b).180 The probe had high selectivity, low toxicity, and good cell permeability and sensitivity. Probe 36 emitted strong fluorescence at 519 nm via UV excitation. Hu and colleagues reported a novel fluorescent Probe 37 for the detection of β-gal activity (Fig. 15b).181 The probe was based on a hydroxyphenylquinazolinone (HPQ) chromophore with AIE and ESIPT effects. The β-galactopyranoside group bond of Probe 37 quenched the fluorescence owing to breaking the intramolecular hydrogen bond and preventing the ESIPT process. After adding β-gal, an efficient enzymatic hydrolysis occurred, releasing the green chromophore. HPQ exhibited enhanced fluorescence because of ESIPT between the phenolic hydroxyl group and the imine moiety. Owing to its high sensitivity, good selectivity, and large Stokes shift, this probe can well image intracellular β-gal activity.
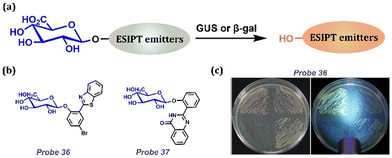 |
| | Fig. 15 (a) General procedure for detecting GUS/β-gal with ESIPT-based probes. (b) Probes for GUS detection. (c) The strains grown for 18 h at 37 °C in the air were observed under visible light (left), and 375 nm UV flashlight excitation (right), dying by Probe 36. (c) Reprinted (adapted) from ref. 180 with permission from Elsevier, copyright 2022. | |
Because β-gal is involved in the significant process in coli, we focus on coli imaging, which is an important application for fluorescent probes (Fig. 15c). Probe 36 was designed as a new type of chromogenic fluorescent medium (C–F E. coli agar) by observing the activity of β-gal in Escherichia coli (E. coli O157:H7) and non-O157:H7 E. coli for effective monitoring and separation. The chromogenic substrates 5-bromo-6-chloro-3-indolyl-β-D-galactoside (Magenta-Gal) and 5-bromo-4-chloro-3-indolyl-β-D-glucoside (X-Glu) were added to C–F E. coli agar. Then, E. coli O157:H7 ATCC 35150, E. coli ATCC 25922, Enterobacter sakazakii ATCC 29544, and Salmonella typhimurium ATCC 14028 were inoculated at the upper right, upper left, lower left and lower right of the C–F E. coli agar plates, respectively. As shown in Fig. 15c, these species were distinguished by excitation with visible light and 375 nm UV light.
3.4 Phosphatase (ALP)
Alkaline Phosphatase (ALP) is a widespread hydrolase in living organisms that can regulate phosphate metabolism. Abnormal fluctuations in ALP serum levels are important for diagnosing diseases, including diabetes, bone disease, liver dysfunction, and prostate cancer (Table 1). By replacing the phenol with phosphate, the probes could effectively respond to ALP through a simple enzymatic reaction (Fig. 16a). Zhang et al. reported a new fluorescent Probe 38 for the detection of ALP activity in aqueous buffers and various cell lines.182 ALP selectively reacted with the phosphate group, and then the hydroxyl group was released to form a fluorescent compound SA (salicylaldazine), which had the characteristics of AIE + ESIPT. Probe 38 selectively exhibited a fast and 240-fold turn-on ratio in response to ALP. It presents the ability to identify and visualize endogenous ALP activity in various cell lineages, including human osteoblasts, mouse melanoma cells, and macrophages. Yang et al. employed the ESIPT fluorophore N-(3-(benzo[d]thiazol-2-yl)-4-hydroxyphenyl) benzylamide to design a ratiometric Probe 39 for ALP detection.183 By enzymatic dephosphorylation, Probe 39 exhibited a large red shift (120 nm) and high sensitivity for ALP sensing at 0.004 mU mL−1. Enzymatic dephosphorylation converted the probe to fluorophore, followed by conspicuous fluorescence changes. Thus, the probe exhibited high sensitivity to ALP, with an extremely low detection limit (0.004 mU mL−1). Then, this probe was applied to monitor endogenous ALP in living cells (HeLa cells) and exhibited excellent biocompatibility. By regulating the protection and deprotection of hydroxyl groups, Ding et al. announced a novel mitochondrial-targeted ratiometric fluorescent Probe 40 with ESIPT characteristics for ALP recognition and visualization.184 The probe was highly soluble in water and emitted green fluorescence in aqueous buffers by blocking ESIPT. As the ALP increased, the fluorescence changed from green to red (650 nm), along with ESIPT recovery. The probe could quantify ALP activity from 0 to 60 mU mL−1 with a minimum LOD of 0.072 mU mL−1. It could also be used to detect ALP in serum samples, HeLa and A549 cell lines, and tumor-bearing BALB/c nude mice.
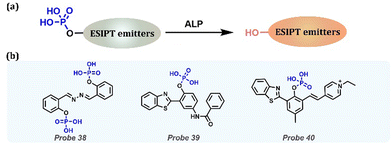 |
| | Fig. 16 (a) General procedure for detecting ALP with ESIPT-based probes. (b) Probes for ALP detection. | |
3.5 Esterase
Esterase catalyzes the esterification reaction, which plays an important role in human physiological processes, such as gene expression, ester metabolism, material transport, and detoxification. As an important biological enzyme, its deletion or abnormal expression is directly related to many diseases, such as Wollman's disease, obesity, atherosclerosis, cancer, hyperlipidemia, and fatty liver. (Table 1). Normally, an ester bond is introduced to block the ESPIT process. Once reacted with specific esterase, the ester group is cleaved by turning on the ESIPT process (Fig. 17a). Tian et al. developed a fluorescent Probe 41 that distinguished living and dead cells in two-color mode by esterification of 3-hydroxyflavones (Fig. 17b).185 They proved that the probe could be degraded to 3-hydroxyflavonoid by active esterase, and the ESIPT process was restored, with an orange fluorescence peak at 570 nm in living cells. In dead cells, the esterase was inactivated with blue emission peaks at 440 nm, preventing the ESIPT process. Thus, this probe is promising for distinguishing between living and dead cells because it can provide accurate information in biological applications. Chen et al. designed and synthesized three fluorescent probes based on HBT derivatives (Probes 42, 43 and 44) to detect esterase (Fig. 17b).186 Based on the ESIPT effect, acetoxymethyl (AM) ether was introduced on HBT as the recognition group. Under esterase catalysis, the recognition group was converted to a hydroxyl group, which led to the release of the fluorescence signal. Moreover, dipstick experiments showed that the probes recognized esterase accompanied by significant color changes. Importantly, this kind of probe is more suitable for detecting and visualizing endogenous esterases in biological environments.
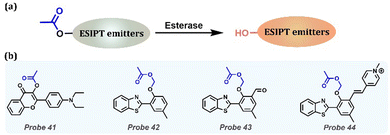 |
| | Fig. 17 (a) General procedure for detecting esterase with ESIPT-based probes. (b) Probes for esterase detection. | |
AChE, a special esterase and the target enzyme for nerve agents, is a biological enzyme involved in the metabolic processes of the human body (Table 1). Once bound to the nerve agent, it inhibits the hydrolysis of the neurotransmitter acetylcholine, leading to neurological problems, such as salivation, convulsions, tremors, and even death. In general, butyrylcholinesterase was selected as the recognition group, which was bound by esterification to fluorophores, turning off the fluorescence (Fig. 18a). Meng et al. reported the fluorescence sensor Probe 45, which can selectively detect the activity of real nerve agents and AChE (Fig. 18b).187 Given that this probe had a dual recognition of nerve agents and AChE, Probe 45 changed from no fluorescence to blue fluorescence when exposed to nerve agent and produced orange fluorescence when exposed to AChE under excitation at 365 nm. Benefiting from the excellent properties of the probe, it was used to monitor the concentration of nerve agent and AChE activity conditions of Probe 45 in living cells. They also used Probe 45 to diagnose neurotoxin intoxication by identifying the AChE activity in the entire blood.
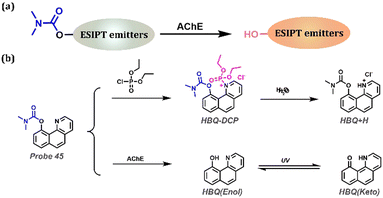 |
| | Fig. 18 (a) General procedure for detecting AChE with ESIPT-based probes. (b) Probe 45 for AChE and nerve agent detection based on ESIPT. | |
3.6 Leucine aminopeptidase (LAP)
As a catalytic enzyme, LAP is involved in many important physiological processes in the human body, and disorders of catalytic function are accompanied by diseases, such as hepatic dysfunction or liver cancer (Table 1). Based on the specificity of the enzymatic reaction, leucine is an ideal substrate for the recognition of LAP (Fig. 19a). Zeng et al. developed the first tetraarylimidazole-based AIE fluorophore by substituting different aryl groups at the 4,5 positions (Fig. 19b).188 Leucine was elaborately introduced to replace the hydrogen bond donor and prohibited ESIPT. Therefore, only short-wavelength emission belonging to the Enol form was perceived. In the presence of LAP, the blocking moiety Leucine was cleaved from the fluorophore along with the release of fluorescence attributed to the AIE and ESIPT. The HBT backbone enabled the fluorophore to exhibit AIE-assisted ESIPT emission at 554 nm, and the obtained Probe 46 was the first probe to combine the AIE with the ESIPT mechanism for precise LAP ratio tracking. Probe 46 possessed a promising ability for sensitive and selective LAP detection in hepatoma cells and zebrafish liver lesions with a large Stokes shift and considerable biocompatibility.
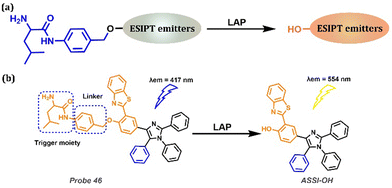 |
| | Fig. 19 (a) General procedure for LAP detection with ESIPT-based probes. (b) Probe 46 and its detection of LAP. | |
3.7 Serum albumin (HAS)
Serum albumin (HSA), which is particularly abundant in the human circulatory system, is closely related to various physiological processes, including the maintenance of colloidal osmotic pressure, the regulation of free radicals, and the transport of nutrients, fatty acids, and metabolites. Additionally, abnormal HSA levels in humans can exhibit various symptoms of the disease, including cardiovascular disease, kidney disease, hypertension, liver cirrhosis, and even cancers (Table 1). Owing to the Lewis acidity from hydrogen and Lewis basicity from the carbonyl group of amide, serum albumin could be sensed by method 2, as depicted in Fig. 2. Huang et al. designed a new fluorescent Probe 47 (Fig. 20).189 The hydroxyl substituent at the specific site in the probe could change the binding site from DS1 (drug site 1) to DS2 (drug site 2) and then restore the ESIPT process of the probe, thus showing a unique dual emission response to HSA. The binding mode of Probe 47 to HSA was confirmed by site-specific competition experiments and molecular docking techniques, and the specific binding of flavonoids to DS2 was confirmed for the first time.
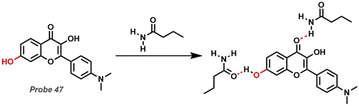 |
| | Fig. 20 Structural description of fluorescent Probe 47 and reaction mechanism with arginine. | |
4. Conclusions and future work
In this review, we investigated the function of some significant biomarkers, possible diseases closely related to them, and the corresponding probes for biomarker detection. Considering some features, including high efficiency, low background interference, and good structural adjustability, ESIPT probes explored in the past five years are emphasized. To functionally monitor specific biomarkers, three major design strategies for ESIPT probes are generally utilized: (1) replacing hydrogen of hydrogen bond donor, (2) destroying hydrogen bond by acid or base, and 3) combining with other electron transfer or energy transfer processes, such as FRET-ESIPT, PET-ESIPT, and ICT-ESIPT. From this perspective, we carefully summarize and classify a series of ESIPT-based probes using deserve strategies. Simultaneously, attributed to the limitation of organizational penetration, NIR ESIPT probes, and their design strategies, such as introducing electron-withdrawing groups, are all highlighted. Moreover, in addition to in vitro experiments, some biological applications are mentioned, including cells, bacteria, and animal experiments.
Through this feature article, we have emphasized recent progress made in the development of ESIPT-based fluorescent probes. Despite these successes, exploring NIR probes with a high quantum yield is still of vital significance to increase the depth of tissue penetration. Additionally, ESIPT probes with a long lifetime might be another rising field, including ESIPT-based chemiluminescence and Thermal Activation Delayed Fluorescence (TADF) probes, showing advantages in observing long-term biological processes.190–193 It is definite that the ESIPT-based fluorescent probes for biomarker detection will still be a challenging and flourishing area with developing measurement techniques and pluralistic biomarker probes, assisted by improved targeting properties, efficient recognizing groups, and excellent photophysical properties. We hope this feature article will inspire researchers to construct more novel, powerful and not-yet-involved probes.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was supported by the National Key R & D Program of China (2021YFC2103600), the National Natural Science Foundation of China (22278224, 21978131, and 21878156), the Natural Science Foundation of Jiangsu Province (BK20200691), the Project of Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the State Key Laboratory of Materials-Oriented Chemical Engineering (KL21-08).
Notes and references
- T. Powles, M. Kockx, A. Rodriguez-Vida, I. Duran, S. J. Crabb, M. S. Van der Heijden, B. Szabados, A. F. Pous, G. Gravis, U. A. Herranz, A. Protheroe, A. Ravaud, D. Maillet, M. J. Mendez, C. Suarez, M. Linch, A. Prendergast, P. J. van Dam, D. Stanoeva, S. Daelemans, S. Mariathasan, J. S. Tea, K. Mousa, R. Banchereau and D. Castellano, Nat. Med., 2019, 25, 1706–1714 CrossRef CAS PubMed.
- A. Sveen, S. Kopetz and R. A. Lothe, Nat. Rev. Clin. Oncol., 2020, 17, 11–32 CrossRef PubMed.
- V. S. LeBleu and R. Kalluri, Trends in Cancer, 2020, 6, 767–774 CrossRef CAS PubMed.
- D. B. Doroshow, S. Bhalla, M. B. Beasley, L. M. Sholl, K. M. Kerr, S. Gnjatic, I. I. Wistuba, D. L. Rimm, M. S. Tsao and F. R. Hirsch, Nat. Rev. Clin. Oncol., 2021, 18, 345–362 CrossRef CAS PubMed.
- J. J. Lee, H. J. Kim, M. Ceko, B. Y. Park, S. A. Lee, H. Park, M. Roy, S. G. Kim, T. D. Wager and C. W. Woo, Nat. Med., 2021, 27, 174–182 CrossRef CAS PubMed.
- Y. F. Liu, Y. F. Li, J. K. Zang, T. Y. Zhang, Y. J. Li, Z. F. Tan, D. Ma, T. Zhang, S. Y. Wang, Y. S. Zhang, L. Huang, Y. S. Wu, X. L. Su, Z. A. Weng, D. Deng, C. K. Tsang, A. D. Xu and D. Lu, Circ. Res., 2022, 130, 907–924 CrossRef CAS PubMed.
- H. Kim, M. Yang, N. Kwon, M. Cho, J. Han, R. Wang, S. Qi, H. Li, V.-N. Nguyen, X. Li, H.-B. Cheng and J. Yoon, Bull. Korean Chem. Soc., 2022, 1 DOI:10.1002/bkcs.12655.
- S. Taniselass, M. K. M. Arshad and S. C. B. Gopinath, Biosens. Bioelectron., 2019, 130, 276–292 CrossRef CAS PubMed.
- L. Y. Wang, S. L. Xie, Z. Y. Wang, F. Liu, Y. F. Yang, C. Q. Tang, X. Y. Wu, P. Liu, Y. J. Li, H. Saiyin, S. Zheng, X. M. Sun, F. Xu, H. B. Yu and H. S. Peng, Nat. Biomed. Eng., 2020, 4, 159–171 CrossRef CAS PubMed.
- H. Karimi-Maleh, Y. Orooji, F. Karimi, M. Alizadeh, M. Baghayeri, J. Rouhi, S. Tajik, H. D. Beitollahi, S. Agarwal, V. K. Gupta, S. Rajendran, A. Ayati, L. Fu, A. L. Sanati, B. Tanhaei, F. Sen, M. Shabani-nooshabadi, P. N. Asrami and A. Al-Othman, Biosens. Bioelectron., 2021, 184, 113252–113267 CrossRef CAS PubMed.
- J. R. Sempionatto, M. Y. Lin, L. Yin, E. de la Paz, K. Pei, T. Sonsa-Ard, A. N. D. Silva, A. A. Khorshed, F. Y. Zhang, N. Tostado, S. Xu and J. S. Wang, Nat. Biomed. Eng., 2021, 5, 737–748 CrossRef CAS PubMed.
- G. Trudel, N. Shahin, T. Ramsay, O. Laneuville and H. Louati, Nat. Med., 2022, 28, 59–62 CrossRef CAS PubMed.
- H. W. Liu, L. L. Chen, C. Y. Xu, Z. Li, H. Y. Zhang, X. B. Zhang and W. H. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC.
- L. Y. Wu, Y. Ishigaki, Y. X. Hu, K. Sugimoto, W. H. Zeng, T. Harimoto, Y. D. Sun, J. He, T. Suzuki, X. Q. Jiang, H. Y. Chen and D. J. Ye, Nat. Commun., 2020, 11, 446–459 CrossRef CAS PubMed.
- L. L. Wu, J. G. Huang, K. Y. Pu and T. D. James, Nat. Rev. Chem., 2021, 5, 406–421 CrossRef CAS.
- H. B. Xiao, P. Li and B. Tang, Coord. Chem. Rev., 2021, 427, 213582–213613 CrossRef CAS.
- R. Kotani, S. Yokoyama, S. Nobusue, S. Yamaguchi, A. Osuka, H. Yabu and S. Saito, Nat. Commun., 2022, 13, 303–311 CrossRef CAS PubMed.
- X. T. Ren, F. Wang, J. Lv, T. W. Wei, W. Zhang, Y. Wang and X. Q. Chen, Dyes Pigm., 2016, 129, 156–162 CrossRef CAS.
- Y. Qi, Y. Huang, B. W. Li, F. Zeng and S. Z. Wu, Anal. Chem., 2018, 90, 1014–1020 CrossRef CAS PubMed.
- T. Rasheed and F. Nabeel, Coord. Chem. Rev., 2019, 401, 213065–213086 CrossRef CAS.
- D. Wu, L. Y. Chen, Q. L. Xu, X. Q. Chen and J. Y. Yoon, Acc. Chem. Res., 2019, 52, 2158–2168 CrossRef CAS PubMed.
- H. H. Han, H. Tian, Y. Zang, A. C. Sedgwick, J. Li, J. L. Sessler, X. P. He and T. D. James, Chem. Soc. Rev., 2021, 50, 9391–9429 RSC.
- Y. N. Liu, Y. A. Yu, Q. Y. Zhao, C. H. Tang, H. Y. Zhang, Y. C. Qin, X. H. Feng and J. M. Zhang, Coord. Chem. Rev., 2021, 427, 213601–213622 CrossRef CAS PubMed.
- D. X. Cao, Z. Q. Liu, P. Verwilst, S. Koo, P. Jangjili, J. S. Kim and W. Y. Lin, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS PubMed.
- Y. Li, Z. Cai, S. Liu, H. Zhang, S. T. H. Wong, J. W. Y. Lam, R. T. K. Kwok, J. Qian and B. Z. Tang, Nat. Commun., 2020, 11, 1255–1264 CrossRef CAS PubMed.
- Z. L. Zeng, S. S. Liew, X. Wei and K. Y. Pu, Angew. Chem., Int. Ed., 2021, 60, 26454–26475 CrossRef CAS PubMed.
- H. D. Li, H. Kim, F. Xu, J. J. Han, Q. C. Yao, J. Y. Wang, K. Y. Pu, X. J. Peng and J. Yoon, Chem. Soc. Rev., 2022, 51, 1795–1835 RSC.
- R. D. Healey, E. M. Saied, X. J. Cong, G. Karsai, L. Gabellier, J. Saint-Paul, E. Del Nero, S. Jeannot, M. Drapeau, S. Fontanel, D. Maurel, S. Basu, C. Leyrat, J. Golebiowski, G. Bossis, C. Bechara, T. Hornemann, C. Arenz and S. Granier, Angew. Chem., Int. Ed., 2022, 61, e202109967 CrossRef CAS PubMed.
- C. Huang, C. Liang, T. Sadhukhan, S. Banerjee, Z. X. Fan, T. X. Li, Z. L. Zhu, P. Y. Zhang, K. Raghavachari and H. Y. Huang, Angew. Chem., Int. Ed., 2021, 60, 9474–9479 CrossRef CAS PubMed.
- C. R. Hoopes, F. J. Garcia, A. M. Sarkar, N. J. Kuehl, D. T. Barkan, N. L. Collins, G. E. Meister, T. R. Bramhall, C. H. Hsu, M. D. Jones, M. Schirle and M. T. Taylor, J. Am. Chem. Soc., 2022, 144, 6227–6236 CrossRef CAS PubMed.
- B. Yan, Inorg. Chem. Front., 2021, 8, 201–233 RSC.
- Y. F. Zhao, H. Zeng, X. W. Zhu, W. G. Lu and D. Li, Chem. Soc. Rev., 2021, 50, 4484–4513 RSC.
- L. L. Wu, J. H. Liu, P. Li, B. Tang and T. D. James, Chem. Soc. Rev., 2021, 50, 702–734 RSC.
- L. L. Wu, A. C. Sedgwick, X. L. Sun, S. D. Bull, X. P. He and T. D. James, Acc. Chem. Res., 2019, 52, 2582–2597 CrossRef CAS PubMed.
- J. Ochi, K. Tanaka and Y. Chujo, Angew. Chem., Int. Ed., 2020, 59, 9841–9855 CrossRef CAS PubMed.
- H. Tian, A. C. Sedgwick, H. H. Han, S. Sen, G. R. Chen, Y. Zang, J. L. Sessler, T. D. James, J. Li and X. P. He, Coord. Chem. Rev., 2021, 427, 213577–213590 CrossRef CAS.
- B. F. E. Curchod and T. J. Martinez, Chem. Rev., 2018, 118, 3305–3336 CrossRef CAS PubMed.
- P. Zhou and K. Han, Acc. Chem. Res., 2018, 51, 1681–1690 CrossRef CAS PubMed.
- H. Q. Yin and X. B. Yin, Acc. Chem. Res., 2020, 53, 485–495 CrossRef CAS PubMed.
- X. Tian, L. C. Murfin, L. Wu, S. E. Lewis and T. D. James, Chem. Sci., 2021, 12, 3406–3426 RSC.
- C.-L. Chen, Y.-T. Chen, A. P. Demchenko and P.-T. Chou, Nat. Rev. Chem., 2018, 2, 131–143 CrossRef CAS.
- Z.-Y. Liu, J.-W. Hu, C.-H. Huang, T.-H. Huang, D.-G. Chen, S.-Y. Ho, K.-Y. Chen, E. Y. Li and P.-T. Chou, J. Am. Chem. Soc., 2019, 141, 9885–9894 CrossRef CAS PubMed.
- Z.-Y. Liu, Y.-C. Wei and P.-T. Chou, J. Phys. Chem. A, 2021, 125, 6611–6620 CrossRef CAS PubMed.
- W. Shao, J. Hao, H. Jiang, P. M. Zimmerman and J. Kim, Adv. Funct. Mater., 2022, 32, 2201256–2201264 CrossRef CAS.
- C.-H. Wang, Z.-Y. Liu, C.-H. Huang, C.-T. Chen, F.-Y. Meng, Y.-C. Liao, Y.-H. Liu, C.-C. Chang, E. Y. Li and P.-T. Chou, J. Am. Chem. Soc., 2021, 143, 12715–12724 CrossRef CAS PubMed.
- C. Xue, Y. Jiang, H.-X. Wang, C. Du, L. Xu, T. Li and M. Liu, Angew. Chem., Int. Ed., 2022, 61, e20220563–e20220569 Search PubMed.
- H. Miyabayashi, K. Fujii, T. Watanabe, Y. Matano, T. Endo and Y. Kimura, J. Phys. Chem. B, 2021, 125, 5373–5386 CrossRef CAS PubMed.
- A. C. Sedgwick, L. Wu, H.-H. Han, S. D. Bull, X.-P. He, T. D. James, J. L. Sessler, B. Z. Tang, H. Tian and J. Yoon, Chem. Soc. Rev., 2018, 47, 8842–8880 RSC.
- Y. Chen, Y. Fang, H. Gu, J. Qiang, H. Li, J. Fan, J. Cao, F. Wang, S. Lu and X. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 55094–55106 CrossRef CAS PubMed.
- P. W. Zhou and K. L. Han, Aggregate, 2022, 3, e160–e182 CrossRef.
- G.-J. Zhao and K.-L. Han, Acc. Chem. Res., 2012, 45, 404–413 CrossRef CAS PubMed.
- J. Z. Zhao, S. M. Ji, Y. H. Chen, H. M. Guo and P. Yang, Phys. Chem. Chem. Phys., 2012, 14, 8803–8817 RSC.
- L. W. He, B. L. Dong, Y. Liu and W. Y. Lin, Chem. Soc. Rev., 2016, 45, 6449–6461 RSC.
- L. Guo, M. Tian, Z. Zhang, Q. Lu, Z. Liu, G. Niu and X. Yu, J. Am. Chem. Soc., 2021, 143, 3169–3179 CrossRef CAS PubMed.
- S. Chen, Y. Hong, Y. Liu, J. Liu, C. W. T. Leung, M. Li, R. T. K. Kwok, E. Zhao, J. W. Y. Lam, Y. Yu and B. Z. Tang, J. Am. Chem. Soc., 2014, 136, 11196 CrossRef CAS.
- Z. Yang, J. Cao, Y. He, J. H. Yang, T. Kim, X. Peng and J. S. Kim, Chem. Soc. Rev., 2014, 43, 4563–4601 RSC.
- C. Corbet and O. Feron, Nat. Rev. Cancer, 2017, 17, 577–593 CrossRef CAS PubMed.
- S. R. Pillai, M. Damaghi, Y. Marunaka, E. P. Spugnini, S. Fais and R. J. Gillies, Cancer Metastasis Rev., 2019, 38, 205–222 CrossRef CAS PubMed.
- G. Song, D. Jiang, L. Wang, X. Sun, H. Liu, Y. Tian and M. Chen, Chin. Chem. Lett., 2022, 33, 339–343 CrossRef CAS.
- B. Suetrong and K. R. Walley, Chest, 2016, 149, 252–261 CrossRef PubMed.
- A. N. Harris, P. R. Grimm, H. W. Lee, E. Delpire, L. J. Fang, J. W. Verlander, P. A. Welling and I. D. Weiner, J. Am. Soc. Nephrol., 2018, 29, 1411–1425 CrossRef CAS PubMed.
- J. Harambat, K. Kunzmann, K. Azukaitis, A. K. Bayazit, N. Canpolat, A. Doyon, A. Duzova, A. Niemirska, B. Sozeri, D. Thurn-Valsassina, A. Anarat, L. Bessenay, C. Candan, A. Peco-Antic, A. Yilmaz, S. Tschumi, S. Testa, A. Jankauskiene, H. Erdogan, A. Rosales, H. Alpay, F. Lugani, K. Arbeiter, F. Mencarelli, A. Kiyak, O. Donmez, D. Drozdz, A. Melk, U. Querfeld, F. Schaefer and C. S. Consortium, Kidney Int., 2017, 92, 1507–1514 CrossRef CAS PubMed.
- L. Y. Wu and R. Wang, Pharmacol. Rev., 2005, 57, 585–630 CrossRef CAS PubMed.
- R. Motterlini and L. E. Otterbein, Nat. Rev. Drug Discov., 2010, 9, 728–743 CrossRef CAS PubMed.
- Z. Q. Meng, Z. H. Yang, J. L. Li and Q. X. Zhang, Chemosphere, 2012, 89, 579–584 CrossRef CAS PubMed.
- D. M. Woodford, R. E. Coutu and E. A. Gaensler, Respiration, 1979, 38, 238–245 CrossRef CAS PubMed.
- H. Vally, N. L. A. Misso and V. Madan, Clin. Exp. Allergy, 2009, 39, 1643–1651 CrossRef CAS PubMed.
- K. R. B. Silva, I. M. Raimundo, I. F. Gimenez and O. L. Alves, J. Agric. Food Chem., 2006, 54, 8697–8701 CrossRef CAS PubMed.
- F. Yan, J. Cui, C. Wang, X. Tian, D. Li, Y. Wang, B. Zhang, L. Feng, S. Huang and X. Ma, Chin. Chem. Lett., 2022, 33, 4219–4222 CrossRef CAS.
- C.-X. Yin, K.-M. Xiong, F.-J. Huo, J. C. Salamanca and R. M. Strongin, Angew. Chem., Int. Ed., 2017, 56, 13188–13198 CrossRef CAS PubMed.
- V. Agouridas, O. El Mahdi, V. Diemer, M. Cargoet, J.-C. M. Monbaliu and O. Melnyk, Chem. Rev., 2019, 119, 7328–7443 CrossRef CAS PubMed.
- R. Zhang, J. Yong, J. Yuan and Z. P. Xu, Coord. Chem. Rev., 2020, 408, 213182–213222 CrossRef CAS.
- X. Ren, L. Liao, Z. Yang, H. Li, X. Li, Y. Wang, Y. Ye and X. Song, Chin. Chem. Lett., 2021, 32, 1061–1065 CrossRef CAS.
- M. J. Cao, H. Y. Chen, D. Chen, Z. Q. Xu, S. H. Liu, X. Q. Chen and J. Yin, Chem. Commun., 2016, 52, 721–724 RSC.
- Y. S. Guo, H. Wang, Y. S. Sun and B. Qu, Chem. Commun., 2012, 48, 3221–3223 RSC.
- X. Y. Han, X. Y. Song, F. B. Yu and L. X. Chen, Chem. Sci., 2017, 8, 6991–7002 RSC.
- X. Li, C. Liu, N. Gao, W. Sheng and B. Zhu, Chin. Chem. Lett., 2022, 33, 2527–2531 CrossRef CAS.
- M. Tian, M. Yang, Y. Liu and F.-L. Jiang, ACS Appl. Bio Mater., 2019, 2, 4503–4514 CrossRef CAS PubMed.
- H. J. Xiang, H. P. Tham, M. D. Nguyen, S. Z. F. Phua, W. Q. Lim, J. G. Liu and Y. L. Zhao, Chem. Commun., 2017, 53, 5220–5223 RSC.
- H. Jiang, G. Yin, Y. Gan, T. Yu, Y. Zhang, H. Li and P. Yin, Chin. Chem. Lett., 2022, 33, 1609–1612 CrossRef CAS.
- X. Li, H. Wang, Y. Zhang, Q. Cao and Y. Chen, Chin. Chem. Lett., 2021, 32, 1541–1544 CrossRef CAS.
- M. Reth, Nat. Immunol., 2002, 3, 1129–1134 CrossRef CAS PubMed.
- R. S. Balaban, S. Nemoto and T. Finkel, Cell, 2005, 120, 483–495 CrossRef CAS PubMed.
- M. Ushio-Fukai, Cardiovasc. Res., 2006, 71, 226–235 CrossRef CAS PubMed.
- H. Ischiropoulos and J. S. Beckman, J. Clin. Invest., 2003, 111, 163–169 CrossRef CAS PubMed.
- P. Sarchielli, F. Galli, A. Floridi, A. Floridi and V. Gallai, Amino Acids, 2003, 25, 427–436 CrossRef CAS PubMed.
- D. A. Wink, Y. Vodovotz, J. Laval, F. Laval, M. W. Dewhirst and J. B. Mitchell, Carcinogenesis, 1998, 19, 711–721 CrossRef CAS PubMed.
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- A. R. Lippert, G. C. V. De Bittner and C. J. Chang, Acc. Chem. Res., 2011, 44, 793–804 CrossRef CAS PubMed.
- G. C. Van de Bittner, C. R. Bertozzi and C. J. Chang, J. Am. Chem. Soc., 2013, 135, 1783–1795 CrossRef CAS PubMed.
- E. R. Stadtman, Free Radic. Res., 2006, 40, 1250–1258 CrossRef CAS PubMed.
- R. M. Touyz and E. L. Schiffrin, Histochem. Cell Biol., 2004, 122, 339–352 CrossRef CAS PubMed.
- H. Cai, Cardiovasc. Res., 2005, 68, 26–36 CrossRef CAS PubMed.
- K. J. Barnham, C. L. Masters and A. I. Bush, Nat. Rev. Drug Discov., 2004, 3, 205–214 CrossRef CAS PubMed.
- E. R. Stadtman, Science, 1992, 257, 1220–1224 CrossRef CAS PubMed.
- E. W. Childs, K. F. Udobi, J. G. Wood, F. A. Hunter, D. M. Smalley and L. Y. Cheung, Shock, 2002, 18, 423–427 CrossRef PubMed.
- X. Q. Chen, F. Wang, J. Y. Hyun, T. W. Wei, J. Qiang, X. T. Ren, I. Shin and J. Yoon, Chem. Soc. Rev., 2016, 45, 2976–3016 RSC.
- S. K. Yoo, T. W. Starnes, Q. Deng and A. Huttenlocher, Nature, 2011, 480, 109–U278 CrossRef CAS PubMed.
- A. Gomes, E. Fernandes and J. Lima, J. Biochem. Biophys. Methods, 2005, 65, 45–80 CrossRef CAS PubMed.
- K. J. Wang, D. Z. Xi, C. T. Liu, Y. H. Chen, H. Gu, L. Jiang, X. Q. Chen and F. Wang, Chin. Chem. Lett., 2020, 31, 2955–2959 CrossRef CAS.
- S. Wang, B. Zhu, B. Wang, X. Cao, L. Zhu, J.-T. Hou and L. Zeng, Chin. Chem. Lett., 2021, 32, 1795–1798 CrossRef CAS.
- J. Chiche, M. C. Brahimi-Horn and J. Pouyssegur, J. Cell. Mol. Med, 2010, 14, 771–794 CrossRef CAS PubMed.
- X. Weng, C. H. Chon, H. Jiang and D. Q. Li, Food Chem., 2009, 114, 1079–1082 CrossRef CAS.
- Y. Liu, Y. Yuan, X. Y. Lei, H. Yang, S. A. Ibrahim and W. Huang, Food Chem., 2013, 138, 2174–2179 CrossRef CAS PubMed.
- A. J. Kleinnijenhuis, Y. C. M. Staal, E. Duistermaat, R. Engel and R. A. Woutersen, Food Chem. Toxicol., 2013, 52, 105–112 CrossRef CAS PubMed.
- J. Lu, J. Y. Miao, T. Su, Y. Liu and R. Q. He, Biochim. Biophys. Acta Gen. Subjects, 2013, 1830, 4102–4116 CrossRef CAS PubMed.
- Z. X. Lin, W. H. Luo, H. Li and Y. Zhang, Toxicol. Lett., 2005, 159, 134–143 CrossRef CAS PubMed.
- Z. Q. Tong, J. L. Zhang, W. H. Luo, W. S. Wang, F. X. Li, H. Li, H. J. Luo, J. Lu, J. N. Zhou, Y. Wan and R. Q. He, Neurobiol. Aging, 2011, 32, 31–41 CrossRef CAS PubMed.
- J. H. Kim, L. B. Vinh, M. Hur, S. C. Koo, W. T. Park, Y. H. Moon, Y. J. Lee, Y. H. Kim, Y. C. Huh and S. Y. Yang, J. Microbiol. Biotechnol., 2021, 31, 1576–1582 CrossRef CAS PubMed.
- H. S. Seo and H. J. Kim, Bull. Korean Chem. Soc., 2017, 38, 1134–1137 CrossRef CAS.
- H. Jung, J. K. Kim and S. H. Ha, J. Korean Soc. Appl. Biol. Chem., 2011, 54, 678–684 CrossRef CAS.
- J. E. Coleman, Annu. Rev. Biophys. Biomol. Struct., 1992, 21, 441–483 CrossRef CAS PubMed.
- C. Liu, S. Yang, Y. Qiao, Y. Zhao, W. Wang, M. Jia, Y. He, Y. Zhou and L. Duan, Chin. Chem. Lett., 2021, 32, 1809–1813 CrossRef CAS.
- K. Ooi, K. Shiraki, Y. Morishita and T. Nobori, J. Clin. Lab. Anal., 2007, 21, 133–139 CrossRef CAS PubMed.
- R. E. Gyurcsanyi, A. Bereczki, G. Nagy, M. R. Neuman and E. Lindner, Analyst, 2002, 127, 235–240 RSC.
- E. Mutch, R. Nave, N. McCracken, K. Zech and F. M. Williams, Biochem. Pharmacol., 2007, 73, 1657–1664 CrossRef CAS PubMed.
- J. Creighton, B. Zhu, M. Alexeyev and T. Stevens, J. Cell Sci, 2008, 121, 110–119 CrossRef CAS PubMed.
- J. Wang, L. Wang, Z. Li, Y. M. Wang, K. Zhu and G. Mu, Eye Contact Lens, 2020, 46, 306–309 CrossRef PubMed.
- L. Feng, Z. M. Liu, J. Hou, X. Lv, J. Ning, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2015, 65, 9–15 CrossRef CAS PubMed.
- S. Kim, H. Kim, Y. Choi and Y. Kim, Chem. – Eur. J., 2015, 21, 9645–9649 CrossRef CAS PubMed.
- L. Peng, S. D. Xu, X. K. Zheng, X. M. Cheng, R. Y. Zhang, J. Liu, B. Liu and A. J. Tong, Anal. Chem., 2017, 89, 3162–3168 CrossRef CAS PubMed.
- X. J. Wang, H. Liu, J. W. Li, K. G. Ding, Z. L. Lv, Y. G. Yang, H. Chen and X. M. Li, Chem. Asian J., 2014, 9, 784–789 CrossRef CAS PubMed.
- D. Y. Li, Z. Li, W. H. Chen and X. B. Yang, J. Agric. Food Chem., 2017, 65, 4209–4215 CrossRef CAS PubMed.
- Q. Jin, L. Feng, D. D. Wang, Z. R. Dai, P. Wang, L. W. Zou, Z. H. Liu, J. Y. Wang, Y. Yu, G. B. Ge, J. N. Cui and L. Yang, ACS Appl. Mater. Interfaces, 2015, 7, 28474–28481 CrossRef CAS PubMed.
- C. B. Xiang, M. Dirak, Y. Luo, Y. L. Peng, L. T. Cai, P. Gong, P. F. Zhang and S. Kolemen, Mater. Chem. Front., 2022, 6, 1515–1521 RSC.
- N. Amend, J. Langgartner, M. Siegert, T. Kranawetvogl, M. Koller, H. John, C. Pfluegler, C. Moegele-Schmid, F. Worek, H. Thiermann and T. Wille, Arch. Toxicol., 2020, 94, 2239–2247 CrossRef CAS PubMed.
- S. He, S. Zhang, X. Zhao, X. Zhu, L. Chen and J. Cui, Chin. Chem. Lett., 2022, 33, 4233–4237 CrossRef CAS.
- M. Tsujimoto, Y. Goto, M. Maruyama and A. Hattori, Heart Fail. Rev., 2008, 13, 285–291 CrossRef CAS PubMed.
- M. Matsui, J. H. Fowler and L. L. Walling, Biol. Chem, 2006, 387, 1535–1544 CAS.
- T. Yamazaki, T. Akada, O. Niizeki, T. Suzuki, H. Miyashita and Y. Sato, Blood, 2004, 104, 2345–2352 CrossRef CAS PubMed.
- J. J. Reichling and M. M. Kaplan, Dig. Dis. Sci., 1988, 33, 1601–1614 CrossRef CAS PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209–215 CrossRef CAS PubMed.
- M. Bernardi, C. Maggioli and G. Zaccherini, Crit Care, 2012, 16, 211–217 CrossRef PubMed.
- H.-M. Lv, D.-H. Yuan, W. Liu, Y. Chen, C.-T. Au and S.-F. Yin, Sens. Actuators B Chem., 2016, 233, 173–179 CrossRef CAS.
- A. C. Sedgwick, W.-T. Dou, J.-B. Jiao, L. Wu, G. T. Williams, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, X.-P. He and T. D. James, J. Am. Chem. Soc., 2018, 140, 14267–14271 CrossRef CAS PubMed.
- M. L. Odyniec, S.-J. Park, J. E. Gardiner, E. C. Webb, A. C. Sedgwick, J. Yoon, S. D. Bull, H. M. Kim and T. D. James, Chem. Sci., 2020, 11, 7329–7334 RSC.
- E. M. Santos, W. Sheng, R. E. Salmani, S. T. Nick, A. Ghanbarpour, H. Gholami, C. Vasileiou, J. H. Geiger and B. Borhan, J. Am. Chem. Soc., 2021, 143, 15091–15102 CrossRef CAS PubMed.
- S. Tsuchiya, K. I. Sakai, K. Kawano, Y. Nakane, T. Kikuchi and T. Akutagawa, Chem. – Eur. J., 2018, 24, 5868–5875 CrossRef CAS PubMed.
- S. Zheng, Y. Fang, Y. Chen, Q. Kong, F. Wang and X. Chen, Spectrochim. Acta, Part A, 2022, 267, 120616–120623 CrossRef CAS PubMed.
- A. P. Demchenko, K.-C. Tang and P.-T. Chou, Chem. Soc. Rev., 2013, 42, 1379–1408 RSC.
- Y. Y. Ning, M. L. Zhu and J. L. Zhang, Coord. Chem. Rev., 2019, 399, 213028–213046 CrossRef CAS.
- H. D. Li, D. Kim, Q. C. Yao, H. Y. Ge, J. Chung, J. L. Fan, J. Y. Wang, X. J. Peng and J. Yoon, Angew. Chem., Int. Ed., 2021, 60, 17268–17289 CrossRef CAS PubMed.
- Z. H. Lei and F. Zhang, Angew. Chem., Int. Ed., 2021, 60, 16294–16308 CrossRef CAS PubMed.
- B. Feng, Y. Zhu, J. Wu, X. Huang, R. Song, L. Huang, X. Feng and W. Zeng, Chin. Chem. Lett., 2021, 32, 3057–3060 CrossRef CAS.
- K. Wang, B. Feng, G. Wang, J. Cui, L. Yang, K. Jiang and H. Zhang, Chem. Commun., 2022, 58, 2894–2897 RSC.
- K. Li, Q. Feng, G. Niu, W. Zhang, Y. Li, M. Kang, K. Xu, J. He, H. Hou and B. Z. Tang, ACS Sens., 2018, 3, 920–928 CrossRef CAS PubMed.
- X. Liu, N. Li, M. Li, H. Chen, N. N. Zhang, Y. L. Wang and K. B. Zheng, Coord. Chem. Rev., 2020, 404, 213109–213129 CrossRef CAS.
- R. Blieck, M. Taillefer and F. Monnier, Chem. Rev., 2020, 120, 13545–13598 CrossRef CAS PubMed.
- M. Liu, R. Xiao, B. Feng, D. Fan, S. Huang, A. Bi, S. Zhong, X. Feng, S. Liu and W. Zeng, Sens. Actuators B Chem., 2021, 342, 130038–130047 CrossRef CAS.
- J. Liang, B. Tang and B. Liu, Chem. Soc. Rev., 2015, 44, 2798–2811 RSC.
- M. H. Chua, H. Zhou, Q. Zhu, B. Z. Tang and J. W. Xu, Mater. Chem. Front., 2021, 5, 659–708 RSC.
- X. Tian, L. C. Murfin, L. L. Wu, S. E. Lewis and T. D. James, Chem. Sci., 2021, 12, 3406–3426 RSC.
- W. Feng, S. Feng and G. Feng, Anal. Chem., 2019, 91, 8602–8606 CrossRef CAS PubMed.
- D.-P. Li, X.-J. Han, Z.-Q. Yan, Y. Cui, J.-Y. Miao and B.-X. Zhao, Dyes Pigm., 2018, 151, 95–101 CrossRef CAS.
- D. Dahal, S. Pokhrel, L. McDonald, K. Bertman, S. Paruchuri, M. Konopka and Y. Pang, ACS Appl. Bio Mater., 2019, 2, 4037–4043 CrossRef CAS PubMed.
- Y. Liu, J. Nie, J. Niu, W. Wang and W. Lin, J. Mater. Chem. B, 2018, 6, 1973–1983 RSC.
- L. Wu, Y. Wang, M. Weber, L. Liu, A. C. Sedgwick, S. D. Bull, C. Huang and T. D. James, Chem. Commun., 2018, 54, 9953–9956 RSC.
- L. Wu, X. Tian, D. J. Lee, J. Yoon, C. S. Lim, H. M. Kim and T. D. James, Chem. Commun., 2021, 57, 11084–11087 RSC.
- L. Wu, H.-H. Han, L. Liu, J. E. Gardiner, A. C. Sedgwick, C. Huang, S. D. Bull, X.-P. He and T. D. James, Chem. Commun., 2018, 54, 11336–11339 RSC.
- G. L. Zeng, Z. H. Liang, X. Jiang, T. T. Quan and T. S. Chen, Chem. – Eur. J., 2022, 28, e202103241–e202103249 CAS.
- X. Liu, H. Tian, L. Yang, Y. Su, M. Guo and X. Song, Sens. Actuators B Chem., 2018, 255, 1160–1165 CrossRef CAS.
- L. Tang, M. Tian, H. Chen, X. Yan, K. Zhong and Y. Bian, Dyes Pigm., 2018, 158, 482–489 CrossRef CAS.
- M. Chen, Z. Liang, G. Zeng, Y. Wang, Z. Mai, X. Chen, G. Wu and T. Chen, Dyes Pigm., 2022, 198, 109995–110002 CrossRef CAS.
- L. He, X. Liu, Y. Zhang, L. Yang, Q. Fang, Y. Geng, W. Chen and X. Song, Sens. Actuators B Chem., 2018, 276, 247–253 CrossRef CAS.
- L. Wu, Q. Yang, L. Liu, A. C. Sedgwick, A. J. Cresswell, S. D. Bull, C. Huang and T. D. James, Chem. Commun., 2018, 54, 8522–8525 RSC.
- Y. Yang, F. Qiu, Y. Wang, Y. Feng, X. Song, X. Tang, G. Zhang and W. Liu, Sens. Actuators B Chem., 2018, 260, 832–840 CrossRef CAS.
- Y. Zhou, J. Yan, N. Zhang, D. Li, S. Xiao and K. Zheng, Sens. Actuators B Chem., 2018, 258, 156–162 CrossRef CAS.
- A. Bi, M. Liu, S. Huang, F. Zheng, J. Ding, J. Wu, G. Tang and W. Zeng, Chem. Commun., 2021, 57, 3496–3499 RSC.
- G. X. Yin, T. T. Niu, Y. B. Gan, T. Yu, P. Yin, H. M. Chen, Y. Y. Zhang, H. T. Li and S. Z. Yao, Angew. Chem., Int. Ed., 2018, 57, 4991–4994 CrossRef CAS PubMed.
- R. Zhang, J. X. Yong, J. L. Yuan and Z. P. Xu, Coord. Chem. Rev., 2020, 408, 213182–213222 CrossRef CAS.
- Y. Pan, L. Ban, J. Li, M. Liu, L. Tang and X. Yan, Dyes Pigm., 2022, 203, 110305–1103012 CrossRef CAS.
- H. Ren, F. Huo, Y. Zhang, S. Zhao and C. Yin, Sens. Actuators B Chem., 2020, 319, 128248–128255 CrossRef CAS.
- Y. Chen, X. Zhong, X. Yang, S. Zhu, Y. Jiang and C. Jin, Chem. Commun., 2021, 57, 8198–8201 RSC.
- M. Chang, C. Yan, L. Shi, D. Li, W. Fu and Z. Guo, Chin. Chem. Lett., 2022, 33, 762–766 CrossRef CAS.
- Y. Long, J. Liu, D. Tian, F. Dai, S. Zhang and B. Zhou, Anal. Chem., 2020, 92, 14236–14243 CrossRef CAS PubMed.
- L. Zhao, X. He, Y. Huang, J. Li, Y. Li, S. Tao, Y. Sun, X. Wang, P. Ma and D. Song, Sens. Actuators B Chem., 2019, 296, 126571–126576 CrossRef CAS.
- B. Feng, Y. Liu, S. Huang, X. Huang, L. Huang, M. Liu, J. Wu, T. Du, S. Wang, X. Feng and W. Zeng, Sens. Actuators B Chem., 2020, 325, 128786–128795 CrossRef CAS.
- H. Ren, F. Huo, T. Shen, X. Liu and C. Yin, Anal. Chem., 2021, 93, 12801–12807 CrossRef CAS PubMed.
- S. Zhang, D. Wu, X. Jiang, F. Xie, X. Jia, X. Song and Y. Yuan, Sens. Actuators B Chem., 2019, 290, 691–697 CrossRef CAS.
- X. Wei, Q. Wu, Y. Feng, M. Chen, S. Zhang, M. Chen, J. Zhang, G. Yang, Y. Ding, X. Yang, Q. Ye, Y. Zhang, Q. Gu, J. Wang, S. Wu, R. Pang and Y. Li, Sens. Actuators B Chem., 2020, 304, 127242–127249 CrossRef CAS.
- W. Yang, X. Zhao, J. Zhang, Y. Zhou, S. Fan, H. Sheng, Y. Cao and Y. Hu, Dyes Pigm., 2018, 156, 100–107 CrossRef CAS.
- Y. He, J. Yu, X. Hu, S. Huang, L. Cai, L. Yang, H. Zhang, Y. Jiang, Y. Jia and H. Sun, Chem. Commun., 2020, 56, 13323–13326 RSC.
- Y. Yang, C. Zhang, R. Pan, S. Zhang, S. Yao, Y. Tang, W. Zhu, L. Wang, W. Zhu, Y. Xu and X. Qian, Chin. Chem. Lett., 2020, 31, 125–128 CrossRef.
- P. Zhang, C. Fu, Q. Zhang, S. Li and C. Ding, Anal. Chem., 2019, 91, 12377–12383 CrossRef CAS PubMed.
- M. Tian, J. Sun, Y. Tang, B. Dong and W. Lin, Anal. Chem., 2018, 90, 998–1005 CrossRef CAS PubMed.
- Q. Kong, J. Wang, Y. Chen, S. Zheng, X. Chen, Y. Wang and F. Wang, Dyes Pigm., 2021, 191, 109349–109354 CrossRef CAS.
- W. Meng, Z. Pei, Y. Wang, M. Sun, Q. Xu, J. Cen, K. Guo, K. Xiao and Z. Li, J. Hazard. Mater., 2021, 410, 124811–124820 CrossRef CAS PubMed.
- X. Huang, Q. Lei, S. Huang, H. Zeng, B. Feng, Q. Zeng, Y. Hu and W. Zeng, Chem. Commun., 2021, 57, 6608–6611 RSC.
- Y. Huang, T. Lv, T. Qin, Z. Xu, L. Wang and B. Liu, Chem. Commun., 2020, 56, 11094–11097 RSC.
- J. Huang, P. Cheng, C. Xu, S. S. Liew, S. He, Y. Zhang and K. Pu, Angew. Chem., Int. Ed., 2022, 61, e202203235 CAS.
- P.-Y. Fu, B.-N. Li, Q.-S. Zhang, J.-T. Mo, S.-C. Wang, M. Pan and C.-Y. Su, J. Am. Chem. Soc., 2022, 144, 2726–2734 CrossRef CAS PubMed.
- J. Xue, Q. Liang, R. Wang, J. Hou, W. Li, Q. Peng, Z. Shuai and J. Qiao, Adv. Mater., 2019, 31, 1808242–1808250 CrossRef PubMed.
- D. M. Mayder, R. Hojo, W. L. Primrose, C. M. Tonge and Z. M. Hudson, Adv. Funct. Mater., 2022, 32, 2204087 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  *
*
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N,
N, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S, and –C
S, and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).45–47 Upon irradiation, ESIPT emitters absorb the energy to achieve E* (excited state of Enol form). Then, ESIPT, a photoisomerization process, appears to form K* (excited state of Keto form) in less than 1 ps.48,49 The decay of K* emits a much longer fluorescence than E* owing to the inner ICT effect arising from the electron-withdrawing property of ketone. Finally, the ground state of the Enol form (E) changes to its original state (ground state of the Keto form, K) called GSIPT (Ground-state Intramolecular Proton Transfer) (Fig. 1).49,50 Benefitting from the transfer from the Enol form to the Keto form, the ESIPT emitters with two emissions could assist in obtaining ratiometric sensing, which could reduce the interference from the background.48,50,51 Simultaneously, compared with traditional probes, such as fluorescein and rhodamine, annoying self-reabsorption and inner-filter effects are prevented in ESIPT emitters.52–54 Consequently, ESIPT emitters are extensively used in biomarker detection.
O).45–47 Upon irradiation, ESIPT emitters absorb the energy to achieve E* (excited state of Enol form). Then, ESIPT, a photoisomerization process, appears to form K* (excited state of Keto form) in less than 1 ps.48,49 The decay of K* emits a much longer fluorescence than E* owing to the inner ICT effect arising from the electron-withdrawing property of ketone. Finally, the ground state of the Enol form (E) changes to its original state (ground state of the Keto form, K) called GSIPT (Ground-state Intramolecular Proton Transfer) (Fig. 1).49,50 Benefitting from the transfer from the Enol form to the Keto form, the ESIPT emitters with two emissions could assist in obtaining ratiometric sensing, which could reduce the interference from the background.48,50,51 Simultaneously, compared with traditional probes, such as fluorescein and rhodamine, annoying self-reabsorption and inner-filter effects are prevented in ESIPT emitters.52–54 Consequently, ESIPT emitters are extensively used in biomarker detection.









![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N in the excited state contributed to Probe 33 emitting double emission (Enol and Keto forms). Conversely, the presence of GSH prevented the formation of the hydrogen bond between “C
N in the excited state contributed to Probe 33 emitting double emission (Enol and Keto forms). Conversely, the presence of GSH prevented the formation of the hydrogen bond between “C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N” and “COOH”, which blocked ESIPT and stabilized the conformation of the probe-GSH adduct. Consequently, this process assisted in emitting the blue-shifted emission arising from the ICT process.
N” and “COOH”, which blocked ESIPT and stabilized the conformation of the probe-GSH adduct. Consequently, this process assisted in emitting the blue-shifted emission arising from the ICT process.


















