Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quorum sensing communication between lipid-based artificial cells†
Antoni
Llopis-Lorente
 ab,
Bastiaan C.
Buddingh'
ab,
Bastiaan C.
Buddingh'
 a,
R.
Martínez-Máñez
a,
R.
Martínez-Máñez
 b,
Jan C. M.
van Hest
b,
Jan C. M.
van Hest
 *a and
Loai K. E.
Abdelmohsen
*a and
Loai K. E.
Abdelmohsen
 *a
*a
aDepartment of Chemical Engineering and Chemistry, Department of Biomedical Engineering, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Het Kranenveld 14, 5600 MB, Eindhoven, The Netherlands. E-mail: J.C.M.v.Hest@tue.nl; l.k.e.a.abdelmohsen@tue.nl
bInstituto Interuniversitario de Investigación de Reconocimiento Molecular y Desarrollo Tecnológico (IDM), Universitat Politècnica de València, Universitat de València; CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Insituto de Salud Carlos III, Spain
First published on 9th December 2022
Abstract
Population behavior based on quorum sensing communication is a key property of living microorganisms. Here, we show quorum sensing behavior in an artificial cell population consisting of giant lipid vesicles loaded with sender–receiver machinery (enzymes and responsive biomolecules). Our system allows the examination of the collective output based on cell density, fuel concentration and proximity, which are important factors controlling natural quorum sensing behavior.
Living organisms employ chemical communication, via the exchange of chemical messengers, to transmit information between cells and to induce collective behavior. One prominent example of chemical communication is the collective behavior of bacteria via quorum sensing.1 For quorum sensing, individual bacterial cells produce signaling molecules that freely diffuse across their membrane to the external milieu. At the same time, bacteria sense the signaling molecules secreted by peers in their surroundings. An increase in the local cell density results in an increase in the concentration of signaling molecules, which upon reaching a threshold, triggers the activation of certain functions across the bacterial population. Thus, quorum sensing allows individual bacteria to sense their relative population density in the environment and act accordingly. For instance, Vibrio fischeri bacteria produce collective luminescence by population-dependent quorum sensing. Across the bacterial world, the outputs of quorum sensing are diverse, e.g. ranging from swarm motility to virulence and biofilm formation.2
Inspired by living systems, the design of artificial compartments (such as liposomes, coacervates, polymersomes, etc.)3 which can mimic cellular processes (such as protein expression, motion, adaptivity)4 has attracted much attention over the last few years, due to fundamental interest and technological potential. Notwithstanding, research has been mostly directed toward the utility of single entities; whereas the ability of artificial particles to autonomously communicate in a collective fashion remains underexplored.5 Some recent studies presented artificial cells able to communicate with another population of artificial cells or with bacteria.6–8 For instance, Mansy and co-workers prepared liposomes loaded with protein-expression machinery able to sense and produce bacterial quorum signaling molecules.9 However, lipid-based artificial cells capable of communication with their artificial peers that results in quorum sensing behavior have not been reported. Harnessing such behavior in artificial cell populations would pave the way for the systematic study and engineering of collective behavior in artificial cell communities.
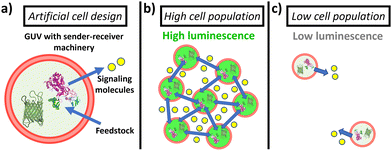
Here, we establish a general strategy for quorum communication and synergistic sensing in consortia of artificial cells. Specifically, we constructed giant unilamellar vesicles (GUVs) equipped with (i) sender machinery (able to produce diffusive signaling molecules in response to a stimulus) and (ii) receiver machinery (able to recognize the signaling molecules and produce an output response). As sender machinery, we opted for the use of enzymes, as they selectively recognize the presence of their substrate and transform it into new products that can act as signaling molecules. In particular, in this study, we selected to encapsulate urease as it catalyzes the conversion of urea into CO2 and NH3 (which acts as membrane-permeable signaling molecule, and raises the pH (Fig. S1, ESI†)). On the other hand, as receiver machinery, we opted for the use of different pH-responsive (bio)molecules that exhibit a fluorescent output as a result of the communication process. In this way, the assembled GUVs containing sender and receiver machinery in their lumen show behavior reminiscent to bacterial quorum sensing communication—GUVs in highly populated consortia display an upregulation of their individual luminescence as a result of communication via the secretion of signaling molecules; in turn, the activation is significantly lower when the number of peers is reduced (Fig. 1). Such platform allows us to study the collective response depending on their population density, fuel concentration and spatial proximity.
For the assembly of GUVs, we followed a methodology based on the previously reported droplet transfer technique.10 Briefly, a 10 mM lipid solution (DOPC/POPC/Chol in 35/35/30 molar ratio) in paraffin was emulsified upon addition of an aliquot of aqueous solution (containing the vesicle inner phase in acetate buffer at pH 5) – thus, creating water-in-oil droplets stabilized by a monolayer of lipids. Next, the emulsion was placed on top of an aqueous solution, and centrifuged—the water droplets passed through the interface covered with phospholipids, yielding the loaded GUVs at the bottom of the tube, which were then washed by centrifugation. We found that incorporating a pegylated lipid (1 mol%) is key for preventing unwanted GUV aggregation during the washing step (Fig. S2, ESI†). This process generated GUVs with a size of 12 ± 5 μm (Fig. S3, ESI†). Successful loading of cargo and removal of unencapsulated material was confirmed by incorporating a fluorescent marker in the loading solution (Fig. S4, ESI†).
Green fluorescent protein (GFP) emission is typically used in the read out of quorum sensing communication in bacteria.11 Inspired by this, we first aimed at developing GUVs containing recombinant GFP that could respond with an upregulation of luminescence to an increase in pH. As observed, GFP exhibited a steep linear increase in emission in the pH 5 to pH 6.5 range (Fig. S5, ESI†), and a clear difference before (at pH 5) and after NH3 (5 mM) addition (Fig. S6, ESI†). Such response is ascribed to structural changes in the hydrogen bonds of GFP's amino acid chains with its phenolate chromophore.12 Next, GUVs encapsulating both urease (as sender machinery) and recombinant GFP (as receiver machinery) were assembled; whilst incorporating DOPE-RhB as a membrane marker. Upon addition of NH3 (signaling molecule) to the medium, a clear increase of GFP luminescence in the GUV lumen was observed (Fig. S7, ESI†), thus, confirming the ability of GFP-loaded GUVs to respond to NH3. As indicated in previous studies, NH3 and H+ (in the form of acetic acid) have a high permeability (1 × 10−4 and 6 × 10−5 m s−1, respectively) and rapidly cross through lipid membranes, in a matter of (milli)seconds.13
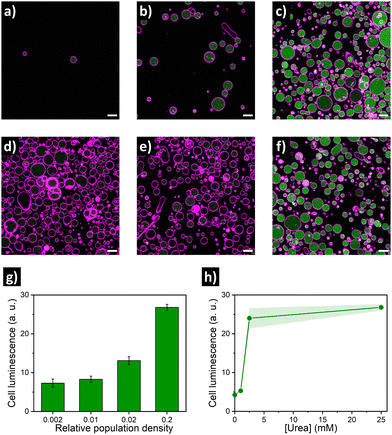
Encouraged by these findings, we then aimed at establishing the quorum sensing model of communication between artificial cells. For these experiments, a batch of urease/GFP-loaded GUVs was divided into several aliquots with different dilution factors (5-, 50-, 100- and 500-fold dilution; referred below as population density (PD) of 0.2, 0.02, 0.01 and 0.002, respectively), and then samples were simultaneously placed in microscope chambers with 25 mM of urea (Fig. 2a–c). In the highly populated consortia (PD = 0.2), microscope images of the membrane marker channel revealed the coexistence of a high number of GUVs in close proximity. In turn, an increase in distance between individual GUVs and a decrease of the number of GUVs per frame upon increasing the dilution factor was observed. Interestingly, visualization of the GFP channel suggested an activation of lumen luminescence for GUVs in the presence of urea in highly populated consortia (Fig. 2c)—in contrast, GFP signal was noticeably smaller for diluted GUV samples (Fig. 2a and b). As shown in Fig. 2g, quantification of GFP luminescence (using ImageJ) revealed a ca. 4-fold increase in the mean cell luminescence under highly populated conditions (PD = 0.2) as compared to the lowest population density (PD = 0.002), with intermediate values at medium population densities.
As the quorum sensing-induced activation should occur in response to the presence of neighboring individuals, the response of the consortia is dependent on the presence of feedstock molecules. Therefore, we parallelly evaluated the effect of varying urea concentration (as fuel of the metabolic process). To this end, additional aliquots of highly populated GUV samples (PD = 0.2) were transferred to different microscope chambers and their media were supplemented with different concentrations of urea. A very low GFP activation signal was observed for GUVs in the absence of urea (Fig. 2d), and addition of 1 mM urea did not produce a significant increase (Fig. 2e). In contrast, a noticeable increase occurred in the presence of 2.5 mM (Fig. 2f), reaching values comparable to 25 mM urea (Fig. 2h) – which indicates that a minimum of fuel concentration should be present for triggering quorum sensing; if fuel concentration is low, cell activation does not happen even in highly populated consortia. Altogether these results validated the establishment of quorum sensing-like communication between artificial cells – the engineered GUVs demonstrated the ability to transform urea from their environment into signaling molecules, inducing a collective increase in luminescence when forming part of highly populated consortia.
After validating this model system using GFP as a reporter molecule, we decided to explore the possibility of using this concept of quorum sensing communication to induce the structural rearrangement of DNA in the interior of artificial cells. DNA nanoswitches are stimulus-responsive DNA-based nanostructures whose structural rearrangement can regulate functions such as cargo loading and release, and transcription–translation processes.14,15 In particular, we employed a Cy3–Cy5-labelled DNA responsive sequence that forms a triplex structure at acidic pH, and rearranges to duplex at basic pH (Fig. 3 and Fig. S8, ESI†), as previously reported by Ricci et al.16 Upon excitation of the Cy3 fluorophore at 530 nm, a relatively low Cy3 signal and a high Cy5 signal were observed at pH 5 due to Cy3–Cy5 proximity in the triplex structure inducing FRET transfer. Upon addition of NH3 (signaling molecule), the nanostructure opened, and a clear increase in emission in the Cy3 channel was observed, as the Cy5-labelled end separated.
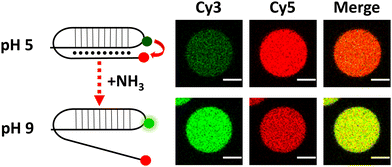 | ||
| Fig. 3 Schematic of DNA reorganization (left) and corresponding changes in confocal fluorescence emission (right) of DNA-loaded GUVs. Scale bar = 5 μm. | ||
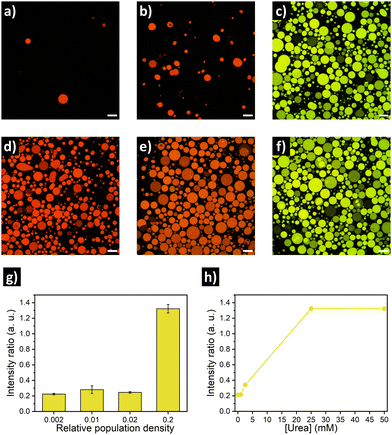
Similar to the experiments with GFP, rearrangement of the DNA nanoswitch as output of quorum sensing was evaluated by placing urease/DNA-loaded GUVs in microscope chambers under different dilution factors (Fig. 4a–c, g). A clear fluoresence activation (indicating opening of the DNA nanostructure) was observed in highly populated consortia (Fig. 4c). In turn, intermediate population density (PD = 0.02) displayed no activation, just as the low populated consortia (PD = 0.002). This resulted in a ca. 6-fold increase in Cy3–Cy5 intensity ratio in highly populated consortia (Fig. 4g). When we evaluated the response depending on the concentration of urea, no activation was registered at 1 mM urea – and 2.5 mM produced minimal activation, far from the activation reached at 25 mM and 50 mM of urea (Fig. 4h). Thus, DNA-GUVs required more urea than GFP-GUVs to reach full activation – this difference can be ascribed to the different pH-responsiveness of the DNA nanoswitch (Fig. S8, ESI†) compared to GFP (Fig. S5, ESI†). These results, together with the programability of DNA nanostructures,17,18 suggest the possibility to engineer artificial cell consortia capable of responding to specific conditions. Reorganization of the DNA nanostructure is a dynamic process, which in our system implied a fluorescence change with a sharp transition after ca. 60 min of incubation (under quorum sensing communication conditions), while the pH raised to ca. 9 (Fig. S9, ESI†).
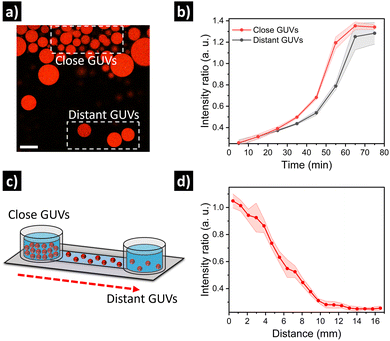
Finally, we set out to assess if the relative spatial location of GUVs in quorum sensing communities could have an impact on their individual output. For this, we prepared GUVs loaded with urease and dextran-RhB-FITC (as pH-responsive probe, Fig. S10, ESI†). In a first approximation, an aliquot of GUVs was carefully placed in the center of a microscope chamber (5-fold dilution) containing 25 mM of urea, and then we compared GUVs located at the edge of the cluster versus GUVs located further away in the same field of view (Fig. 5a). Interestingly, we observed a delayed response in GUVs located further away from the cluster (Fig. 5b and Fig. S11, ESI†). This can be ascribed to higher local production of NH3 by GUVs in close proximity, that diffuses to more distant GUVs after some minutes. In a second approximation, in order to have better control over the spatial distribution of GUVs and the diffusion of signal molecules, GUVs were loaded in a microfluidic channel device (17 mm) under diluted conditions with 25 mM of urea, and then a highly populated area was created by depositing concentrated GUVs at the end of the channel (Fig. 5c). After incubation (12 h), the signal from GUVs located along the channel was registered by acquiring micrographs of subsequent positions. Interestingly, a clear activation was observed for GUVs located in the highly populated area indicating quorum sensing, while activation diminished with distance (Fig. 5d and Fig. S12, ESI†). Thus, the highly populated consortia is able to produce a signal gradient that diffuses through the channel, creating a spatial activation pattern according to the position of distant GUVs.
In conclusion, we demonstrated the engineering of quorum sensing communication between artificial cells, based on GUVs loaded with sender–receiver machinery. As a model system, we prepared urease-loaded GUVs equipped with different responsive biomolecules (such as GFP or DNA nanostructures) that are able to (i) exhibit tuneable behaviour depending on their population density and fuel concentration in the medium, and (ii) display spatiotemporal activation patterns depending on their relative distance. We think this is a versatile approach for the design and investigation of collective behavior in artificial cell communities, and may inspire further advances in the areas of chemical communication and bioinspired materials.
The authors acknowledge funding from the Dutch Ministry of Education, Culture and Science (Gravitation program 024.001.035 and Spinoza premium), ERC Advanced Grant Artisym (694120), ERC Advanced Grant Edison (101052997); the Spanish Ministry of Science and Innovation, AEI, and FEDER-EU (project PID2021-126304OB-C41), and the GVA (Project CIPROM/2021/007). A. L.-L. thanks the MSCA Cofund project oLife (847675), which has received funding from the EU's Horizon 2020 research and innovation program; and the Spanish Ministry of Universities for his María Zambrano fellowship funded by Next Generation EU from the European Union. Dr W. Altenburg is thanked for supplying GFP. Fig. 1 was created using Biorender.com.
Conflicts of interest
There are no conflicts to declare.References
- K. Stephens and W. E. Bentley, Trends Microbiol., 2020, 28, 633–643 CrossRef CAS PubMed
.
- M. E. Taga and B. L. Bassler, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14549–14554 CrossRef CAS PubMed
.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC
.
- B. C. Buddingh and J. C. M. Van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS PubMed
.
- B. De Luis, A. Llopis-Lorente, F. Sancenón and R. Martínez-Máñez, Chem. Soc. Rev., 2021, 50, 8829–8856 RSC
.
- B. C. Buddingh, J. Elzinga and J. C. M. van Hest, Nat. Commun., 2020, 11, 1652 CrossRef CAS PubMed
.
- Y. Elani, Angew. Chem., Int. Ed., 2021, 11, 5602–5611 CrossRef PubMed
.
- G. Rampioni, F. D’Angelo, M. Messina, A. Zennaro, Y. Kuruma, D. Tofani, L. Leoni and P. Stano, Chem. Commun., 2018, 54, 2090–2093 RSC
.
- R. Lentini, N. Y. Martín, M. Forlin, L. Belmonte, J. Fontana, M. Cornella, L. Martini, S. Tamburini, W. E. Bentley, O. Jousson and S. S. Mansy, ACS Cent. Sci., 2017, 3, 117–123 CrossRef CAS PubMed
.
- S. Pautot, B. J. Frisken and D. A. Weitz, Langmuir, 2003, 19, 2870–2879 CrossRef CAS
.
- Y. Bai, S. N. Patil, S. D. Bowden, S. Poulter, J. Pan, G. P. C. Salmond, M. Welch, W. T. S. Huck and C. Abell, Int. J. Mol. Sci., 2013, 14, 10570–10581 CrossRef PubMed
.
- M. A. Elsliger, R. M. Wachter, G. T. Hanson, K. Kallio and S. J. Remington, Biochemistry, 1999, 38, 5296–5301 CrossRef CAS PubMed
.
-
(a) Y. Miele, Z. Medveczky, G. Holló, B. Tegze, I. Derényi, Z. Hórvölgyi, E. Altamura, I. Lagzi and F. Rossi, Chem. Sci., 2020, 11, 3228–3235 RSC
; (b) S. Liu, P. C. Hu and N. Malmstadt, Biophys. J., 2011, 101, 700–708 CrossRef PubMed
.
- E. Del Grosso, G. Ragazzon, L. J. Prins and F. Ricci, Angew. Chem., Int. Ed., 2019, 58, 5582–5586 CrossRef CAS PubMed
.
- L. Heinen and A. Walther, Chem. Sci., 2017, 8, 4100–4107 RSC
.
- T. Patiño, A. Porchetta, A. Jannasch, A. Lladó, T. Stumpp, E. Schäffer, F. Ricci and S. Sánchez, Nano Lett., 2019, 19, 3440–3447 CrossRef PubMed
.
- A. Idili, A. Vallée-Bélisle and F. Ricci, J. Am. Chem. Soc., 2014, 136, 5836–5839 CrossRef CAS PubMed
.
- R. Rubio-Sanchez, G. Fabrini, P. Cicuta and L. Di Michele, Chem. Commun., 2021, 57, 12725–12740 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods, and supplementary figures. See DOI: https://doi.org/10.1039/d2cc05367e |
| This journal is © The Royal Society of Chemistry 2023 |