DOI:
10.1039/D3BM01119D
(Paper)
Biomater. Sci., 2023,
11, 7062-7066
Cationic copolymer and crowding agent have a cooperative effect on a Na+-dependent DNAzyme†
Received
4th July 2023
, Accepted 29th August 2023
First published on 30th August 2023
Abstract
DNAzymes are promising agents for theranostics and biosensors. Sodium dependent DNAzymes have been developed for sensing and imaging of Na+, but these DNAzymes have low catalytic activity. Herein, we demonstrate that a molecular crowded environment containing 10 to 40 wt% PEG enhanced the catalytic activity of a Na+-dependent DNAzyme, EtNa, although dextran did not. The cationic copolymer poly(L-lysine)-graft-poly(ethylene glycol) at 0.03 wt% (0.3 g L−1) enhanced the reaction rate of EtNa by 10-fold, which is similar to the acceleration induced by 15 wt% (150 g L−1) PEG. A cooperative impact of the copolymer and crowding agent was observed: the combination resulted in an impressive 46-fold acceleration effect. Thus, the use of a cationic copolymer and a crowding agent is a promising strategy to improve the activity of Na+-dependent DNAzyme-based nanomachines, biosensors, and theranostics, especially in environments lacking divalent metal ions.
1. Introduction
DNAzymes are DNA molecules that can catalyze various reactions such as site-specific RNA cleavage.1–3 Due to their programmability, excellent stability, and low cost, DNAzymes are used in nanotechnology applications,4–7 biosensors,8–11 and theranostics.12–14 The RNA cleavage by a DNAzyme involves the formation of a DNAzyme/substrate (ES) complex, chemical cleavage, and product dissociation. Like ribozymes, DNAzymes usually require divalent metal ions such as Mg2+,15 Pb2+,16 and Zn2+ ac cofactors.17 Sodium-dependent DNAzymes have also been identified for sensing of Na+;18–21 however, low catalytic activities of these DNAzymes have limited their applications.
The environment within a living cell differs significantly from that of aqueous solution. High concentrations (20%–40%) of various macromolecules, such as proteins and nucleic acids, create a molecularly crowded cellular environment that is abundant in nucleic acid–protein interactions,22,23 which impacts various biological processes including enzymatic reactions. In vitro experiments are usually conducted in dilute aqueous solutions that do not accurately mimic the cellular environment. Molecular crowding can affect the kinetic and thermodynamic properties of nucleic acids, thereby improving the catalytic efficiency of nucleic acid enzymes. For example, the presence of a molecular crowding agent such as polyethylene glycol (PEG) was shown to increase the reaction rate of a hairpin ribozyme by enhancing the stability of the docked, active state, which is a compact conformation.24 The addition of PEG also increased the catalytic activity of the 17E DNAzyme by enhancing the stability of the active conformation.25
Our group has studied the interactions between nucleic acids and the cationic copolymer, poly(L-lysine)-graft-dextran (PLL-g-Dex), which is composed of a cationic main chain and a hydrophilic side chain (Fig. 1A, for a recent review see ref. 26). PLL-g-Dex enhances the catalytic activities of two kinds of divalent metal ion-dependent DNAzymes, 8–17 and 10–23, by promoting the DNAzyme/substrate association under multiple-turnover conditions.27,28 Herein, we describe our investigation of the effects of cationic copolymers and molecular crowding on the Na+-dependent DNAzyme EtNa. The EtNa DNAzyme, reported by Liu and coworkers, is highly specific for Na+, and its activity is significantly promoted in an alcoholic environment.19 We found that the cationic copolymer and molecular crowding effectively accelerated the reaction rate of EtNa, and a cooperative effect was observed.
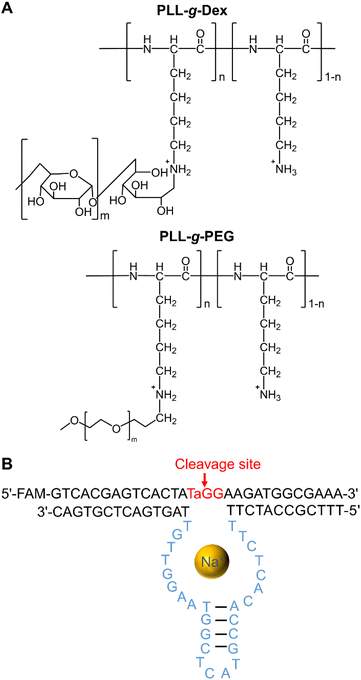 |
| | Fig. 1 (A) Chemical structures of cationic copolymers, PLL-g-Dex and poly(L-lysine)-graft-poly(ethylene glycol) (PLL-g-PEG). (B) Secondary structure of EtNa DNAzyme. | |
2. Materials and methods
2.1 Materials
Poly(L-lysine hydrobromide) (Mw = 1.5 × 104) and PEG were purchased from Sigma-Aldrich. Dextran (Mn = 8.0 × 103–1.2 × 104) was provided by Funakoshi Co. NaCl was obtained from Nacalai Tesque, Inc. MES was purchased from DOJINDO Laboratories. All oligonucleotides were HPLC-grade and purchased from Fasmac Co., Ltd. The cationic copolymers were synthesized by reductive amination according to our previous publication.29 Synthesis is described in detail in the Experimental Methods section of the ESI.† PLL-g-Dex with 90 wt% grafted dextran and PLL-g-PEG with 80 wt% grafted PEG were used in this study.
2.2 Gel electrophoresis-based activity assays
A reaction mixture containing 2 μM DNAzyme and 1 μM FAM-labeled substrate was incubated in MES buffer (50 mM MES, 120 mM NaCl, pH 6.0) at 25 °C in the absence or presence of polymers. For the reactions in the presence of the cationic copolymer (PLL-g-Dex or PLL-g-PEG), the molar ratio of positively charged amino groups of the copolymer to negatively charged phosphate groups of DNA was fixed at 2. For the reactions with PEG or Dex, mixed reaction solutions were prepared at different concentrations by weight (10–40%). For the reaction with ethanol (EtOH), the mixed reaction solution was prepared at different concentrations by volume (10–54%). After indicated incubation times, 5 μL of the reaction mixture was added to 20 μL formamide quenching buffer (95% formamide, 5% TrackIT Cyan/Yellow Loading Buffer). For the reactions in the presence of the cationic copolymer, 5 μL of 1 mM anionic poly(vinylsulfonic acid) was added to the sample. Then 10 μL of sample was analyzed by 15% denaturing PAGE at 100 V for 70 min. Gels were visualized using a PharoFX molecular imager (Bio-Rad) and quantified using ImageJ. The observed reaction rate (kobs) was calculated by fitting the percentage of the cleaved substrate and reaction time (in hours) to the following equation:
where P∞ is the reaction plateau, and Pt is the cleavage percentage at time t.
2.3 Measurement of melting temperature (Tm)
The full-DNA substrate in which the deoxy A nucleotide replaced the ribo A nucleotide was used to avoid the cleavage reaction. The EtNa DNAzyme (1 μM) and the full-DNA substrate (1 μM) were mixed in MES buffer (50 mM MES, 120 mM NaCl, pH6.0) without any additives or with PLL-g-PEG, PLL-g-Dex, 10% PEG or 10% EtOH as an additive, respectively. The mixture was incubated at 20 °C for 30 min to form the ES complex. The Tm was determined by measuring the UV melting curves on a V730 UV-visible spectrometer (Jasco).
2.4 Measurement of circular dichroism (CD) spectra
Samples containing a 2 μM EtNa/full-DNA substrate complex without or with additives were analyzed on a J-820 CD spectrometer (Jasco) using a 1 cm quartz cell and a scan rate of 100 nm min−1 at 25 °C. The CD spectra was obtained by averaging over 5 accumulations for each experiment. The CD spectra for two independent experiments were measured and averaged.
3. Results and discussion
3.1 Effect of molecular crowding on DNAzyme activity
The catalytic core of the EtNa DNAzyme is composed of a hairpin loop and two bulges (Fig. 1B). Its substrate strand contains a single riboadenine (rA) linkage, which is the cleavage site. This Na+-dependent DNAzyme has a reaction rate of 1.138 h−1 in the presence of 54% ethanol (Fig. S1†).19 In aqueous solution with 120 mM NaCl, the activity of EtNa was very low with a kobs of 0.010 h−1 (Fig. 2A, red line), which is consistent with a previous report.19 The effect of molecular crowding on EtNa was examined using PEG or Dex as the crowding agent under single-turnover conditions with 2 μM EtNa and 1 μM substrate. The reaction rates in 20% PEGs with different molecular weights (PEG550, PEG2000, and PEG5000) were almost the same (Fig. S2†), indicating that the molecular weight of the crowding agent does not influence the activity. PEG2000 was used in the subsequent experiments. The activity of EtNa was dependent on the concentration of PEG, as higher concentrations of PEG led to an increase in DNAzyme activity (Fig. 2A). Rate enhancement was minimal when Dex was employed as the crowding agent (Fig. 2B). The analysis of Tm and CD spectra showed that the presence of PEG did not influence the stability and conformation of the ES complex (Fig. S3 and S4†). However, the presence of crowding agents alters the physical properties, including the dielectric constant and viscosity, of a solution.30,31 The electrostatic binding of metal ions to nucleic acids is enhanced when the dielectric constant is reduced. A 20% solution of PEG2000 has a relative dielectric constant of ∼60, the relative dielectric constant of 20% Dex is ∼65, and that of pure water is ∼80 at 25 °C.32 Thus, the presence of the crowding agent might increase the amount of the catalytically active ES complex. Moreover, PEG has higher immiscibility than Dex with DNA.33–35 This property likely contributes to a stronger excluded-volume effect of PEG compared to that of Dex, making PEG a better inducer of the assembly of DNA molecules.
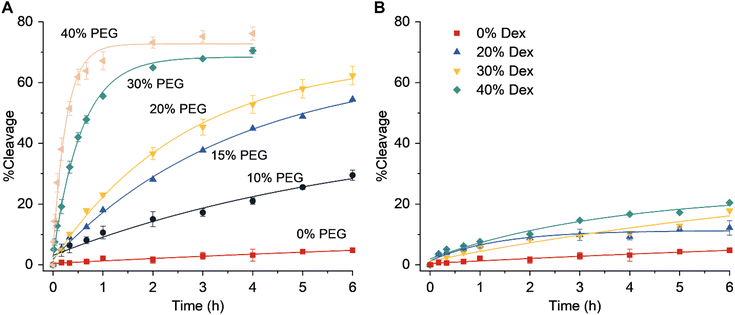 |
| | Fig. 2 Effect of molecular crowding on DNAzyme activity. The dependence of the DNAzyme activity on the amount of (A) PEG or (B) Dex. Error bars were derived from two independent experiments. | |
3.2 Effects of cationic copolymers on DNAzyme activity
Next, we investigated the effects of two kinds of cationic copolymers, PLL-g-PEG and PLL-g-Dex (Fig. 1A), on the reaction catalyzed by EtNa. The cleavage reaction of EtNa in the presence of 0.03 wt% PLL-g-PEG had a kobs of 0.123 h−1, which is more than 10-fold faster than the rate without a copolymer (Fig. 3) and comparable to that in a solution containing 15 wt% PEG (0.134 h−1) (Fig. 4). This result demonstrated that the cationic copolymer more effectively enhanced the cleavage activity of EtNa DNAzyme than the crowding agent PEG. A similar rate enhancement was observed upon the addition of PLL-g-Dex (Fig. 3), indicating that the copolymer accelerated the DNAzyme activity by a mechanism that differed from the enhancement due to PEG. The electrostatic interaction of the cationic backbone of the copolymer with DNA likely played a role in the acceleration, since the two copolymers have the same cationic main chain.36 We previously reported that PLL-g-Dex improved the activity of the 10–23 DNAzyme under multiple-turnover conditions.27 However, PLL-g-Dex does not promote the chemical cleavage step of 10–23 since the addition of PLL-g-Dex does not enhance the reaction rate under single-turnover conditions.27 In contrast, the activity of EtNa was enhanced ∼10-fold by copolymers under single-turnover conditions.
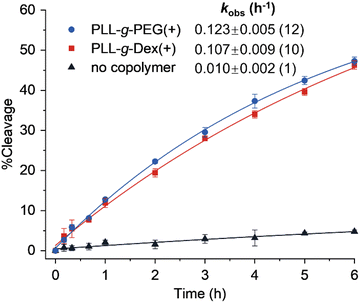 |
| | Fig. 3 Cleavage kinetics of EtNa DNAzyme in the absence or presence of a cationic copolymer. Error bars were derived from two independent experiments. | |
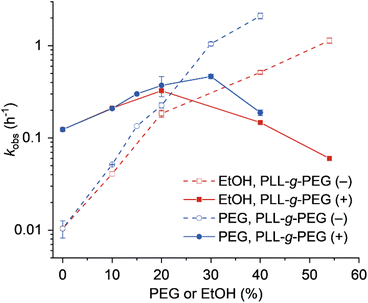 |
| | Fig. 4 Plot of kobs values against the amount of PEG or EtOH in the absence or presence of PLL-g-PEG. Error bars were derived from two independent experiments. | |
The Tm of the ES complex was 53 °C in the absence of a copolymer, and the addition of the cationic copolymers increased the Tm to more than 65 °C (Fig. S3†), suggesting that the copolymer stabilized the ES complex. The reaction temperature (25 °C) was much lower than the Tm values, indicating thorough association of DNAzymes with substrates under our reaction conditions. Therefore, the copolymers promoted the chemical cleavage step catalyzed by EtNa. The CD spectra of the ES complex exhibited a negative band at ∼250 nm and a positive band at ∼280 nm, and the presence of the copolymers did not alter the CD spectra (Fig. S4†), indicating that the conformation of the ES complex was not significantly influenced by the interactions of the copolymers. However, the CD spectra were not sensitive enough for the determination of the local structures of the ES complex. It is possible that the binding of the copolymer to EtNa alters the local microenvironment of the ES complex, thereby promoting the cleavage reaction.
3.3 Cooperativity of cationic copolymers with crowding agents
As the mechanisms of copolymer-mediated enhancement and crowding agent-mediated enhancement of EtNa activity likely differ, we speculated that the copolymer and the crowding agent might act cooperatively. To test this, we analyzed the EtNa-catalyzed reaction in the presence of PLL-g-PEG and PEG or EtOH. The reaction rate was increased by the addition of PLL-g-PEG when the concentration of PEG or EtOH was lower than 30% or 20%, respectively (Fig. 4). The reaction rate reached maxima in the presence of PLL-g-PEG with 30% PEG (kobs of 0.464 h−1) and with 20% EtOH (kobs of 0.325 h−1). This indicated that the cooperativity of the copolymer and crowding agents resulted in a maximum acceleration effect of 46-fold compared with that in the aqueous solution. Further increase in the concentration of the crowding agent, however, resulted in the decrease of the reaction rate. This might be due to the excessively enhanced interactions of the cationic copolymer with DNA in the solution with a low dielectric constant. Consistently, the lysine homopolymer that has stronger interactions with DNA than the cationic copolymer inhibited the cleavage reaction of EtNa (Fig. S5†).
4. Conclusion
In this study, we investigated the impacts of molecular crowding and cationic copolymer on a Na+-dependent RNA-cleaving DNAzyme, EtNa. Compared with its activity in aqueous solution, the cleavage activity of EtNa DNAzyme was enhanced in the molecular crowded environment containing PEG, likely due to the reduced dielectronic constant. The catalytic activity of EtNa increased as the amount of PEG was increased. The cationic copolymer enhanced the DNAzyme activity through its electrostatic interaction with DNA. The copolymer at 0.03 wt% accelerated the reaction rate of EtNa by 10-fold, similar to the acceleration observed in the solution containing 15 wt% PEG. The cationic copolymer and crowding agent act cooperatively to enhance the reaction to a greater extent than that observed with either additive alone. This finding suggested that both the copolymer and crowding agents should be evaluated when optimizing DNAzyme reaction conditions. Crowding agents may have a distinct effect when the DNAzyme associated with protein, a type of interaction usually stabilized by electrostatic interactions. The use of the copolymer and crowding agent may expand the application of DNAzyme into divalent metal ion-deficient environments such as blood and the extracellular environment.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was financially supported by a Grants-in-Aid for Transformative Research Areas “Molecular Cybernetics” (20A40), Promotion of Distinctive Joint Research Center Program Grant (JPMXP 0621467946) from MEXT, the Cooperative Research Program of “Network Joint Research Center for Materials and Devices”, and JSPS KAKENHI (21H03816). J. W. was supported by a Chinese Scholarship Council fellowship.
References
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Biochemistry, 1998, 37, 13330–13342 CrossRef CAS PubMed.
- S. K. Silverman, Trends Biochem. Sci., 2016, 41, 595–609 CrossRef CAS PubMed.
- Y. Chen, M. Wang and C. Mao, Angew. Chem., Int. Ed., 2004, 43, 3554–3557 CrossRef CAS PubMed.
- Z. Wang, J. Yang, G. Qin, C. Zhao, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2022, 61, e202204291 CrossRef CAS PubMed.
- M. N. Stojanovic, T. E. Mitchell and D. Stefanovic, J. Am. Chem. Soc., 2002, 124, 3555–3561 CrossRef CAS PubMed.
- Q. Li, Z. Tong, Y. Cao and H. Gu, Chem, 2021, 7, 2556–2568 CAS.
- X. Wang, G. Kim, J. L. Chu, T. Song, Z. Yang, W. Guo, X. Shao, M. L. Oelze, K. C. Li and Y. Lu, J. Am. Chem. Soc., 2022, 144, 5812–5819 CrossRef CAS PubMed.
- E. Mokany, S. M. Bone, P. E. Young, T. B. Doan and A. V. Todd, J. Am. Chem. Soc., 2010, 132, 1051–1059 CrossRef CAS PubMed.
- P. Zhang, Z. He, C. Wang, J. Chen, J. Zhao, X. Zhu, C.-Z. Li, Q. Min and J.-J. Zhu, ACS Nano, 2015, 9, 789–798 CrossRef CAS PubMed.
- Y. Gao, S. Zhang, C. Wu, Q. Li, Z. Shen, Y. Lu and Z.-S. Wu, ACS Nano, 2021, 15, 19211–19224 CrossRef CAS PubMed.
- Q. Wang, K. Tan, H. Wang, J. Shang, Y. Wan, X. Liu, X. Weng and F. Wang, J. Am. Chem. Soc., 2021, 143, 6895–6904 CrossRef CAS PubMed.
- H. Wang, Y. Chen, H. Wang, X. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2019, 58, 7380–7384 CrossRef CAS PubMed.
- A. I. Taylor, C. J. K. Wan, M. J. Donde, S.-Y. Peak-Chew and P. Holliger, Nat. Chem., 2022, 14, 1–11 CrossRef PubMed.
- J. Borggräfe, J. Victor, H. Rosenbach, A. Viegas, C. G. W. Gertzen, C. Wuebben, H. Kovacs, M. Gopalswamy, D. Riesner, G. Steger, O. Schiemann, H. Gohlke, I. Span and M. Etzkorn, Nature, 2021, 1–6 Search PubMed.
- H. Liu, X. Yu, Y. Chen, J. Zhang, B. Wu, L. Zheng, P. Haruehanroengra, R. Wang, S. Li, J. Lin, J. Li, J. Sheng, Z. Huang, J. Ma and J. Gan, Nat. Commun., 2017, 8, 2006 CrossRef PubMed.
- J. Li, W. Zheng, A. H. Kwon and Y. Lu, Nucleic Acids Res., 2000, 28, 481–488 CrossRef CAS PubMed.
- S.-F. Torabi, P. Wu, C. E. McGhee, L. Chen, K. Hwang, N. Zheng, J. Cheng and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5903–5908 CrossRef CAS PubMed.
- W. Zhou, R. Saran, Q. Chen, J. Ding and J. Liu, ChemBioChem, 2016, 17, 159–163 CrossRef CAS PubMed.
- L. Ma and J. Liu, ChemBioChem, 2019, 20, 537–542 CrossRef CAS PubMed.
- L. Ma, S. Kartik, B. Liu and J. Liu, Nucleic Acids Res., 2019, 47, 8154–8162 CrossRef CAS PubMed.
- R. J. Ellis, Trends Biochem. Sci., 2001, 26, 597–604 CrossRef CAS PubMed.
- N. Chowdhury and A. Bagchi, Curr. Chem. Biol., 2015, 9, 73–83 CrossRef CAS.
- B. P. Paudel and D. Rueda, J. Am. Chem. Soc., 2014, 136, 16700–16703 CrossRef CAS PubMed.
- S. Nakano, M. Horita, M. Kobayashi and N. Sugimoto, Catalysts, 2017, 7, 355 CrossRef.
- O. Hanpanich and A. Maruyama, Polym. J., 2019, 51, 935–943 CrossRef CAS.
- J. Gao, N. Shimada and A. Maruyama, Biomater. Sci., 2015, 3, 308–316 RSC.
- K. Rudeejaroonrung, O. Hanpanich, K. Saito, N. Shimada and A. Maruyama, Biomater. Sci., 2020, 8, 3812–3818 RSC.
- A. Maruyama, M. Katoh, T. Ishihara and T. Akaike, Bioconjugate Chem., 1997, 8, 3–6 CrossRef CAS PubMed.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS PubMed.
- S. Nakano and N. Sugimoto, Biophys. Rev., 2016, 8, 11–23 CrossRef CAS PubMed.
- S. Nakano, H. Hirayama, D. Miyoshi and N. Sugimoto, J. Phys. Chem. B, 2012, 116, 7406–7415 CrossRef CAS PubMed.
- A. Alberga, M. Ferrez and E. E. Baulieu, FEBS Lett., 1976, 61, 223–226 CrossRef CAS PubMed.
- P.-G. Lantz, M. Matsson, T. Wadström and P. Rådström, J. Microbiol. Methods, 1997, 28, 159–167 CrossRef CAS.
- H. Barbosa, A. V. Hine, S. Brocchini, N. K. H. Slater and J. C. Marcos, J. Chromatogr. A, 2008, 1206, 105–112 CrossRef CAS PubMed.
- Y. Sato, Y. Kobayashi, T. Kamiya, H. Watanabe, T. Akaike, K. Yoshikawa and A. Maruyama, Biomaterials, 2005, 26, 703–711 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2023 |
 and
Atsushi
Maruyama
and
Atsushi
Maruyama
 *
*




