Recent nanotechnology-based strategies for interfering with the life cycle of bacterial biofilms
Jiahe
Wu
ab,
Bo
Zhang
a,
Nengming
Lin
*a and
Jianqing
Gao
 *b
*b
aKey Laboratory of Clinical Cancer Pharmacology and Toxicology Research of Zhejiang Province, Affiliated Hangzhou First People's Hospital, Zhejiang University School of Medicine, Hangzhou 310006, China. E-mail: lnm1013@zju.edu.cn
bInstitute of Pharmaceutics, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China. E-mail: gaojianqing@zju.edu.cn
First published on 13th January 2023
Abstract
Biofilm formation plays an important role in the resistance development in bacteria to conventional antibiotics. Different properties of the bacterial strains within biofilms compared with their planktonic states and the protective effect of extracellular polymeric substances contribute to the insusceptibility of bacterial cells to conventional antimicrobials. Although great effort has been devoted to developing novel antibiotics or synthetic antibacterial compounds, their efficiency is overshadowed by the growth of drug resistance. Developments in nanotechnology have brought various feasible strategies to combat biofilms by interfering with the biofilm life cycle. In this review, recent nanotechnology-based strategies for interfering with the biofilm life cycle according to the requirements of different stages are summarized. Additionally, the importance of strategies that modulate the bacterial biofilm microenvironment is also illustrated with specific examples. Lastly, we discussed the remaining challenges and future perspectives on nanotechnology-based strategies for the treatment of bacterial infection.
1 Introduction
Persistent bacterial infection has become a life-threatening challenge worldwide. Particularly, the emergence of drug-resistant pathogens prompts the over-consumption of antibiotics, which in turn exacerbates the rapid development of drug resistance.1 In light of the current dilemma, great effort has been devoted to discovering new antibiotics or synthesizing novel antimicrobial drugs,2,3 but their efficiency may not be high enough to keep up with the rise of drug resistance.4 Some bacteria with intrinsic resistance are naturally insensitive to antimicrobial agents. Interestingly, drug resistance can also be acquired from other bacterial strains through specific biochemical routes, including gene mutation and gene transfer.According to extensive research in the past few decades, biofilm formation is considered a vital pathway for the revolutionary development of resistance.5 Biofilm formation is a mechanism for bacteria to protect them against the host immune system and hazardous environmental challenges.6 The properties of the bacteria inside the biofilm are distinct from their planktonic counterparts, especially the susceptibility of pathogenic bacteria to conventional antibiotics.7 Bacterial cells within the biofilm usually stay in slow growth states which enable the tolerance to the antimicrobial drugs that target physiological processes occurring during growth.8 Moreover, the extracellular polymeric substance (EPS), as well as the biofilm microenvironment, plays vital roles in hindering the susceptibility of bacteria to antibiotics.9 There is a diffusion-reaction inhibition effect exerted by EPS molecules during the penetration of antimicrobials in the biofilm, which limits the diffusion extent in the biofilm and quenches the activity of antimicrobial drugs. Additionally, attributed to the local metabolic activity and host immune response, there is an inherent acidic microenvironment (pH values of 4.5–6.5)10–14 prevailing in a biofilm, leading to the disabling of antibacterial agents.6 Unfortunately, numerous persistent infectious diseases are related to biofilm formation, such as caries, fibrosis pneumonia, and endocarditis.15,16 Apart from infectious diseases, the urgent need to defeat biofilms also exists in various fields, including biomedical implants and food packaging.17 Consequently, antibiofilm strategies, either preventing biofilm formation or eradicating them, have been a topic of great interest over the past few decades.
A typical life cycle of planktonic bacteria to biofilms is comprised of four stages, including the planktonic bacteria adhesion on either biological or inert surfaces, bacterial growth and EPS secretion, mature biofilm formation, and detachment of planktonic bacteria from the mature biofilm.18 The irreversible attachment initiates various changes in the attached bacteria at the genetic level, boosting the biosynthesis of the matrix and tolerance to conventional antibiotics. Then, bacterial cells grow and aggregate with EPSs into a more complex three-dimensional structure. With the formation of mature biofilms, EPS, as a natural physical barrier, can not only protect the resident bacteria as described above, but also separate the biofilm cells from nutrients, thus leading to the emergence of subpopulations to adapt to the ever-changing microenvironment accompanied by cell crowding. Besides, bacterial cells can be detached from the biofilm and return to the planktonic mode which can initiate a new cycle for biofilm formation after their colonization on a new surface. In light of the biofilm formation life cycle and the pathways endowing biofilm bacteria with drug resistance, strategies that interfere with the biofilm life cycle, including inhibiting initial bacterial adhesion, inhibiting the biofilm formation, eradicating mature biofilms, and killing planktonic bacterial cells, have been widely explored. However, these strategies cannot be carried out with barely conventional antibiotics due to their limited biofilm penetration, insufficient bactericidal efficacy, poor targeting ability, developed drug resistance of bacteria, and barely any effect on the EPS.
Recent advances in nanotechnology have furnished promising alternative tools to combat biofilms and great effort has been devoted to leveraging nanomaterials for interfering with the biofilm life cycle. At the nanoscale, materials exhibit special and unique features. Moreover, diverse functionalities can be imparted for enhanced drug delivery, such as prolonged systemic circulation, improved drug solubility, enlarged drug loading capacity, targeted drug delivery and controlled drug release. Many approaches using nanotechnology-based drug delivery systems, including liposomes,19–22 polymeric nano-vehicles,23,24 and mesoporous silica nanoparticles,25–28 have been verified and found to be feasible for combating drug-resistant bacterial strains.29 Apart from being utilized as drug carriers, some nanomaterials with intrinsic antimicrobial effects, such as noble metal (e.g. gold,30 silver31) nanomaterials, metallic oxide (e.g. iron oxide,32 vanadium pentoxide33) nanomaterials, and carbon nanomaterials,34–36 have also been developed as antibiotic-free antimicrobial agents that kill pathogens through various pathways (e.g. toxic metal ion leaching, generation of reactive oxygen species (ROS), and physical interactions) simultaneously.4
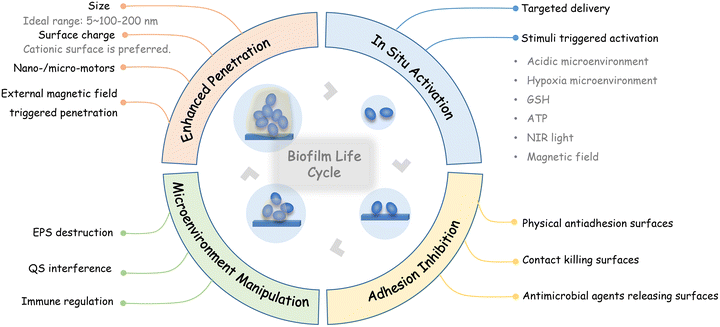
In this review, the current nanotechnology-based strategies for interfering with the biofilm life cycle, which includes inhibiting initial bacterial adhesion, promoting antimicrobial penetration into biofilms, and in situ activation of antimicrobial activities, as well as modulating the infection microenvironment, are summarized according to the requirements of different stages (Fig. 1). Finally, perspectives on the further development and challenges of nanotechnology-based antibiofilm strategies are discussed.
2 Engineered surfaces for adhesion inhibition
In the biofilm life cycle, the adhesion of planktonic bacteria plays a pivotal role in initiating biofilm formation.18 Therefore, modified surfaces that prevent the non-specific attachment of planktonic bacterial cells and sterilize the adherent bacteria through a contact-killing effect or by releasing antibacterial agents, are highly desirable to avoid subsequent biofilm formation.37,38 The properties of these surfaces are summarized in Table 1. In particular, biomimicry surfaces that are inspired by plants or animals are also highly feasible for preventing microbial adhesion.39,40 The special nanostructured textures in nature (e.g. the nanopillar geometry of insect wings, nanogroove geometry of shark skin) play important roles in endowing an antimicrobial effect.41| Surface type | Characteristics and antiadhesion mechanism | Advantages | Disadvantages |
|---|---|---|---|
| Physical antiadhesion surfaces | • Electrostatic repulsion between the negative surfaces and the negative bacterial membrane | Inhibit the interaction of bacteria with the surfaces directly | Remain risks of bacterial attachment and initiation of biofilm formation |
| • Superhydrophobic surfaces capture stable air cushions | |||
| • Superhydrophilic surfaces form a stable hydration layer | |||
| Contact-killing surfaces | Bacterial physical attachment on the surfaces: | Deactivate the adhered bacterial cells | Rely on the physical attachment of bacterial cells and kill bacteria in a limited range |
| • Tethering positively charged molecules on surfaces to interfere with the integrity of the bacterial membrane | |||
| • Decorating bactericidal peptides and nanoparticles on surfaces to kill pathogens directly | |||
| • Manufacturing special nanostructured textures on surfaces to trap bacteria or rupture bacterial cells | |||
| Antimicrobial agents releasing surfaces | Decorating antibiotic-loaded nanocarriers, antimicrobial metal ions or ions-leaching metal nanoparticles on the surfaces | Deactivate both adhered bacteria cells and planktonic bacterial cells | Require rational design for controlled release to eliminate probable toxicity |
2.1 Physical antiadhesion surfaces
To develop surfaces interdicting adhesion, surface topographical features, including charge, roughness, and wettability, have to be taken into consideration.4Due to the inherent negative surface charge of bacterial cells, surfaces with negative charge are capable of suppressing bacterial adhesion through electrostatic repulsion. For example, Cheng Zhu et al. decorated negatively charged carbon nanodots on polymeric coatings, enabling a robust antiadhesive effect and effective inhibition of biofilm formation.42 Similarly, graphene–silicone elastomer composite materials that were tailored with strong electronegativity could significantly expel both Gram-negative bacteria and Gram-positive bacteria through physical interaction.43 On the other hand, although the positive surface usually cannot resist bacterial adhesion, it can interfere with the integrity of the bacterial membrane and lead to a contact killing effect,44 which will be introduced in section 2.2.1.
Regarding roughness, bacterial adhesive abilities differ from micron-scale roughness and nanoscale roughness.45 Although it has been well recognized that nanoscale roughness on surfaces generally provides better anti-adhesion properties, some studies verified that bacterial adhesion occurs more easily on rough surfaces in the range of nanoparticles.28,46 For instance, Hao Song et al. constructed mesoporous silica nanoparticles with rough surfaces, which exhibited enhanced adhesion to the bacterial surface compared with mesoporous silica nanoparticles with smooth surfaces.28
Wettability is an important indicator of bacterial adhesion on a surface. It has been well recognized that the initial bacterial adhesion force on hydrophobic surfaces is higher than it on hydrophilic surfaces due to the hydrophobic interactions. On the other hand, there are some studies indicating that hydrogen bonding with hydrophilic surfaces can also help bacterial adhesion.45 Consequently, surfaces with moderate wettability allow bacterial adhesion through hydrogen bonding or hydrophobic interactions,47–49 which depends on not only the surface wettability but also the bacterial species.50,51 However, superwettable surfaces can resist bacterial adhesion through the formation of natural barriers.52 Superhydrophobic surfaces can capture stable air cushions, while superhydrophilic surfaces can form a stable hydration layer.53,54 The surface of the lotus leaf is a typical natural superhydrophobic surface with self-cleaning and antifouling properties, which is attributed to its micro/nanostructure.55 It is also worth noting that surface chemistry and surface roughness can be adjusted to influence the surface wettability.55,56 For example, Gege Wang et al. endowed polylactic acid (PLA) surfaces with micron-/nanoscale roughness via an adjusted exfoliation process to form superhydrophobic surfaces.57 The results turned out that the PLA surfaces with a higher roughness value (Ra = 302 nm) presented higher water contact angles, while pristine PLA surfaces were smoother (Ra = 7.155 nm) and exhibited hydrophilicity.
It has been reported that a conditioning film that is formed by the deposition of organic and inorganic molecules present in aqueous environments is a critical parameter for bacterial adhesion.58 Components in the conditioning films with various properties, such as ionic strength, hydrophilicity and charge, can alter the physiochemical features of the surface.59,60 Consequently, modifying the conditioning films can also influence bacterial adhesion. For example, Kim Doiron et al. utilized snow crab peptides that are natural organic matter in seawater to alter the nature of the conditioning film on mild steel, which consequently limits the biofilm formation and reduces the microbial-induced corrosion.61
2.2 Bactericidal surfaces
Although physical antiadhesion surfaces can largely repel bacteria, there remain risks of bacterial attachment and initiation of biofilm formation. To this end, strategies that use bactericidal surfaces have been proposed to inhibit biofilm formation. According to the bactericidal regime, these surfaces can be generally categorized as contact-killing surfaces and antimicrobial agent releasing surfaces. Additionally, the physical antiadhesive ability can also be integrated into bactericidal surfaces to maximize the inhibition of biofilm formation.62,63Apart from tethering antibacterial groups/molecules, polymers and peptides, some nanoparticles with antimicrobial capability can also be grafted or loaded to the surface for contact killing. Additionally, the decoration of nanoparticles onto the surface may change the surface topographical features as well. Guanidine-based compounds with bacteriostatic properties can be utilized to fabricate antibacterial nanomaterials.79 Hua Han et al. utilized guanidine-based compounds to construct nanogels by copolymerization.73 The as-fabricated nanogels presented a strong contact-killing effect by destroying the cell membrane and causing bacterial lysis. Then the nanogels were grafted on the surface of cotton fibers, which converted the surface from hydrophilicity to hydrophobicity. Consequently, the cotton grafted with nanogels was able to repel most bacteria and further kill the immobilized bacteria.
Inspired by the special nanostructured textures in nature, surfaces with specific nanopatterns may trap bacteria or rupture bacterial cells and thus can be developed for contact-killing surfaces. Among them, nanopillars are a nanopattern that has been widely investigated. For instance, Martyna Michalska et al. fabricated a nanopillar geometry inspired by insect wings on black silicon.80 The as-fabricated biomimicry surfaces presented tunable antibacterial effects. The longer (up to 7 μm) and exceedingly sharp pillars presented a strong bactericidal effect on various bacterial species, while the shorter (<2 μm) and blunt pillars appeared effective for specific bacterial species. They illustrated that the strong adhesion effect between bacterial cells and nanopillars, which was dependent on the topography of the nanopillar array and the properties of bacterial cells, was the main mechanism for the bactericidal effect of as-fabricated nanopillar patterned surfaces.80 Similarly, Joshua Jenkins et al. also explored the morphological effects of mimetic titanium nanopillars on bacteria.81 They found that nanopillars with topography that mimic the surface of dragonfly wings could deform and penetrate the bacterial envelope rather than rupturing or lysing bacterial cells. Interestingly, such mimetic titanium nanopillars performed physiological effects on bacteria, including inhibiting cell division and increasing intracellular oxidative stress.81 Besides, integrating bactericidal compounds onto the nanopillar array can obtain a synergistic antibacterial effect as evidenced by Goro Choi et al., who fabricated a polymeric nanopillar array covered by an ionic polymer layer containing quaternary ammonium compounds.82
Some antimicrobial metal ions (e.g., Cu2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 84 and Ag+
84 and Ag+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 85) and ion-leaching metal nanoparticles (e.g., silver nanoparticles (AgNPs),86 metal–organic framework (MOF) nanoparticles87) are easily decorated on biomaterial surfaces and thus usually utilized to fabricate antimicrobial agent releasing surfaces. AgNPs have been well-recognized and widely adopted as bactericidal nanomaterials. Ag+ leaching from the surface of AgNPs plays a major antimicrobial role. Therefore, AgNPs can be deemed as a carrier of Ag+. Hossein Yazdani-Ahmadabadi et al. used AgNP assemblies to form silver coatings that provided a sustained release of Ag+ ions.86 Consequently, the fabricated silver coatings exerted a long-term anti-adhesion effect (>30 days) and a long-term antibacterial effect in vitro on various bacterial species for up to 28 days. MOFs consisting of crystalline porous coordination polymers have emerged as antibacterial materials in recent years. Similarly, MOFs can sustainably release the stored metal ions and thus achieve a long-term antibacterial effect.88 Xiaoxue Yao et al. prepared omniphobic porous hydrogels loaded with zinc imidazolate framework 8 (ZIF–8) nanoparticles as wound dressings.38 Anchoring of ZIF–8 nanoparticles endowed hydrogels with a reentrant architecture and increased roughness, which was beneficial for eliminating the adhesion of pathogens. Bactericidal Zn2+ could be gradually released from ZIF-8 nanoparticles. With long-term antiadhesion and antibacterial effects, the ZIF-8 nanoparticle loaded hydrogel successfully accelerated wound closure in vivo.
85) and ion-leaching metal nanoparticles (e.g., silver nanoparticles (AgNPs),86 metal–organic framework (MOF) nanoparticles87) are easily decorated on biomaterial surfaces and thus usually utilized to fabricate antimicrobial agent releasing surfaces. AgNPs have been well-recognized and widely adopted as bactericidal nanomaterials. Ag+ leaching from the surface of AgNPs plays a major antimicrobial role. Therefore, AgNPs can be deemed as a carrier of Ag+. Hossein Yazdani-Ahmadabadi et al. used AgNP assemblies to form silver coatings that provided a sustained release of Ag+ ions.86 Consequently, the fabricated silver coatings exerted a long-term anti-adhesion effect (>30 days) and a long-term antibacterial effect in vitro on various bacterial species for up to 28 days. MOFs consisting of crystalline porous coordination polymers have emerged as antibacterial materials in recent years. Similarly, MOFs can sustainably release the stored metal ions and thus achieve a long-term antibacterial effect.88 Xiaoxue Yao et al. prepared omniphobic porous hydrogels loaded with zinc imidazolate framework 8 (ZIF–8) nanoparticles as wound dressings.38 Anchoring of ZIF–8 nanoparticles endowed hydrogels with a reentrant architecture and increased roughness, which was beneficial for eliminating the adhesion of pathogens. Bactericidal Zn2+ could be gradually released from ZIF-8 nanoparticles. With long-term antiadhesion and antibacterial effects, the ZIF-8 nanoparticle loaded hydrogel successfully accelerated wound closure in vivo.
For antimicrobial agent releasing surfaces, antibacterial agents are incorporated and usually released in a controlled manner.38 Moreover, to improve the antibacterial efficacy and reduce the potential toxicity, stimuli-responsive moieties are usually introduced to allow stimuli-triggered drug release for enhanced specificity to pathogens.10,89 At the site of infection, the acidic microenvironment as well as different types of molecules secreted by the bacteria can be used as triggers.90 For instance, a pH-responsive grafted bilayer constructed by Hyun-Su Lee et al.10 could responsively release loaded antibiotics in the acidic microenvironment to perform a significantly enhanced antibacterial activity.
3 Tailored nanomaterials for enhanced biofilm penetration
After the formation of biofilm architectures, the EPS acts as a natural fortress to protect the bacterial cells within the biofilm. The susceptibility of conventional antibiotics and the toxic metal ions to the enwrapped bacteria might be diminished by chelation or enzymatic degradation during the diffusion process. Thus the bacterial strains within the biofilm can survive the challenge of antimicrobials with sublethal concentrations, which can further promote resistance to the exposed antimicrobials. Therefore, it is a desirable strategy to combat biofilms by improving the penetration capacity and retaining the sterilization activity in the meanwhile. Utilizing nanocarriers to deliver conventional antibiotics or toxic metal ions is an alternative to maintain their disinfecting activity. Additionally, it has been noted that the physicochemical properties of nanomaterials, such as size, surface charge, and surface ligands, can alter their penetration extent in biofilms.3.1 Size
Regarding size, the ideal diameter for nanomaterials to treat biofilms has been arrived at as the range between 5 and 100–200 nm and no larger than 500 nm.6 Specifically, compared to large nanoparticles of limited biofilm penetration capability, small nanoparticles with a size of <20 nm have been reported to penetrate biofilms to the deep layers.23,91,92 For instance, Xiaokai Chen et al. synthesized epoxy group-functionalized organosilica nanodots (OSiNDs) with a high photoluminescence quantum yield.93 Due to the ultrasmall size (≈6.4 nm), OSiNDs were able to penetrate biofilms and thus could be used as a universal platform for imaging or therapy. Considering that small-sized nanoparticles can be readily cleared from the body and larger nanoparticles have prolonged circulation time and are liable to accumulate at the infection site due to the host immune response, size transition triggered by the external or internal stimuli might favor both accumulation and penetration.94–96 Our group fabricated pH-responsive silver nanoassemblies composed of ultrasmall AgNPs with a diameter of about 4 nm and pH-sensitive polymeric ligands.97 The as-fabricated pH-responsive silver nanoassemblies could undergo structural transformation in the acidic microenvironment in the biofilm and disassembled into dispersed AgNPs. The resulting size transition from ∼150 nm to ∼8 nm enabled enhanced penetration and further bactericidal effect activation.973.2 Surface charge
Surface charge is also a crucial factor affecting the penetration capacity.23,91,92,98 Yong Liu et al. found that the micellar nanocarriers with surface-adaptive charge reversal properties could perform enhanced penetration while those micellar nanocarriers with a negative charge could only penetrate to a limited extent.23 Xiaoning Li et al. compared the penetration capacity of quantum dots (QDs) with different surface modifications.98 Their results demonstrated that cationic QDs could readily penetrate fully into biofilms while neutral or anionic QDs could hardly penetrate or accumulate into biofilms. Therefore, nanomaterials with a positive charge tend to have better penetration capacity.99,100 Although cationic nanocarriers are preferred for penetration, their surfaces easily absorb proteins and form a corona, which may cause an enlarged size and hinder their penetration to biofilm. Based on the fact that poly(ethylene oxide) (PEO) with a negative charge can resist protein adhesion, Yuejing Xi et al. utilized PEO to construct nanocarriers to avoid the blocking of EPS in the biofilms.101 With the further incorporation of positively charged polymer and loading of antibiotics, the as-fabricated nanoformulation could reduce dental plaque as verified in a rat periodontitis model. Depending on the different requirements in blood circulation and biofilms, biofilm microenvironment-responsive charge-reversal strategies are also often used to improve the penetration and accumulation of antimicrobials in bacterial biofilms while reducing their effects on normal tissues.102–104 For example, AgNPs functionalized with carboxyl betaine groups presented an acidic-responsive charge reversal from negative to positive, which benefited their antibacterial effects deep in the biofilm and reduced their possible toxicity to healthy tissues.1053.3 Others
Apart from altering the physicochemical properties of nanomaterials, enhanced penetration can also be achieved by exerting special physical and mechanical features of nanomaterials. Magnesium-based micromotors developed by Liangfang Zhang's group presented efficient motor propulsion under acidic conditions and were of great potential for antibiotic delivery with enhanced penetration to the bacterial biofilm.106 Similarly, Tingting Cui et al. fabricated a self-propelled nanoswimmer for biofilm penetration using mesoporous silica asymmetrically functionalized with gold nanoparticles (AuNPs).107 Upon exposure to near-infrared (NIR) light irradiation, the photothermal conversion of AuNPs triggered the thermophoretic motion of the nanoswimmers and resulted in a rapid penetration into the biofilm. Moreover, magnetic materials with the assistance of an external magnetic field can reach the deep side of biofilms.108,109 Naside Gozde Durmus et al. prepared silver-conjugated superparamagnetic iron oxide nanoparticles for the eradication of antibiotic-resistant biofilms.110 In the presence of an external magnetic field, the antibacterial effect of silver was further enhanced, which might be attributed to the improved penetration of the nanoparticles within the biofilm. Besides, as verified by Kecheng Quan et al., the distribution of magnetic nanoparticles in biofilms was dependent on the exposure time of the external magnetic field.111 They identified that there was an optimal exposure time for magnetic nanoparticles to distribute homogeneously in biofilms rather than accumulating in their top or bottom layer. Apart from magnetic force-triggered penetration, the photochemical internalization effect has also been demonstrated beneficial for biofilm permeability.112Some studies also observed that some biomaterials presented inherent biofilm penetration ability. For example, galactose can bind to LecA in the EPS and thus can be incorporated with bactericidal agents to promote their penetration.113,114 Yiwen Zhu synthesized a series of tunable block ratio diblock copolymers PαGal50-b-PGRBn that could self-assemble into micelles spontaneously (Fig. 2).114 Galactose moieties in the polymers were used to target Pseudomonas aeruginosa (P. aeruginosa) biofilm due to the carbohydrate–protein interaction between galactose and LecA in the EPS. The penetration and dispersion of micelles in the biofilm enabled their effective photodynamic bactericidal activities inside the biofilm.114
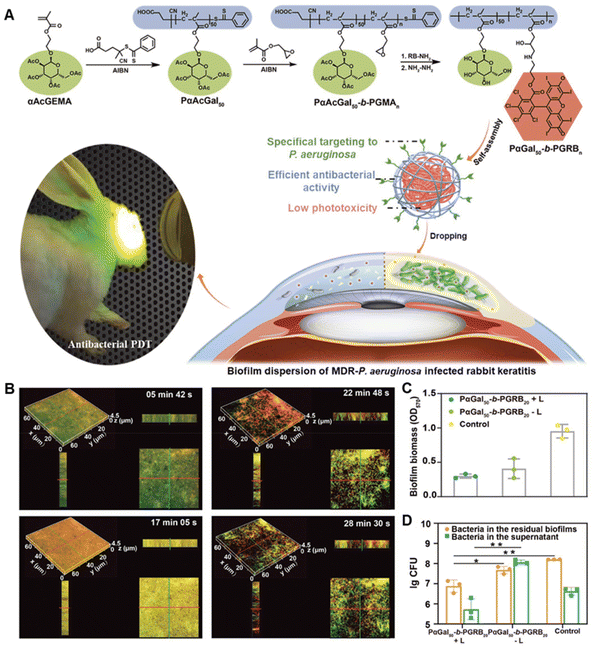 | ||
| Fig. 2 (A) Schematic illustration of the synthesis of biofilm-targeted copolymers PαGal50-b-PGRBn and their antibacterial PDT against P. aeruginosa biofilm. (B) The penetration of PαGal50-b-PGRB20 in the mature P. aeruginosa biofilm. (C) Biofilm eradication effect of PαGal50-b-PGRB20. (D) Bactericidal effect of PαGal50-b-PGRB20 on the biofilm bacteria. Adapted with permission from ref. 114, copyright 2022, Wiley. | ||
4 Stimuli-responsive nanomaterials for in situ activation
To treat superficial bacterial infections such as diabetic wounds, a local antimicrobial delivery strategy is usually adopted to create high localized concentrations for biofilm prevention.6,39 However, for deep-seated infectious diseases, unspecific distribution of antimicrobial agents upon systemic administration may not only cause severe side effects but also be invalid to treat the biofilm-related infection. An infection site-targeted delivery system is an alternative to decrease the unspecific distribution. Sazid Hussain et al.25 used a cyclic 9-amino-acid peptide CARGGLKSC, a Staphylococcus aureus (S. aureus) targeting peptide screened via phage display in the S. aureus-induced pneumonia mice model, to modify vancomycin-loaded porous silicon nanoparticles. The as-constructed antibiotic delivery system presented specific accumulation in the S. aureus-infected tissues and minimized systemic side effects with a reduced dose. Additionally, the targeted systems exhibited better therapeutic effects on S. aureus-induced pneumonia than the untargeted delivery systems or free vancomycin with equivalent doses. Moreover, due to the possible diffusion inhibition effect of the EPS in the biofilm, a higher concentration of antimicrobials is required for the bactericidal purpose.115 Therefore, it is highly desirable to develop stimuli-responsive nanoformulations with in situ activation features to improve antibiofilm efficacy and eliminate systemic toxicity.1164.1 Acidic microenvironment
Due to the bacterial metabolic activity, there is an inherent acidic microenvironment in the biofilm. Hence, the acidic microenvironment of a biofilm is distinguished from the normal physiological conditions, which can be utilized to develop pH-responsive drug delivery systems for in situ activation.117 Benjamin Horev et al. developed polymeric nanocarriers to deliver the antibacterial drug farnesol to oral biofilms.24 In the acidic microenvironment, the nanocarrier underwent core destabilization, which induced the release of farnesol in situ and facilitated its therapeutic effect. Some antibacterial peptides can also be assembled and fabricated into pH-responsive nanoparticles. The acidic microenvironment may cause their structural transformation and thus enable a biofilm-specific activity.95,118 Similar strategies are also adopted for the photosensitizer delivery for selective antibacterial photodynamic therapy (PDT). For example, Lei Xia et al. constructed pH-responsive supramolecular-based nanoparticles through the host–guest recognition of carboxylatopillar[5]arene (CP5) and a quaternary ammonium group functionalized tetrafluorophenyl porphyrin (TFPP-QA).119 Under acidic conditions, the supramolecule-based nanoparticles dissociated and the exposed positive TFPP-QA could disrupt the bacterial membrane. Moreover, upon light irradiation, the anchoring of TFPP-QA on the bacterial surface benefited the PDT.However, the acidic microenvironment might quench the activity of some conventional antibiotics. In contrast, some metal ion-leaching-based antimicrobial agents, such as AgNPs and MOF, can be promoted in the acidic conditions and thus enable the in situ amplification of the bactericidal effect.97,105,120 Typically, due to the structural characteristics of MOFs, antibiotics can be loaded and released along with acid-induced degradation.121 Consequently, leaching metal ions and antibiotics can synergistically combat pathogens. Moreover, due to the ion-leaching efficiency being related to the specific surface area, smaller nanoparticles with a higher specific surface area generally exhibited accelerated ion leaching. Therefore, combined with the pH-responsive size-transition strategy, the antimicrobial effect based on metal ion-leaching can be further amplified.97
4.2 Other endogenous stimuli
A biofilm is a heterologous structure with steep gradients of not only pH values but also nutrients, oxygen, electron acceptors and donors, and redox conditions.8,122 These characteristics can also be utilized as endogenous stimuli for in situ activation in the biofilm microenvironment (Table 2).| Endogenous stimuli | Nanoformulations | Outcomes | Ref. |
|---|---|---|---|
| pH | Nanoassembly of chitosan–polyethylene glycol–peptide conjugate | Enhanced penetration to the biofilm. Expose the α-helical peptide at the bacterial cell membrane. | 95 |
| pH | Nanoassembly composed of AgNPs and imidazole-containing polymer | Enhanced penetration to the biofilm. Accelerated Ag+ leaching in the biofilm. | 97 |
| pH and lipid | Peptide nanoparticles (pHly-1 NPs) | Coil-helix conformational transition for killing cariogenic bacteria. | 118 |
| pH | Supramolecular nanoformulation composed of CP5 and TFPP-QA | Release photosensitizer in situ. | 119 |
| pH | Au/Ag hybrid nanoparticles by coating gold nanorods with silver | Acid-triggered Ag+ release and in situ photoacoustic imaging. | 120 |
| pH | ZIF-8 coated with polydopamine loading vancomycin | Release antimicrobials in situ. | 121 |
| Azoreductase (hypoxia) | Supramolecular nanoformulation self-assembled by lactose-modified LacAC4A | Hypoxia-responsive antibiotic delivery. | 124 |
| GSH and pH | Supramolecular nanoformulation composed of α-cyclodextrin conjugated prodrugs and pH-sensitive polypeptide copolymer | Enhanced penetration to the biofilm due to the pH-responsive charge reversal. GSH-responsive drug release. GSH consumption benefits the PDT. | 102 |
| ATP | IAA carrying ZIF-8 and HRP co-loaded in the polyacrylamide hydrogel microspheres | ATP-triggered release of IAA enabling the cascade amplification of antibacterial effect by the reaction of IAA and HRP. | 127 |
Due to the fast growth of bacteria and rapid oxygen consumption, bacteria at a depth in biofilms experience hypoxic conditions.123 Based on overexpression of azoreductase by bacteria in the hypoxic biofilm microenvironment, Juan-Juan Li et al. synthesized lactose-modified azocalix[4]arene (LacAC4A) and fabricated supramolecular nanoformulation using LacAC4A for hypoxia-responsive antibiotic delivery.124
Besides, overexpression of glutathione (GSH) was also observed in the bacterial biofilm microenvironment.125 Dengfeng Hu et al. conjugated GSH-sensitive α-cyclodextrin to nitric oxide (NO) and chlorin e6 (Ce6) respectively to obtain GSH-activated antibacterial prodrugs.102 These prodrugs together with pH-sensitive polymers were then fabricated into supramolecular nanoformulations through host–guest recognition. The supramolecular nanoformulations underwent charge reversal in the acidic biofilm microenvironment and thus successfully penetrated the biofilm. Then these GSH-prodrugs were activated by the overexpressed GSH in the biofilm and released bactericidal NO and a photosensitizer. During the activation process, GSH consumption was beneficial for the Ce6-mediated PDT as well.102
Living bacteria can secret adenosine triphosphate (ATP) and thus the extracellular ATP is abundant in biofilms.126 Based on the affinity of ATP with Zn2+, Yuhao Weng et al. designed an ATP-activated platform for the co-delivery of indole-3-acetic acid (IAA) and horseradish peroxidase (HRP) to biofilms (Fig. 3).127 IAA carrying ZIF-8 and HRP were co-loaded in polyacrylamide hydrogel microspheres. IAA was loaded to ZIF-8 that was self-assembled by Zn2+ and 2-methylimidazole to avoid premature interaction with HRP before arriving at the bacterial biofilm. Due to the stronger affinity between ATP and Zn2+ than it between Zn2+ and 2-methylimidazole, ZIF-8 dissociated in the biofilm and released IAA. ROS generated from the reaction of IAA and HRP damaged the bacterial membrane and triggered more ATP leakage, resulting in cascade amplification of the antibacterial effect.127
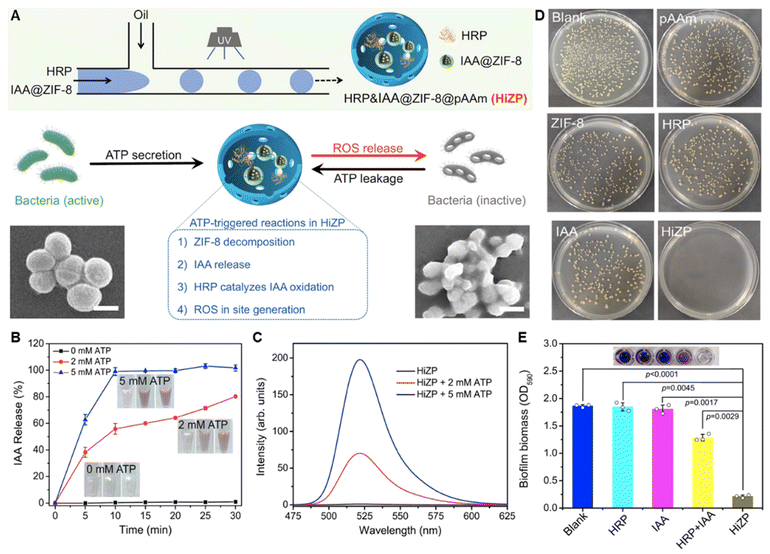 | ||
| Fig. 3 (A) Schematic illustration of the construction and antibacterial mechanism of HiZP. (B) The ATP-responsive release profiles of IAA from IAA@ZIF-8. (C) ATP-responsive ROS generation of HiZP. (D) Antibacterial effect of HiZP on S. aureus. (E) Biofilm eradication effect of HiZP. Adapted with permission from ref. 127, copyright 2019, Springer. | ||
4.3 NIR light
Light with short wavelengths (ultraviolet or near-ultraviolet visible light) presents low penetration with high phototoxicity. As feasible non-invasive external stimuli, red light and NIR light with deep penetration capacity have been widely used in various biomedical applications, such as antitumor and infection treatment. Zhiqiang Shen et al. developed coumarin-based nitrogen oxide donors that could be activated by red-light irradiation through a photoredox catalysis reaction.113 With a rational design, NIR light irradiation can also be utilized to trigger the release of antibacterial molecules in biofilms by increased temperature via the photothermal effect.107,128,129 For instance, Xile Hu et al. fabricated glycosheets self-assembled by the galactose- and fucose-based ligands on a molybdenum disulfide thin-layer.130In situ thermal release of antibiotics triggered by NIR light together with white light-triggered ROS generation enabled the effective therapy of multidrug-resistant bacterial infection on wounds. Besides, the photoacoustic effect can also be adopted for enhanced penetration into biofilms as well as controlled release in situ. As verified by Bing Cao et al., NIR laser irradiation could trigger the gasification of perfluorohexane that was loaded in nanodroplets and resulted in photoacoustic cavitation and the release of loaded rifampicin in the biofilm.131Apart from the NIR light-triggered antimicrobial release, nanomaterials with intrinsic NIR-activated properties are also utilized for in situ activation.34,132 By incorporating photosensitizer molecules (e.g. IR780,133 indocyanine green,134 tetrafluorophenyl porphyrin,119 Ce6,112 Rose bengal114), PDT within the biofilm can occur upon NIR light application. Specifically, Gram-negative bacterial strains are more susceptible to PDT. However, PDT requires sufficient oxygen to generate destructive ROS while biofilm bacteria experience hypoxic conditions, especially for those deep in the biofilm. Insufficient oxygen may hinder the outcome of PDT. To this end, Xiaolin Sun et al. incorporated a MnO2 nanolayer on the PDT nanocomposite to decompose the generated hydrogen peroxide (H2O2) during PDT and produce oxygen, resulting in promotion of ROS generation and improved PDT efficacy in the biofilm.135 In addition, photothermal therapy (PTT) can also be adopted for in situ activation.34,37,92,136,137 For example, Luoxiao Ran et al. utilized gold–silver alloy nanoparticles to carry gentamicin and wrapped them with polydopamine.138 Upon NIR excitation, both gold–silver alloy nanoparticles and polydopamine performed photothermal effects, triggering the release of gentamicin and Ag+ at the same time. Consequently, increased temperature (47 °C) in situ and released antibacterial molecules exhibited a synergistic antibiofilm effect. Specifically, some nanomaterials can simultaneously perform PDT and PTT for synergistic biofilm irradiation.139,140
As an external trigger, NIR light can be utilized together with both endogenous stimuli103,104,141 and other external triggers109 to achieve a more precise control and to promote the antibiofilm effect.
4.4 External magnetic field
Magnetic nanomaterials can be transported to the infection site upon the application of an external magnetic field. Besides, as evidenced by Aaron Elbourne et al., gallium-based liquid metal droplets could transform their shape and equip themselves with sharp edges in response to a low-intensity rotating magnetic field.142 Such magneto-induced shape transformation consequently resulted in damaged biofilm structure and ruptured bacterial cells. Among various magnetic nanoparticles, iron oxide nanoparticles (IONPs) have intrinsic peroxidase-like properties with the generation of bactericidal free radicals that can perform synergistic antibiofilm effects with magnetic field-triggered physical effects.143,144Moreover, with an alternating magnetic field, local hyperthermia can be achieved by magnetic nanomaterials.145 For instance, Jie Li et al. evaluated the antibiofilm effect of IONPs under different conditions and demonstrated that magnetic hyperthermia could affect the integrity of biofilm structure.145 Similar to the photothermal effect-induced drug release, magnetic hyperthermia can trigger the release of antibacterial molecules in the biofilm as well. For instance, Jiaxing Wang et al. coated nitrosothiols on magnetic CoFe2O4@MnFe2O4 nanoparticles for synergistic biofilm eradication.146 With an alternating magnetic field, magnetic hyperthermia damaged the dense structure of biofilms and enabled the penetration of nanoparticles. Moreover, thermosensitive nitrosothiols release antibacterial NO inside biofilms. The synergetic therapeutic effect was validated in implant-associated infection models.146
5 Nanocomposites for regulating infection microenvironment
Apart from the strategies targeting bacteria directly, manipulating the biofilm microenvironment, including destroying the EPS and inhibiting quorum sensing (QS), also represented a potential way to interfere with the life cycle of biofilms. Besides, invading pathogens can trigger the activation of innate immune reactions. Immune cells can combat bacterial infection through various pathways.147 Therefore, regulating the immune microenvironment around the infection is a feasible approach to participate in antibiofilm.5.1 EPS destruction
The EPS plays an essential role in maintaining the integrity of biofilms and protecting the enwrapped bacterial cells. Therapeutic strategies targeting the EPS have emerged as a topic of intense research attempts. Inhibiting the EPS has been verified as a robust antibiofilm strategy without the development of resistance.148,149 Destroying the EPS in a mature biofilm can not only enable bacterial exposure but also promote the penetration and diffusion of antibacterial agents in the biofilm.Some components exhibit an inherent EPS-disrupting effect and rhamnolipid is one of the typical examples. Rhamnolipid is a bacteria-secreted anionic surfactant that possesses negligible direct antibacterial effects but can impede bacterial adhesion and destroy the EPS in mature biofilms. Incorporating rhamnolipid into nanoparticles can eradicate biofilms effectively, especially for the Helicobacter pylori biofilm.150,151 Some natural enzymes (e.g. dextranase or mutanase) can degrade the polysaccharides and thus cleave the biofilm matrix.152 Inspired by the EPS degradation property of these natural enzymes, Anheng Wang et al. decorated protease alcalase on the surface of ciprofloxacin-loaded nanogels. Degraded EPS and enhanced penetration of ciprofloxacin in the depth of the biofilm could be achieved by the functionalized nanogels.153
It has been widely recognized that ROS benefits matrix degradation. Therefore, apart from the direct bactericidal effect, the EPS as well as the dense structure of biofilms can be destroyed simultaneously by PDT.154 Most recently, taking lessons from natural enzymes,155 some nanomaterials have been found to mimic the activity of natural enzymes (oxidase, peroxidase, deoxyribonuclease), which mainly increase the oxidative stress by generating ROS within biofilms.17,156–161 These artificial nanozymes, based on metal, metal oxide and carbon, have been explored for antibiofilm application.143,162–166 Yanqiu Wang et al. proposed a strategy utilizing nanozymes with peroxidase activity with engineered bacteria generating H2O2 to degrade the biofilm EPS and eradicate the biofilm.162 Using this strategy, significantly reduced bacteria number (5 log reduction), as well as biofilm matrix removal (85% reduction), could be achieved. In addition to the ROS-generating activities, more enzymatic activities of nanozymes have also been discovered and exploited. For example, DNase-mimicking activity is identified in cerium(IV) complexes, which can be utilized to hydrolyze extracellular DNA and thus disrupt established biofilms. Therefore, Zhengwei Liu et al. designed a series of MOF/Ce-based nanozymes to integrate the peroxidase-like activity of MOF and DNase-mimicking activity of cerium(IV) complexes for biofilm eradication (Fig. 4).167 Interestingly, such a design could avoid the recurrence of biofilms as well.
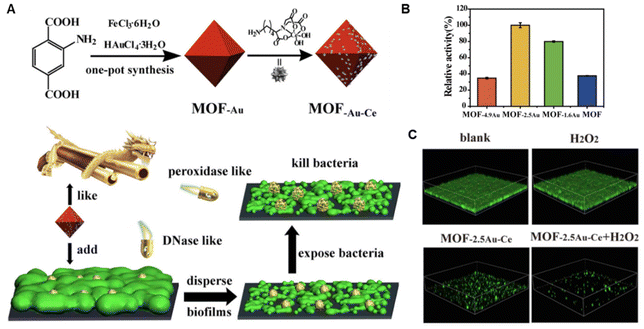 | ||
| Fig. 4 (A) Schematic illustration of construction and antibiofilm mechanism of MOF-Au-Ce. (B) Peroxidase-like activity of various MOFs. (C) Biofilm eradication effect of MOF-2.5Au-Ce. Adapted with permission from ref. 167, copyright 2019, Elsevier. | ||
Recently, a nanohole-boosted electron transport antibiofilm strategy rather than using simple enzymatic activities was proposed by Tonglei Shi et al.168 Nanoholes with atomic vacancies in disulfide nanosheets served as electronic donors while bacterial biofilms served as electronic receptors. Boosted electron transport and redox reaction resulted in destruction of essential components in the EPS. Additionally, the proposed strategy also downregulated expression of biofilm formation-related genes and thus inhibited the EPS.168
5.2 QS Interference
QS is a process whereby bacteria communicate and present coordinated activities mediated by specific signaling molecules.169,170 These molecules can sense the cell density and thus govern biofilm formation. Therefore, interfering with the QS is an effective microenvironment manipulating strategy for antibiofilm application.62,171,172 QS inhibitors, such as acylase, lactonase, and curcumin, can degrade the extracellular QS signals and interfere with the biofilm life cycle, which requires targeted delivery to the infection site especially when there are already mature biofilms.173,174 Aleksandra Ivanova et al. coated AgNPs with aminocellulose and acylase to obtain a nanoplatform for the delivery of antimicrobials and QS inhibitors deep into biofilms.173 The efficient delivery of QS inhibitors could promote the therapeutic effect of antimicrobials with reduced minimum inhibitory concentration. Considering that natural haloperoxidase can also deactivate QS molecules and result in quorum quenching, Zijun Zhou developed cerium-based MOFs that were endowed with haloperoxidase-mimicking activity for an antibacterial effect and inhibiting the formation of antibiofilm.175Apart from direct inhibition of the QS molecules, researchers also observed that modulating the biofilm microenvironment may regulate the QS. Dengfeng Hu et al. constructed perfluorohexane-loaded liposomes to deliver oxygen to the biofilm and tried to relieve the hypoxic conditions that were responsible for the antibiotic resistance.176 The results turned out that such a strategy successfully retained the bacterial sensitivity to commercial antibiotics. Interestingly, the gene sequencing indicated that relieving biofilm hypoxia could restrain the QS as well, which requires further exploration.
5.3 Immune regulation
Macrophages and neutrophils are essential innate immune cells capable of defense against invading microbes. Moreover, both macrophages and neutrophils exhibited heterogeneous regulatory functions in response to various diseases and pathological courses. Therefore, these two cells are attractive targets and are usually paid more attention to when regulating the immune microenvironment for antibiofilm.The polarization outcome of macrophages can be simply designated as pro-inflammatory (M1) and anti-inflammatory (M2) phenotypes. M1 macrophages can perform phagocytic and bactericidal effects while the formation of biofilms inhibits the M1 polarization. It has been reported that magnetic nanoparticles are capable of recruiting macrophages and influencing their polarization. Therefore, magnetic nanoparticles can be utilized not only for the direct antibacterial effect but also for macrophage-mediated immune therapy by promoting M1 polarization.146 In contrast, M2 macrophages can accelerate tissue repair and benefit recovery after antibacterial management. As validated by Weijun Xiu et al., after PDT-activated chemotherapy using hyaluronic acid-Ce6-metronidazole nanoparticles to treat bacterially infected wounds, an increased ratio of M2-like macrophages was observed in the microenvironment.177 Consequently, PDT-activated chemotherapy-triggered M2 polarization promoted wound healing.
Similarly, the bactericidal function of neutrophils can also be suppressed. To eradicate biofilms and reactivate neutrophils for a synergistic effect, Wanbo Zhu et al. synthesized Fe3O4 nanoparticle dotted graphene oxide nanosheets and loaded them into a methacrylated hyaluronic acid microneedle patch.178 Biofilm microenvironment and NIR triggered the disruption of biofilms and intracellular iron overload, which further resulted in a ferroptosis-like death. Interestingly, bactericidal functions were reactivated in iron-nourished neutrophils and consequently eliminated the biofilm synergistically.
Apart from these nanocomposites integrating with antibacterial effect and immune regulation ability, some nanomaterials don't show intrinsic bactericidal functions but can significantly evoke the local innate immunity for immune antibacterial therapy.179 For example, Chuang Yang et al. explored the potential of polyvinylpyrrolidone-modified manganese chalcogenophosphates nanosheets (MPS-PVP) in presurgical neoadjuvant immunotherapy to treat medical implant infections (Fig. 5).180 MPS-PVP generated ROS that destructive the biofilm structure and led to the expression and exposure of bacterial-associated antigens. The following amplified priming of antigen-presenting cells and biofilm-infiltrating B cells enabled biofilm-specific humoral immune and memory responses. Therefore, immune-stimulatory MPS-PVP successfully mitigated residual infections and suppressed the infection recurrence after surgical removal of the infected implants.180
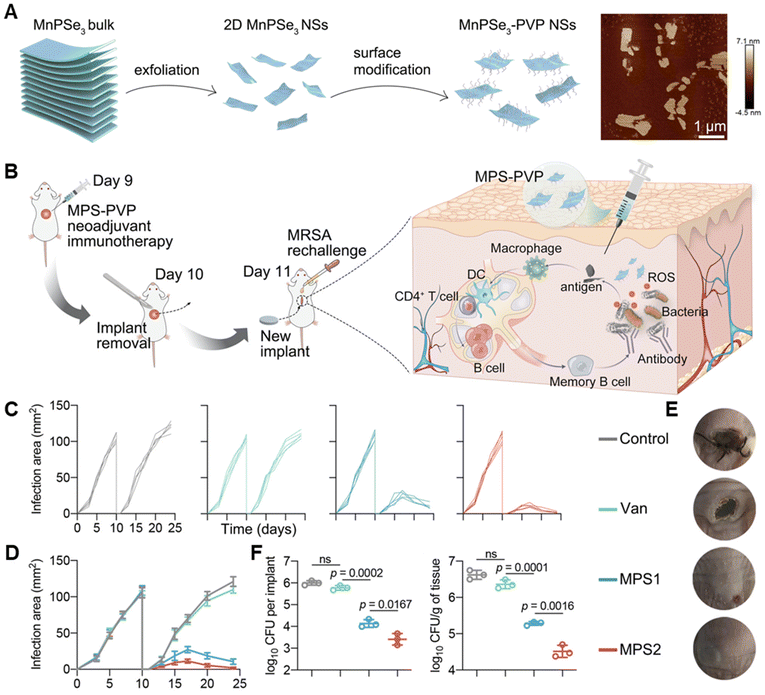 | ||
| Fig. 5 (A) Schematic illustrations of the construction of MPS and its typical AFM images. (B) Schematic illustration of the experimental procedure of the presurgical neoadjuvant immunotherapy mediated by MPS-PVP. (C–E) Changes of individual infection area (C), average infection area (D) and representative pictures (E) of mice after treatments. (F) Statistical analysis of bacterial amounts in extracted implants and peri-implant tissues respectively. Adapted with permission from ref. 180, copyright 2022, Springer. | ||
6 Conclusion and outlook
Immense progress in nanotechnology-based strategies for interfering with the biofilm life cycle has been achieved. According to the requirement of interfering with the biofilm life cycle in different stages, nanotechnology can be feasibly used to effectively inhibit initial bacterial adhesion, promote antimicrobial penetration to biofilms, boost antimicrobial activities in situ, and modulate the infection microenvironment.However, there is still a long way to go for the antibiofilm exploration since a comprehensive understanding of the bacterial biofilm is still lacking.170,181 First of all, the current understanding of the biofilm life cycle is limited and challenged by some new findings. And Karin Sauer et al. recently proposed a revised conceptual model of the biofilm life cycle, encompassing aggregation, growth and disaggregation independently of surfaces.122 According to this model, strategies that combat pathogens directly rather than inhibiting bacterial adhesion are essential for infection management. Secondly, although the planktonic bacteria are considered to be more susceptible to antimicrobial agents and immune regulation, bacteria dissociated from the dispersal of biofilm are distinct from both biofilm bacteria and planktonic cells, which requires further exploration and development of more robust antibacterial strategies accordingly.182
In the current nanotechnology-based strategies for interfering with the biofilm life cycle, evoking immune function to antibacterial agents and antibiofilm is a rather promising future direction. The inherent interaction between nanomaterials and the immune system makes nanomaterials attractive in anti-infection immunotherapy. Moreover, the unique thermogenesis or ROS production of some nanomaterials in response to exogenous or external stimuli has been demonstrated capable of activating the immune system. If the autoimmune function can be fully utilized to treat bacterial infections, it can play a synergistic effect with antimicrobial materials. Consequently, the dosage of antimicrobial agents can be reduced, thereby reducing the exposure to antimicrobial agents and attenuating the progress of bacterial resistance to antimicrobial agents.
Despite the various antibacterial strategies, developing imaging strategies for the monitoring of bacterial infections deserves more attention. If the rapid diagnosis and identification of bacterial infection can be realized, the control of bacterial infection in the early stage can avoid the subsequent formation of bacterial biofilm. Moreover, imaging tools that are capable of indicating bacterial living mode can be used to guide the choice of strategies with rational design for interfering with the life cycle of bacterial biofilm. The diagnostic strategy based on nanotechnology has been demonstrated to be feasible in the theranostics of diverse diseases.183–185 However, bacterial resistance to nanomaterials has been reported.186–189 Therefore, close attention should be paid to the revolution of bacterial resistance to nanomaterials when using nanomaterials to treat or diagnose bacterial infection.
Author contributions
Jiahe Wu: conceptualization, visualization, writing – original draft, and writing – review & editing. Bo Zhang: writing – review & editing. Jianqing Gao: conceptualization, supervision, and writing – review & editing. Nengming Lin: supervision and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Zhejiang Province (grant number LQ22H180009), the Postdoctoral Research Foundation of China (grant number 2021M702876), the Zhejiang Provincial Program for the Cultivation of High-level Leading Health Talents (grant number ZWB-2020-18), and the Clinical Pharmacy of Hangzhou Medical Key Discipline (grant number HWB-2021-21-16).References
- W. Gao, S. Thamphiwatana, P. Angsantikul and L. Zhang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 532–547 CAS.
- W. Kim, W. Zhu, G. L. Hendricks, D. Van Tyne, A. D. Steele, C. E. Keohane, N. Fricke, A. L. Conery, S. Shen and W. Pan, Nature, 2018, 556, 103–107 CrossRef CAS PubMed.
- W. Chin, G. Zhong, Q. Pu, C. Yang, W. Lou, P. F. De Sessions, B. Periaswamy, A. Lee, Z. C. Liang and X. Ding, Nat. Commun., 2018, 9, 917 CrossRef PubMed.
- G. J. Mi, D. Shi, M. Wang and T. J. Webster, Adv. Healthc. Mater., 2018, 7, 1800103 CrossRef PubMed.
- R. Laxminarayan, A. Duse, C. Wattal, A. K. M. Zaidi, H. F. L. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Lancet Infect. Dis., 2013, 13, 1057–1098 CrossRef PubMed.
- Y. Liu, L. Q. Shi, L. Z. Su, H. C. van der Mei, P. C. Jutte, Y. J. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC.
- J. H. Li, K. X. Zhang, L. Ruan, S. F. Chin, N. Wickramasinghe, H. B. Liu, V. Ravikumar, J. H. Ren, H. W. Duan, L. Yang and M. B. Chan-Park, Nano Lett., 2018, 18, 4180–4187 CrossRef CAS PubMed.
- H. C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS PubMed.
- A. L. Furst, M. J. Smith and M. B. Francis, Biochemistry, 2018, 57, 3017–3026 CrossRef CAS PubMed.
- H. S. Lee, S. S. Dastgheyb, N. J. Hickok, D. M. Eckmann and R. J. Composto, Biomacromolecules, 2015, 16, 650–659 CrossRef CAS PubMed.
- M. Wilton, L. Charron-Mazenod, R. Moore and S. Lewenza, Antimicrob. Agents Chemother., 2016, 60, 544–553 CrossRef CAS PubMed.
- W. Siala, M. P. Mingeot-Leclercq, P. M. Tulkens, M. Hallin, O. Denis and F. Van Bambeke, Antimicrob. Agents Chemother., 2014, 58, 6385–6397 CrossRef PubMed.
- L. K. Jennings, K. M. Storek, H. E. Ledvina, C. Coulon, L. S. Marmont, I. Sadovskaya, P. R. Secor, B. S. Tseng, M. Scian, A. Filloux, D. J. Wozniak, P. L. Howell and M. R. Parsek, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11353–11358 CrossRef CAS PubMed.
- A. Gupta, R. Das, G. Y. Tonga, T. Mizuhara and V. M. Rotello, ACS Nano, 2018, 12, 89–94 CrossRef CAS PubMed.
- O. Ciofu, T. Tolker-Nielsen, P. O. Jensen, H. Z. Wang and N. Hoiby, Adv. Drug Delivery Rev.Adv Drug Deliver Rev, 2015, 85, 7–23 CrossRef CAS PubMed.
- D. Lebeaux, A. Chauhan, O. Rendueles and C. Beloin, Pathogens, 2013, 2, 288–356 CrossRef PubMed.
- Z. W. Chen, Z. Z. Wang, J. S. Ren and X. G. Qu, Acc. Chem. Res.Accounts Chem Res, 2018, 51, 789–799 CrossRef CAS PubMed.
- B. S. Gomes, B. Simoes and P. M. Mendes, Nat. Rev. Chem., 2018, 2, 0120 CrossRef.
- D. Pornpattananangkul, L. Zhang, S. Olson, S. Aryal, M. Obonyo, K. Vecchio, C. M. Huang and L. Zhang, J. Am. Chem. Soc., 2011, 133, 4132–4139 CrossRef CAS PubMed.
- C. M. Huang, C. H. Chen, D. Pornpattananangkul, L. Zhang, M. Chan, M. F. Hsieh and L. Zhang, Biomaterials, 2011, 32, 214–221 CrossRef CAS PubMed.
- S. Thamphiwatana, W. Gao, M. Obonyo and L. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 17600–17605 CrossRef CAS PubMed.
- X. Pang, Q. Xiao, Y. Cheng, E. Ren, L. Lian, Y. Zhang, H. Gao, X. Wang, W. Leung and X. Chen, ACS Nano, 2019, 13, 2427–2438 CAS.
- Y. Liu, H. J. Busscher, B. Zhao, Y. Li, Z. Zhang, H. C. van der Mei, Y. Ren and L. Shi, ACS Nano, 2016, 10, 4779–4789 CrossRef CAS PubMed.
- B. Horev, M. I. Klein, G. Hwang, Y. Li, D. Kim, H. Koo and D. S. W. Benoit, ACS Nano, 2015, 9, 2390–2404 CrossRef CAS PubMed.
- S. Hussain, J. Joo, J. Kang, B. Kim, G. B. Braun, Z. G. She, D. Kim, A. P. Mann, T. Molder, T. Teesalu, S. Carnazza, S. Guglielmino, M. J. Sailor and E. Ruoslahti, Nat. Biomed. Eng., 2018, 2, 95–103 CrossRef CAS PubMed.
- Y. Wang, Y. A. Nor, H. Song, Y. N. Yang, C. Xu, M. H. Yu and C. Z. Yu, J. Mater. Chem. B, 2016, 4, 2646–2653 RSC.
- Y. A. Nor, H. W. Zhang, S. Purwajanti, H. Song, A. K. Meka, Y. Wang, N. Mitter, D. Mahony and C. Z. Yu, J. Mater. Chem. B, 2016, 4, 7014–7021 RSC.
- H. Song, Y. A. Nor, M. H. Yu, Y. N. Yang, J. Zhang, H. W. Zhang, C. Xu, N. Mitter and C. Z. Yu, J. Am. Chem. Soc., 2016, 138, 6455–6462 CrossRef CAS PubMed.
- R. H. Fang and L. F. Zhang, Adv. Healthc. Mater., 2018, 7, 1800392 CrossRef CAS PubMed.
- S. K. Boda, J. Broda, F. Schiefer, J. Weber-Heynemann, M. Hoss, U. Simon, B. Basu and W. Jahnen-Dechent, Small, 2015, 11, 3183–3193 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- Y. A. Nor, L. Zhou, A. K. Meka, C. Xu, Y. T. Niu, H. W. Zhang, N. Mitter, D. Mahony and C. Z. Yu, Adv. Funct. Mater., 2016, 26, 5408–5418 CrossRef CAS.
- F. Natalio, R. Andre, A. F. Hartog, B. Stoll, K. P. Jochum, R. Wever and W. Tremel, Nat. Nanotechnol., 2012, 7, 530–535 CrossRef CAS PubMed.
- Y. Huang, W. T. Dou, F. Xu, H. B. Ru, Q. Gong, D. Wu, D. Yan, H. Tian, X. P. He and Y. Mai, Angew. Chem., Int. Ed., 2018, 57, 3366–3371 CrossRef CAS PubMed.
- Q. Xin, H. Shah, A. Nawaz, W. Xie, M. Z. Akram, A. Batool, L. Tian, S. U. Jan, R. Boddula, B. Guo, Q. Liu and J. R. Gong, Adv. Mater., 2019, 31, 1804838 CrossRef CAS PubMed.
- J. Zhang, J. Xu, H. Ma, H. Bai, L. Liu, C. Shu, H. Li, S. Wang and C. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 14597–14607 CrossRef CAS PubMed.
- Y.-Q. Zhao, Y. Sun, Y. Zhang, X. Ding, N. Zhao, B. Yu, H. Zhao, S. Duan and F.-J. Xu, ACS Nano, 2020, 14, 2265–2275 CrossRef CAS PubMed.
- X. Yao, G. Zhu, P. Zhu, J. Ma, W. Chen, Z. Liu and T. Kong, Adv. Funct. Mater., 2020, 30, 1909389 CrossRef CAS.
- H. Koo, R. N. Allan, R. P. Howlin, P. Stoodley and L. Hall-Stoodley, Nat. Rev. Microbiol., 2017, 15, 740–755 CrossRef CAS PubMed.
- E. Chee and A. C. Brown, Biomater. Sci., 2020, 8, 1089–1100 RSC.
- P. K. Sahoo, R. Janissen, M. P. Monteiro, A. Cavalli, D. M. Murillo, M. V. Merfa, C. L. Cesar, H. F. Carvalho, A. A. De Souza and E. P. Bakkers, Nano Lett., 2016, 16, 4656–4664 CrossRef CAS PubMed.
- C. Zhu, H. Li, H. B. Wang, B. W. Yao, H. Huang, Y. Liu and Z. H. Kang, Small, 2019, 15, 1900007 CrossRef PubMed.
- W. Bing, L. M. Tian, Y. J. Wang, H. C. Jin, L. Q. Ren and S. Y. Dong, Adv. Mater. Technol., 2019, 4, 1800480 CrossRef.
- L. H. Gao, X. X. Liu, M. X. Xu, G. Sun, S. J. Xu, T. Zou, L. T. M. Wang, F. J. Wang, J. Da, Y. W. Wang and L. Wang, Small, 2021, 17, 2006815 CrossRef CAS PubMed.
- K. Yang, J. R. Shi, L. Wang, Y. Z. Chen, C. Y. Liang, L. Yang and L. N. Wang, J. Mater. Sci. Technol., 2022, 99, 82–100 CrossRef.
- F. Cao, L. Zhang, H. Wang, Y. You, Y. Wang, N. Gao, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2019, 58, 16236–16242 CrossRef CAS PubMed.
- F. Song, L. L. Zhang, R. R. Chen, Q. Liu, J. Y. Liu, J. Yu, P. L. Liu, J. Z. Duan and J. Wang, ACS Appl. Mater. Interfaces, 2021, 13, 33417–33426 CrossRef CAS PubMed.
- J. Lin, X. F. Cai, Z. L. Liu, N. Liu, M. Xie, B. P. Zhou, H. Q. Wang and Z. H. Guo, Adv. Funct. Mater., 2020, 30, 2000398 CrossRef CAS.
- S. Dhingra, V. Gaur, V. Saini, K. Rana, J. Bhattacharya, T. Loho, S. Ray, A. Bajaj and S. Saha, Biomater. Sci., 2022, 10, 3856–3877 RSC.
- G. M. Bruinsma, H. C. van der Mei and H. J. Busscher, Biomaterials, 2001, 22, 3217–3224 CrossRef CAS PubMed.
- L. Caro-Lara, E. Ramos-Moore, I. T. Vargas, M. Walczak, C. Fuentes, A. V. Gomez, N. P. Barrera, J. Castillo and G. Pizarro, Colloids Surf., B, 2021, 202, 111656 CrossRef CAS PubMed.
- W. L. Li, E. S. Thian, M. Wang, Z. Y. Wang and L. Ren, Adv. Sci., 2021, 8, 2100368 CrossRef CAS PubMed.
- Y. Zhan, S. Yu, A. Amirfazli, A. R. Siddiqui and W. Li, Adv. Eng. Mater., 2022, 24, 2101053 CrossRef CAS.
- Y. M. Wang, F. Wang, H. Zhang, B. Yu, H. L. Cong and Y. Q. Shen, Appl. Mater. Today, 2021, 25, 101192 CrossRef.
- H. Fan and Z. Guo, Biomater. Sci., 2020, 8, 1502–1535 RSC.
- L. Liu, H. Shi, H. Yu, S. Yan and S. Luan, Biomater. Sci., 2020, 8, 4095–4108 RSC.
- G. Wang, C. Yang, M. Shan, H. Jia, S. Zhang, X. Chen, W. Liu, X. Liu, J. Chen and X. Wang, Langmuir, 2022, 38, 8987–8998 CrossRef CAS PubMed.
- C. Berne, C. K. Ellison, A. Ducret and Y. V. Brun, Nat. Rev. Microbiol., 2018, 16, 616–627 CrossRef CAS PubMed.
- G. Hwang, S. Kang, M. G. El-Din and Y. Liu, Colloids Surf., B, 2012, 91, 181–188 CrossRef CAS PubMed.
- S. BinAhmed, A. Hasane, Z. Wang, A. Mansurov and S. R. V. Castrillon, Environ. Sci. Technol., 2018, 52, 162–172 CrossRef CAS PubMed.
- K. Doiron, L. Beaulieu, R. St-Louis and K. Lemarchand, Colloids Surf., B, 2018, 167, 524–530 CrossRef CAS PubMed.
- Y. Zou, C. Liu, H. Zhang, Y. Wu, Y. Lin, J. Cheng, K. Lu, L. Li, Y. Zhang and H. Chen, Acta Biomater., 2022, 151, 254–263 CrossRef CAS PubMed.
- Y. X. Zhang, W. Jiang, L. L. Lei, Y. Wang, R. N. Xu, L. Qin and Q. B. Wei, Langmuir, 2022, 38, 7157–7167 CrossRef CAS PubMed.
- A. F. Radovic-Moreno, T. K. Lu, V. A. Puscasu, C. J. Yoon, R. Langer and O. C. Farokhzad, ACS Nano, 2012, 6, 4279–4287 CrossRef CAS PubMed.
- Y.-M. Bai, J. Mao, D.-X. Li, X.-J. Luo, J. Chen, F. R. Tay and L.-N. Niu, Acta Biomater., 2019, 85, 229–240 CrossRef CAS PubMed.
- P. P. Kalelkar, Z. Geng, M. Finn and D. M. Collard, Biomacromolecules, 2019, 20, 3366–3374 CrossRef CAS PubMed.
- J. Lin, X. Chen, C. Chen, J. Hu, C. Zhou, X. Cai, W. Wang, C. Zheng, P. Zhang, J. Cheng, Z. Guo and H. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 6124–6136 CrossRef CAS PubMed.
- R. Wang, Y. Li, Y. Si, F. Wang, Y. Liu, Y. Ma, J. Yu, X. Yin and B. Ding, Nanoscale Adv., 2019, 1, 1948–1956 RSC.
- N. Nazi, V. Humblot and C. Debiemme-Chouvy, Langmuir, 2020, 36, 11005–11014 CrossRef CAS PubMed.
- Z. Huang, S. Nazifi, P. Jafari, A. Karim and H. Ghasemi, ACS Appl. Bio Mater., 2020, 3, 911–919 CrossRef CAS PubMed.
- Y. He, X. Wan, K. Xiao, W. Lin, J. Li, Z. Li, F. Luo, H. Tan, J. Li and Q. Fu, Biomater. Sci., 2019, 7, 5369–5382 RSC.
- R. Gharibi and S. Agarwal, ACS Appl. Bio Mater., 2021, 4, 4629–4640 CrossRef CAS PubMed.
- H. Han, J. Zhu, D. Q. Wu, F. X. Li, X. L. Wang, J. Y. Yu and X. H. Qin, Adv. Funct. Mater., 2019, 29, 1806594 CrossRef.
- Y. Ma, J. Li, Y. Si, K. Huang, N. Nitin and G. Sun, ACS Appl. Mater. Interfaces, 2019, 11, 17814–17822 CrossRef CAS PubMed.
- L. Segev-Zarko, R. Saar-Dover, V. Brumfeld, M. L. Mangoni and Y. Shai, Biochem. J., 2015, 468, 259–270 CrossRef CAS PubMed.
- A. Khan, M. Xu, T. Wang, C. You, X. Wang, H. Ren, H. Zhou, A. Khan, C. Han and P. Li, Biosci. Rep., 2019, 39, BSR20190504 CrossRef CAS PubMed.
- M. Xu, A. Khan, T. Wang, Q. Song, C. Han, Q. Wang, L. Gao, X. Huang, P. Li and W. Huang, ACS Appl. Bio Mater., 2019, 2, 3329–3340 CrossRef CAS PubMed.
- L. H. Peng, Y. F. Huang, C. Z. Zhang, J. Niu, Y. Chen, Y. Chu, Z. H. Jiang, J. Q. Gao and Z. W. Mao, Biomaterials, 2016, 103, 137–149 CrossRef CAS PubMed.
- S. Guo, Q. Huang, Y. Chen, J. Wei, J. Zheng, L. Wang, Y. Wang and R. Wang, Angew. Chem., Int. Ed., 2021, 60, 618–623 CrossRef CAS PubMed.
- M. Michalska, F. Gambacorta, R. Divan, I. S. Aranson, A. Sokolov, P. Noirot and P. D. Laible, Nanoscale, 2018, 10, 6639–6650 RSC.
- J. Jenkins, J. Mantell, C. Neal, A. Gholinia, P. Verkade, A. H. Nobbs and B. Su, Nat. Commun., 2020, 11, 1626 CrossRef CAS PubMed.
- G. Choi, Y. Song, H. Lim, S. H. Lee, H. K. Lee, E. Lee, B. G. Choi, J. J. Lee, S. G. Im and K. G. Lee, Adv. Healthc. Mater., 2020, 9, 2000447 CrossRef CAS PubMed.
- R. M. Mendes, A. P. Francisco, F. A. Carvalho, M. Dardouri, B. Costa, A. F. Bettencourt, J. Costa, L. Goncalves, F. Costa and I. A. C. Ribeiro, Colloids Surf., B, 2021, 208, 112057 CrossRef CAS PubMed.
- Z. Zhu, Q. Gao, Z. Long, Q. Huo, Y. Ge, N. Vianney, N. A. Daliko, Y. Meng, J. Qu and H. Chen, Bioact. Mater., 2021, 6, 2546–2556 CrossRef CAS PubMed.
- S. Zhang, L. Wang, X. Liang, J. Vorstius, R. Keatch, G. Corner, G. Nabi, F. Davidson, G. M. Gadd and Q. Zhao, ACS Biomater. Sci. Eng., 2019, 5, 2804–2814 CrossRef CAS PubMed.
- H. Yazdani-Ahmadabadi, D. F. Felix, K. Yu, H. H. Yeh, H. D. Luo, S. Khoddami, L. E. Takeuchi, A. Alzahrani, S. Abbina, Y. Mei, L. Fazli, D. Grecov, D. Lange and J. N. Kizhakkedathu, ACS Cent. Sci., 2022, 8, 546–561 CrossRef CAS PubMed.
- A. Arenas-Vivo, G. Amariei, S. Aguado, R. Rosal and P. Horcajada, Acta Biomater., 2019, 97, 490–500 CrossRef CAS PubMed.
- K. Huang, J. Wang, Q. Zhang, K. Yuan, Y. Yang, F. Li, X. Sun, H. Chang, Y. Liang and J. Zhao, Adv. Funct. Mater., 2022, 2204906 CrossRef CAS.
- T. Wei, Q. Yu and H. Chen, Adv. Healthc. Mater., 2019, 8, 1801381 CrossRef CAS PubMed.
- W. Ahmed, Z. Zhai and C. Gao, Mater. Today Bio, 2019, 2, 100017 CrossRef CAS PubMed.
- K. Giri, L. R. Yepes, B. Duncan, P. K. Parameswaran, B. Yan, Y. Jiang, M. Bilska, D. F. Moyano, M. A. Thompson and V. M. Rotello, RSC Adv., 2015, 5, 105551–105559 RSC.
- D. Hu, H. Li, B. Wang, Z. Ye, W. Lei, F. Jia, Q. Jin, K. F. Ren and J. Ji, ACS Nano, 2017, 11, 9330–9339 CrossRef CAS PubMed.
- X. Chen, X. Zhang, F. Lin, Y. Guo and F. G. Wu, Small, 2019, 15, 1901647 CrossRef PubMed.
- Q. Deng, P. Sun, L. Zhang, Z. Liu, H. Wang, J. Ren and X. Qu, Adv. Funct. Mater., 2019, 29, 1903018 CrossRef.
- X. Ju, J. Chen, M. Zhou, M. Zhu, Z. Li, S. Gao, J. Ou, D. Xu, M. Wu and S. Jiang, ACS Appl. Mater. Interfaces, 2020, 12, 13731–13738 CrossRef CAS PubMed.
- Y. Gao, J. Wang, M. Chai, X. Li, Y. Deng, Q. Jin and J. Ji, ACS Nano, 2020, 14, 5686–5699 CrossRef CAS PubMed.
- J. Wu, F. Li, X. Hu, J. Lu, X. Sun, J. Gao and D. Ling, ACS Cent. Sci., 2019, 5, 1366–1376 CrossRef CAS PubMed.
- X. Li, Y.-C. Yeh, K. Giri, R. Mout, R. F. Landis, Y. Prakash and V. M. Rotello, Chem. Commun., 2015, 51, 282–285 RSC.
- K. R. Sims Jr., J. P. Maceren, Y. Liu, G. R. Rocha, H. Koo and D. S. Benoit, Acta Biomater., 2020, 115, 418–431 CrossRef PubMed.
- M. Chai, Y. Gao, J. Liu, Y. Deng, D. Hu, Q. Jin and J. Ji, Adv. Healthc. Mater., 2020, 9, 1901542 CrossRef CAS PubMed.
- Y. Xi, Y. Wang, J. Gao, Y. Xiao and J. Du, ACS Nano, 2019, 13, 13645–13657 CrossRef CAS PubMed.
- D. Hu, Y. Deng, F. Jia, Q. Jin and J. Ji, ACS Nano, 2019, 14, 347–359 CrossRef PubMed.
- H. Han, Y. Gao, M. Chai, X. Zhang, S. Liu, Y. Huang, Q. Jin, A. Grzybowski, J. Ji and K. Yao, J. Controlled Release, 2020, 327, 676–687 CrossRef CAS PubMed.
- S. Wu, C. Xu, Y. Zhu, L. Zheng, L. Zhang, Y. Hu, B. Yu, Y. Wang and F. J. Xu, Adv. Funct. Mater., 2021, 31, 2103591 CrossRef CAS.
- Z. Qiao, Y. Yao, S. Song, M. Yin and J. Luo, J. Mater. Chem. B, 2019, 7, 830–840 RSC.
- B. E. de Avila, P. Angsantikul, J. Li, M. A. Lopez-Ramirez, D. E. Ramirez-Herrera, S. Thamphiwatana, C. Chen, J. Delezuk, R. Samakapiruk, V. Ramez, M. Obonyo, L. Zhang and J. Wang, Nat. Commun., 2017, 8, 272 CrossRef PubMed.
- T. Cui, S. Wu, Y. Sun, J. Ren and X. Qu, Nano Lett., 2020, 20, 7350–7358 CrossRef CAS PubMed.
- C. Zhang, C. Du, J.-Y. Liao, Y. Gu, Y. Gong, J. Pei, H. Gu, D. Yin, L. Gao and Y. Pan, Biomater. Sci., 2019, 7, 2833–2840 RSC.
- A. A. Balhaddad, Y. Xia, Y. Lan, L. Mokeem, M. S. Ibrahim, M. D. Weir, H. H. Xu and M. A. S. Melo, ACS Nano, 2021, 15, 19888–19904 CrossRef CAS PubMed.
- N. G. Durmus and T. J. Webster, Adv. Healthc. Mater., 2013, 2, 165–171 CrossRef CAS PubMed.
- K. Quan, Z. Zhang, Y. Ren, H. J. Busscher, H. C. van der Mei and B. W. Peterson, ACS Biomater. Sci. Eng., 2019, 6, 205–212 CrossRef PubMed.
- J. Zhu, J. Tian, C. Yang, J. Chen, L. Wu, M. Fan and X. Cai, Small, 2021, 17, 2101495 CrossRef CAS PubMed.
- Z. Shen, S. Zheng, S. Xiao, R. Shen, S. Liu and J. Hu, Angew. Chem., Int. Ed., 2021, 60, 20452–20460 CrossRef CAS PubMed.
- Y. Zhu, S. Wu, Y. Sun, X. Zou, L. Zheng, S. Duan, J. Wang, B. Yu, R. Sui and F. J. Xu, Adv. Funct. Mater., 2022, 32, 2111066 CrossRef CAS.
- W. Gao, Y. Chen, Y. Zhang, Q. Zhang and L. Zhang, Adv. Drug Delivery Rev., 2018, 127, 46–57 CrossRef CAS PubMed.
- Y. Hu, X. Ruan, X. Lv, Y. Xu, W. Wang, Y. Cai, M. Ding, H. Dong, J. Shao and D. Yang, Nano Today, 2022, 46, 101602 CrossRef CAS.
- Y. Shi, Y. Cao, J. Cheng, W. Yu, M. Liu, J. Yin, C. Huang, X. Liang, H. Zhou and H. Liu, Adv. Funct. Mater., 2022, 2111148 CrossRef CAS.
- P. Zhang, S. Wu, J. Li, X. Bu, X. Dong, N. Chen, F. Li, J. Zhu, L. Sang and Y. Zeng, Theranostics, 2022, 12, 4818 CrossRef CAS PubMed.
- L. Xia, J. Tian, T. Yue, H. Cao, J. Chu, H. Cai and W. Zhang, Adv. Healthc. Mater., 2022, 11, 2102015 CrossRef CAS PubMed.
- T. Kim, Q. Zhang, J. Li, L. Zhang and J. V. Jokerst, ACS Nano, 2018, 12, 5615–5625 CrossRef CAS PubMed.
- Y. Xiao, M. Xu, N. Lv, C. Cheng, P. Huang, J. Li, Y. Hu and M. Sun, Acta Biomater., 2021, 122, 291–305 CrossRef CAS PubMed.
- K. Sauer, P. Stoodley, D. M. Goeres, L. Hall-Stoodley, M. Burmolle, P. S. Stewart and T. Bjarnsholt, Nat. Rev. Microbiol., 2022, 20, 608–620 CrossRef CAS PubMed.
- O. Ciofu, C. Moser, P. O. Jensen and N. Hoiby, Nat. Rev. Microbiol., 2022, 20, 621–635 CrossRef CAS PubMed.
- J. J. Li, Y. Hu, B. Hu, W. Wang, H. Xu, X. Y. Hu, F. Ding, H. B. Li, K. R. Wang, X. Zhang and D. S. Guo, Nat. Commun., 2022, 13, 6279 CrossRef CAS PubMed.
- Z. Wang, X. Liu, Y. Duan and Y. Huang, Biomaterials, 2022, 280, 121249 CrossRef CAS PubMed.
- M. Proietti, L. Perruzza, D. Scribano, G. Pellegrini, R. D'Antuono, F. Strati, M. Raffaelli, S. F. Gonzalez, M. Thelen, W. D. Hardt, E. Slack, M. Nicoletti and F. Grassi, Nat. Commun., 2019, 10, 250 CrossRef PubMed.
- Y. Weng, H. Chen, X. Chen, H. Yang, C. H. Chen and H. Tan, Nat. Commun., 2022, 13, 4712 CrossRef CAS PubMed.
- H. Yu, X. Xu, Z. Xie, X. Huang, L. Lin, Y. Jiao and H. Li, ACS Appl. Mater. Interfaces, 2022, 14, 36947–36956 CrossRef CAS PubMed.
- S. Yu, G. Li, P. Zhao, Q. Cheng, Q. He, D. Ma and W. Xue, Adv. Funct. Mater., 2019, 29, 1905697 CrossRef CAS.
- X. L. Hu, L. Chu, X. Dong, G. R. Chen, T. Tang, D. Chen, X. P. He and H. Tian, Adv. Funct. Mater., 2019, 29, 1806986 CrossRef.
- B. Cao, X. Lyu, C. Wang, S. Lu, D. Xing and X. Hu, Biomaterials, 2020, 262, 120341 CrossRef CAS PubMed.
- Z. H. Yu, X. Li, F. Xu, X. L. Hu, J. Yan, N. Kwon, G. R. Chen, T. Tang, X. Dong and Y. Mai, Angew. Chem., 2020, 132, 3687–3693 CrossRef.
- L. Tan, J. Li, X. Liu, Z. Cui, X. Yang, S. Zhu, Z. Li, X. Yuan, Y. Zheng and K. W. Yeung, Adv. Mater., 2018, 30, 1801808 CrossRef PubMed.
- Z. Yuan, B. Tao, Y. He, C. Mu, G. Liu, J. Zhang, Q. Liao, P. Liu and K. Cai, Biomaterials, 2019, 223, 119479 CrossRef CAS PubMed.
- X. Sun, J. Sun, Y. Sun, C. Li, J. Fang, T. Zhang, Y. Wan, L. Xu, Y. Zhou and L. Wang, Adv. Funct. Mater., 2021, 31, 2101040 CrossRef CAS.
- Y. Qiao, F. Ma, C. Liu, B. Zhou, Q. Wei, W. Li, D. Zhong, Y. Li and M. Zhou, ACS Appl. Mater. Interfaces, 2018, 10, 193–206 CrossRef CAS PubMed.
- H. Tang, Y. Liu, B. Li, B. Shang, J. Yang, C. Zhang, L. Yang, K. Chen, W. Wang and J. Liu, Bioact. Mater., 2021, 6, 4758–4771 CrossRef CAS PubMed.
- L. Ran, B. Lu, H. Qiu, G. Zhou, J. Jiang, E. Hu, F. Dai and G. Lan, Bioact. Mater., 2021, 6, 2956–2968 CrossRef CAS PubMed.
- X. Zhang, G. Zhang, M. Chai, X. Yao, W. Chen and P. K. Chu, Bioact. Mater., 2021, 6, 12–25 CrossRef CAS PubMed.
- H. Hu, H. Wang, Y. Yang, J. F. Xu and X. Zhang, Angew. Chem., Int. Ed., 2022, e202200799 CAS.
- X. Wei, H. Sun, Y. Bai, Y. Zhang, Z. Ma, J. Li and X. Zhang, Biomater. Sci., 2020, 8, 6912–6919 RSC.
- A. Elbourne, S. Cheeseman, P. Atkin, N. P. Truong, N. Syed, A. Zavabeti, M. Mohiuddin, D. Esrafilzadeh, D. Cozzolino and C. F. McConville, ACS Nano, 2020, 14, 802–817 CrossRef CAS PubMed.
- Y. Dong, L. Wang, K. Yuan, F. Ji, J. Gao, Z. Zhang, X. Du, Y. Tian, Q. Wang and L. Zhang, ACS Nano, 2021, 15, 5056–5067 CrossRef CAS PubMed.
- Y. Liu, Y. Huang, D. Kim, Z. Ren, M. J. Oh, D. P. Cormode, A. T. Hara, D. T. Zero and H. Koo, Nano Lett., 2021, 21, 9442–9449 CrossRef CAS PubMed.
- J. Li, R. Nickel, J. Wu, F. Lin, J. van Lierop and S. Liu, Nanoscale, 2019, 11, 6905–6915 RSC.
- J. Wang, L. Wang, J. Pan, J. Zhao, J. Tang, D. Jiang, P. Hu, W. Jia and J. Shi, Adv. Sci., 2021, 8, 2004010 CrossRef CAS PubMed.
- J. Wu, T. Ma, M. Zhu, T. Huang, B. Zhang, J. Gao and N. Lin, Nano Today, 2022, 46, 101577 CrossRef CAS.
- L. Dieltjens, K. Appermans, M. Lissens, B. Lories, W. Kim, E. V. Van der Eycken, K. R. Foster and H. P. Steenackers, Nat. Commun., 2020, 11, 107 CrossRef CAS PubMed.
- M. Wei, J. Wu, H. Sun, B. Zhang, X. Hu, Q. Wang, B. Li, L. Xu, T. Ma, J. Gao, F. Li and D. Ling, Small, 2022, 19, 2205471 CrossRef PubMed.
- P. Li, X. Chen, Y. Shen, H. Li, Y. Zou, G. Yuan, P. Hu and H. Hu, J. Controlled Release, 2019, 300, 52–63 CrossRef CAS PubMed.
- Y. Shen, Y. Zou, X. Chen, P. Li, Y. Rao, X. Yang, Y. Sun and H. Hu, J. Controlled Release, 2020, 328, 575–586 CrossRef CAS PubMed.
- R. Otsuka, S. Imai, T. Murata, Y. Nomura, M. Okamoto, H. Tsumori, E. Kakuta, N. Hanada and Y. Momoi, Microbiol. Immunol., 2015, 59, 28–36 CrossRef CAS PubMed.
- A. Wang, P. J. Weldrick, L. A. Madden and V. N. Paunov, ACS Appl. Mater. Interfaces, 2021, 13, 22182–22194 CrossRef CAS PubMed.
- H. Zhang, Y. Zhu, Y. Li, X. Qi, J. Yang, H. Qi, Q. Li, Y. Ma, Y. Zhang and X. Zhang, Adv. Funct. Mater., 2021, 31, 2104799 CrossRef CAS.
- F. Vatansever, W. C. de Melo, P. Avci, D. Vecchio, M. Sadasivam, A. Gupta, R. Chandran, M. Karimi, N. A. Parizotto, R. Yin, G. P. Tegos and M. R. Hamblin, FEMS Microbiol. Rev., 2013, 37, 955–989 CrossRef CAS PubMed.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- B. Yang, Y. Chen and J. Shi, Adv. Mater., 2019, 31, e1901778 CrossRef PubMed.
- D. P. Cormode, L. Gao and H. Koo, Trends Biotechnol., 2018, 36, 15–29 CrossRef CAS PubMed.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- M. Huo, L. Wang, H. Zhang, L. Zhang, Y. Chen and J. Shi, Small, 2019, 15, e1901834 CrossRef PubMed.
- Y. Wang, X. Shen, S. Ma, Q. Guo, W. Zhang, L. Cheng, L. Ding, Z. Xu, J. Jiang and L. Gao, Biomater. Sci., 2020, 8, 2447–2458 RSC.
- G. Hwang, A. J. Paula, E. E. Hunter, Y. Liu, A. Babeer, B. Karabucak, K. Stebe, V. Kumar, E. Steager and H. Koo, Sci. Robot., 2019, 4, eaaw2388 CrossRef PubMed.
- P. C. Naha, Y. Liu, G. Hwang, Y. Huang, S. Gubara, V. Jonnakuti, A. Simon-Soro, D. Kim, L. Gao and H. Koo, ACS Nano, 2019, 13, 4960–4971 CrossRef CAS PubMed.
- L. Gao, Y. Liu, D. Kim, Y. Li, G. Hwang, P. C. Naha, D. P. Cormode and H. Koo, Biomaterials, 2016, 101, 272–284 CrossRef CAS PubMed.
- Y. Tao, E. Ju, J. Ren and X. Qu, Adv. Mater., 2015, 27, 1097–1104 CrossRef CAS PubMed.
- Z. Liu, F. Wang, J. Ren and X. Qu, Biomaterials, 2019, 208, 21–31 CrossRef CAS PubMed.
- T. L. Shi, X. Hou, S. Q. Guo, L. Zhang, C. H. Wei, T. Peng and X. G. Hu, Nat. Commun., 2021, 12, 493 CrossRef CAS PubMed.
- J. M. V. Makabenta, A. Nabawy, C.-H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- H.-C. Flemming, E. D. van Hullebusch, T. R. Neu, P. H. Nielsen, T. Seviour, P. Stoodley, J. Wingender and S. Wuertz, Nat. Rev. Microbiol., 2023, 21, 70–86 CrossRef CAS PubMed.
- F. Sedlmayer, T. Jaeger, U. Jenal and M. Fussenegger, Nano Lett., 2017, 17, 5043–5050 CrossRef CAS PubMed.
- Y. Sun, H. Qin, Z. Yan, C. Zhao, J. Ren and X. Qu, Adv. Funct. Mater., 2019, 29, 1808222 CrossRef.
- A. Ivanova, K. Ivanova, A. Tied, T. Heinze and T. Tzanov, Adv. Funct. Mater., 2020, 30, 2001284 CrossRef CAS.
- G. Zhang, Y. Yang, J. Shi, X. Yao, W. Chen, X. Wei, X. Zhang and P. K. Chu, Biomaterials, 2021, 269, 120634 CrossRef CAS PubMed.
- Z. Zhou, S. Li, G. Wei, W. Liu, Y. Zhang, C. Zhu, S. Liu, T. Li and H. Wei, Adv. Funct. Mater., 2022, 2206294 CrossRef CAS.
- D. Hu, L. Zou, W. Yu, F. Jia, H. Han, K. Yao, Q. Jin and J. Ji, Adv. Sci., 2020, 7, 2000398 CrossRef CAS PubMed.
- W. Xiu, L. Wan, K. Yang, X. Li, L. Yuwen, H. Dong, Y. Mou, D. Yang and L. Wang, Nat. Commun., 2022, 13, 3875 CrossRef CAS PubMed.
- W. Zhu, J. Mei, X. Zhang, J. Zhou, D. Xu, Z. Su, S. Fang, J. Wang, X. Zhang and C. Zhu, Adv. Mater., 2022, 2207961 CrossRef CAS PubMed.
- C. Yang, J. Li, C. Zhu, Q. Zhang, J. Yu, J. Wang, Q. Wang, J. Tang, H. Zhou and H. Shen, Acta Biomater., 2019, 89, 403–418 CrossRef CAS PubMed.
- C. Yang, Y. Luo, H. Shen, M. Ge, J. Tang, Q. Wang, H. Lin, J. Shi and X. Zhang, Nat. Commun., 2022, 13, 4866 CrossRef CAS PubMed.
- J. Jo, A. Price-Whelan and L. E. P. Dietrich, Nat. Rev. Microbiol., 2022, 20, 593–607 CrossRef CAS PubMed.
- K. P. Rumbaugh and K. Sauer, Nat. Rev. Microbiol., 2020, 18, 571–586 CrossRef CAS PubMed.
- L. E. Low, J. Wu, J. Lee, B. T. Tey, B.-H. Goh, J. Gao, F. Li and D. Ling, J. Controlled Release, 2020, 324, 69–103 CrossRef CAS PubMed.
- L. Li, R. He, H. Yan, Z. Leng, S. Zhu and Z. Gu, Nano Today, 2022, 47, 101654 CrossRef CAS.
- H. Wu, F. Xia, L. Zhang, C. Fang, J. Lee, L. Gong, J. Gao, D. Ling and F. Li, Adv. Mater., 2022, 34, 2108348 CrossRef CAS PubMed.
- C. Zhang, R. Sun and T. Xia, Nano Today, 2020, 34, 100909 CrossRef CAS.
- C. Gunawan, C. P. Marquis, R. Amal, G. A. Sotiriou, S. A. Rice and E. J. Harry, ACS Nano, 2017, 11, 3438–3445 CrossRef CAS PubMed.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek, O. Tomanec and R. Zbořil, Nat. Nanotechnol., 2018, 13, 65–71 CrossRef PubMed.
- L. M. Stabryla, K. A. Johnston, N. A. Diemler, V. S. Cooper, J. E. Millstone, S.-J. Haig and L. M. Gilbertson, Nat. Nanotechnol., 2021, 16, 996–1003 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |