Modulating liquid–liquid phase separation of FUS: mechanisms and strategies
Yanglimin
Ji
 ab,
Fen
Li
ab,
Fen
Li
 ab and
Yan
Qiao
ab and
Yan
Qiao
 *ab
*ab
aBeijing National Laboratory for Molecular Sciences (BNLMS), Laboratory of Polymer Physics and Chemistry, CAS Research/Education Center for Excellence in Molecular Sciences, Institute of Chemistry, Chinese Academy of Sciences, Beijing, 100190, China. E-mail: yanqiao@iccas.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing, 100049, China
First published on 21st October 2022
Abstract
Liquid–liquid phase separation (LLPS) of biomolecules inspires the construction of protocells and drives the formation of cellular membraneless organelles. The resulting biomolecular condensates featuring dynamic assembly, disassembly, and phase transition play significant roles in a series of biological processes, including RNA metabolism, DNA damage response, signal transduction and neurodegenerative disease. Intensive investigations have been conducted for understanding and manipulating intracellular phase-separated disease-related proteins (e.g., FUS, tau and TDP-43). Herein, we review current studies on the regulation strategies of intracellular LLPS focusing on FUS, which are categorized into physical stimuli, biochemical modulators, and protein structural modifications, with summarized molecular mechanisms. This review is expected to provide a sketch of the modulation of FUS LLPS with its pros and cons, and an outlook for the potential clinical treatments of neurodegenerative diseases.
1. Introduction
Liquid–liquid phase separation (LLPS) underlies the formation of coacervate protocells and intracellular membraneless organelles.1,2 Coacervate microdroplets based on LLPS were hypothesized to be relevant to the origin of life on the Earth by Oparin early in the 1930s.3 In the past decade, coacervates have been studied extensively as protocells/synthetic cells due to their abilities of compartmentalization,4,5 molecular enrichment,6,7 reaction localization and enhancement,8–10 active growth and division,11–14 communication,15,16 as well as artificial predation and phagocytosis,17,18 protocell–natural cell interaction,19,20 intracellular delivery,21–23 protocell-based microactuator translocation,24 and network-driven signal processing.25 On the other side, the membraneless organelles based on intracellular condensation were known since the discovery of liquid-like P granules in dividing embryos of Caenorhabditis elegans by Hyman and Brangwynne et al.26 These liquid-like organelles without membranes as a typical intracellular compartmentalization form, such as nucleoli, Cajal bodies, stress granules, and P bodies,27–30 are widely associated with cellular environmental sensing,31 transcription regulation,32,33 DNA damage repairing,34 RNA processing and transportation,35 neurotransmitter release,36,37 innate immunity,38 and the episode of neurodegenerative diseases in organisms.39Membraneless organelles are demonstrated to be formed by the LLPS of proteins and nucleic acids with multivalent interactions, which can be categorized into three cases.40,41 First, proteins that contain low complexity (LC) domains, also named intrinsically disordered regions (IDRs), have high potential to undergo phase separation, such as fused in sarcoma (FUS), LAF1 and Ddx4. This phase separation is driven by the dynamic and multivalent interactions from the inherent disordered structures and multiple binding sites, including cation–π, electrostatic, hydrophobic interactions, etc.42 Second, specific protein interactions can likewise induce LLPS, and are identified in the condensates of NCK and N-WASP proteins with their repeating SH3 domains and PRM domains, respectively.43 Third, nucleic acid-mediated phase separation is driven by unspecific electrostatic attraction or specific DNA/RNA–protein binding.44
FUS as a representative RNA-binding protein distributes in nuclear and cytoplasmic condensates, involving in the formation of stress granules and modulation of RNA processes.45 With a high intrinsic propensity to self-assemble, FUS undergoes LLPS and further liquid–solid phase transition (LSPT) when pathological cells form irreversible amyloid fibers under environmental stress, which have been closely associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).46,47 These neurodegenerative diseases can be avoided by preventing persistent accumulation of biocondensates.48 In this review, we focus on the modulation of LLPS using the FUS system as a representative. By introducing the structure of FUS, clarifying the driving forces of LLPS and summarizing the physical, chemical and biological tools for phase separation regulation, we intend to evaluate these general strategies to leverage our understanding for the development of potential preventing methods for neurodegenerative diseases and provide a basic research pathway for LLPS-related proteins and diseases.
2. Structure and phase separation of FUS
Full-length FUS containing 526 residues is usually divided into seven parts, that is, a disordered N-terminal domain (NTD) of LC enriched in serine, tyrosine, glycine and glutamine (QGSY) residues, three arginine–glycine–glycine-rich domains (RGG), an RNA recognition motif (RRM), a zinc-finger domain (ZnF) and a proline–tyrosine nuclear localization sequence (PY-NLS) (Fig. 1a).49 FUS LC (residues 1–163 or 1–214) and RGG are the crucial regions that drive phase separation (Fig. 1b).50,51 The former has few charged residues but is rich in hydrophobic residues (such as tyrosine), driving LLPS and amyloid aggregation independently. The latter mediates self-association and nucleic acid binding through charged arginine residues.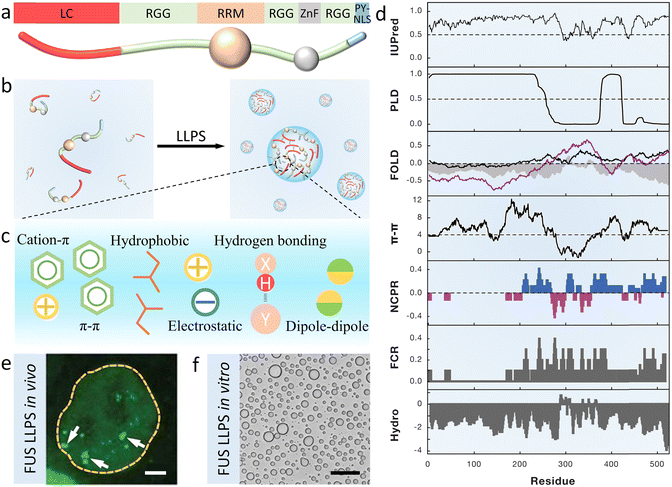 | ||
| Fig. 1 Structure and phase separation of FUS. (a) Schematic image of full-length FUS domain architecture, including a QGSY-rich LC domain, three arginine–glycine-rich RGG regions, an RRM, a ZnF domain and a PY-NLS. (b) Cartoon showing LLPS of FUS driven by multivalent interaction containing cation–π, π–π, electrostatic, and hydrophobic interaction, hydrogen bonding and dipole–dipole interaction (c). (d) Bioinformatic analysis of the FUS sequence including predictions of the intrinsic disorder region (IUPred), prion-like region (PLD), fold index (FOLD, gray filling), π–π interaction (π–π), net charge per residue (NCPR), fraction of charged residues (FCR) and hydrophobicity (HYDRO). Reproduced from ref. 52 with permission, Copyright Elsevier (2019). (e) Fluorescence microscopy image of HeLa cells showing green coacervate droplets formed by GFP-FUS in vivo. Scale bar, 2 μm. Reproduced from ref. 53 with permission, Copyright Science (2018). (f) Optical microscopy image of FUS LC condensates in vitro. Scale bar, 50 μm. Reproduced from ref. 51 with permission, Copyright Elsevier (2017). | ||
Cation–π interaction between arginine and tyrosine is the main driving force for the LLPS of FUS.50 Moreover, other non-covalent interactions including the π–π interaction, hydrophobic interaction, electrostatic interaction, hydrogen bonding and dipole–dipole interaction are also considered to promote condensation (Fig. 1c). To better understand these interactions, sequence analysis tools with algorithms are used to predict the physicochemical properties of FUS (Fig. 1d), including the predictions of the intrinsic disorder region (IUPred), prion-like region (PLD), fold index (FOLD), π–π and hydrophobicity interaction (π–π and HYDRO), distribution of charged residues of net charge per residue (NCPR) and fraction of charged residues (FCRs). These methods have been well reviewed by Alberti, Gladfelter and Mittag.52
The formation of FUS coacervates can be observed in vivo by overexpressing the GFP-tagged FUS in HeLa cells (Fig. 1e).53 Not only in mammalian cells but also in eukaryotic fungus yeast and prokaryotic bacteria E. coli, FUS are expressed and purified.54,55 FUS condensates are investigated in vitro and in vivo due to the common mechanism of phase separation. As shown in Fig. 1f, purified FUS can also undergo LLPS in certain buffers.51 Furthermore, fluorescence recovery after photobleaching (FRAP) and droplet fusing experiments are widely used to identify the liquid-like properties of FUS condensates.46,56–58
3. Modulation of FUS phase separation by physical stimulation
The physical conditions of the external environment in which organisms live have significant impacts on the physiological processes. Cells respond to environmental stimuli by viscoadaptation,59 ion transportation,60 membrane potential alteration,61 and membraneless organelle formation.31 Intracellular dynamic phase separation of FUS implicates in RNA metabolic pathways including transcription, pre-mRNA splicing and miRNA processing,45 which can be modulated by the environmental factors, e.g., temperature, pressure and light. These physical stimuli regulate the LLPS of FUS by changing the inter- and intramolecular interactions, protein conformations and modular aggregations, whose process can also be utilized to study the kinetics, mechanisms and applications of condensates. Temperature and pressure are outstanding for their insignificant alteration of the original compositions of the systems, although they may bring side effects to cellular functions, while light modulation is relatively complicated for the requirement of protein restructuring and genetic modification.3.1 Temperature
FUS protein exhibits thermal-responsive LLPS, whose temperature–concentration phase diagram defines one- and two-phase regimes with the phase boundary exhibiting an upper critical solution temperature (UCST) (Fig. 2a).52 A decrease in temperature favours the condensation of FUS since the contribution of increased molecular interactions at lower temperatures is greater than that of entropic reduction during phase separation.40 The temperature-induced FUS phase separation is commonly characterized by turbidity and optical/fluorescence microscopy due to the generation of micron-sized spherical droplets.58,62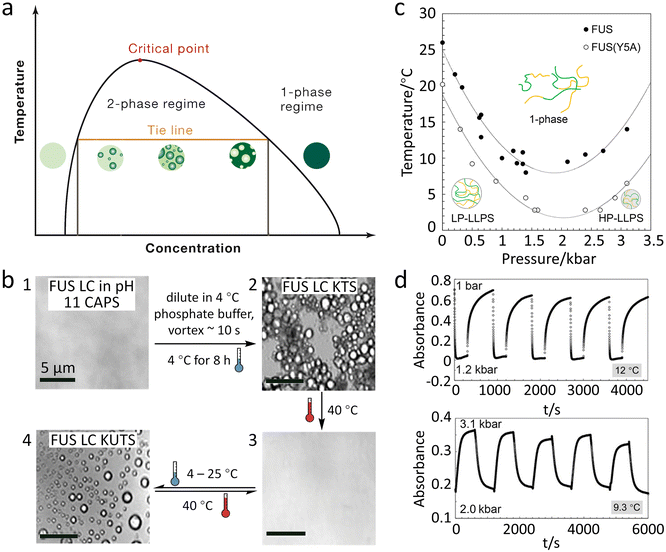 | ||
| Fig. 2 Reversible LLPS of FUS under temperature and pressure stimuli. (a) Schematic phase diagram of the FUS system modulated by temperature showing an UCST of LLPS. Reproduced from ref. 52 with permission, Copyright Elsevier (2019). (b) Microscope images presenting the formation of KTS (1 to 2) and KUTS (2 to 4) of FUS LC condensates. There are reversible temperature responses of KUTS condensates by altering temperature (3 to 4). Reproduced from ref. 63 with permission, Copyright John Wiley and Sons (2022). (c) Pressure–temperature phase diagram for wild-type FUS (solid circles) and FUS with the Y5A variant (open circles), showing that LP- and HP-LLPS are formed at low and high pressure, respectively. (d) Pressure jump relaxation study of FUS indicates reversible condensation under pressure cycles at 12 °C (upper) and 9.3 °C (lower). Reproduced from ref. 66 with permission, Copyright American Chemical Society (2021). | ||
Although the reversible formation of FUS coacervates by temperature cycling has been widely observed, the thermodynamic processes are less discussed. Parekh et al. reported that FUS LC condensates generated through different thermal treatments have two states of molecular organization, i.e., the kinetically trapped state (KTS) and untrapped state (KUTS).63 FUS condensates in the KTS are obtained by incubation at 4 °C for 8 h and show distinct boundaries when they come in contact with one another (Fig. 2b, 1 to 2), while the KUTS condensates are formed by an irreversible annealing process of heating KTS condensates to 40 °C followed by cooling to room temperature (Fig. 2b, 2 to 4). Unlike KTS condensates, KUTS condensates can fuse and reversibly form under temperature cycles (Fig. 2b, 3 to 4). Thermal annealing of KTS condensates allows FUS LC proteins to escape the kinetic trap and reconfigure a more favourable thermodynamic state. The authors find that temperature alteration during the condensation process can significantly affect the distribution of amino acids in the condensates. As a result, proteins in KTS condensates expose more hydrophobic residues and have increased β-sheet conformation toward the aqueous buffer than those in KUTS condensates, leading to the formation of more hydrophobic interfaces and inhibition of fusion. Moreover, tyrosines in KTS condensates show increased hydrogen bonding with other tyrosines, resulting in stronger protein–protein interactions and more crowded internal environments.
3.2 Pressure
Pressure modulates the phase separation of FUS towards the thermodynamic equilibrium state in which protein favours the folding structure with the lowest Gibbs free energy, leading to redistribution of amino acids.64 The behaviours of FUS under varying hydrostatic pressure can be explained using a molar excess function of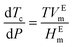 , where Tc is the critical solution temperature and VEm and HEm are the molar excess volume and the molar excess enthalpy of the mixture, respectively.65 FUS as a phase separation protein with UCST has positive HEm.
, where Tc is the critical solution temperature and VEm and HEm are the molar excess volume and the molar excess enthalpy of the mixture, respectively.65 FUS as a phase separation protein with UCST has positive HEm. 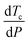 and VEm of FUS are negative on the low-pressure side and positive on the high-pressure side. Thus, the LLPS of FUS is first inhibited and then promoted with increasing hydrostatic pressure as shown in the pressure–temperature phase diagram (Fig. 2c), resulting in low-pressure LLPS (LP-LLPS) and high-pressure LLPS (HP-LLPS).
and VEm of FUS are negative on the low-pressure side and positive on the high-pressure side. Thus, the LLPS of FUS is first inhibited and then promoted with increasing hydrostatic pressure as shown in the pressure–temperature phase diagram (Fig. 2c), resulting in low-pressure LLPS (LP-LLPS) and high-pressure LLPS (HP-LLPS).
Pressure not only changes the packing structure of the protein in coacervates but also adjusts molecular interactions. For example, increasing pressure reduces the electrostatic interactions, and meanwhile enhances the cation–π and hydrophobic interactions, generating coacervates with more condensed configurations. Moreover, the formation and vanishing cycling of FUS coacervates can be realized by pressure-jump at certain temperatures in vitro.66 As shown in Fig. 2d, the turbidity jumps reflect the reversible formation of LP- and HP-LLPS condensates between 0.001–1.2 kbar and 2.0–3.1 kbar, respectively. The vanishing half-life time (t1/2) of HP-LLPS is 20-fold longer than that of LP-LLPS, which is consistent with the result that HP-LLPS showing stronger molecular interactions and self-association.
3.3 Light
Optogenetic tools have been developed to dynamically modulate intracellular protein interactions, which enable the spatiotemporal control of condensation by light within living cells. Essentially, light-controlled LLPS of FUS proteins is achieved by varying the strength of intermolecular interactions by fusing them with light-induced associative/dissociative modules.67 These optogenetic platforms are generally classified into light-induced assembly (e.g., optoDroplets and Corelet systems) and light-induced disassembly systems (e.g., PixELLs system), which have been utilized in the fields of artificial organelle construction,68 biosensing,69 transcription regulation,70 metabolic adjustment71,72 and pathological mechanism studies.73,74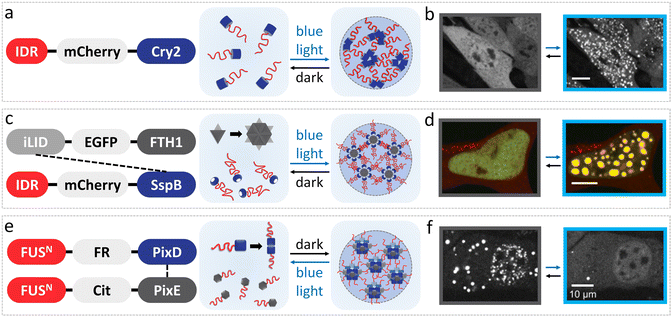 | ||
| Fig. 3 LLPS modulation of FUS under light stimuli in vivo. (a) OptoDroplet system of FUS IDR-Cry2 protein induces LLPS under blue light stimuli. (b) Fluorescence microscopy images display light-activated assembly of optoFUS condensates in 293T cells before and after exposure for 225 s under 488 nm light. Reproduced from ref. 56 with permission, Copyright Elsevier (2017). (c) Corelet system facilitates phase separation of FUS through 24 IDR modules captured by each core upon blue light illumination. (d) Confocal microscopy images show photo-activated FUS Corelet-expressing HEK293 cells before and after irradiation for 150 s under 488 nm blue light. Reproduced from ref. 75 with permission, Copyright Elsevier (2018). (e) PixELLs system composed of FUS-PixD and FUS-PixE proteins that form clusters in dark and dissociate under blue light. (f) Representative fluorescence microscopy images show NIH-3T3 cells with the PixELLs system before and after 450 nm blue light irradiation. Reproduced from ref. 76 with permission, Copyright Elsevier (2018). | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PixE stoichiometry of 10
PixE stoichiometry of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 or 10
4 or 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and dissociate upon blue light irradiation into dimers of FUSN-PixD and monomers of FUSN-PixE within seconds (Fig. 3e and f).76 With this methodology, light-induced disassembly of condensates with an asymmetric distribution of spatial patterns in cells can be established in minutes of light exposure, and it persisted for hours after removing the stimulus. Thus, the PixELLs system offers a long-term spatial memory compared with optoDroplets and Corelet systems, which have been applied in metabolic scenarios in living cells, such as cytoplasmic flow and cytoskeletal assembly/disassembly to drive asymmetric phenotypes, showing great potential for biological regulation.76
5 and dissociate upon blue light irradiation into dimers of FUSN-PixD and monomers of FUSN-PixE within seconds (Fig. 3e and f).76 With this methodology, light-induced disassembly of condensates with an asymmetric distribution of spatial patterns in cells can be established in minutes of light exposure, and it persisted for hours after removing the stimulus. Thus, the PixELLs system offers a long-term spatial memory compared with optoDroplets and Corelet systems, which have been applied in metabolic scenarios in living cells, such as cytoplasmic flow and cytoskeletal assembly/disassembly to drive asymmetric phenotypes, showing great potential for biological regulation.76
4. Modulation of FUS phase separation by molecular modulators
Molecular modulators are able to influence the protein–protein interactions by involving in FUS condensation as guest molecules. The readily designed molecular structures that are widely manufactured in industries endow molecular modulators with significant clinical potential, which circumvents the complex operation of genetic modification. The LLPS of FUS is (i) inhibited by 1,6-hexanediol, molecular chaperones and adenosine triphosphate (ATP) and (ii) promoted by certain crowing agents. Moreover, FUS undergoes (iii) biphasic separation regulated by H+/OH−, ATP, nucleic acids and organic small molecules and (iv) reentrant phase separation with the addition of inorganic salts.4.1 1,6-Hexanediol
1,6-Hexanediol has been widely used to disassemble biomolecular condensates probably by affecting the conformational ensemble of natural disordered proteins through breaking the hydrophobic interactions and hydrogen bonds (Fig. 4a).66,77,78 Alberti and his coauthors proved that liquid-like FUS coacervates are rapidly dissolved with the addition of hexanediol while solid-like protein aggregates are resistant in vitro (Fig. 4e).79 Also, 1,6-hexanediol is widely used to examine the liquid-like properties of membraneless organelles.77 More recently, it has been demonstrated that other alkanediols (such as 2,5-hexanediol and 1,5-pentanediol) with different lengths and configurations also impact protein solvation to disassemble the condensates of FUS LC.80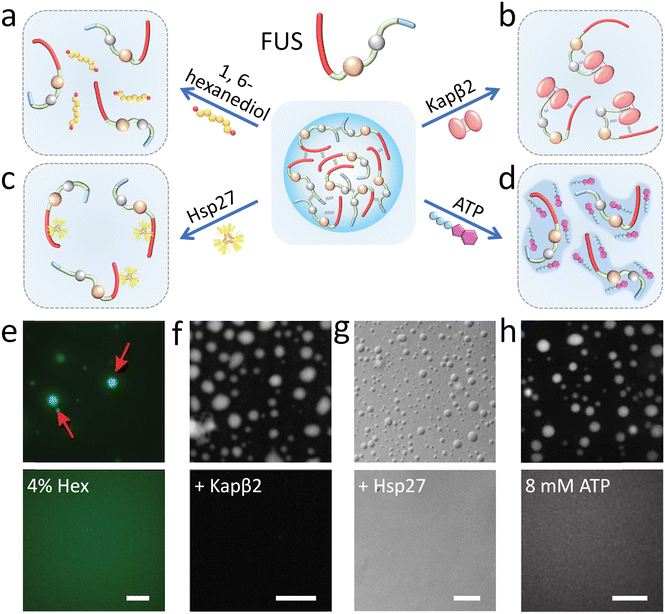 | ||
| Fig. 4 Modulation of FUS condensation droplets by molecular modulators. (a–d) Illustrative scheme of the inhibition mechanism of FUS phase separation with (a) 1,6-hexanediol affecting the conformational ensemble, (b) Kapβ2 interacting with the PY-NLS domain, (c) Hsp27 binding with a wide region of FUS LC, and (d) ATP acting as a hydrotrope. (e–h) Microscopy images show that FUS coacervate droplets (top) disassemble (down) with 4% 1,6-hexanediol (e), Kapβ2 (f), Hsp27 (g) and 8 mM ATP (h) in vitro. Scale bars, 10 μm. Reproduced with permissions from ref. 79, Copyright ScienceMatters AG (2017); ref. 84, Copyright Elsevier (2018); ref. 87, Copyright Springer Nature (2020); and ref. 96, Copyright Science (2017), respectively. | ||
4.2 Molecular chaperones
Molecular chaperones play a major role in maintaining proteostasis by assisting the refolding of proteins to ensure their function or by promoting the degradation of terminally misfolded proteins.81 Specifically, they may serve as inhibitors of FUS phase separation through preventing misfolded or abnormal aggregated proteins to modulate stress granules.82 Nuclear import receptors and small heat shock proteins (Hsps) are two kinds of typical molecular chaperone proteins for the regulation of the FUS phase behaviour.4.3 Crowding agents
Crowding agents such as PEG,46 dextran,89 BSA90 and Ficoll91 are commonly used in FUS LLPS studies in vitro for promoting condensation by decreasing the Cs. The solutions with crowding agents were also thought to better reproduce the crowded internal environment of cells. The probable mechanisms underpinning that have been proposed with three effects, including the excluded volume effect, the specific interaction between macromolecules and proteins, and the decreased protein solubility in crowded environments.92 Moreover, the transition of FUS assembly from a viscous fluid to a viscoelastic gel-like state was realized in a crowding-dependent manner, which can be explained by the increased intermolecular interactions in RGG and PLD domains caused by volume exclusion.934.4 H+/OH−
The concentrations of H+ and OH− are usually more stable in vivo than in vitro. For intercellular experiments, they are adjustable with an influence on the solvability of proteins. The pH of the system affects the protonation/deprotonation of protein residues, regulating the strength of intermolecular interactions and further influencing the phase separation. Generally, full-length FUS and FUS LC can form coacervates in acidic and alkalescent buffers with a pH range from 3.5 to 8.0.58,78,87,944.5 ATP
ATP has well-characterized functions in providing energy for biochemical reactions in vivo.95 Recent studies show that a high concentration of ATP in cells may also prevent protein aggregation.96 ATP possesses amphiphilicity due to the hydrophilic tripolyphosphate group and the hydrophobic adenosine group and is capable of dissolving hydrophobic proteins as a hydrotrope. Thus, ATP can inhibit the formation of condensates in full-length FUS (Fig. 4d and h).The Song laboratory demonstrated that ATP displays different regulatory effects for NTD, C-terminal domains (CTDs) and full-length of FUS.94,97 For the NTD, ATP acts as a hydrotrope that monotonically disassembles condensates through specifically binding between triphosphate chains and RGG residues, which is consistent with the results of full-length FUS, while for the CTD lacking the intrinsic capacity, ATP acts as a bivalent binder to modulate its LLPS in a concentration-dependent way. At low concentrations, ATP enhances the LLPS by specific binding with the arginine/lysine residues on CTDs. In contrast, excess ATP molecules disassemble condensates at high concentrations.
4.6 Organic small molecules
Organic small molecules with highly hydrophobic moieties and negatively charged groups, such as 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (bis-ANS) and Congo red, have been developed as biphasic modulators for biocondensates.78 The bivalence of sulfonate groups allows them to act as small intermolecular scaffolds or transient, weak cross-linkers between protein molecules, which promote FUS LLPS at low concentrations. Meanwhile, the hydrophobic naphthalene groups synergistically induce condensation through enhancing π–π stacking. At high concentrations, negatively charged sulfonate groups are essential for the regulation, which drives decondensation by electrostatic repulsion.4.7 Nucleic acids
ssDNA is proven to inhibit the LLPS of FUS NTD while acting as a biphasic modulator for the CTD, which shows similar properties to ATP. Importantly, ssDNA shows a much higher binding affinity to FUS RGG residues than ATP, which means that lower concentration of ssDNA is sufficient to replace the same effect of ATP.97 The complex coacervation of FUS and RNA is driven through non-specific electrostatic interaction and specific binding between diverse functional domains. The specific binding has been deciphered by Allain et al. who showed that RRM and ZnF regions are the structural basis for the recognition.98,99Additionally, the stoichiometry and structure (sequence and length) of RNA are the key to regulate FUS phase separation. First, when [FUS]:[RNA] > 1, LLPS is facilitated where RNA acts as scaffolds for the multimerization of FUS in vitro,58 while when [FUS]:[RNA] < 1, the excess RNA prevent FUS LLPS by suppressing interactions (Fig. 5a). In cells, nuclear RNA concentration is normally high enough to keep FUS soluble, which can be proved by the immediate condensation of FUS after microinjecting RNase A into the nucleus of HeLa cells.100 Second, the sequence and length of RNA also greatly influence the LLPS of FUS.101 The specific binding motifs within the RNA sequence enhance multivalent interactions and drive synergistic phase separation. For example, the middle domain of NEAT1_2 contains multiple binding sites for FUS.102 Typically, short RNA is more potent in disassembling FUS condensates, while the long RNA requires much higher concentration to inhibit phase separation (Fig. 5b).53 The length dependence of RNA in controlling the binding of FUS has been investigated systematically by Myong et al. through the smFRET assay. The results indicate that long ssRNA allows the formation of FUS condensates by acting as scaffolds, while short one simply binds to single FUS monomers.103
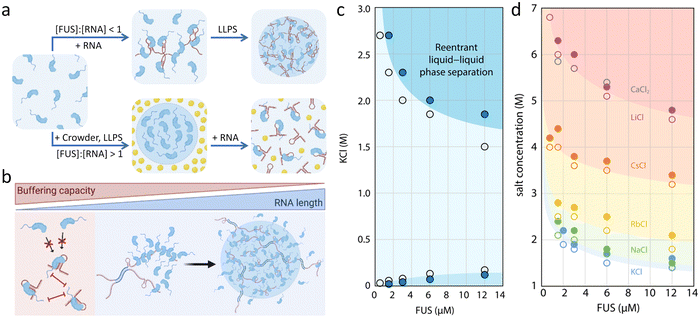 | ||
Fig. 5 Biphasic regulation of FUS LLPS by salts and RNA. (a) Influence of the [FUS]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [RNA] ratio on FUS LLPS. The LLPS of FUS is promoted when [FUS] [RNA] ratio on FUS LLPS. The LLPS of FUS is promoted when [FUS]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [RNA] > 1, while it is prevented when [FUS] [RNA] > 1, while it is prevented when [FUS]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [RNA] < 1. (b) Buffering capacity of RNA is strongly related to the length of RNA as the shorter one has stronger capability to dissolve FUS droplets. Reproduced from ref. 101 with permission, Copyright Elsevier (2021). (c) Phase diagram of reentrant phase separation of FUS adjusted by KCl. The LLPS of FUS happens in specific KCl concentration ranges (below 125 mM or above 1.5 M). (d) Phase diagram showing the Hofmeister effect in the high-salt phase separation region of FUS. The phase boundaries shift to higher salt concentrations in the order of Hofmeister series. Reproduced from ref. 77 with permission, Copyright Springer Nature (2021). [RNA] < 1. (b) Buffering capacity of RNA is strongly related to the length of RNA as the shorter one has stronger capability to dissolve FUS droplets. Reproduced from ref. 101 with permission, Copyright Elsevier (2021). (c) Phase diagram of reentrant phase separation of FUS adjusted by KCl. The LLPS of FUS happens in specific KCl concentration ranges (below 125 mM or above 1.5 M). (d) Phase diagram showing the Hofmeister effect in the high-salt phase separation region of FUS. The phase boundaries shift to higher salt concentrations in the order of Hofmeister series. Reproduced from ref. 77 with permission, Copyright Springer Nature (2021). | ||
The nucleic acid-mimicking biopolymer poly(ADP-ribose) (PAR) with similar properties to RNA also participates in the regulation of nucleic acid-related phase separation in cells. The PAR synthesized from NAD+ by the catalysis of poly(ADP-ribose) polymerase (PARP) interacts with the RGG2 domain of FUS, which drives LLPS of FUS to DNA damage sites to recruit the components for promoting double-strand break repair.104,105 On the other side, Myong et al. revealed that PAR seeds the homotypic FUS–FUS multimerization via a transient multivalent interaction with RGG domains without participation in coacervate formation. In this case, PAR runs a length-dependent catalyst-like mechanism that PAR chains with lengths comparable to those of RNA show one thousand times higher potency for promoting condensation than RNA in vitro.106
4.8 Inorganic salts
Hydrophobic interaction is important to the LLPS of FUS due to the hydrophobic tyrosine residues, which make up 15% of sequence involving in most intermolecular or intramolecular hydrophobic contacts.107 An increase in salt concentration facilitates the phase separation of FUS.58 Hofmeister series are utilized to evaluate the effect of salts on the hydration of proteins.107 Generally, Hofmeister cations show little effect on LLPS of FUS LC, while monovalent Hofmeister anions follow the trend.Reentrant phase separation of FUS mediated by inorganic salts was reported by the Knowles group, who showed that the condensates form at relatively low and high salt concentration while disappear in the mild-concentration region.77 The molecular interactions in the high-salt regime are fundamentally different from those driving LLPS in the low-salt regime. At high salt concentration, hydrophobic and non-ionic interactions are the main driving forces of phase separation, where electrostatics are screened out. At low salt concentrations without charge shielding, phase separation is driven by both electrostatic and hydrophobic interactions. The results were mapped out as a function of KCl concentration in the phase diagram (Fig. 5c). FUS forms condensates at a KCl concentration below ∼125 mM and above 1.5 M, otherwise a well-mixed single phase is formed. Furthermore, the phase separation of FUS in the high-salt regime with the chloride salts shows that higher salt concentration is needed with cations later in the Hofmeister series (Fig. 5d).
FUS-derived protein by fusing a hexhistidine tag to the N-terminal of FUS LC (6His-FUS LC) exhibits a specific recognition of metal ions reported by our group recently.108 The dynamic LLPS of 6His-FUS LC is triggered by metal ion–histidine coordination and further inhibited by the competitive binding of chelators (e.g., EDTA and imidazole). The regulation of 6His-FUS LC by metal ions displays universality to nickel, zinc, cupric and cobalt ions, which have been described to coordinate to histidine residues. Moreover, the metal ions with varying binding affinities can be used to adjust internal organization and molecular diffusivity within protein condensates.
5. Modulation of the FUS phase behaviour by altering protein structures
Posttranslational modifications (PTMs) and mutations of amino acid sequences serve as on/off switches of LLPS.39,109 More than half of proteins in eukaryotic cells undergo PTM to adjust the charge distribution, spatial conformation, intracellular localization, catalytic activity and protein–protein interaction.110 PTMs of FUS mainly include phosphorylation, methylation, and acetylation. Phosphorylation and methylation are mostly studied due to their ubiquitousness in natural cells and close combination with the modulation of phase separation to further affect the episode of neurodegenerative disease. The ALS/FTD-related PTM positions are summarized in Fig. 6a.111 Sequence mutation in FUS may lead to mislocalization, aggregation and RNA-binding disruption, and may also be a possible pathogenesis of ALS/FTD.112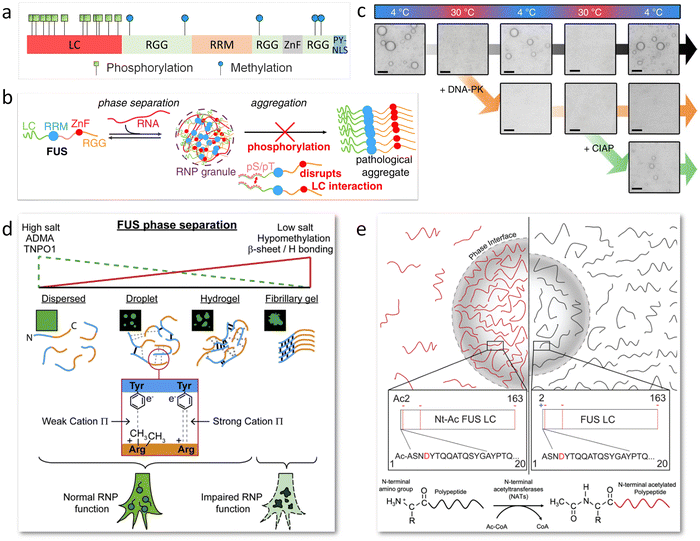 | ||
| Fig. 6 PTM of FUS for phase separation adjustment. (a) Scheme showing phosphorylation and methylation sites on FUS associated with neurodegenerative diseases. (b) Schematic image indicating phosphorylation of FUS LC regulates FUS/RNA phase separation and disrupts pathological aggregation by discouraging LC–LC intermolecular interactions. Reproduced from ref. 54 with permission, Copyright John Wiley and Sons (2017). (c) Optical microscopy images displaying the reversible condensation of FUS LC in temperature cycles of 4 °C and 30 °C, which is further adjusted with phosphorylation by DNA-PK and dephosphorylation by CIAP. Scale bars, 5 μm. Reproduced from ref. 62 with permission, Copyright Springer Nature (2018). (d) Schematic diagram illustrates that methylation weakens the cation–π interactions between arginine at the C terminal and tyrosine at the N terminal to modulate FUS phase transition. Reproduced from ref. 115 with permission, Copyright Elsevier (2018). (e) Illustrative diagram of FUS LC phase separation impacted by N-terminal acetylation. Acetylation removes the positive charge of the FUS NTD, promoting phase separation while disrupting followed LSPT. Reproduced from ref. 55 with permission, Copyright John Wiley and Sons (2021). | ||
5.1 Phosphorylation
The FUS LC domain is rich in serine and tyrosine, which are prone to undergo phosphorylation by phosphoinositide 3-kinase-like kinases (PIKKs)113 and DNA-dependent protein kinase (DNA-PK).114 Phosphorylation regulates phase transition of FUS coacervates by altering the charge without changing the disordered structure. The phosphorylated FUS presents a significant increase in the number of negatively charged residues, which disrupts LC–LC intermolecular interactions, RNA-FUS interactions, and FUS abnormal aggregation (Fig. 6b).54 Besides, the reversible condensation of FUS LC is realized in temperature cycles of 4 °C and 30 °C and further regulated with phosphorylation by DNA-PK and dephosphorylation by calf intestinal alkaline phosphatase (CIAP) (Fig. 6c).625.2 Methylation
Methylation usually acts on arginine residues in the FUS RGG motifs by protein arginine methyltransferases (PRMTs), which reduces the cation–π interactions between arginine and tyrosine in FUS NTD to modulate phase separation (Fig. 6d).115 It has been proved that hypomethylation of FUS is associated with FTD due to the transition of condensates into intermolecular β-sheet-rich hydrogel, which disrupts the RNP granule function and impairs protein synthesis. The degree of methylation of FUS displays a critical impact on the electrostatic combination with molecular chaperones.116 Unmethylated and monomethylated FUS show a higher binding affinity than dimethylated FUS to transportin-1 (TNPO1, i.e., Kapβ2), which electrostatically binds the RGG3. Therefore, regulating FUS methylation combined with TNPO1 levels to a normal state in vivo may lead to new therapies for ALS/FTD.915.3 Acetylation
The N terminal of FUS can be acetylated by co-expressing with an N-terminal acetyltransferase (NatA complex) and co-factor acetyl-CoA (Fig. 6e).55 The acetylation results in a small change in the net charge of FUS LC from −2 to −3, while it does not change its secondary structure or local motions. Because of the increase in negative charges, the LLPS of FUS LC is slightly promoted. Moreover, the acetylated FUS LC shows lower tendency of solid-like aggregation. It is also noted that N-terminal acetylated full-length FUS cannot be obtained by co-expression of the NatA complex, which means that the rule of FUS LC acetylation may not apply to full-length FUS.5.4 Sequence mutation
The mutation of tyrosine residues which participate in the cation–π interaction is used to modulate the FUS LLPS.117 Rosen et al. found that the number of tyrosine residues rather than the distribution has a crucial effect on the CS of LLPS. The aromaticity of tyrosine is critical since the threshold of phase separation significantly increases when replacing tyrosine with non-aromatic hydrophilic or hydrophobic amino acids, while less increase for the replacement of tyrosine with other aromatic amino acids. Moreover, computational methods combining a genetic algorithm with a sequence-dependent coarse-grained model and an all-atom model are also used for directional variations of amino acid sequence to promote or inhibit FUS LLPS.118Besides the abovementioned influence on the ability of FUS phase separation, the sequence mutations are also closely associated with the occurrence of neurodegenerative diseases.112 ALS/FTD-linked FUS mutations in arginine (e.g., R216C, R244C, R514G, R521C, and R521G) and glycine (e.g., G156E, G187S, G225V, G230C, and G399V) residues have been demonstrated to lead the aggregation propensity in disease.103 For arginine mutants, loss of the positively charged arginine leads to defective binding of FUS and RNA, forming an altered configuration that has more potential to aggregate or grow into aberrantly large condensates with less fluidity, while for glycine mutants, the decrease in the positive charges induces the generation of condensates showing accelerated loss of fluidity. Mutation of the FUS sequence also affects the protein–protein interaction.119 For example, glycine mutants can hardly interact with wild-type FUS, while arginine mutants form mixed condensates with wild-type FUS to restore the mutant defects.
6. Conclusions and perspective
Biocondensates formed by LLPS participate in a variety of cellular activities. Although our understanding of the dynamic assembly and physiological functions of condensates is still in its infancy, the investigations of intra- and intercellular modulation of LLPS have shed light on it. In this review, we summarized the approaches for regulating LLPS of FUS from three practical perspectives: physical stimuli, biochemical modulators and protein structure changes. Physical stimuli show insignificant alteration of the original composition of the system while may bring side effects to cellular functions. Molecular modulators as flexible tools adjust the LLPS and LSPT by affecting multivalent interactions, which are promising for clinical application to prevent diseases. PTM and protein subsequence mutation are the inherent LLPS regulatory mechanisms in living systems, which alter the structure, charge, aromaticity, and hydrophobicity and influence the combination of molecular modulators with proteins.Aberrant phase separation and solid-like transition of biomolecules have been implicated in the neurodegenerative diseases. Systematically summarizing the regulation methods of FUS LLPS provides a basic pathway for phase separation research and will be further extended to other LLPS-related proteins and diseases. Despite that these phase separation proteins obey distinct molecular grammars, regulation modes, aggregation forms, and have distinct phase behaviours, novel strategies for drug development and therapeutic treatment including identification of molecules that modulate phase separation and transition and alteration of enzymes that associate with PTM have been provided. In the future, these regulatory methods can be systematically integrated with proteomic, transcriptomic, imaging, genetic, and epidemiologic data to explore the pathogenesis of abnormal phase transitions, propose potential therapeutic targets and prevent and treat these diseases, and to guide drug screening strategies.
Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (22072159).References
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed.
- A. I. Oparin, The Origin of Life, New York: Macmillan, 1938 Search PubMed.
- T. Z. Jia, K. Chandru, Y. Hongo, R. Afrin, T. Usui, K. Myojo and H. J. Cleaves 2nd, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 15830–15835 CrossRef CAS PubMed.
- L. Jia, Z. Ji, Y. M. Ji, C. Zhou, G. W. Xing and Y. Qiao, ChemSystemsChem, 2020, 2, e2000044 CrossRef.
- Y. Ji, W. Mu, H. Wu and Y. Qiao, Adv. Sci., 2021, 8, e2101187 CrossRef PubMed.
- Z. Liu, Y. Ji, W. Mu, X. Liu, L. Y. Huang, T. Ding and Y. Qiao, Chem. Commun., 2022, 58, 2536–2539 RSC.
- Y. Zhang, Y. Chen, X. Yang, X. He, M. Li, S. Liu, K. Wang, J. Liu and S. Mann, J. Am. Chem. Soc., 2021, 143, 2866–2874 CrossRef CAS PubMed.
- R. R. Poudyal, R. M. Guth-Metzler, A. J. Veenis, E. A. Frankel, C. D. Keating and P. C. Bevilacqua, Nat. Commun., 2019, 10, 490 CrossRef CAS PubMed.
- W. Mu, Z. Ji, M. Zhou, J. Wu, Y. Lin and Y. Qiao, Sci. Adv., 2021, 7, eabf9000 CrossRef CAS PubMed.
- K. K. Nakashima, M. H. I. van Haren, A. A. M. Andre, I. Robu and E. Spruijt, Nat. Commun., 2021, 12, 3819 CrossRef CAS PubMed.
- E. Te Brinke, J. Groen, A. Herrmann, H. A. Heus, G. Rivas, E. Spruijt and W. T. S. Huck, Nat. Nanotechnol., 2018, 13, 849–855 CrossRef CAS PubMed.
- A. Ianeselli, D. Tetiker, J. Stein, A. Kuhnlein, C. B. Mast, D. Braun and T. Y. Dora Tang, Nat. Chem., 2022, 14, 32–39 CrossRef CAS PubMed.
- P. Adamski, M. Eleveld, A. Sood, Á. Kun, A. Szilágyi, T. Czárán, E. Szathmáry and S. Otto, Nat. Rev. Chem., 2020, 4, 386–403 CrossRef.
- T. Mashima, M. van Stevendaal, F. R. A. Cornelissens, A. F. Mason, B. Rosier, W. J. Altenburg, K. Oohora, S. Hirayama, T. Hayashi, J. C. M. van Hest and L. Brunsveld, Angew. Chem., Int. Ed., 2022, 61, e202115041 CrossRef CAS PubMed.
- Y. Qiao, M. Li, D. Qiu and S. Mann, Angew. Chem., Int. Ed., 2019, 58, 17758–17763 CrossRef CAS PubMed.
- Y. Qiao, M. Li, R. Booth and S. Mann, Nat. Chem., 2017, 9, 110–119 CrossRef CAS PubMed.
- N. Gao, C. Xu, Z. Yin, M. Li and S. Mann, J. Am. Chem. Soc., 2022, 144, 3855–3862 CrossRef CAS PubMed.
- S. Liu, Y. Zhang, M. Li, L. Xiong, Z. Zhang, X. Yang, X. He, K. Wang, J. Liu and S. Mann, Nat. Chem., 2020, 12, 1165–1173 CrossRef CAS PubMed.
- C. Zhao, J. Li, S. Wang, Z. Xu, X. Wang, X. Liu, L. Wang and X. Huang, ACS Nano, 2021, 15, 10048–10057 CrossRef PubMed.
- Y. Sun, S. Y. Lau, Z. W. Lim, S. C. Chang, F. Ghadessy, A. Partridge and A. Miserez, Nat. Chem., 2022, 14, 274–283 CrossRef CAS PubMed.
- W. Xiao, M. D. Jakimowicz, I. Zampetakis, S. Neely, F. Scarpa, S. A. Davis, D. S. Williams and A. W. Perriman, Adv. Biosyst., 2020, 4, e2000101 CrossRef PubMed.
- Y. Zhang, S. Liu, Y. Yao, Y. Chen, S. Zhou, X. Yang, K. Wang and J. Liu, Small, 2020, 16, e2002073 CrossRef PubMed.
- N. Gao, M. Li, L. Tian, A. J. Patil, B. Pavan Kumar and S. Mann, Nat. Chem., 2021, 13, 868–879 CrossRef CAS PubMed.
- A. Samanta, M. Horner, W. Liu, W. Weber and A. Walther, Nat. Commun., 2022, 13, 3968 CrossRef CAS PubMed.
- C. P. Brangwynne, C. R. Eckmann, D. S. Courson, A. Rybarska, C. Hoege, J. Gharakhani, F. Julicher and A. A. Hyman, Science, 2009, 324, 1729–1732 CrossRef CAS PubMed.
- C. P. Brangwynne, T. J. Mitchison and A. A. Hyman, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4334–4339 CrossRef CAS PubMed.
- M. Strzelecka, A. C. Oates and K. M. Neugebauer, Nucleus, 2010, 1, 96–108 CrossRef PubMed.
- S. Hennig, G. Kong, T. Mannen, A. Sadowska, S. Kobelke, A. Blythe, G. J. Knott, K. S. Iyer, D. Ho, E. A. Newcombe, K. Hosoki, N. Goshima, T. Kawaguchi, D. Hatters, L. Trinkle-Mulcahy, T. Hirose, C. S. Bond and A. H. Fox, J. Cell Biol., 2015, 210, 529–539 CrossRef CAS PubMed.
- A. Molliex, J. Temirov, J. Lee, M. Coughlin, A. P. Kanagaraj, H. J. Kim, T. Mittag and J. P. Taylor, Cell, 2015, 163, 123–133 CrossRef CAS PubMed.
- T. M. Franzmann, M. Jahnel, A. Pozniakovsky, J. Mahamid, A. S. Holehouse, E. Nuske, D. Richter, W. Baumeister, S. W. Grill, R. V. Pappu, A. A. Hyman and S. Alberti, Science, 2018, 359, eaao5654 CrossRef PubMed.
- B. R. Sabari, A. Dall’Agnese, A. Boija, I. A. Klein, E. L. Coffey, K. Shrinivas, B. J. Abraham, N. M. Hannett, A. V. Zamudio, J. C. Manteiga, C. H. Li, Y. E. Guo, D. S. Day, J. Schuijers, E. Vasile, S. Malik, D. Hnisz, T. I. Lee, I. I. Cisse, R. G. Roeder, P. A. Sharp, A. K. Chakraborty and R. A. Young, Science, 2018, 361, 379 CrossRef CAS PubMed.
- A. Boija, I. A. Klein, B. R. Sabari, A. Dall'Agnese, E. L. Coffey, A. V. Zamudio, C. H. Li, K. Shrinivas, J. C. Manteiga, N. M. Hannett, B. J. Abraham, L. K. Afeyan, Y. E. Guo, J. K. Rimel, C. B. Fant, J. Schuijers, T. I. Lee, D. J. Taatjes and R. A. Young, Cell, 2018, 175, 1842–1855 CrossRef CAS PubMed.
- S. Kilic, A. Lezaja, M. Gatti, E. Bianco, J. Michelena, R. Imhof and M. Altmeyer, EMBO J., 2019, 38, e101379 CrossRef PubMed.
- V. H. Ryan, G. L. Dignon, G. H. Zerze, C. V. Chabata, R. Silva, A. E. Conicella, J. Amaya, K. A. Burke, J. Mittal and N. L. Fawzi, Mol. Cell, 2018, 69, 465–479 CrossRef CAS PubMed.
- X. Wu, M. Ganzella, J. Zhou, S. Zhu, R. Jahn and M. Zhang, Mol. Cell, 2021, 81, 13–24 CrossRef CAS PubMed.
- D. Milovanovic, Y. Wu, X. Bian and P. De Camilli, Science, 2018, 361, 604–607 CrossRef CAS PubMed.
- M. Du and Z. J. Chen, Science, 2018, 361, 704–709 CrossRef CAS PubMed.
- A. Zbinden, M. Perez-Berlanga, P. De Rossi and M. Polymenidou, Dev. Cell, 2020, 55, 45–68 CrossRef CAS PubMed.
- E. W. Martin and T. Mittag, Biochemistry, 2018, 57, 2478–2487 CrossRef CAS PubMed.
- C. Y. Shi, Q. Zhang, H. Tian and D. H. Qu, SmartMat, 2020, 1, e1012 CrossRef.
- V. N. Uversky, Curr. Opin. Struct. Biol., 2017, 44, 18–30 CrossRef CAS PubMed.
- P. Li, S. Banjade, H. C. Cheng, S. Kim, B. Chen, L. Guo, M. Llaguno, J. V. Hollingsworth, D. S. King, S. F. Banani, P. S. Russo, Q. X. Jiang, B. T. Nixon and M. K. Rosen, Nature, 2012, 483, 336–340 CrossRef CAS PubMed.
- J. A. Joseph, J. R. Espinosa, I. Sanchez-Burgos, A. Garaizar, D. Frenkel and R. Collepardo-Guevara, Biophys. J., 2021, 120, 1219–1230 CrossRef CAS PubMed.
- S. Reber, D. Jutzi, H. Lindsay, A. Devoy, J. Mechtersheimer, B. R. Levone, M. Domanski, E. Bentmann, D. Dormann, O. Muhlemann, S. M. L. Barabino and M. D. Ruepp, Nucleic Acids Res., 2021, 49, 7713–7731 CrossRef CAS PubMed.
- A. Patel, H. O. Lee, L. Jawerth, S. Maharana, M. Jahnel, M. Y. Hein, S. Stoynov, J. Mahamid, S. Saha, T. M. Franzmann, A. Pozniakovski, I. Poser, N. Maghelli, L. A. Royer, M. Weigert, E. W. Myers, S. Grill, D. Drechsel, A. A. Hyman and S. Alberti, Cell, 2015, 162, 1066–1077 CrossRef CAS PubMed.
- Y. Lu, L. Lim and J. Song, Sci. Rep., 2017, 7, 1043 CrossRef PubMed.
- A. Aguzzi and M. Altmeyer, Trends Cell Biol., 2016, 26, 547–558 CrossRef CAS PubMed.
- S. N. Rhoads, Z. T. Monahan, D. S. Yee and F. P. Shewmaker, Int. J. Mol. Sci., 2018, 19, 886 CrossRef PubMed.
- A. C. Murthy, W. S. Tang, N. Jovic, A. M. Janke, D. H. Seo, T. M. Perdikari, J. Mittal and N. L. Fawzi, Nat. Struct. Mol. Biol., 2021, 28, 923–935 CrossRef CAS PubMed.
- D. T. Murray, M. Kato, Y. Lin, K. R. Thurber, I. Hung, S. L. McKnight and R. Tycko, Cell, 2017, 171, 615–627 CrossRef CAS PubMed.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- S. Maharana, J. Wang, D. K. Papadopoulos, D. Richter, A. Pozniakovsky, I. Poser, M. Bickle, S. Rizk, J. Guillen-Boixet, T. M. Franzmann, M. Jahnel, L. Marrone, Y. T. Chang, J. Sterneckert, P. Tomancak, A. A. Hyman and S. Alberti, Science, 2018, 360, 918–921 CrossRef CAS PubMed.
- Z. Monahan, V. H. Ryan, A. M. Janke, K. A. Burke, S. N. Rhoads, G. H. Zerze, R. O'Meally, G. L. Dignon, A. E. Conicella, W. Zheng, R. B. Best, R. N. Cole, J. Mittal, F. Shewmaker and N. L. Fawzi, EMBO J., 2017, 36, 2951–2967 CrossRef CAS PubMed.
- A. S. Bock, A. C. Murthy, W. S. Tang, N. Jovic, F. Shewmaker, J. Mittal and N. L. Fawzi, Protein Sci., 2021, 30, 1337–1349 CrossRef CAS PubMed.
- Y. Shin, J. Berry, N. Pannucci, M. P. Haataja, J. E. Toettcher and C. P. Brangwynne, Cell, 2017, 168, 159–171 CrossRef CAS PubMed.
- T. Murakami, S. Qamar, J. Q. Lin, G. S. Schierle, E. Rees, A. Miyashita, A. R. Costa, R. B. Dodd, F. T. Chan, C. H. Michel, D. Kronenberg-Versteeg, Y. Li, S. P. Yang, Y. Wakutani, W. Meadows, R. R. Ferry, L. Dong, G. G. Tartaglia, G. Favrin, W. L. Lin, D. W. Dickson, M. Zhen, D. Ron, G. Schmitt-Ulms, P. E. Fraser, N. A. Shneider, C. Holt, M. Vendruscolo, C. F. Kaminski and P. St George-Hyslop, Neuron, 2015, 88, 678–690 CrossRef CAS PubMed.
- K. A. Burke, A. M. Janke, C. L. Rhine and N. L. Fawzi, Mol. Cell, 2015, 60, 231–241 CrossRef CAS PubMed.
- L. B. Persson, V. S. Ambati and O. Brandman, Cell, 2020, 183, 1572–1585 CrossRef CAS PubMed.
- S. Shabala and L. Shabala, Biomol. Concepts, 2011, 2, 407–419 CAS.
- S. Szmelcman and J. Adler, Proc. Natl. Acad. Sci. U. S. A., 1976, 73, 4387–4391 CrossRef CAS PubMed.
- F. Luo, X. Gui, H. Zhou, J. Gu, Y. Li, X. Liu, M. Zhao, D. Li, X. Li and C. Liu, Nat. Struct. Mol. Biol., 2018, 25, 341–346 CrossRef CAS PubMed.
- S. Chatterjee, Y. Kan, M. Brzezinski, K. Koynov, R. M. Regy, A. C. Murthy, K. A. Burke, J. J. Michels, J. Mittal, N. L. Fawzi and S. H. Parekh, Adv. Sci., 2022, 9, e2104247 CrossRef PubMed.
- K. Akasaka, R. Kitahara and Y. O. Kamatari, Arch. Biochem. Biophys., 2013, 531, 110–115 CrossRef CAS PubMed.
- S. Li, T. Yoshizawa, R. Yamazaki, A. Fujiwara, T. Kameda and R. Kitahara, J. Phys. Chem. B, 2021, 125, 6821–6829 CrossRef CAS PubMed.
- R. Kitahara, R. Yamazaki, F. Ide, S. Li, Y. Shiramasa, N. Sasahara and T. Yoshizawa, J. Am. Chem. Soc., 2021, 143, 19697–19702 CrossRef CAS PubMed.
- C. D. Reinkemeier and E. A. Lemke, Curr. Opin. Chem. Biol., 2021, 64, 174–181 CrossRef CAS PubMed.
- E. M. Zhao, N. Suek, M. Z. Wilson, E. Dine, N. L. Pannucci, Z. Gitai, J. L. Avalos and J. E. Toettcher, Nat. Chem. Biol., 2019, 15, 589–597 CrossRef CAS PubMed.
- M. Marsafari, J. Ma, M. Koffas and P. Xu, Curr. Opin. Biotechnol., 2020, 64, 175–182 CrossRef CAS PubMed.
- M. Wei, X. Fan, M. Ding, R. Li, S. Shao, Y. Hou, S. Meng, F. Tang, C. Li and Y. Sun, Sci. Adv., 2020, 6, eaay6515 CrossRef CAS PubMed.
- D. W. Sanders, N. Kedersha, D. S. W. Lee, A. R. Strom, V. Drake, J. A. Riback, D. Bracha, J. M. Eeftens, A. Iwanicki, A. Wang, M. T. Wei, G. Whitney, S. M. Lyons, P. Anderson, W. M. Jacobs, P. Ivanov and C. P. Brangwynne, Cell, 2020, 181, 306–324 CrossRef CAS PubMed.
- N. Treen, S. F. Shimobayashi, J. Eeftens, C. P. Brangwynne and M. Levine, Nat. Commun., 2021, 12, 1561 CrossRef CAS PubMed.
- P. Zhang, B. Fan, P. Yang, J. Temirov, J. Messing, H. J. Kim and J. P. Taylor, eLife, 2019, 8, e39578 CrossRef PubMed.
- S. Basu, S. D. Mackowiak, H. Niskanen, D. Knezevic, V. Asimi, S. Grosswendt, H. Geertsema, S. Ali, I. Jerkovic, H. Ewers, S. Mundlos, A. Meissner, D. M. Ibrahim and D. Hnisz, Cell, 2020, 181, 1062–1079 CrossRef CAS PubMed.
- D. Bracha, M. T. Walls, M. T. Wei, L. Zhu, M. Kurian, J. L. Avalos, J. E. Toettcher and C. P. Brangwynne, Cell, 2018, 175, 1467–1480 CrossRef CAS PubMed.
- E. Dine, A. A. Gil, G. Uribe, C. P. Brangwynne and J. E. Toettcher, Cell Syst., 2018, 6, 655–663 CrossRef CAS PubMed.
- G. Krainer, T. J. Welsh, J. A. Joseph, J. R. Espinosa, S. Wittmann, E. de Csillery, A. Sridhar, Z. Toprakcioglu, G. Gudiskyte, M. A. Czekalska, W. E. Arter, J. Guillen-Boixet, T. M. Franzmann, S. Qamar, P. S. George-Hyslop, A. A. Hyman, R. Collepardo-Guevara, S. Alberti and T. P. J. Knowles, Nat. Commun., 2021, 12, 1085 CrossRef CAS.
- W. M. Babinchak, B. K. Dumm, S. Venus, S. Boyko, A. A. Putnam, E. Jankowsky and W. K. Surewicz, Nat. Commun., 2020, 11, 5574 CrossRef CAS PubMed.
- S. Kroschwald, S. Maharana and A. Simon, Matters, 2017 DOI:10.19185/matters.201702000010.
- T. M. Perdikari, A. C. Murthy and N. L. Fawzi, bioRxiv, 2022, preprint DOI:10.1101/2022.05.05.490812.
- Y. E. Kim, M. S. Hipp, A. Bracher, M. Hayer-Hartl and F. U. Hartl, Annu. Rev. Biochem., 2013, 82, 323–355 CrossRef CAS PubMed.
- J. Tittelmeier, E. Nachman and C. Nussbaum-Krammer, Front. Aging Neurosci., 2020, 12, 581374 CrossRef CAS PubMed.
- C. E. Springhower, M. K. Rosen and Y. M. Chook, Curr. Opin. Cell Biol., 2020, 64, 112–123 CrossRef CAS PubMed.
- T. Yoshizawa, R. Ali, J. Jiou, H. Y. J. Fung, K. A. Burke, S. J. Kim, Y. Lin, W. B. Peeples, D. Saltzberg, M. Soniat, J. M. Baumhardt, R. Oldenbourg, A. Sali, N. L. Fawzi, M. K. Rosen and Y. M. Chook, Cell, 2018, 173, 693–705 CrossRef CAS PubMed.
- L. Guo, H. J. Kim, H. Wang, J. Monaghan, F. Freyermuth, J. C. Sung, K. O'Donovan, C. M. Fare, Z. Diaz, N. Singh, Z. C. Zhang, M. Coughlin, E. A. Sweeny, M. E. DeSantis, M. E. Jackrel, C. B. Rodell, J. A. Burdick, O. D. King, A. D. Gitler, C. Lagier-Tourenne, U. B. Pandey, Y. M. Chook, J. P. Taylor and J. Shorter, Cell, 2018, 173, 677–692 CrossRef CAS PubMed.
- Z. Li and P. Srivastava, Curr. Protoc. Immunol., 2004 Search PubMed Appendix 1, Appendix 1T.
- Z. Liu, S. Zhang, J. Gu, Y. Tong, Y. Li, X. Gui, H. Long, C. Wang, C. Zhao, J. Lu, L. He, Y. Li, Z. Liu, D. Li and C. Liu, Nat. Struct. Mol. Biol., 2020, 27, 363–372 CrossRef CAS PubMed.
- J. Gu, Z. Liu, S. Zhang, Y. Li, W. Xia, C. Wang, H. Xiang, Z. Liu, L. Tan, Y. Fang, C. Liu and D. Li, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 31123–31133 CrossRef CAS PubMed.
- J. Wang, J. M. Choi, A. S. Holehouse, H. O. Lee, X. Zhang, M. Jahnel, S. Maharana, R. Lemaitre, A. Pozniakovsky, D. Drechsel, I. Poser, R. V. Pappu, S. Alberti and A. A. Hyman, Cell, 2018, 174, 688–699 CrossRef CAS PubMed.
- Y. Lin, D. S. Protter, M. K. Rosen and R. Parker, Mol. Cell, 2015, 60, 208–219 CrossRef CAS PubMed.
- M. Hofweber, S. Hutten, B. Bourgeois, E. Spreitzer, A. Niedner-Boblenz, M. Schifferer, M. D. Ruepp, M. Simons, D. Niessing, T. Madl and D. Dormann, Cell, 2018, 173, 706–719 CrossRef CAS PubMed.
- A. A. M. Andre and E. Spruijt, Int. J. Mol. Sci., 2020, 21, 5908 CrossRef CAS PubMed.
- T. Kaur, I. Alshareedah, W. Wang, J. Ngo, M. M. Moosa and P. R. Banerjee, Biomolecules, 2019, 9, 71 CrossRef CAS PubMed.
- J. Kang, L. Lim and J. Song, Biochem. Biophys. Res. Commun., 2018, 504, 545–551 CrossRef CAS PubMed.
- T. Elston, H. Wang and G. Oster, Nature, 1998, 391, 510–513 CrossRef CAS PubMed.
- A. Patel, L. Malinovska, S. Saha, J. Wang, S. Alberti, Y. Krishnan and A. A. Hyman, Science, 2017, 356, 753–756 CrossRef CAS PubMed.
- J. Kang, L. Lim, Y. Lu and J. Song, PLoS Biol., 2019, 17, e3000327 CrossRef PubMed.
- F. E. Loughlin, P. J. Lukavsky, T. Kazeeva, S. Reber, E. M. Hock, M. Colombo, C. Von Schroetter, P. Pauli, A. Clery, O. Muhlemann, M. Polymenidou, M. D. Ruepp and F. H. Allain, Mol. Cell, 2019, 73, 490–504 CrossRef CAS PubMed.
- J. C. Schwartz, X. Wang, E. R. Podell and T. R. Cech, Cell Rep., 2013, 5, 918–925 CrossRef CAS PubMed.
- J. Aarum, C. P. Cabrera, T. A. Jones, S. Rajendran, R. Adiutori, G. Giovannoni, M. R. Barnes, A. Malaspina and D. Sheer, EMBO Rep., 2020, 21, e49585 CrossRef CAS PubMed.
- J. R. Mann and C. J. Donnelly, Neuron, 2021, 109, 2663–2681 CrossRef CAS PubMed.
- T. Yamazaki, S. Souquere, T. Chujo, S. Kobelke, Y. S. Chong, A. H. Fox, C. S. Bond, S. Nakagawa, G. Pierron and T. Hirose, Mol. Cell, 2018, 70, 1038–1053 CrossRef CAS PubMed.
- A. G. Niaki, J. Sarkar, X. Cai, K. Rhine, V. Vidaurre, B. Guy, M. Hurst, J. C. Lee, H. R. Koh, L. Guo, C. M. Fare, J. Shorter and S. Myong, Mol. Cell, 2020, 77, 82–94 CrossRef CAS PubMed.
- A. S. Mastrocola, S. H. Kim, A. T. Trinh, L. A. Rodenkirch and R. S. Tibbetts, J. Biol. Chem., 2013, 288, 24731–24741 CrossRef CAS PubMed.
- M. Altmeyer, K. J. Neelsen, F. Teloni, I. Pozdnyakova, S. Pellegrino, M. Grofte, M. D. Rask, W. Streicher, S. Jungmichel, M. L. Nielsen and J. Lukas, Nat. Commun., 2015, 6, 8088 CrossRef CAS PubMed.
- K. Rhine, M. Dasovich, J. Yoniles, M. Badiee, S. Skanchy, L. R. Ganser, Y. Ge, C. M. Fare, J. Shorter, A. K. L. Leung and S. Myong, Mol. Cell, 2022, 82, 969–985 CrossRef CAS PubMed.
- A. C. Murthy, G. L. Dignon, Y. Kan, G. H. Zerze, S. H. Parekh, J. Mittal and N. L. Fawzi, Nat. Struct. Mol. Biol., 2019, 26, 637–648 CrossRef CAS PubMed.
- F. Li, Y. Lin and Y. Qiao, Chin. J. Polym. Sci., 2022, 40(9), 1043–1049 CrossRef CAS.
- F. G. Quiroz and A. Chilkoti, Nat. Mater., 2015, 14, 1164–1171 CrossRef CAS PubMed.
- M. Mann and O. N. Jensen, Nat. Biotechnol., 2003, 21, 255–261 CrossRef CAS PubMed.
- B. Wang, L. Zhang, T. Dai, Z. Qin, H. Lu, L. Zhang and F. Zhou, Signal Transduction Targeted Ther., 2021, 6, 290 CrossRef PubMed.
- H. Deng, K. Gao and J. Jankovic, Nat. Rev. Neurol., 2014, 10, 337–348 CrossRef CAS PubMed.
- M. Gardiner, R. Toth, F. Vandermoere, N. A. Morrice and J. Rouse, Biochem. J., 2008, 415, 297–307 CrossRef CAS PubMed.
- Q. Deng, C. J. Holler, G. Taylor, K. F. Hudson, W. Watkins, M. Gearing, D. Ito, M. E. Murray, D. W. Dickson, N. T. Seyfried and T. Kukar, J. Neurosci., 2014, 34, 7802–7813 CrossRef CAS PubMed.
- S. Qamar, G. Wang, S. J. Randle, F. S. Ruggeri, J. A. Varela, J. Q. Lin, E. C. Phillips, A. Miyashita, D. Williams, F. Strohl, W. Meadows, R. Ferry, V. J. Dardov, G. G. Tartaglia, L. A. Farrer, G. S. Kaminski Schierle, C. F. Kaminski, C. E. Holt, P. E. Fraser, G. Schmitt-Ulms, D. Klenerman, T. Knowles, M. Vendruscolo and P. St George-Hyslop, Cell, 2018, 173, 720–734 CrossRef CAS PubMed.
- D. Dormann, T. Madl, C. F. Valori, E. Bentmann, S. Tahirovic, C. Abou-Ajram, E. Kremmer, O. Ansorge, I. R. Mackenzie, M. Neumann and C. Haass, EMBO J., 2012, 31, 4258–4275 CrossRef CAS PubMed.
- Y. Lin, S. L. Currie and M. K. Rosen, J. Biol. Chem., 2017, 292, 19110–19120 CrossRef CAS PubMed.
- S. M. Lichtinger, A. Garaizar, R. Collepardo-Guevara and A. Reinhardt, PLoS Comput. Biol., 2021, 17, e1009328 CrossRef CAS PubMed.
- K. Rhine, M. A. Makurath, J. Liu, S. Skanchy, C. Lopez, K. F. Catalan, Y. Ma, C. M. Fare, J. Shorter, T. Ha, Y. R. Chemla and S. Myong, Mol. Cell, 2020, 80, 666–681 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |



