Open Access Article
Open Access ArticleEngineering microbes to synthesize functionalized biopolymers
Kang
Zhou
*
Department of Chemical and Biomolecular Engineering, National University of Singapore, Singapore. E-mail: kang.zhou@nus.edu.sg
First published on 31st August 2022
Abstract
Engineering the metabolism of microbes has allowed simultaneous co-production of functional small molecules and biopolymers. This perspective briefly introduces its working principles and summarizes my views on how this approach could lead to consolidated fermentation processes for producing functionalized biopolymers. The potential applications of the polymer products and the foreseen challenges are also discussed.
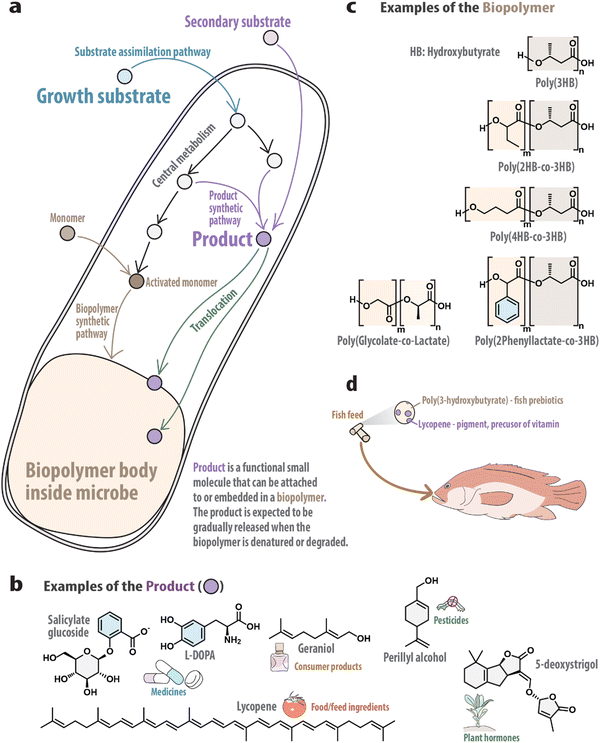
Bacteria can be engineered to accumulate polymers intracellularly using cheap and renewable substrates, such as sugars.1 The substrates can be transformed into a large number of building blocks through the native metabolism. Through genetic engineering, new enzymes can be expressed in the cells to convert some of these building blocks into suitable activated monomers and polymerize them into homo- or hetero-polymers (Fig. 1a). Through decades of research and development, the number and type of polymers that can be made in this way have increased substantially.1–4
During the same period, the same bacteria have also been engineered to transform their native metabolites or exogenously added substrates into functional small molecules (Fig. 1a), which may be used as drugs,5,6 pesticides7 and food/feed ingredients8 (Fig. 1b).
The above two processes may compete in the same cell for substrates, energy and/or reducing equivalents, but they are not exclusive. Recently, it has been demonstrated that one cell can be engineered to produce both a polymer and functional small molecules.9 Moreover, the functional small molecules could have interactions with the polymer.9 This opens opportunities for developing a consolidated fermentation process for producing functionalized polymers from low-cost substrates. In this article, I will share my views on this approach and the potential applications.
As a hypothetical example, Escherichia coli can be engineered to synthesize L-DOPA (l-3,4-dihydroxyphenylalanine, a frontline drug used to treat Parkinson's disease5) and a biopolymer from glucose (Fig. 1a). L-DOPA could be encapsulated in/on the biopolymer due to hydrophobic or ionic interactions. After the L-DOPA-containing biopolymer is purified from the cells, it may be administered to a patient, in which L-DOPA is released in a controlled manner when the biopolymer is denatured or degraded in the digestive tract. A slow and sustained release profile may lead to prolonged relief of the patient's symptoms and less side effects. Such fermentation processes, when developed, could lead to simpler and more cost-effective ways of producing the formulated drug.
Gram per liter of L-DOPA has been produced from glucose using engineered E. coli.10 Systematic research will be needed to evaluate the biopolymers that currently can be produced in large quantities by E. coli, with the objective of identifying a polymer that has (1) sufficient affinity with L-DOPA and (2) suitable properties needed by controlled release applications. Poly(3-hydroxybutyrate) (P3HB) is naturally produced by many microorganisms to store excessive carbon substrates.11 Such a polymer produced by the cells can easily occupy >50% of the cell's mass. Usually, L-DOPA-producing cells secrete almost all the L-DOPA to the extracellular space. If the cells were found to accumulate a larger fraction of L-DOPA intracellularly after they were engineered to produce a biopolymer, it is possible that L-DOPA has been co-localized with the biopolymer. Advanced characterizations would be needed to confirm the hypothesis and to reveal more details, such as whether L-DOPA are embedded into the biopolymer during their synthesis or simply attached to the surface of the biopolymer. New methods and tools will be needed to complete these characterizations. Such simultaneous co-production of the drug and the polymer may also lead to encapsulation patterns that cannot be achieved using the mainstream formulation methods.
Bioengineers have developed strategies to incorporate many monomers when polyester is synthesized in E. coli (Fig. 1c). The monomers include 2-hydroxybutyrate,2 4-hydroxybutyrate,4 lactate,1 glycolate1 and phenyllactate.3 These exciting advances provide a large space to explore to meet the two requirements described above, because they (1) confer different hydrophobicity for interacting with L-DOPA and (2) could moderate the polymer degradation properties for achieving the desired controlled release profile. This search space will be further expanded into new dimensions when bioengineers tune the content, molecular weight, size and surface properties of the polymers. The content refers to the ratio of the biopolymer mass to the dry cell mass. It can be changed by metabolic engineering, such as changing the rate of supplying of acetyl-CoA to PHB biosynthesis. The molecular weight and size of the biopolymer may be changed by controlling the expression level of the polymerase. The surface of these polyesters is at least partly covered by proteins,12 which can be genetically modified to include new residues for changing the hydrophobicity, charge and biodegradation rate of the polymer.
Theoretically, it should be possible to replace L-DOPA in the above example with other therapeutic small molecules. One example molecule to highlight is salicylate-2-O-β-D-glucose (SAG), a plant natural product with similar efficacy to aspirin (acetylsalicylate).6 SAG has been produced from glucose at gram per liter level by engineered E. coli. Plant natural products are an important arsenal in the combat against infectious diseases, pains, and cancers. da Silva Antonio et al. summarized the molecules that have potential in treating COVID-1913 and many of them can be produced in engineered microbes. Opioids and cannabinoids play critical roles in pain management and complete microbial synthesis of some molecules within these families has been demonstrated in the past decade, despite the fact that the product titers are still low.14,15 Paclitaxel is widely used in treating various cancers and has been extensively studied in the drug delivery field. Although constructing its biosynthesis in microbes is not yet completed, I am optimistic that it will be achieved in one to two decades. Co-production with a biopolymer may facilitate these efforts because many of the involved enzymes (such as cytochrome P450s) have structural support in plant cells and such support may be provided by polymer bodies in microbes.16
A challenge in the applications articulated above is the safety of the functionalized polymers. There could be other cellular components also encapsulated on/in the biopolymers. Such impurities may be toxic or immunogenic.17 A direction to address this concern is host selection and engineering. Escherichia coli may be replaced by a GRAS (Generally Regarded As Safe) host species, which could be further genetically modified to deactivate the synthesis of the contaminating molecules, or to disrupt the undesired interactions between these molecules and the biopolymers. If the biopolymer is immunogenic, human proteins (e.g., tropoelastin) may be recombinantly expressed and targeted onto the biopolymer for improving its compatibility, especially in tissue engineering applications. Suitable separation techniques will be needed to isolate the polymer bodies from the rest of the biomass at a reasonable cost.
If the above concern over safety cannot be adequately addressed, the focus may be shifted to applications outside the healthcare sector. A few designs are shared below to attract experts from various fields to investigate this approach with material scientists and metabolic engineers. Such interdisciplinary collaborations would be the key to transform designs into reality.
Many volatile natural products can be efficiently synthesized from cheap substrates using microbes.18 If they can be co-produced with and encapsulated in biopolymers, the resulting product may be used to gradually release the volatiles to the gas phase. If the volatile is a natural pesticide (such as perillyl alcohol7,19), the cells may be directly added into the soil and programmed to lyse themselves under the environment; if the volatile is a fragrance molecule (e.g., geraniol18), the functionalized polymer may be applied on cloth as a new perfume design. In the agricultural example, if we replace the volatile with plant hormones (e.g., 5-deoxystrigol20), these small molecules can be slowly released to positively regulate plant growth. When any of these applications can be safely implemented without purifying the polymer from the cells, the economic feasibility of the application would be improved substantially.
Besides being a carrier, the biopolymer could have additional functions. For example, P3HB was found to be a prebiotic for farmed fish: the fish growth rate was 20% higher than the control when the diet contained 5% of PHB (by weight).8 If lycopene-containing P3HB is included in fish feed, lycopene could be slowly released for boosting fish nutritional value and color development, while P3HB contributed to improving fish growth (Fig. 1d).
As a final note, this article has focused on polyesters because of the author's prior experiences. There are other families of polymers that can be synthesized in large quantities in cells, such as polysaccharides, polyphosphates, polyamides (including proteins), and nucleic acids. It could also be possible to co-localize functional small molecules with these biopolymers after they are synthesized in vivo, which could generalize the biopolymer production method discussed in this article.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The research projects related to this article were supported by Singapore National Research Foundation through a Singapore-MIT Alliance for Research and Technology (SMART) Program and a Competitive Research Project Program (Grant identifiers: R-279-000-587-592, R-279-000-512-281).References
- S. Y. Choi, S. J. Park, W. J. Kim, J. E. Yang, H. Lee, J. Shin and S. Y. Lee, Nat. Biotechnol., 2016, 34, 435–440 CrossRef CAS PubMed.
- S. Mizuno, Y. Enda, A. Saika, A. Hiroe and T. Tsuge, J. Biosci. Bioeng., 2018, 125, 295–300 CrossRef CAS PubMed.
- J. E. Yang, S. J. Park, W. J. Kim, H. J. Kim, B. J. Kim, H. Lee, J. Shin and S. Y. Lee, Nat. Commun., 2018, 9, 79 CrossRef PubMed.
- Y. Wang, H. Wu, X. Jiang and G. Q. Chen, Metab. Eng., 2014, 25, 183–193 CrossRef CAS PubMed.
- X. Ma, G. Gozaydin, H. Yang, W. Ning, X. Han, N. Y. Poon, H. Liang, N. Yan and K. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 7719–7728 CrossRef CAS.
- M. K. Ahmadi, L. Fang, N. Moscatello and B. A. Pfeifer, Metab. Eng., 2016, 38, 382–388 CrossRef CAS PubMed.
- W. P. Mayeku, N. I. Omollo, O. J. Odalo and A. Hassanali, Med. Vet. Entomol., 2014, 28, 253–256 CrossRef CAS PubMed.
- M. Dawood, M. Abdel-Tawwab and H. Abdel-Latif, Rev. Aquac., 2020, 12, 2511–2526 CrossRef.
- Y. Liu, Z. J. Low, X. Ma, H. Liang, A. J. Sinskey, G. Stephanopoulos and K. Zhou, Metab. Eng., 2020, 61, 206–214 CrossRef CAS.
- A. J. Munoz, G. Hernandez-Chavez, R. de Anda, A. Martinez, F. Bolivar and G. Gosset, J. Ind. Microbiol. Biotechnol., 2011, 38, 1845–1852 CrossRef CAS.
- R. Tang, C. Weng, X. Peng and Y. Han, Metab. Eng., 2020, 61, 11–23 CrossRef CAS PubMed.
- A. Sznajder, D. Pfeiffer and D. Jendrossek, Appl. Environ. Microbiol., 2015, 81, 1847–1858 CrossRef.
- A. da Silva Antonio, L. Wiedemann and V. Veiga-Junior, RSC Adv., 2020, 10, 23379–23393 RSC.
- X. Luo, M. A. Reiter, L. d'Espaux, J. Wong, C. M. Denby, A. Lechner, Y. Zhang, A. T. Grzybowski, S. Harth, W. Lin, H. Lee, C. Yu, J. Shin, K. Deng, V. T. Benites, G. Wang, E. E. K. Baidoo, Y. Chen, I. Dev, C. J. Petzold and J. D. Keasling, Nature, 2019, 567, 123–126 CrossRef CAS PubMed.
- S. Galanie, K. Thodey, I. J. Trenchard, M. Filsinger Interrante and C. D. Smolke, Science, 2015, 349, 1095–1100 CrossRef CAS PubMed.
- A. Gerber, M. Kleser, R. Biedendieck, R. Bernhardt and F. Hannemann, Microb. Cell Fact., 2015, 14, 107 CrossRef PubMed.
- M. F. Moradali and B. H. A. Rehm, Nat. Rev. Microbiol., 2020, 18, 195–210 CrossRef CAS PubMed.
- Y. Liu, X. Ma, H. Liang, G. Stephanopoulos and K. Zhou, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab065 CrossRef CAS PubMed.
- J. Alonso-Gutierrez, R. Chan, T. S. Batth, P. D. Adams, J. D. Keasling, C. J. Petzold and T. S. Lee, Metab. Eng., 2013, 19, 33–41 CrossRef CAS PubMed.
- S. Wu, X. Ma, A. Zhou, A. Valenzuela, K. Zhou and Y. Li, Sci. Adv., 2021, 7, eabh4048 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |