Molecular imprinting as a simple way for the long-term maintenance of the stemness and proliferation potential of adipose-derived stem cells: an in vitro study†
Abolfazl
Nazbar
a,
Saeed
Samani
 b,
Sepideh
Yazdian Kashani
b,
Sepideh
Yazdian Kashani
 *c,
Amir
Amanzadeh
a,
Shahram
Shoeibi
*c,
Amir
Amanzadeh
a,
Shahram
Shoeibi
 d and
Shahin
Bonakdar
*a
d and
Shahin
Bonakdar
*a
aNational Cell Bank Department, Pasteur Institute of Iran, Tehran, Iran. E-mail: sh_bonakdar@pasteur.ac.ir
bDepartment of Tissue Engineering and Applied Cell Sciences, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran
cDepartment of Chemical Engineering, Amirkabir University of Technology (Tehran Polytechnic), Tehran, Iran. E-mail: ykashani@aut.ac.ir
dFood and Drug Laboratory Research Center (FDLRC), Iran Food and Drug Administration (IFDA), MOH & ME, Tehran, Iran
First published on 16th June 2022
Abstract
Cells are smart creatures that respond to every signal after isolation and in vitro culture. Adipose-derived stem cells (ADSCs) gradually lose their characteristic spindle shape, multi-lineage differentiation potential, and self-renewal ability, and enter replicative senescence after in vitro expansion. This loss of cellular function is a serious impediment to clinical applications that require huge numbers of cells. It has been proven that substrates with cell imprints can be applied for stem cells' differentiation into desired cells or to re-culture any cell type while maintaining its ordinary activity. This study demonstrated the application of cell-imprinted substrates as a novel method in the long-term expansion of ADSCs while maintaining their stemness. Here we used molecular imprinting of stem cells as a physical signal to maintain stem cells' stemness. First, ADSCs were isolated and cultured on the tissue culture plate. Then, cells were fixed, and stem cell-imprinted substrates were fabricated using PDMS. Afterward, ADSCs were cultured on these substrates and subjected to osteogenic and adipogenic differentiation signals. The results were compared with ADSCs cultured on a polystyrene tissue culture plate and non-patterned PDMS. Morphology analysis with optical and fluorescence microscopy and SEM images illustrated that ADSCs seeded on imprinted substrates kept ADSC morphology. Alizarin Red S and Oil Red O staining, flow cytometry, and qPCR results showed that ADSC-imprinted substrates could reduce the differentiation of stem cells in vitro even if the differentiating stimulations were applied. Also, cell cycle analysis revealed that ADSCs could maintain their proliferation potential. So this method can maintain stem cells' stemness for a long time and reduce the unwanted stem cell differentiation that occurs in conventional cell culture on tissue culture plates.
1. Introduction
In recent years the application of adult and embryonic stem cells has received special attention in cell therapy and tissue engineering; in many cases, it is impossible to obtain a suitable cell substitute for adult tissues from the patient's adult cells to repair damaged tissues.1–3 When a stem cell divides in the human body, it may not change its nature or differentiate into a specific cell, such as muscle, red blood, or brain cells. So, stem cells in many tissues serve as a regeneration system to renew damaged cells. Chemical, mechanical, electrical, and surface factors can affect stem cell fate. Despite the conditions causing differentiation into desirable cells, the conditions under which proliferation occurs without differentiation are also necessary. Many researchers have been interested in mesenchymal stem cells (MSCs) to overcome the constraints of employing adult cells in regenerative medicine by considering self-renewal capacity, multi-lineage differentiation potential, and immunosuppressive ability. So, because of their unique characteristics, such as availability, differentiation potential, immune cell evasion, and easy in vitro proliferation, MSCs are a promising cell type for regenerative medicine and stem cell therapy.4 Also, the advantages of using allogeneic instead of autologous cells are essential factors to be considered in cell transplantation methods that can be achieved using MSCs.5–7Intravenous injection of MSCs requires minimal effective dosages of about 70–190 million cells per patient per dose, and around 100 million cells are necessary to treat a person weighing 70 kg. However, on the one hand, clinical trials are hampered by the deficient availability of MSCs in isolated samples. For example, MSCs make up just 0.001–0.01% of nucleated cells aspirated from bone marrow.8,9 Therefore, extensive in vitro passaging is needed to expand freshly homogenous MSC populations before clinical applications.5 On the other hand, isolated adult stem cells may inadvertently differentiate into undesirable cell lineages during in vitro continuously culturing in conventional tissue culture plates (TCP),5 leading to the loss of their normal phenotype.10 Indeed, biochemical and biophysical cues in the surrounding environment influence cell proliferation and differentiation,11 and regulate the MSCs' differentiation ability into a specific lineage.4
The in vivo microenvironment provides numerous regulating stimuli not available in conventional cell culture methods.12 In native tissues, cells do not live alone and continually sense and interact with the extracellular matrix (ECM).13 The ECM supports most cellular functions chemically, mechanically, and physically,13,14 and can determine the fate of stem cells.15 For large-scale clinical stem cell applications requiring large numbers of stem cells, loss or alteration of cell function is a significant barrier.9 Although many studies have been performed to mimic the ECM, preparation of an acceptable artificial ECM is not facile.16
Because complex topographical niches, such as ECM shape, govern cell activities in vivo, the topographic surfaces at the micro and nanoscale can regulate stem cell fate.17 Previous studies have shown that stem cells can sense changes in nanoscale topographical features, highlighting the role of nanostructured biomaterials' surface free energy in regulating cell fate.18 For example, for osteogenic differentiation of stem cells, Shi et al. made micropatterned PDA-coated parafilms with varied groove widths providing topographic cues for cell control on PDA-coated parafilms.19 These results support the hypothesis that besides the mechanosensing characteristics such as rigidity or elasticity, cells recognize the topography of their substrates, and surface topography affects stem cell differentiation and fate.20–26
Adipose tissue can be obtained from by-products of certain processes, such as esthetic surgery, which was previously considered as biological waste. Thus, using adipose-derived mesenchymal stem cells (ADSCs) avoids many ethical problems associated with bone marrow-derived stem cells.27In vitro expansion of ADSCs changes their characteristic spindle shape, self-renewal ability, and multi-lineage differentiation potential causing their replicative senescence.9 For clinical applications, strategies that allow the generation of large numbers of ADSCs parallel to the long-term maintenance of their stemness are required.28
The importance of culture substrate properties, such as substrate nanostructures, topography, and stiffness on cell fate, has been evaluated in various studies.29–34 The most often used substrates for in vitro cell culture are tissue culture plates that increase the likelihood of osteogenic differentiation because of the substrate rigidity.35 So, in stem cell-based regenerative medicine, suitable cell culture platforms providing well-defined biophysical and biochemical properties are needed to mimic the in vivo microenvironments,12 and simulate identical cell proliferation and differentiation conditions.36 Nonetheless, designing a substrate that will give the required biological cues to drive cells still is challenging.37
One of the fundamental areas in regenerative medicine is the responsiveness of stem cells to topographical cues, including micro- and nanoscaled patterns influencing cell fate. Although patterns are frequently employed to enhance cell functions, their mechanisms are not known completely. Furthermore, the concerns of chemical cues, such as growth factors' expensiveness, their short half-life in vivo, and the possibility of harmful effects from overdosing, have been solved in topographical patterns.37
Molecular recognition is essential in most biological processes because it allows selective and intelligent distinguishing of individual molecules in complex surroundings.38 Molecular imprinting is a synthetic method that creates biomimetic materials for protein recognition.39 Molecularly imprinted polymers (MIPs) have been proven to promote cell adhesion and proliferation, are stable in various chemical and physiological conditions and can be used multiple times.40 MIPs are a popular alternative to natural molecular recognition elements,38,41 and can mimic the antigen-binding properties of antibodies.39 Surface imprinting is one of the leading fabrication strategies of MIPs, including different approaches such as soft lithography. In soft lithography, an elastomeric stamp is made by casting a pre-polymer solution, typically polydimethylsiloxane (PDMS) with a relatively low elastic modulus, onto the template. After polymerization and stamp production, specific geometries can be made from nanoscale (30 nm) to micro-scale (100 μm).36,37
PDMS has been employed in the imprinting procedure because of its simplicity of fabrication, gas permeability, optical transparency, and low chemical reactivity.42 Also, against conventional TCP, PDMS has many other advantages such as nontoxicity, adjustable physico-mechanical properties, easy moldability, precision at micro-scale and nanoscale, and low manufacturing costs, which make it desirable as a cell culture substrate.12 As a substrate to study cell behavior, PDMS provides a better understanding of cell behavior toward stretching,43 stiffness,44 mechanical stimulation,45 and topography.46–49
However, the surface hydrophobicity of PDMS has a negative effect on cell adhesion and, consequently, normal cell behaviors and functions.12 Additionally, because cells and especially stem cells do not intrinsically attach to any surface, some surface modifications are mandatory for PDMS to improve cell attachment and spreading.50 So, many approaches have tried to alter the PDMS surface somehow to overcome its limitations,51,52 which can be done by plasma treatment, ECM protein coating, or other molecular coatings. Despite better cell adhesion and spreading acquired via these modifications, they are not practical for all cell lines, can be influenced by variable conditions, and cannot ensure the expected demand for the qualified cell source for human therapeutic use.12
Some studies have focused on using cellular patterns as biophysical stimulating factors to direct cellular functions in a specific direction. For the first time, Mahmoudi et al. prepared a cell-imprinted substrate by PDMS casting and polymerization on mature chondrocytes to induce chondrogenic differentiation in rabbit adipose tissue-derived mesenchymal stem cells.49 Furthermore, chondrocyte spherical shape-imprinted substrates were used to induce differentiation, redifferentiation, and transdifferentiation. It was discovered that a chondrocyte template might successfully shift MSC and semifibroblast spindle morphology toward spherical morphology. By manipulating the cellular shape, any cells with chondrogenic differentiation potential could be differentiated into chondrocytes.47 In other studies, osteogenic,53 keratinogenic,48 neurogenic,54–56 and tenogenic57 differentiation of stem cells was achieved by the imprinting method. Also, the imprinting approach for induced pluripotent stem cells can improve cardiomyogenic differentiation efficiency.29 Cell imprinting can also reduce or eliminate the requirement for lengthy patient sample analysis.58 Overall, molecular and cellular imprinting may be a promising way to manipulate cell phenotypes and functions without any chemical cues.
Rather than stem cell differentiation studied in previous reports, this study reveals a novel role of cell-imprinted substrates for maintaining stemness and proliferation potential of ADSCs during their in vitro long-term culture for future clinical and tissue engineering applications. Based on published findings of the positive effects of nanotopographies on cell fate, we concluded and expected that the patterns of stem cells could influence cellular behaviors better. Thus we developed ADSC-patterned and non-patterned PDMS substrates and cultured ADSCs on them. Then, we evaluated the effect of the ADSC pattern on maintaining stemness and decreasing osteogenic and adipogenic differentiation for a long time. We hope that our observations encourage other researchers to perform new studies to understand stemness maintenance better.
2. Experimental
2.1. ADSC isolation
The experiments were carried out at the Pasteur Institute of Iran and were approved by the institute's ethics committee (code: IR.PII.REC.1399.005). After signing the consent forms by all patients, adipose tissue was obtained from healthy individuals having a cesarean delivery and preserved at room temperature in DMEM (GIBCO, Scotland) containing 10% (v/v) fetal bovine serum (FBS, GIBCO, Scotland), 100 U mL−1 penicillin (Sigma-Aldrich, USA), and 100 μg mL−1 streptomycin (Sigma-Aldrich, USA) (complete culture medium).Based on Iran National Cell Bank's protocols and previously published studies, harvested adipose tissue was rinsed three times with DMEM containing an antibiotic/antimycotic solution (1%, Invitrogen, USA). Then connective tissues and blood vessels were separated, the adipose fragments were washed with phosphate-buffered saline (PBS) solution supplemented by antibiotic/antimycotic agents (1%, Invitrogen, USA), and treated with collagenase type I (0.02 mg mL−1, GIBCO, USA) for 45 minutes at 37 °C. After centrifuging the homogenate at 2000 rpm for five minutes, the deposited cell pellet was suspended and cultured in DMEM supplemented with 10% FBS and 1% Pen/Strep under common cell culture conditions (37 °C, 5% CO2, in a humidified atmosphere).2,47,59
2.2. ADSC characterization
The isolated ADSCs were characterized by flow cytometry with positive and negative CD markers. Additionally, the ADSCs were cultured in osteogenic and adipogenic mediums. Alizarin Red S and Oil Red O were used to stain the fixed cells after 21 days to confirm the osteogenic and adipogenic differentiation potential, respectively.60 The detailed procedures are as below.2.3. Developing cell-imprinted substrates
Freshly isolated ADSCs at a density of 3 × 105 cells per well were seeded in the 6-well tissue culture plate and incubated for one week. After reaching 90% confluence, the cells were fixed with 4% glutaraldehyde solution for 24 hours and washed with PBS three times. The PDMS casting procedure was performed by pouring the prepared elastomer solution onto the fixed ADSCs and kept at 37 °C for 48 hours. PDMS-based substrate platforms were fabricated by blending PDMS (Sylgard 184 Silicon Elastomer Kit, Dow Corning) with different curing agent weight ratios (curing agent/base polymer = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 1
30, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, and 1
40, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). Then the mold was peeled off from the tissue culture plate and rinsed thoroughly with 1 M NaOH solution for 30 minutes to eliminate any residual cell debris and existing chemicals from the imprinted substrates.2,49,54,55,59
50). Then the mold was peeled off from the tissue culture plate and rinsed thoroughly with 1 M NaOH solution for 30 minutes to eliminate any residual cell debris and existing chemicals from the imprinted substrates.2,49,54,55,59
2.4. Surface characterization of PDMS substrates
Scanning Electron Microscopy (SEM, Philips XL30, Netherlands) images of the substrates were evaluated based on previously published protocols.2,552.5. ADSC culture on the cell-imprinted substrates
ADSCs were proliferated on the cell-imprinted PDMS, non-patterned PDMS, and tissue culture plate (TCP) as the control, which were called ADSCI, ADSCP, and ADSCT, respectively. The cells (at a density of 4 × 104 cells per sample) were seeded onto the fabricated PDMS-based culture platforms placed in the 6-well plates and tissue culture plates as well. Fresh DMEM supplemented with 10% FBS was added, and exchanged every three days, and the cells were passaged every seven days. Finally, appropriate analyses were carried out considering suitably designed intervals.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL onto the fabricated PDMS-based culture platforms placed in the 24-well plates and TCP for the mentioned time points. The culture medium was removed slowly, and the cells were rinsed gently with PBS to avoid cell detachment, and treated for four hours with a 9
000 cells per mL onto the fabricated PDMS-based culture platforms placed in the 24-well plates and TCP for the mentioned time points. The culture medium was removed slowly, and the cells were rinsed gently with PBS to avoid cell detachment, and treated for four hours with a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) dilution of Alamar Blue in FBS-free DMEM. After incubation, three 150 μL replicates of Alamar Blue-containing solutions were pipetted into a blank 96-well plate, and optical densities at 560 nm excitation and 590 nm emission were read by an ELx808 ELISA reader (BioTek, USA).
1 (v/v) dilution of Alamar Blue in FBS-free DMEM. After incubation, three 150 μL replicates of Alamar Blue-containing solutions were pipetted into a blank 96-well plate, and optical densities at 560 nm excitation and 590 nm emission were read by an ELx808 ELISA reader (BioTek, USA).
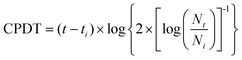 | (1) |
| Primer | Sequence (5′ →3′) | |
|---|---|---|
| GAPDH | Forward | GAGTCCACTGGCGTCTTCA |
| Reverse | TCTTGAGGCTGTTGTCATACTTC | |
| Collagen I | Forward | CGATGGCTGCACGAGTCA |
| Reverse | GGTTCAGTTTGGGTTGCTTGTC | |
| PPRγ | Forward | ACGAAGACATTCCATTCACAAG |
| Reverse | CTCCACAGACACGACATTCAAT | |
| Osteocalcin | Forward | CAGCGAGGTAGTGAAGAGACC |
| Reverse | TCTGGAGTTTATTTGGGAGCAG | |
| CD34 | Forward | GCCCAGATCAGCTCCTAACCC |
| Reverse | GATCCCTGCTCAACCCCT | |
| CD105 | Forward | GCATCCTTCGTGGAGCTACC |
| Reverse | GAGGAGTGGTCTGGATCGG | |
| CD90 | Forward | ATGAAGGTCCTCTACTTATCCGC |
| Reverse | GCACTGTGACGTTCTGGGA | |
2.6. Statistical analysis
All results are presented as mean ± standard deviation. For statistical comparisons between multiple groups, one-way ANOVA on Ranks followed by Tukey's post hoc test was done at the significance level of 0.05.3. Results and discussion
3.1. ADSC characterization
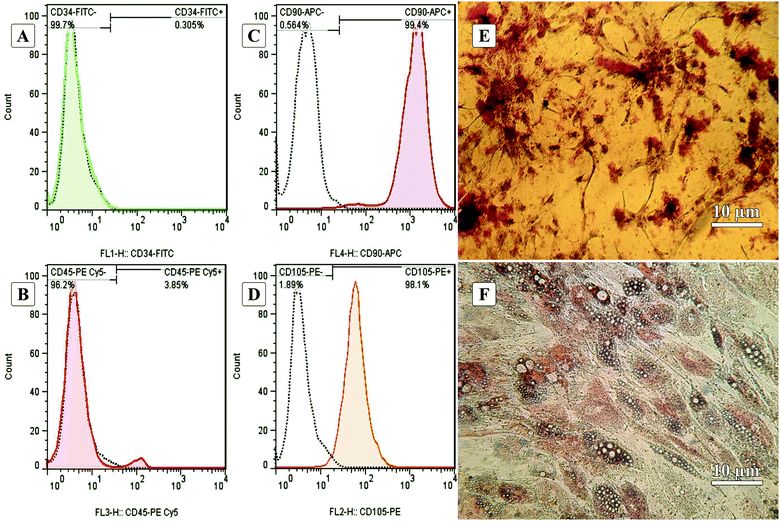
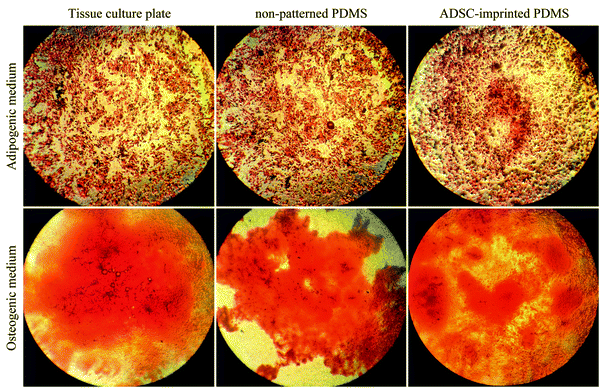
Fig. 1A–D show the characterization of stem cells by flow cytometry with positive and negative CD markers. Flow cytometry analysis revealed that the ADSCs at the first passage could express the MSC-specific markers (CD105 and CD90) well, and the expressions of the hematopoietic markers (CD45 and CD34) were at a low level.5,62Fig. 1E and F show the Alizarin Red S and Oil Red O staining after 21-days of culturing in osteogenic and adipogenic mediums. These results confirmed the capability of the ADSCs for differentiating into osteoblast- and adipocyte-like cells demonstrated by deposited calcium compound (Fig. 1E) and lipid droplets (Fig. 1F), respectively.63 After isolation, to reach the appropriate confluence in a short time, the ADSCs were counted and transferred to culture plates in the desired number to maintain the cells in ideal conditions, less affected by growth and proliferation conditions in tissue culture plates. After culturing for one week in a tissue culture plate, the ADSCs were fixed with 4% glutaraldehyde before pouring PDMS on them. This fixation maintains the cell morphology during the cell-imprinting process.3.2. Characterization of ADSC-imprinted substrates
Fig. 2 shows the SEM images of imprinted PDMS substrates from different surfaces, including air (PDMS with no imprint), TCP, and ADSCs. SEM evaluations of the cell-imprinted PDMS substrates showed that the imprinting process was performed well, and ADSCs' topography was transferred to the PDMS substrate.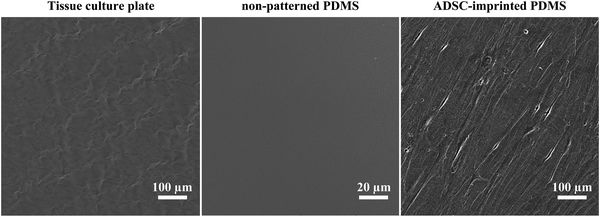 | ||
| Fig. 2 Scanning electron micrographs of tissue culture polystyrene plate (TCP)-imprinted PDMS, PDMS with no imprint (air surface), and ADSC-imprinted PDMS. | ||
To gain the topography of ADSCs precisely, PDMS must flow slowly during the molding and curing processes, which is dependent on the curing agent/PDMS ratio. It has been shown that there is a relationship between substrate stiffness and stem cell potency. A soft substrate retains the proliferative and differentiating potential of mesenchymal stem cells during long-term expansion.9,64 Overall, mixtures of different curing agent/PDMS ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 1
30, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, and 1
40, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) were evaluated to adjust the surface stiffness and better molding. A ratio of 1
50) were evaluated to adjust the surface stiffness and better molding. A ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 was chosen in this study because it could perform the cell-imprinting process well and could be easily peeled from the plate surface after curing. Diluted mixtures (1
30 was chosen in this study because it could perform the cell-imprinting process well and could be easily peeled from the plate surface after curing. Diluted mixtures (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and 1
40 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratios) were not appropriate for the cell-imprinting process because they could not be peeled easily after curing and were torn.
50 ratios) were not appropriate for the cell-imprinting process because they could not be peeled easily after curing and were torn.
Also, washing the surface of the cell-imprinted substrate with 1 M NaOH removes cell debris and proteins after mold casting, eliminates the effect of these residues on cell fate, and restricts the results and observations to the cellular imprinted template. The X-ray photoelectron spectroscopy (XPS) elemental analysis of the surface of cell-imprinted PDMS substrate confirmed the absence of detectable nitrogen atoms after chemical treatment by NaOH, proving the absence of detectable protein on the surface.54
According to the Mahmoudi et al. study in which high-resolution AFM analysis was performed for cell-imprinted replicas of different cells, the PDMS-based imprinting method assures a high-quality mold surface by replicating a surface roughness of less than 2 nm.49 Also, Kamgouyan and coworkers fabricated substrates with the imprinted patterns of five spindle-like cell types and compared their nanotopography. They analyzed the substrates by AFM, obtained roughness profiles at the nanoscale, and confirmed the difference between the nanotopographies of the cellular imprints.65 These findings can guarantee the usage of the PDMS imprinting method to copy and save the molecular pattern of the cell membrane precisely for future clinical regulation of cell behaviors.
Studies have shown that surface-specific nanotopograpies can affect the cell fate.17,34 Cells respond to nanoscale surface features by actin- and fibril-based filopodia, by which cells explore the surface and understand it. Although the morphology of ADSCs at the micro-scale is similar to some primary cells such as osteoblasts, microscopic characteristics cannot solely stimulate the cells to exhibit a specific behavior.53 Since cell-imprinted substrates can mimic the nanoscale properties, cell templates are promising for cell culture applications.
3.3. Evaluation of adhesion and morphology of ADSCs
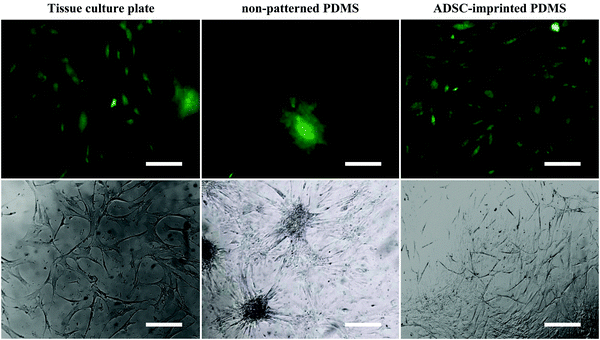
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68) and better adhesion conditions. ADSCs on non-patterned PDMS (ADSCP sample) had poor adhesion and spherical shape in aggregated colonies because of the inherent hydrophobicity of PDMS and its low stiffness. However, ADSCs cultured on the cell-imprinted PDMS surface (ADSCI sample) had an expected spindle shape for these cells. Despite the inherent hydrophobicity and low stiffness of the PDMS surface, ADSCI had better cell adhesion compared to ADSCP, which could be attributed to the presence of irregular nanotopographies; because the cells grew randomly after isolation and before the fixation stage, creating an irregular ADSC-specific pattern was inevitable.
68) and better adhesion conditions. ADSCs on non-patterned PDMS (ADSCP sample) had poor adhesion and spherical shape in aggregated colonies because of the inherent hydrophobicity of PDMS and its low stiffness. However, ADSCs cultured on the cell-imprinted PDMS surface (ADSCI sample) had an expected spindle shape for these cells. Despite the inherent hydrophobicity and low stiffness of the PDMS surface, ADSCI had better cell adhesion compared to ADSCP, which could be attributed to the presence of irregular nanotopographies; because the cells grew randomly after isolation and before the fixation stage, creating an irregular ADSC-specific pattern was inevitable.
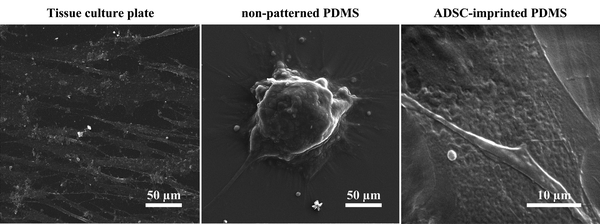 | ||
| Fig. 3 Scanning electron micrographs of ADSCs cultured on TCP, non-patterned PDMS, and ADSC-imprinted substrate. ADSC stands for adipose-derived stem cell. | ||
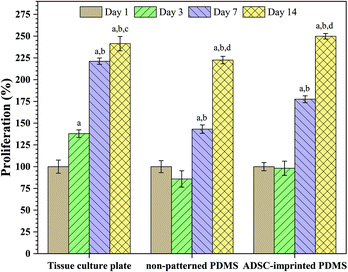
Fig. 4 shows the effect of surface characteristics on cell proliferation after 1, 3, 7, and 14 days for ADSCs cultured on TCP (ADSCT), non-patterned PDMS (ADSCP), and cell-imprinted PDMS (ADSCI). Over time, it was found that the number of cells attached to the surface increased significantly in each group, such that the proliferation percentage for ADSCT, ADSCP, and ADSCI increased by 141.4, 122.5, and 149.9%, respectively, after 14 days. Although a high proliferation percent on TCP was expected because of its favorable surface characteristics for cell adhesion and proliferation, the presence of the ADSC template in the ADSCI sample could largely eliminate the negative effect of the surface hydrophobicity of PDMS, causing a significant increase in the proliferation percent by 12.3% compared to ADSCP (p < 0.001). Moderate proliferation on the surface of PDMS with an ADSC membrane pattern reflects a moderate adhesion state that can provide an ADSC mechanism for self-renewal, differentiation suppression, and long-term in vitro culture. Adhesion strength to culture substrate determines the cell fate, such that stronger adhesion leads the stem cell to hard tissue phenotypes and weaker adhesion causes differentiation into soft tissues. So, creating a moderate surface stiffness associated with physical anchoring sites through cell-specific patterns can provide suitable cell adhesion and proliferation.
3.4. Cell population doubling time and cell cycle evaluation
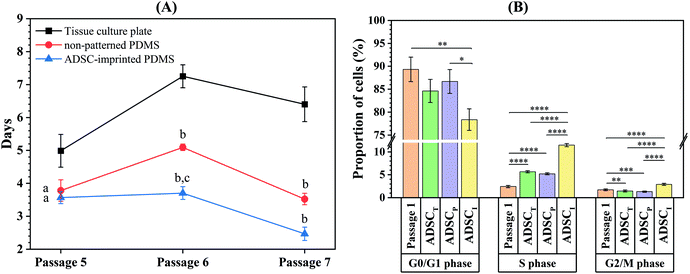
Isolated ADSCs were subcultured every seven days up to 21 days on the surface of TCP (ADSCT sample), non-patterned PDMS (ADSCP sample), and ADSC-imprinted PDMS substrate (ADSCI sample) and counted at the end of each time point (passages 5, 6, and 7) to calculate cell population doubling time (CPDT). At passage 5, the CPDT of ADSCP and ADSCI was significantly lower than that of ADSCT by 24.2 and 28.5%, respectively (Fig. 6A). Despite similar reduction trends in CPDTs of ADSCP and ADSCI at passage 6, the presence of the ADSC pattern had a more significant effect on CPDT reduction (p < 0.01, 109%) compared to ADSCP (29.7%). Although CPDT of ADSCI did not change at passage 6 and significantly decreased at passage 7 (p < 0.05), CPDTs of ADSCT and ADSCP increased significantly at passage 6 (p < 0.0001 and p < 0.01 for ADSCT and ADSCP, respectively), and decreased significantly at passage 7 for ADSCP (p < 0.001). At passage 7, the CPDTs of ADSCP and ADSCI were significantly lower than that of ADSCT by 45 and 61.2%, respectively. Despite the non-significant difference between the CPDTs of PDMS-based substrates, the presence of an ADSC pattern caused 30% in the CPDT compared to non-patterned PDMS (ADSCP sample).Evaluation of the cell cycle at passage 8 showed that the number of ADSCs for the ADSCI sample in the S and G2/M phases increased more significantly than those of ADSCT and ADSCP samples (102.5 and 119.9% in the S phase, 96.6 and 118.9% in the G2/M phase) (Fig. 6B and Fig. S1, ESI†). In addition, a decreased G1 phase in the ADSCI sample compared to ADSCT and ADSCP samples (by 310 and 132%, respectively) was parallel to the decrease in doubling time and proved the prevention of ADSC differentiation.
Mesenchymal stem cells are in the stationary phase in tissue homeostatic conditions to protect against DNA damage. These cells are thought to become a functional phenotype for responding to tissue demands by producing rapidly proliferating progenitor cells before entering the area needing regeneration.66 When mesenchymal stem cells are isolated from their native microenvironment in vitro, they enter an active form whose imprinted patterns save the features of that form at the micro- and nanoscale. So, the cell population rate increases, and the G1 phase decreases.
3.5. Effect of ADSC-imprinted substrates on ADSC stemness
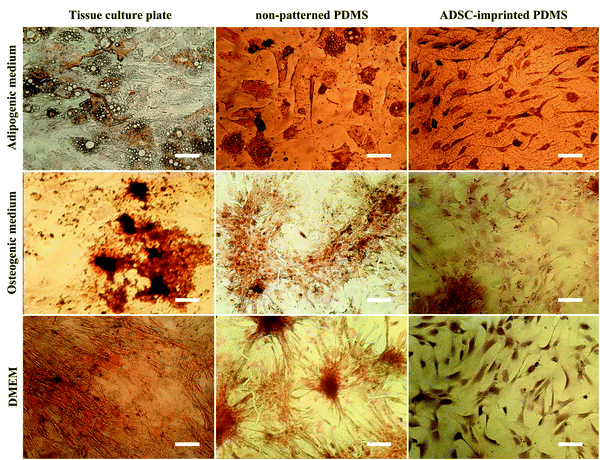
ADSCT cultured in the osteogenic and adipogenic medium was differentiated into osteocyte- and adipocyte-like cells indicated by calcium precipitation and lipid droplet formation, respectively. Also, a little calcium precipitation was observed for ADSCT cultured in DMEM, which could be because of the TCP stiffness effect on stem cells.
ADSCP cultured in differentiating mediums showed osteogenic and adipogenic differentiation. However, ADSCP in adipogenic medium had spherical morphology due to the hydrophobicity and weak surface adhesion of non-patterned PDMS. By culturing in DMEM, calcium precipitation increased for ADSCP, which might be because of the pressure between cellular aggregates and changes in gene expression.
However, the staining of ADSCI cultured in osteogenic and adipogenic medium revealed that ADSCs received the least differentiating effects from the mediums, resulting in low calcium deposition and rare lipid droplets. Furthermore, ADSCI could maintain its spindle-shaped morphology compared to ADSCP, which had spherical morphology. Since it has been reported that the surface topography of a culture substrate can even maintain stem cells' stemness,69 ADSC-specific surface topography can significantly reduce the influence of the differentiating mediums and maintain the stemness of stem cells for a long time.
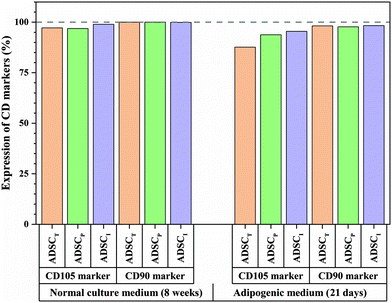
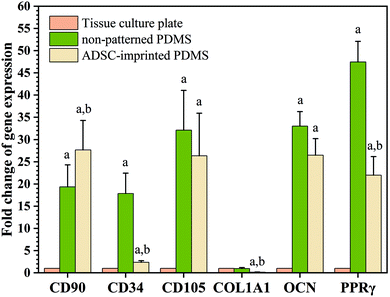
To better understand the effect of ADSC-imprinting on ADSC fate, the cells were cultured in the adipogenic differentiating medium for 21 days. As can be seen in Fig. 8, the expression of CD105 marker ADSCI increased compared to the other two groups, though CD90 expression did not show a significant difference between all groups.
Although expressions of CD34 (as a negative marker for ADSC) in ADSCI and particularly in ADSCP were significantly higher than that of ADSCT, the fold change of CD34 for ADSCI was 2.36 times greater than ADSCT only. Furthermore, the influence of the ADSC template (ADSCI sample) on CD34 downregulation by ∼87% was significant (p < 0.05) compared to ADSCP, proving the more positive effect of the ADSC template on stemness maintenance.
The PPRγ analysis demonstrated significant upregulation in ADSCP and ADSCI compared to ADSCT, which was inevitable due to the effect of lower surface stiffness on adipogenic differentiation. Also, the lower strength of cell adhesion can promote the expression of adipocyte-specific genes.66 Although ADSCP and ADSCI had similar surface stiffness because of the same chemical composition and fabrication process, the presence of an ADSC-specific template in the ADSCI sample could decrease PPRγ expression by ∼53.7% supporting the exact application of ADSC-imprinting to produce a better cell culture substrate. Various studies have shown that increasing the expression of CD90 reduces the ability of stem cells to differentiate into adipose cells.70,71 Therefore, the presence of an ADSC-specific pattern could protect ADSCs against adipogenesis, thereby maintaining stemness long-term.
Analysis of specific genes involved in the osteogenesis pathway showed that Col1A1 underwent significant downregulation in ADSCI compared to ADSCT and ADSCP (by ∼85% for both samples), confirming the positive effect of the ADSC template on protecting ADSCs against osteogenic differentiation. Although the OCN gene was significantly upregulated in ADSCP and ADSCI compared to ADSCT, the positive presence of the ADSC template caused a ∼20% decrease in OCN level compared to ADSCP.
The qPCR results were truly conformed to Alizarin Red S and Oil Red O staining of ADSCs cultured on different substrates under osteogenic and adipogenic conditions (Fig. 7). Despite higher cytoplasmic expression of some osteogenic and adipogenic genes in PDMS-based substrates, other results demonstrating morphological or functional characteristics (microscopic imaging, staining, and flow cytometry) confirmed that PDMS-based substrates, particularly ADSC-imprinted ones, did not follow differentiation pathways, and the ADSC-specific pattern had a noteworthy influence on long-term stemness maintenance.
Examination of several commercial TCPs has shown that their surface topography is different (ranged 1–7 nm), resulting in various protein adsorption and population doubling.72 Also, spontaneous differentiation of MSCs by culturing them on common TCP causes rapid diminution resulting in a heterogeneous population of predominantly fibroblastic phenotype.73 Although the differentiation potential of TCP and ADSC-imprinted PDMS has similarities to some extent, it is probable that using commercial TCPs in each clinical center and the possibility of changes in laboratory equipment may cause heterogeneous consequences for clinical therapies. So, creating a culture substrate having cell-specific, cell-friendly, and maybe reproducible patterns can increase success in cell-based studies and therapies.
4. Conclusions
The application of stem cells to heal various diseases has received more attention in the last few decades. To control the differentiation of stem cells and form a homogeneous population in vitro, biomaterial–cell interactions and making a substrate that can control cellular functions without using chemical agents have become an interesting topic recently. However, the produced substrate should have features such as easy fabrication, cheapness, large-scale production, easy use for different users, and availability of raw materials and equipment.Although common tissue culture plates (TCPs) can be used for many stem cell applications, it is expected that using commercial TCPs may cause heterogeneous consequences for clinical therapies throughout the world because of their variety of surface topography. So, creating a culture substrate with a cell-friendly pattern can ensure success in cell-based studies and therapies. Cell-imprinted substrates can also be autoclaved for multiple usages, which reduces costs and creates appropriate maintenance conditions for in vitro cell culture.
In this study, we discussed a novel approach to making a suitable culture substrate for the long-term expansion of stem cells. We isolated adipose-derived stem cells (ADSCs), developed a PDMS-based substrate having an ADSC-specific pattern and performed some evaluations to analyze the capability of the ADSC-imprinted PDMS substrate to delay cell differentiation and maintain stemness after long-term cell culture. In comparison to common TCP and non-patterned PDMS, ADSC-imprinted PDMS had better cell attachment and the highest proliferation percentage due to the presence of ADSC patterns. It could capture the ADSCs' topography well and stimulate cultured ADSCs to present their original spindle-like morphology compared to other substrates. The presence of an ADSC-specific pattern significantly decreased the cell population doubling time compared to TCP and influenced phases of the cell cycle, resulting in more rapid cell division. Furthermore, the ADSC-specific pattern could largely prevent the differentiation of ADSCs cultured on the substrate, confirming the positive effect of specific surface topography on stemness maintenance. After long-term cell culturing, ADSC-specific surface markers were highly expressed on the cell membrane underlying stemness preservation. It can be concluded that biomimetic ADSC-imprinted substrates delay stem cell differentiation in vitro while maintaining their proliferative potential even when differentiating stimulations are used.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Pasteur Institute of Iran [grant no. 1707]; and Iran Food and Drug Administration (IFDA) [grant no. Pr977302].References
- S. Lopa, C. Mondadori, V. L. Mainardi, G. Talò, M. Costantini, C. Candrian, W. Swieszkowski and M. Moretti, Translational application of microfluidics and bioprinting for stem cell-based cartilage repair, Stem Cells Int., 2018, 2018, 6594841, DOI:10.1155/2018/6594841.
- S. Yazdian Kashani, M. Keshavarz Moraveji, M. Taghipoor, R. Kowsari-Esfahan, A. A. Hosseini, L. Montazeri, M. M. Dehghan, H. Gholami, S. Farzad-Mohajeri, M. Mehrjoo, M. Majidi, P. Renaud and S. Bonakdar, An integrated microfluidic device for stem cell differentiation based on cell-imprinted substrate designed for cartilage regeneration in a rabbit model, Mater. Sci. Eng., C, 2021, 121, 111794, DOI:10.1016/j.msec.2020.111794.
- Y. Liu, G. Zhou and Y. Cao, Recent Progress in Cartilage Tissue Engineering—Our Experience and Future Directions, Engineering, 2017, 3, 28–35, DOI:10.1016/J.ENG.2017.01.010.
- M. Mohammed, T. S. Lai and H. C. Lin, Substrate stiffness and sequence dependent bioactive peptide hydrogels influence the chondrogenic differentiation of human mesenchymal stem cells, J. Mater. Chem. B, 2021, 9, 1676–1685, 10.1039/d0tb02008g.
- Y. H. K. Yang, C. R. Ogando, C. Wang See, T. Y. Chang and G. A. Barabino, Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro, Stem Cell Res. Ther., 2018, 9, 1–14, DOI:10.1186/s13287-018-0876-3.
- C. Li, H. Zhao, L. Cheng and B. Wang, Allogeneic vs. autologous mesenchymal stem/stromal cells in their medication practice, Cell Biosci., 2021, 11, 1–21, DOI:10.1186/s13578-021-00698-y.
- W. Zakrzewski, M. Dobrzyński, M. Szymonowicz and Z. Rybak, Stem cells: Past, present, and future, Stem Cell Res. Ther., 2019, 10, 68, DOI:10.1186/s13287-019-1165-5.
- M. Kabat, I. Bobkov, S. Kumar and M. Grumet, Trends in mesenchymal stem cell clinical trials 2004-2018: Is efficacy optimal in a narrow dose range, Stem Cells Transl. Med., 2020, 9, 17–27, DOI:10.1002/sctm.19-0202.
- S. K. Kureel, P. Mogha, A. Khadpekar, V. Kumar, R. Joshi, S. Das, J. Bellare and A. Majumder, Soft substrate maintains proliferative and adipogenic differentiation potential of human mesenchymal stem cells on long-term expansion by delaying senescence, Biol. Open, 2019, 8(4), bio039453, DOI:10.1242/bio.039453.
- I. Jun, H. S. Han, J. W. Lee, K. Lee, Y. C. Kim, M. R. Ok, H. K. Seok, Y. J. Kim, I. S. Song, H. Shin, J. R. Edwards, K. Y. Lee and H. Jeon, On/off switchable physical stimuli regulate the future direction of adherent cellular fate, J. Mater. Chem. B, 2021, 9, 5560–5571, 10.1039/d1tb00908g.
- K. Ishihara, M. Kaneyasu, K. Fukazawa, R. Zhang and Y. Teramura, Induction of mesenchymal stem cell differentiation by co-culturing with mature cells in double-layered 2-methacryloyloxyethyl phosphorylcholine polymer hydrogel matrices, J. Mater. Chem. B, 2022, 10, 2561–2569, 10.1039/d1tb01817e.
- F. Etezadi, M. N. T. Le, H. Shahsavarani, A. Alipour, N. Moazzezy, S. Samani, A. Amanzadeh, S. Pahlavan, S. Bonakdar, M. A. Shokrgozar and K. Hasegawa, Optimization of a PDMS-Based Cell Culture Substrate for High-Density Human-Induced Pluripotent Stem Cell Adhesion and Long-Term Differentiation into Cardiomyocytes under a Xeno-Free Condition, ACS Biomater. Sci. Eng., 2022, 8(5), 2040–2052, DOI:10.1021/acsbiomaterials.2c00162.
- F. Han, C. Zhu, Q. Guo, H. Yang and B. Li, Cellular modulation by the elasticity of biomaterials, J. Mater. Chem. B, 2016, 4, 9–26, 10.1039/c5tb02077h.
- E. S. Place, N. D. Evans and M. M. Stevens, Complexity in biomaterials for tissue engineering, Nat. Mater., 2009, 8, 457–470, DOI:10.1038/nmat2441.
- C. Y. Yang, W. Y. Huang, L. H. Chen, N. W. Liang, H. C. Wang, J. Lu, X. Wang and T. W. Wang, Neural tissue engineering: The influence of scaffold surface topography and extracellular matrix microenvironment, J. Mater. Chem. B, 2021, 9, 567–584, 10.1039/d0tb01605e.
- A. Steier, A. Muñiz, D. Neale and J. Lahann, Emerging Trends in Information-Driven Engineering of Complex Biological Systems, Adv. Mater., 2019, 31(26) DOI:10.1002/adma.201806898.
- A. Higuchi, Q. D. Ling, S. S. Kumar, Y. Chang, A. A. Alarfaj, M. A. Munusamy, K. Murugan, S. T. Hsu and A. Umezawa, Physical cues of cell culture materials lead the direction of differentiation lineages of pluripotent stem cells, J. Mater. Chem. B, 2015, 3, 8032–8058, 10.1039/c5tb01276g.
- K. Metavarayuth, E. Villarreal, H. Wang and Q. Wang, Surface topography and free energy regulate osteogenesis of stem cells: effects of shape-controlled gold nanoparticles, Biomater. Transl., 2021, 2, 165–173, DOI:10.12336/biomatertransl.2021.02.006.
- L. Jia, F. Han, H. Wang, C. Zhu, Q. Guo, J. Li, Z. Zhao, Q. Zhang, X. Zhu and B. Li, Polydopamine-assisted surface modification for orthopaedic implants, J. Orthop. Transl., 2019, 17, 82–95, DOI:10.1016/j.jot.2019.04.001.
- R. S. Fischer, K. A. Myers, M. L. Gardel and C. M. Waterman, Stiffness-controlled three-dimensional extracellular matrices for high-resolution imaging of cell behavior, Nat. Protoc., 2012, 7, 2056–2066, DOI:10.1038/nprot.2012.127.
- W. Wang, H. Cui, P. Zhang, J. Meng, F. Zhang and S. Wang, Efficient Capture of Cancer Cells by Their Replicated Surfaces Reveals Multiscale Topographic Interactions Coupled with Molecular Recognition, ACS Appl. Mater. Interfaces, 2017, 9, 10537–10543, DOI:10.1021/acsami.7b01147.
- X. Zhou, J. Shi, F. Zhang, J. Hu, X. Li, L. Wang, X. Ma and Y. Chen, Reversed cell imprinting, AFM imaging and adhesion analyses of cells on patterned surfaces, Lab Chip, 2010, 10, 1182–1188, 10.1039/b926325j.
- L. Yang, L. Yang, L. Ge, L. Ge, P. Van Rijn and P. Van Rijn, Synergistic Effect of Cell-Derived Extracellular Matrices and Topography on Osteogenesis of Mesenchymal Stem Cells, ACS Appl. Mater. Interfaces, 2020, 12, 25591–25603, DOI:10.1021/acsami.0c05012.
- D. Hernandez, E. T. Ritschdorff, J. L. Connell and J. B. Shear, In Situ Imprinting of Topographic Landscapes at the Cell-Substrate Interface, J. Am. Chem. Soc., 2018, 140, 14064–14068, DOI:10.1021/jacs.8b09226.
- S. Kumari, S. Vermeulen, B. Van Der Veer, A. Carlier, J. De Boer and D. Subramanyam, Shaping Cell Fate: Influence of Topographical Substratum Properties on Embryonic Stem Cells, Tissue Eng., Part B, 2018, 24, 255–266, DOI:10.1089/ten.teb.2017.0468.
- P. Bhattacharjee, B. L. Cavanagh and M. Ahearne, Effect of substrate topography on the regulation of human corneal stromal cells, Colloids Surf., B, 2020, 190, 110971, DOI:10.1016/j.colsurfb.2020.110971.
- B. Bellei, E. Migliano, M. Tedesco, S. Caputo, F. Papaccio, G. Lopez and M. Picardo, Adipose tissue-derived extracellular fraction characterization: Biological and clinical considerations in regenerative medicine, Stem Cell Res. Ther., 2018, 9, 1–18, DOI:10.1186/s13287-018-0956-4.
- J. Liu, Y. Ding, Z. Liu and X. Liang, Senescence in Mesenchymal Stem Cells: Functional Alterations, Molecular Mechanisms, and Rejuvenation Strategies, Front. Cell Dev. Biol., 2020, 8, 258, DOI:10.3389/fcell.2020.00258.
- P. P. S. S. Abadi, J. C. Garbern, S. Behzadi, M. J. Hill, J. S. Tresback, T. Heydari, M. R. Ejtehadi, N. Ahmed, E. Copley, H. Aghaverdi, R. T. Lee, O. C. Farokhzad and M. Mahmoudi, Engineering of Mature Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Using Substrates with Multiscale Topography, Adv. Funct. Mater., 2018, 28, 1707378, DOI:10.1002/adfm.201707378.
- G. Abagnale, A. Sechi, M. Steger, Q. Zhou, C. C. Kuo, G. Aydin, C. Schalla, G. Müller-Newen, M. Zenke, I. G. Costa, P. van Rijn, A. Gillner and W. Wagner, Surface Topography Guides Morphology and Spatial Patterning of Induced Pluripotent Stem Cell Colonies, Stem Cell Rep., 2017, 9, 654–666, DOI:10.1016/j.stemcr.2017.06.016.
- M. Ali and J. B. Shear, Real time remodeling of cellular morphology using optical imprinting of cell-culture substrates, Biomed. Phys. Eng. Express, 2019, 5, 035029, DOI:10.1088/2057-1976/aafc8e.
- E. Costa, C. González-García, J. L. Gómez Ribelles and M. Salmerón-Sánchez, Maintenance of chondrocyte phenotype during expansion on PLLA microtopographies, J. Tissue Eng., 2018, 9, 1–10, DOI:10.1177/2041731418789829.
- B. Liu, C. Tao, Z. Wu, H. Yao and D.-A. Wang, Engineering strategies to achieve efficient in vitro expansion of haematopoietic stem cells: development and improvement, J. Mater. Chem. B, 2022, 10, 1734–1753, 10.1039/d1tb02706a.
- J. O. Abaricia, N. Farzad, T. J. Heath, J. Simmons, L. Morandini and R. Olivares-Navarrete, Control of innate immune response by biomaterial surface topography, energy, and stiffness, Acta Biomater., 2021, 133, 58–73, DOI:10.1016/j.actbio.2021.04.021.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix Elasticity Directs Stem Cell Lineage Specification, Cell, 2006, 126, 677–689, DOI:10.1016/j.cell.2006.06.044.
- A. F. Bonatti, C. De Maria and G. Vozzi, Molecular imprinting strategies for tissue engineering applications: A review, Polymers, 2021, 13, 1–20, DOI:10.3390/polym13040548.
- K. Zhang, X. Xiao, X. Wang, Y. Fan and X. Li, Topographical patterning: Characteristics of current processing techniques, controllable effects on material properties and co-cultured cell fate, updated applications in tissue engineering, and improvement strategies, J. Mater. Chem. B, 2019, 7, 7090–7109, 10.1039/c9tb01682a.
- W. Zhao, B. Li, S. Xu, X. Huang, J. Luo, Y. Zhu and X. Liu, Electrochemical protein recognition based on macromolecular self-assembly of molecularly imprinted polymer: A new strategy to mimic antibody for label-free biosensing, J. Mater. Chem. B, 2019, 7, 2311–2319, 10.1039/c9tb00220k.
- A. K. Venkataraman, J. R. Clegg and N. A. Peppas, Polymer composition primarily determines the protein recognition characteristics of molecularly imprinted hydrogels, J. Mater. Chem. B, 2020, 8, 7685–7695, 10.1039/d0tb01627f.
- M. Komiyama, T. Mori and K. Ariga, Molecular imprinting: materials nanoarchitectonics with molecular information, Bull. Chem. Soc. Jpn., 2018, 91, 1075–1111 CrossRef CAS.
- T. Saeki, E. Takano, H. Sunayama, Y. Kamon, R. Horikawa, Y. Kitayama and T. Takeuchi, Signalling molecular recognition nanocavities with multiple functional groups prepared by molecular imprinting and sequential post-imprinting modifications for prostate cancer biomarker glycoprotein detection, J. Mater. Chem. B, 2020, 8, 7987–7993, 10.1039/d0tb00685h.
- S. Yazdian Kashani, A. Afzalian, F. Shirinichi and M. Keshavarz Moraveji, Microfluidics for core–shell drug carrier particles – a review, RSC Adv., 2020, 11, 229–249, 10.1039/d0ra08607j.
- Y. Shao, J. M. Mann, W. Chen and J. Fu, Global architecture of the F-actin cytoskeleton regulates cell shape-dependent endothelial mechanotransduction, Integr. Biol., 2014, 6, 300–311, 10.1039/c3ib40223a.
- R. C. Lyon, F. Zanella, J. H. Omens and F. Sheikh, Mechanotransduction in cardiac hypertrophy and failure, Circ. Res., 2015, 116, 1462–1476, DOI:10.1161/CIRCRESAHA.116.304937.
- M. Ehrbar, S. C. Rizzi, R. Hlushchuk, V. Djonov, A. H. Zisch, J. A. Hubbell, F. E. Weber and M. P. Lutolf, Enzymatic formation of modular cell-instructive fibrin analogs for tissue engineering, Biomaterials, 2007, 28, 3856–3866, DOI:10.1016/j.biomaterials.2007.03.027.
- H. Kavand, H. Van Lintel, S. Bakhshi Sichani, S. Bonakdar, H. Kavand, J. Koohsorkhi and P. Renaud, Cell-Imprint Surface Modification by Contact Photolithography-Based Approaches: Direct-Cell Photolithography and Optical Soft Lithography Using PDMS Cell Imprints, ACS Appl. Mater. Interfaces, 2019, 11, 10559–10566, DOI:10.1021/acsami.9b00523.
- S. Bonakdar, M. Mahmoudi, L. Montazeri, M. Taghipoor, A. Bertsch, M. A. Shokrgozar, S. Sharifi, M. Majidi, O. Mashinchian, M. H. Sekachaei, P. Zolfaghari and P. Renaud, Cell-Imprinted Substrates Modulate Differentiation, Redifferentiation, and Transdifferentiation, ACS Appl. Mater. Interfaces, 2016, 8, 13777–13784, DOI:10.1021/acsami.6b03302.
- O. Mashinchian, S. Bonakdar, H. Taghinejad, V. Satarifard, M. Heidari, M. Majidi, S. Sharifi, A. Peirovi, S. Saffar, M. Taghinejad, M. Abdolahad, S. Mohajerzadeh, M. A. Shokrgozar, S. M. Rezayat, M. R. Ejtehadi, M. J. Dalby and M. Mahmoudi, Cell-imprinted substrates act as an artificial niche for skin regeneration, ACS Appl. Mater. Interfaces, 2014, 6, 13280–13292, DOI:10.1021/am503045b.
- M. Mahmoudi, S. Bonakdar, M. A. Shokrgozar, H. Aghaverdi, R. Hartmann, A. Pick, G. Witte and W. J. Parak, Cell-imprinted substrates direct the fate of stem cells, ACS Nano, 2013, 7, 8379–8384, DOI:10.1021/nn403844q.
- M. T. Lam and M. T. Longaker, Comparison of several attachment methods for human iPS, embryonic and adipose-derived stem cells for tissue engineering, J. Tissue Eng. Regen. Med., 2012, 6, s80–s86, DOI:10.1002/term.1499.
- J. N. Lee, X. Jiang, D. Ryan and G. M. Whitesides, Compatibility of mammalian cells on surfaces of poly(dimethylsiloxane), Langmuir, 2004, 20, 11684–11691, DOI:10.1021/la048562.
- J. Zhou, A. V. Ellis and N. H. Voelcker, Recent developments in PDMS surface modification for microfluidic devices, Electrophoresis, 2010, 31, 2–16, DOI:10.1002/elps.200900475.
- K. Kamguyan, A. A. Katbab, M. Mahmoudi, E. Thormann, S. Zajforoushan Moghaddam, L. Moradi and S. Bonakdar, An engineered cell-imprinted substrate directs osteogenic differentiation in stem cells, Biomater. Sci., 2018, 6, 189–199, 10.1039/c7bm00733g.
- Z. S. Ghazali, M. Eskandari, S. Bonakdar, P. Renaud, O. Mashinchian, S. Shalileh, F. Bonini, I. Uckay, O. Preynat-Seauve and T. Braschler, Neural priming of adipose-derived stem cells by cell-imprinted substrates, Biofabrication, 2021, 13, 035009, DOI:10.1088/1758-5090/abc66f.
- S. Dadashkhan, S. Irani, S. Bonakdar and B. Ghalandari, P75 and S100 gene expression induced by cell-imprinted substrate and beta-carotene to nerve tissue engineering, J. Appl. Polym. Sci., 2021, 138, 50624, DOI:10.1002/app.50624.
- M. Moosazadeh Moghaddam, S. Bonakdar, M. A. Shokrgozar, A. Zaminy, H. Vali and S. Faghihi, Engineered substrates with imprinted cell-like topographies induce direct differentiation of adipose-derived mesenchymal stem cells into Schwann cells, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 1022–1035, DOI:10.1080/21691401.2019.1586718.
- S. M. A. Haramshahi, S. Bonakdar, M. Moghtadaei, K. Kamguyan, E. Thormann, S. Tanbakooei, S. Simorgh, P. Brouki-Milan, N. Amini, N. Latifi, M. T. Joghataei, A. Samadikuchaksaraei, M. Katebi and M. Soleimani, Tenocyte-imprinted substrate: A topography-based inducer for tenogenic differentiation in adipose tissue-derived mesenchymal stem cells, Biomed. Mater., 2020, 15, 035014, DOI:10.1088/1748-605X/ab6709.
- J. Medlock, A. A. K. Das, L. A. Madden, D. J. Allsup and V. N. Paunov, Cancer bioimprinting and cell shape recognition for diagnosis and targeted treatment, Chem. Soc. Rev., 2017, 46, 5110–5127, 10.1039/c7cs00179g.
- S. Yazdian Kashani, M. Keshavarz Moraveji and S. Bonakdar, Computational and experimental studies of a cell-imprinted-based integrated microfluidic device for biomedical applications, Sci. Rep., 2021, 11, 1–17, DOI:10.1038/s41598-021-91616-2.
- M. A. Shokrgozar, M. Fattahi, S. Bonakdar, I. R. Kashani, M. Majidi, N. Haghighipour, V. Bayati, H. Sanati and S. N. Saeedi, Healing potential of mesenchymal stem cells cultured on a collagen-based scaffold for skin regeneration, Iran. Biomed. J., 2012, 16, 1–9, DOI:10.6091/ibj.1053.2012.
- H. Ghaneialvar, L. Soltani, H. R. Rahmani, A. S. Lotfi and M. Soleimani, Characterization and Classification of Mesenchymal Stem Cells in Several Species Using Surface Markers for Cell Therapy Purposes, Indian J. Clin. Biochem., 2018, 33, 46–52, DOI:10.1007/s12291-017-0641-x.
- S. T. L. Pinto Filho, M. M. Pillat, M. P. Rosa, F. Dalmolin, H. Ulrich and N. L. Pippi, Expression patterns of mesenchymal stem cell-specific proteins in adipose tissue-derived cells: possible immunosuppressing agent in partial allograft for restoring the urinary bladder in rabbits, Pesqui. Veterinária Bras., 2018, 38, 2183–2189 CrossRef.
- F. Aleahmad, S. Ebrahimi, M. Salmannezhad, M. Azarnia, M. Jaberipour, M. Hoseini and T. Talaei-Khozani, Heparin/Collagen 3D Scaffold Accelerates Hepatocyte Differentiation of Wharton's Jelly-Derived Mesenchymal, Stem Cells, Tissue Eng. Regener. Med., 2017, 14, 443–452, DOI:10.1007/s13770-017-0048-z.
- L. R. Smith, S. Cho and D. E. Discher, Stem cell differentiation is regulated by extracellular matrix mechanics, Physiology, 2018, 33, 16–25, DOI:10.1152/physiol.00026.2017.
- K. Kamguyan, S. Z. Moghaddam, A. Nazbar, S. M. A. Haramshahi, S. Taheri, S. Bonakdar and E. Thormann, Cell-imprinted substrates: In search of nanotopographical fingerprints that guide stem cell differentiation, Nanoscale Adv., 2021, 3, 333–338, 10.1039/d0na00692k.
- M. J. Dalby, N. Gadegaard and R. O. C. Oreffo, Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate, Nat. Mater., 2014, 13, 558–569, DOI:10.1038/nmat3980.
- A. J. García, Get a grip: Integrins in cell-biomaterial interactions, Biomaterials, 2005, 26, 7525–7529, DOI:10.1016/j.biomaterials.2005.05.029.
- P. M. Gilbert, K. L. Havenstrite, K. E. G. Magnusson, A. Sacco, N. A. Leonardi, P. Kraft, N. K. Nguyen, S. Thrun, M. P. Lutolf and H. M. Blau, Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture, Science, 2010, 329, 1078–1081, DOI:10.1126/science.1191035.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6, 997–1003, DOI:10.1038/nmat2013.
- C. F. Woeller, C. W. O’Loughlin, S. J. Pollock, T. H. Thatcher, S. E. Feldon and R. P. Phipps, Thy1 (CD90) controls adipogenesis by regulating activity of the Src family kinase, Fyn, FASEB J., 2015, 29, 920–931, DOI:10.1096/fj.14-257121.
- D. A. Moraes, T. T. Sibov, L. F. Pavon, P. Q. Alvim, R. S. Bonadio, J. R. Da Silva, A. Pic-Taylor, O. A. Toledo, L. C. Marti, R. B. Azevedo and D. M. Oliveira, A reduction in CD90 (THY-1) expression results in increased differentiation of mesenchymal stromal cells, Stem Cell Res. Ther., 2016, 7, 97, DOI:10.1186/s13287-016-0359-3.
- A. S. Zeiger, B. Hinton and K. J. Van Vliet, Why the dish makes a difference: Quantitative comparison of polystyrene culture surfaces, Acta Biomater., 2013, 9, 7354–7361, DOI:10.1016/j.actbio.2013.02.035.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. C. Oreffo and M. J. Dalby, Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency, Nat. Mater., 2011, 10, 637–644, DOI:10.1038/nmat3058.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb00279e |
| This journal is © The Royal Society of Chemistry 2022 |