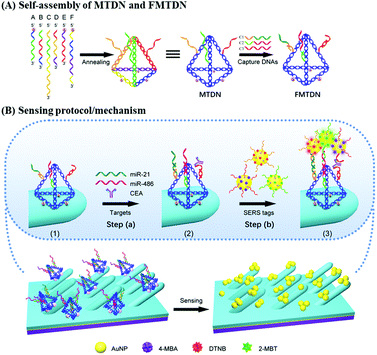
DNA self-assembled Au nanoparticle clusters on silver nanorod arrays for high-sensitive and multiplex detection of cancer-related biomarkers†
Yanjun
Yang
ab,
Chunyuan
Song
 *a,
Jingjing
Zhang
a,
Jie
Chao
a,
Hoang Mai
Luong
c,
Yiping
Zhao
*c and
Lianhui
Wang
*a,
Jingjing
Zhang
a,
Jie
Chao
a,
Hoang Mai
Luong
c,
Yiping
Zhao
*c and
Lianhui
Wang
 *a
*a
aState Key Laboratory for Organic Electronics and Information Displays & Jiangsu Key Laboratory for Biosensors, Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts & Telecommunications, Nanjing 210023, China. E-mail: iamcysong@njupt.edu.cn; iamlhwang@njupt.edu.cn
bSchool of Electrical and Computer Engineering, College of Engineering, University of Georgia, Athens, GA 30602, USA
cDepartment of Physics and Astronomy, University of Georgia, Athens, Georgia 30602, USA. E-mail: zhaoy@uga.edu
First published on 22nd February 2022
Abstract
To sensitively detect multiple and cross-species disease-related targets from a single biological sample in a quick and reliable manner is of high importance in accurately diagnosing and monitoring diseases. Herein, a surface-enhanced Raman scattering (SERS) sensor based on a functionalized multiple-armed tetrahedral DNA nanostructure (FMTDN) immobilized silver nanorod (AgNR) array substrate and Au nanoparticle (AuNP) SERS tags is constructed to achieve both multiplex detection and enhanced sensitivity using a sandwich strategy. The sensor can achieve single, dual, and triple biomarker detections of three lung cancer-related nucleic acid and protein biomarkers, i.e., miRNA-21, miRNA-486 and carcinoembryonic antigen (CEA) in human serum. The enhanced SERS signals in multiplex detections are due to the DNA self-assembled AuNP clusters on the silver nanorod array during the assay, and the experimentally obtained relative enhancement factor ratios, 150 for AuNP dimers and 840 for AuNP trimers, qualitatively agree with the numerically calculated local electric field enhancements. The proposed FMTDN-functionalized AgNR SERS sensor is capable of multiplex and cross-species detection of nucleic acid and protein biomarkers with improved sensitivity, which has great potential for the screening and clinical diagnosis of cancer in the early stage.
Introduction
In practical biomedical applications (e.g., clinical diagnosis), the ability to detect trace multiple disease-related biomarkers from a single biological sample in a quick and reliable manner is of high importance. Many different techniques have been developed to target this goal.1,2 Among them, surface-enhanced Raman spectroscopy (SERS), with the potential to achieve single molecule detection, is very attractive and promising for multiplex detection.3,4 SERS offers a unique “signature” spectral profile with very narrow spectral peaks for an individual analyte, and has been demonstrated to be able to detect trace amounts of biomarkers.5–8 However, in general, SERS faces two critical challenges in improving the sensitivity or lowering the limit of detection (LOD), and achieving multiplex detection. In most studies, these two challenges are targeted separately.To improve the sensitivity, researchers have paid specific attention to developing various methods to construct SERS substrates with high enhancement factors and good reproducibility.9,10 These methods include top-down strategies, such as electron beam lithography,11 optical lithography,12 nanosphere lithography,13 and focused-ion-beam milling;14 as well as bottom-up strategies, such as chemical synthesis,15 and assembly of metallic nano-components.16 For the past 16 years, it has been well-known that aligned silver nanorod (AgNR) arrays, fabricated by the oblique angle deposition method, can act as extremely sensitive, reproducible, and uniform SERS substrates for various analyte detections.17–19 To improve both the biocompatibility and sensitivity, gold or silver nanoparticles (NPs) based SERS tags have been employed. Song et al. reported sensitive sandwich immunoassays using immune-AuNP SERS tags on SERS-active AgNR substrates for human IgG detection, with a LOD of 2.5 fg mL−1.20 It is suggested that using noble metal NPs or NP clusters to decorate AgNRs is one way to improve the sensitivity and lower the LOD, as well as to promote biocompatibility.
Currently there are two strategies to achieve multiplex chip-based SERS sensors for biomarkers such as microRNAs. One is to use an array of SERS sensors functionalized with different capture molecule probes. For example, in our previous work on the detection of multiple lung cancer-related microRNA biomarkers, different molecular beacons were assembled on different locations of an AgNR substrate to construct a sensor array.21 This strategy requires several SERS measurements to be performed on different functionalized regions, and the functionalization process for each probe molecule has to be optimized individually. The other strategy is to immobilize a mixture of multiple probe molecules on one SERS sensor.22 In most biomarker detections, the probe molecules are self-assembled on a SERS substrate. Due to different adsorption and desorption rates, different probe molecules would result in different surface coverages and intermolecular spacings. Such an uncontrollable assembly will affect the target recognition efficiency of the target molecules and reduce the detection sensitivity. Therefore, it remains a major challenge to control the assembly of the capture DNAs on the SERS substrate.
Is there a way to simultaneously deal with the above two challenges, i.e., to achieve both higher sensitivity and multiplex detection of nucleic acids and proteins? Recently, with advances in DNA nanotechnology, self-assembled DNA structures, such as DNA frameworks, DNA origami and DNA tiles, provide a way to concurrently assemble different capture DNAs on the same DNA superstructure with controlled spacing. For example, the tetrahedral DNA nanostructures (TDNs) assembled with several designed DNA strands and probes can intrinsically control the density and orientation of capture DNAs immobilized on the substrate.23,24 Multiple capture DNAs can be easily functionalized on a single TDN for multiplex detection at the same substrate location. Thus, multiplex detection can be achieved with a single SERS measurement, which becomes more efficient for sensing. In addition, TDNs can also be used to assemble AuNP clusters and control the gap between NPs,25 which provides a method to assemble NPs on the AgNR substrate. Therefore, a strategy using multiplex detection and NP self-assembly capabilities of DNA superstructures on AgNR substrates could provide a solution to the above two challenges simultaneously.
Here we present a proof of concept for the aforementioned strategy. A functionalized multiple-armed tetrahedral DNA nanostructure (FMTDN)-modified AgNR array SERS substrate that can assemble AuNP clusters was constructed for the detection of multiple lung cancer-related biomarkers with improved sensitivity. The sensor can achieve single, dual, and triple biomarker detections of three lung cancer-related nucleic acid and protein biomarkers, i.e., miRNA-21, miRNA-486 and carcinoembryonic antigen (CEA) in human serum. The assembly of AuNP clusters on the AgNR array was confirmed by TEM and SEM images. The specificity and enhanced SERS performance during multiplex detection were demonstrated. The SERS enhancement model of this multiplex detection strategy has been established, and the relative SERS enhancement factors qualitatively agreed with the local electric field enhancements calculated by the three-dimensional finite-difference time-domain (FDTD) method.
Experimental section
Materials
4-Mercaptobenzoic acid (4-MBA, 99%), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, 99%) and tris (2-carboxyethyl) phosphine hydrochloride (TCEP, ≥98%) were obtained from Sigma-Aldrich (Shanghai, China). 2-Mercaptobenzothiazole (2-MBT, analytical reagent) and gold nanoparticles (20 nm in diameter) were purchased from Aladdin, British Biocell International (Cardiff, UK), respectively. Carcinoembryonic antigen (CEA) and neuron-specific enolase (NSE) were purchased from Shanghai Lingchao Biological Technology Co., Ltd (Shanghai, China). Human serum and bovine serum albumin (BSA) were obtained from Biosharp (Shenzhen, China). The single-stranded DNAs (ssDNAs), including the 6 nucleotide sequences (A–F) to construct the MTDN, capture strands (C1, C2 and C3), probe strands (P1, P2 and P3), target miRNAs (miR-21 and miR-486), single-base mismatched miRNA (relative to miR-21) and a noncomplementary miRNA (miR-375), as listed in Table S1,† were synthesized and HPLC-purified by Takara Biotechnology (Dalian, China). Unless otherwise specified, all the materials were used without additional purification, and sterilized ultrapure Millipore water (18.2 MΩ cm) was used as the solvent throughout.Several different buffers were prepared and used for the treatment of nucleic acids, including TM buffer (20 mM Tris, 50 mM MgCl2, pH 8.0), TBE buffer (89 mM Tris, 90 mM boric acid, 2 mM EDTA, pH 8.0), TBE-Mg2+ buffer (89 mM Tris, 90 mM boric acid, 2 mM EDTA, 12.5 mM MgCl2), and PBS buffer (10 mM NaH2PO4, 10 mM Na2HPO4, 100 mM NaCl, 2 mM MgCl2, pH 7.4).
Sensing protocol
Fig. 1 shows the general procedures for multiple-armed tetrahedral DNA nanostructure (MTDN) assembly, SERS substrate functionalization, and SERS detection. Six ssDNAs (A–F) are designed to form MTDN structures (Fig. 1A), and the MTDN hybridizes with three capture DNAs (C1, C2, and C3) to form a functionalized multiple-armed tetrahedral DNA nanostructure (FMTDN). The design and self-assembly of FMTDN can be found in section S1 of the ESI.† Then FMTDNs are immobilized onto the AgNR array via the thiol functional groups at the 5′ end of the selected ssDNAs to obtain the SERS-active sensor (state (1) in Fig. 1B). The functionalized AgNR substrate is incubated with the analyte mixtures containing miR-21, miR-486, and CEA, and C1, C2, and C3 on FMTDNs will capture the corresponding biomarkers (step (a) and state (2) in Fig. 1B). Then, equal amounts of three AuNP-based SERS-tags, which also exist probe ssDNAs partially complementary to the captured biomarkers, are mixed and incubated with the substrate (step (b) in Fig. 1B). The three SERS tags are pre-prepared, and each SERS tag is designed to specifically target a particular target biomarker with a different SERS reporter molecule, i.e., the miR-21-SERS tag with the Raman reporter DTNB (main peak wavenumber ΔvDTNB = 1327 cm−1), the miR-486-SERS tag with 4-MBA (ΔvMBA = 1585 cm−1), and the CEA-SERS tag with 2-MBT (ΔvMBT = 1393 cm−1). Detailed preparation and characterization of SERS tags are described in section S3 of the ESI.† After incubation with SERS tags, AuNPs or AuNP clusters (AuNP dimers and trimers) are expected to assemble onto the AgNRs by a FMTDN-mediated target-triggered sandwich assay, and the corresponding characteristic SERS peaks of different reporter molecules (DTNB, 4-MBA, and 2-MBT) can be measured and distinguished. According to the presence of peaks at ΔvDTNB, ΔvMBT, and ΔvMBA and the corresponding peak intensities, multiplex detection can be achieved. It is also expected that the formation of the AuNP clusters can further improve the sensitivity of the detection.FMTDN immobilization
The details of FMTDN formation and AgNR substrate preparation are shown in section S2 of the ESI.† FMTDNs were immobilized onto the AgNR array substrate via S–Ag bonds as illustrated in Fig. 1B. Briefly, 20 μL of 50 nM FMTDN solution was pipetted into each small well (well diameter of 4 mm, depth of 1 mm) patterned on the AgNR array substrate, followed by placing into a thermotank at 25 °C and a humidity of ∼60%. After 3 h, the wells were washed with TM buffer thoroughly, and the FMTDN-modified AgNR array (F-AgNR) SERS sensors were ready for the detection of targets.Preparation of AuNP-based SERS tags
AuNP-based SERS tags were prepared by following our previous reports,26 and the structures of three SERS tags can be seen from the magnified diagrammatic sketch shown in Fig. 1B. The SERS tag 1 specifically binding to miR-21 was prepared by labeling Probe-A-miR-21 and Raman reporter DTNB onto the AuNPs in sequence. Specifically, 500 μL of 2.3 nM colloidal AuNPs (20 nm) was mixed with 50 μL of 10 μM Probe-A-miR-21 in 0.5 × TBE and incubated at 25 °C for more than 4 hours. Then 12.5 μL of 2 M NaCl solution was added slowly into the mixture in 4 times over 2 hours with the final concentration of NaCl reaching 200 mM, followed by aging at 25 °C overnight. The Probe-A-miR-21 functionalized AuNPs were washed with 0.5 × TBE buffer by centrifugation (9000 rpm, 20 min) 3 times and finally re-dispersed in 500 μL. Next, 5 μL of 100 μM DTNB was added into the Probe-A-miR-21 functionalized AuNPs to label the AuNPs with DTNB molecules. After stirring for 3 h, the above mixture was purified by centrifugation three times and the final Probe-A-miR-21 and DTNB labeled AuNPs, i.e., SERS tag 1, were obtained by re-dispersing the precipitates with 50 μL of PBS buffer. The preparation process of SERS tag 2 for miR-486 detection is similar to the protocol of SERS tag 1, by labeling Probe-C-miR-486 and 4-MBA onto the AuNPs in sequence. Specifically, 500 μL of 2.3 nM colloidal AuNPs (20 nm) was mixed with 50 μL of 10 μM Probe-C-miR-486, and 10 μL of 100 μM 4-MBA was added into the Probe-C-miR-486 functionalized AuNPs to label the AuNPs with 4-MBA molecules. The preparation process of SERS tag 3 for CEA detection is similar to the protocol of SERS tag 1, by labeling Probe-E-CEA and 2-MBT onto the AuNPs in sequence. Specifically, 500 μL of 2.3 nM colloidal AuNPs (20 nm) was mixed with 50 μL of 10 μM Probe-E-CEA aptamers, and 7.5 μL of 100 μM 2-MBT was added into the Probe-E-CEA functionalized AuNPs to label the AuNPs with 2-MBT molecules. The details of SERS-tag characterization are shown in section S3 of the ESI.†Biomarker detection
20 μL of the target mixture (100 pM miR-21, 100 pM miR-486 and 100 pM CEA) was added into different F-AgNR wells and incubated in a thermotank (37 °C with a humidity of ∼60%) for 2 h, followed by a thorough PBS wash. Then, 18 μL of mixed SERS tags containing equal-volumes (6 μL) of miR-21-SERS, miR-486-SERS, and CEA-SERS tags were added into each well. After incubation in the thermotank at 37 °C for 3 h, the wells were washed with PBS and water subsequently. After air-drying, 20 SERS measurements at different locations in each well were performed (details see section S2 in the ESI†).FDTD calculations
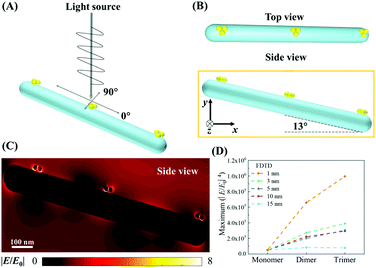
A commercial software package (FDTD Solutions version 8.16.931, Lumerical Solutions Inc.) was used to calculate the localized electric field (E-field) distribution of the assembled AuNP clusters on the AgNR. The schematic of calculations is shown in Fig. 2A, and the entire structure consists of an AgNR and three AuNP clusters (monomer, dimer, and trimer, respectively) placed at the tip, middle, and bottom of the AgNR (Fig. 2B). The entire structure was surrounded by the dielectric environment of vacuum. The sizes and geometric parameters of AuNPs or AgNRs were determined from the corresponding electron microscopy images: the diameter of AuNPs was fixed to be 20 nm while the diameter and length of an AgNR were set to be 100 nm and 1 μm. The gaps between AgNR and AuNP clusters dc were set as 0.5 nm, while the gaps between AuNPs dp were systematically adjusted from 1 nm to 15 nm. Linearly polarized light at a wavelength of 785 nm, with 77° angle between its propagation direction and the long axis of the nanorod, was applied, which is consistent with the SERS measurement configuration. Two polarization directions, one perpendicular (ϕp = 90°) and the other parallel (ϕp = 0°) to the plane of the light propagation direction and the axis direction of the nanorod, were used (Fig. 2A). Perfectly matched layer (PML) absorbing boundaries were used in all directions. To ensure the convergence of the calculations, a mesh size of 1 nm × 1 nm × 1 nm was chosen. The dielectric functions of Ag were taken from the model of Palik,27 while those for Au were taken from Johnson and Christy.28 A monitor of the “frequency-domain field profile” was set up to calculate the localized E-field distributions and the obtained E-fields were normalized to the magnitude of the incident E-fields. The average local E-field of the entire structure, including the AgNR and AuNP clusters (1 μm × 0.35 μm, the orange box in Fig. 2B), was calculated using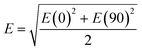 .
.
Results and discussion
Theoretical prediction of SERS enhancement from AuNP clusters.
The mechanism for SERS includes both EM enhancement and chemical enhancement.29 However, the proposed sandwich-based detection shown in Fig. 1, the chemical enhancement can be neglected. Usually, chemical enhancement only occurs when there is a charge transfer between the chemisorbed molecule and the SERS substrate, or there is a severe bond deformation for a chemisorbed molecule.30 This effect is very local. The SERS reporter molecules used in the experiments have already immobilized onto the AuNPs. Therefore, when the AuNPs are attached to the AgNR surface or form clusters, the SERS reporter molecules would remain on the original AuNPs and cannot bond to the AgNR surface or another AuNP. Therefore, it is impossible to induce any chemical enhancement during the AuNP clustering. In addition, one shall note that there are not only EM coupling among AuNPs but also coupling from the nanogaps between the AgNRs and AuNPs. Thus, it is expected that the plasmon resonant condition is not determined by the AuNPs, but by both the AgNRs and AuNPs. In fact, our later results show that there are many AgNRs on the structure, suggesting that AgNRs would dominate the optical response of the AuNP–AgNR system. The typical absorption spectra of the AgNR array substrate indicated a broad adsorption feature from 350 nm to 1200 nm.31 Thus, the calculated field distribution is only the consequence of the experimental excitation wavelength, i.e., 785 nm, instead of the resonance absorption wavelength of the AgNR. In fact, our previous results showed that the 785 nm excitation could produce SERS enhancement as high as 109 for AgNRs.17In order to validate the proposed strategy for improved SERS sensitivity, the local E-field distribution of the assembled AuNP clusters on AgNRs was first evaluated. Fig. 2C shows an example of a typical localized E-field distribution of the assembled AuNP trimers with dp = 1 nm on the AgNR. Multiple hot spots (locations with the highest local E-fields) were formed inside the nanogaps between the AuNP clusters as well as between the AuNP and AgNR. The AuNP cluster at the tip of the AgNR (upper left in Fig. 2C) shows the strongest hot spots, while the cluster at the bottom exhibits the slightly weaker hot spots. The overall SERS enhancement is approximately proportional to |E/E0|4, and Fig. 2D shows a plot of the corresponding maximum |E/E0|4 of AuNP monomers, dimers, and trimers assembled on the AgNR from the 1 μm × 0.35 μm region (Fig. 2B) for different dp values. Two consistent trends are observed: For the same dp, the |E/E0|4 value of the AuNP clusters increases monotonically with the AuNP number in a cluster, i.e., the trimer shows the maximum |E/E0|4; for the same cluster, the |E/E0|4 value decreases monotonically with dp. When dp = 15 nm, the maximum |E/E0|4 only increases 63% from the monomer to dimer, and then 55% to the trimer. When dp = 10 nm, the |E/E0|4 for the trimer is 6 times that of the monomer. When dp = 1 nm, the |E/E0|4 of the dimer and trimer is 13 and 20 times larger than that of the monomer, respectively. Such a small gap among the AuNP cluster is expected. According to the all-atom molecular dynamics simulation by Shen et al., when filled by ssDNAs, the average gap among the assembled AuNP clusters was 0.6 nm.32 The corresponding E-field distributions of free suspended AuNP dimers and trimers are shown in section S4 of the ESI,† and the corresponding local E-field versus dp plot (Fig. S3B†) shows a significantly smaller |E/E0|4 value compared to that of the AuNP clusters on AgNRs. Therefore, based on the FDTD results, the use of FMTDNs to control the assembly of AuNP clusters on the AgNR substrate has the potential to significantly enhance the intensity of the SERS signal.
Characterization of FMTDN formation
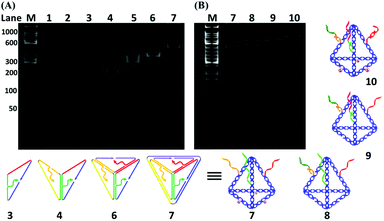
As shown in Fig. 1A, six specially designed ssDNAs (A–F in Table S1†) are self-assembled to form the MTDN. The detailed design description can be found in section S1 in the ESI.† The 5′-ends of A, E and F ssDNAs (Fig. 1A) are modified with thiol groups, while three overhangs are left on three other arms to link capture strands, i.e., C1, C2, and C3, which are complementary to the targets, i.e., miR-21, miR-486, and CEA, respectively. Capture-miR-21 (C1) is divided into two parts, the underlined sequences (21-nt, green color in Table S1†) at the 3′-end is complementary to the first overhang (green color) of the MTDNs on sequence A; the sequence (11-nt) at the 5′-end is specially designed to capture target miR-21. A similar strategy is used to design Capture-miR-486 (C2). For the Capture-CEA (C3), the sequence (18-nt) at the 5′-end is an aptamer, which is specially designed to capture the target CEA protein.33,34 When the MTDNs hybridize with the three specially designed capture DNAs, the functionalized MTDNs (FMTDNs) are formed. The theoretical height of the MTDNs is 8 nm with equal edge lengths of 10 nm. There is a nick in the middle of each edge where the 5′ and 3′-ends of the ssDNA meet. The three overhangs (21-nt) at nick on the edges are partially complementary to the three capture strands. Native polyacrylamide gel electrophoresis (PAGE) analysis (Fig. 3A) demonstrates the stepwise assembly of MTDNs as each strand was added. A distinct band shift was observed from lanes 2 to 7, indicating high-yield MTDN formation (lane 7). The hybridizations of three capture strands (C1, C2 and C3) were also observed from lanes 8 to 10 in the PAGE analysis in Fig. 3B. As the number of capture strands increased, a distinct band shift was observed due to the mobility change. Different overhang designs for site-specific capture strand hybridization allows precise control of the number and orientation of capture strands, and their specificity to different biomarkers are detailed in section S5 of the ESI.†AuNP cluster formation via FMTDN
The formation of AuNP clusters via FMTDNs was demonstrated in solution using three different SERS tags. To assemble AuNP dimers, 10 nM FMTDNs were mixed with 10 nM miR-21 and 10 nM miR-486, and incubated together for 0.5 h at room temperature, followed by adding a mixture of equal amounts of the miR-21-SERS tag and miR-486-SERS tag. The molar ratio of the SERS tag and FMTDNs was 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
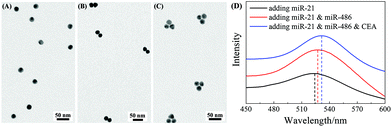
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Fig. 4A and B show the representative TEM images of AuNP monomers and AuNP dimers formed via FMTDN assembly. The yield of the AuNP dimers was estimated to be 72%, and the average dp in the dimers was determined to be 0.5 ± 0.2 nm. The AuNP trimers were assembled using a similar strategy, 10 nM miR-21, 10 nM miR-486, and 10 nM CEA, along with three SERS tags, mixed and incubated. Fig. 4C shows a typical TEM image of AuNP dimers. The yield of AuNP trimers was estimated to be about 30%, and the average dp in trimers was measured to be 1.5 ± 0.3 nm. These measured dp values are consistent with those in the simulation by Shen et al.32 In addition, UV-Vis absorption spectroscopy was used to further confirm the formation of AuNP dimers and trimers. As shown in Fig. 4D, for the AuNP monomer suspension, the localized surface plasmon resonance (LSPR) peak was centered at λp = 523 nm. When dimers were formed, the λp red-shifted to 527 nm (Fig. 4D). With the formation of AuNP trimers, the λp further red-shifted to 531.5 nm. The continuous redshift in λp further confirms the formation of dimer and trimer clusters in suspensions.
1. Fig. 4A and B show the representative TEM images of AuNP monomers and AuNP dimers formed via FMTDN assembly. The yield of the AuNP dimers was estimated to be 72%, and the average dp in the dimers was determined to be 0.5 ± 0.2 nm. The AuNP trimers were assembled using a similar strategy, 10 nM miR-21, 10 nM miR-486, and 10 nM CEA, along with three SERS tags, mixed and incubated. Fig. 4C shows a typical TEM image of AuNP dimers. The yield of AuNP trimers was estimated to be about 30%, and the average dp in trimers was measured to be 1.5 ± 0.3 nm. These measured dp values are consistent with those in the simulation by Shen et al.32 In addition, UV-Vis absorption spectroscopy was used to further confirm the formation of AuNP dimers and trimers. As shown in Fig. 4D, for the AuNP monomer suspension, the localized surface plasmon resonance (LSPR) peak was centered at λp = 523 nm. When dimers were formed, the λp red-shifted to 527 nm (Fig. 4D). With the formation of AuNP trimers, the λp further red-shifted to 531.5 nm. The continuous redshift in λp further confirms the formation of dimer and trimer clusters in suspensions.
Formation of AuNP dimers and trimers on the AgNR array
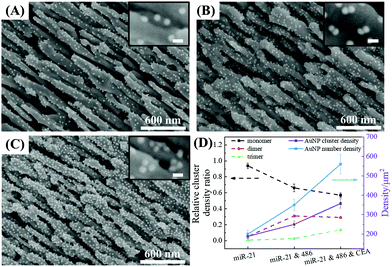
The optimal conditions required to assemble FMTDNs onto the AgNR array for SERS measurements are detailed in section S6 in the ESI† following the detection strategy shown in Fig. 1B. Under these optimal conditions, the formation of different AuNP clusters on AgNRs was investigated by SEM. When only adding miR-21 (100 pM) followed by miR-21-SERS tags (2.3 nM), as shown in Fig. 5A, almost all the AuNPs were randomly distributed and scattered all over the surface of AgNRs, and only a few formed dimers and trimers. The AuNP density is nSAuNP = 200 ± 20 μm−2 and the apparent surface densities of the monomers, dimers, and trimers are nSm = 178 ± 7 μm−2, nSd = 10 ± 3 μm−2, and nSt = 2 ± 1 μm−2, respectively. When two or three kinds of target molecules and the corresponding SERS tags were added, as shown in Fig. 5B and C, the total number of AuNPs increases, and the AuNP dimers or trimers are consistently observed, especially in the zoomed-in images. Based on the three dimers shown in the zoomed-in image of Fig. 5B, the average dp is determined to be 0.6 ± 0.3 nm, which is consistent with the TEM result. The densities of AuNPs, AuNP monomers, dimers, and trimers on the AgNR are estimated to be nDAuNP = 350 ± 30 μm−2, nDm = 170 ± 13 μm−2, nDd = 78 ± 3 μm−2, and nDt = 7 ± 3 μm−2, respectively (Fig. 5B). From the two trimers shown in the zoomed-in image in Fig. 5C, the average dp is 2 ± 1 nm, and the densities of AuNPs, AuNP monomers, dimers and trimers are nTAuNP = 560 ± 50 μm−2, nTm = 200 ± 11 μm−2, nTd = 104 ± 5 μm−2, and nTt = 50 ± 9 μm−2, respectively (Fig. 5C). Comparing Fig. 5A–C, when adding more biomarkers, the total number of AuNPs appearing on the AgNR surface becomes larger, i.e., nDAuNP ∼ 2nSAuNP and nTAuNP ∼ 3nSAuNP. This result indicates that the hybridization probability of these three SERS tags is very similar regardless of the immobilization amounts of different kinds of AuNPs. In fact, Fig. 5D (solid curves) shows the plot of both the total AuNP density and the total AuNP cluster density n0 = nm + nd + nt for three assembly conditions, and a monotonic increase is revealed. Fig. 5D (dashed curves) also shows a plot of the relative cluster density ratio,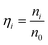 , where i = m, d, and t, for three assembly conditions. From single biomarker assays (only adding miR-21) to triple biomarker assays (adding miR-21, 486, and CEA), the monomer ratio ηm decreases dramatically (black dashed curve), but both ηd and ηt increase (red and green dashed lines), which demonstrates that (1) the FMTDNs on the AgNR surface facilitate the assembly of AuNP clusters and (2) the AuNP clusters can play a more important role in multiplex detection by SERS.
, where i = m, d, and t, for three assembly conditions. From single biomarker assays (only adding miR-21) to triple biomarker assays (adding miR-21, 486, and CEA), the monomer ratio ηm decreases dramatically (black dashed curve), but both ηd and ηt increase (red and green dashed lines), which demonstrates that (1) the FMTDNs on the AgNR surface facilitate the assembly of AuNP clusters and (2) the AuNP clusters can play a more important role in multiplex detection by SERS.
Multiplex SERS sensing performance
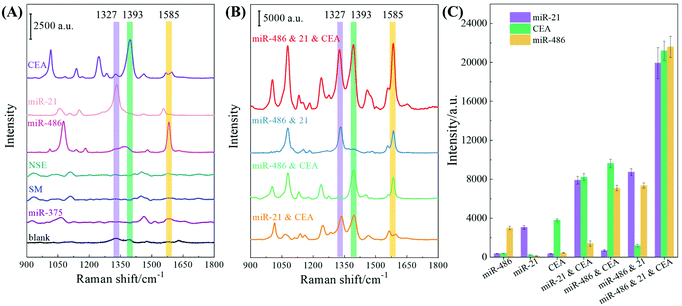
The sensing performance of the F-AgNR SERS sensor was evaluated for the specificity and multiplex detection of the three biomarkers in 80% human serum. The performance of single biomarker detection is shown in Fig. 6A. For this assay, only a single biomarker and a mixture of three AuNP-based SERS tags were added. As a reference, the 80% human serum without spiking the biomarkers was used as the control. The nonspecific assays were carried out using miR-375 (1 nM), a single-base mismatched strand relative to miR-21 (SM) (1 nM), as well as NSE protein (1 nM) in human serum. Fig. 6A shows the corresponding SERS spectra and the purple, green, and orange stripes indicate the characteristic peaks at ΔvDTNB, ΔvMBA, and ΔvMBT. The corresponding SERS peak intensities at ΔvDTNB, ΔvMBA, and ΔvMBT are shown in Fig. 6C. For the blank control spectrum (black curve), a very weak SERS signal is observed at 1324 cm−1 and 1391 cm−1. The peak at Δv = 1324 cm−1 can be assigned to ring-stretching from Adenosine,35,36 while the peak at Δv = 1391 cm−1 may result from unknown contamination from the environment (Fig. S10†). Similarly, weak SERS spectra were recorded in the nonspecific target groups of miR-375, SM, and NSE, suggesting that different non-specific biomolecules (i.e., other miRNAs, proteins, etc.) could not provide interference sensing signals. The variation in the detailed spectral features for these four cases may be due to the nonspecific binding of biomolecules on the F-AgNR SERS sensor, while the probability of nonspecific binding of SERS tags is very small as revealed in the SERS spectra. However, when the F-AgNR SERS sensor was treated with miR-21, miR-486, and CEA, respectively, distinguished SERS peaks at ΔvDTNB, ΔvMBT and ΔvMBA, were observed, as shown in Fig. 6A. After treatment with 100 pM miR-21, the peak at ΔvDTNB = 1327 cm−1 appears in Fig. 6A. Similarly, after treatment with 100 pM miR-486 or 100 pM CEA, distinctive peaks at ΔvMBA or ΔvMBT appear, which indicates the successful recognition of the specific biomarkers and capture corresponding SERS tags, as designed. The intensities plotted in Fig. 6C demonstrate the same trend. Thus, the results in Fig. 6A demonstrate that the F-AgNR SERS sensor is only specific to biomarkers of miR-21, miR-486, and CEA.Dual and triple biomarker assays were carried out and the results are plotted in Fig. 6B. In dual detections, three dual biomarker combinations were used: 100 pM miR-21 and 100 pM CEA, 100 pM miR-486 and 100 pM CEA, as well as 100 pM miR-21 and 100 pM miR-486. According to the design, one expects to observe two distinguished SERS peaks from ΔvDTNB, ΔvMBT and ΔvMBA. In fact, this is true as revealed in Fig. 6B: when the miR-21 and CEA mixture was tested, two SERS tag peaks at ΔvDTNB and ΔvMBT were observed; when the miR-486 and CEA mixture was assayed, two peaks at ΔvMBA and ΔvMBT appear; and for the test of the miR-21 and miR-486 mixture, two peaks at ΔvDTNB and ΔvMBA emerge. The peak intensities of these three peaks under the three different conditions plotted in Fig. 6C confirm the observation. However, compared to the single biomarker detection, the peak intensities at the corresponding SERS tag wavenumbers for dual biomarker detection are roughly almost doubled. The triple biomarker detection was performed using a mixture of 100 pM miR-21, 100 pM miR-486 and 100 pM CEA, and the corresponding SERS spectrum is shown in Fig. 6B (red curve). As expected, all three distinguished SERS peaks at ΔvDTNB, ΔvMBT and ΔvMBA are observed. The corresponding SERS intensity plotted at ΔvDTNB, ΔvMBT and ΔvMBA in Fig. 6C shows that the corresponding intensity in triple biomarker detection is nearly 5–7 times that of the single biomarker detection.
The above results can be briefly understood as follows: the functionalized AuNP is acting as a SERS tag. When the SERS tag is hybridized on the AgNR substrate, the SERS signal of the Raman report molecules will be enhanced due to the enhancement of the AgNR. In addition, during the multiplex detection, the AuNP clusters are formed, and an additional SERS enhancement is achieved as demonstrated in Fig. 6. The SERS responses of the three as-prepared SERS tags were characterized by dispensing 20 μL of the SERS tag suspension into PMDS wells on a silicon substrate respectively and after air-drying SERS measurements were performed and the corresponding Raman spectra are shown in Fig. S2 of the ESI† (785 nm wavelength, 177.2 mW laser power, 5 s acquisition time). The surface AuNP density was estimated to be ∼2.2 × 104 particle per μm2. Then based on Fig. 5A, the AuNP density on the AgNR was estimated to be ∼189 particle per μm2. The SERS measurements were performed with 785 nm wavelength, 9 mW laser power, and 2 s acquisition time. Thus, the SERS signal per AuNP particle on the AgNR was 1164 times as large as that on the Si substrate, indicating a strong enhancement of the AuNP tag by the AgNR substrate.
In addition, other performance parameters of the sensor, such as uniformity and reproducibility, have been characterized as shown in section S7 of the ESI.† The results show that the F-AgNR SERS sensors are highly uniform, the SERS signal variations for all three SERS tags are less than 9%, and 6 batches of AgNR substrates only produce <8.3% variation in different detection signals, i.e., the sensors are highly reproducible. Therefore, based on the above results, the following four conclusions can be drawn: (1) The designed F-AgNR SERS sensor is only specific to biomarkers of miR-21, miR-486, and CEA; (2) The sensor can achieve single, dual and triple biomarker detections; (3) With the same concentration of the biomarker, the SERS intensity of the SERS tag peak with respect to the corresponding biomarker increases significantly when the detection changes from single to dual, and then to triple biomarker detection; and (4) The sensors are very uniform and highly reproducible.
However, due to the additional clustering induced enhancement in multiplex detection, quantitative detection based on this strategy is quite challenging. For example, even the miR-486 concentration will be fixed, and its respective SERS tags produce a certain amount of SERS signal on the AgNR substrate, but when the FMTDN further hybridized with the SERS tags for CEA, or together with SERS tags for CEA and miR-21, the SERS signal for miR-486 would be more than doubled or even as high as 7 times according to Fig. 6. However, the total amount of the SERS tags of miR-486 on the AgNR substrate was not changed. Thus, the quantitation is impossible by just examining the intensity of the characteristic SERS peak. However, machine learning (or deep learning) could be an excellent solution for this, if the spectra of a suitable number of a selective mixture of biomarkers with different mixing ratios and concentrations could be used as a modeling spectral set to adapt an appropriate algorithm.37–39
Understanding the SERS intensity changes due to multiplex detection or AuNP clustering
In the above conclusions, the third point needs to be further understood. Table 1 shows a summary of the intensity ratios for the single biomarker detection at the corresponding ΔvDTNB, ΔvMBT and ΔvMBA values for the single, dual, and triple biomarker detection configurations. For the dual biomarker detection, the corresponding SERS peak intensity is 2.30–2.70 times that of single biomarker detection while this ratio jumps to 5.58–7.22 for triple biomarker detection. Such a significant increase in SERS intensities can only come from two possible sources: the number of SERS tags corresponding to a specific biomarker is increased or the SERS enhancement factor increases significantly. According to Fig. 5D, the AuNP densities nAuNP for single (nSAuNP), dual (nDAuNP), and triple (nTAuNP) biomarker detection satisfy the following relationship, i.e., nDAuNP ≈ 2nSAuNP and nTAuNP ≈ 3nSAuNP. Since each SERS tag only carries one kind of Raman report molecule, it is concluded that when the detection configuration changes from single to dual biomarker detection, the extra AuNPs on the F-AgNR SERS sensor are all coming from different AuNP-based SERS tags. A similar conclusion can be drawn for triple biomarker detection. That is, if the three detection configurations have one of the same biomarkers, then the surface densities of the AuNP-based SERS tag for that biomarker under these three conditions are the same, i.e., the amount of Raman reporter molecules is roughly the same for all three configurations. Therefore, the enhanced intensity shown in Table 1 can only come from the SERS enhancement factor change.| Single detection | Dual detection | Triple detection | |||
|---|---|---|---|---|---|
| Average | |||||
| miR-21 (ΔvDTNB) | 1 | 2.58 | 2.85 | 2.72 | 6.51 |
| miR-486 (ΔvMBA) | 1 | 2.37 | 2.46 | 2.40 | 7.22 |
| CEA (ΔvMBT) | 1 | 2.12 | 2.53 | 2.30 | 5.58 |
For a fixed biomarker in single, dual, and triple biomarker detection configurations, the corresponding SERS peak intensity can be written as,
| IS ∝ EmnSd + 2EdnSd + 3EtnSt, | (1) |
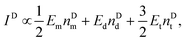 | (2) |
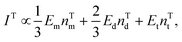 | (3) |
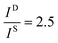 and
and 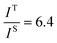 , and taking the data for the cluster density from Fig. 5D, we obtain that
, and taking the data for the cluster density from Fig. 5D, we obtain that 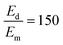 and
and 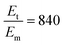 . This result shows that the AuNP cluster on the AgNR substrate not only provides a way to achieve multiplex detection, but also significantly enhances the SERS signal. In fact, recalling the FDTD results from Fig. 2D, for dp = 1 nm, the |E/E0|4 values of the dimer and trimer are 13 and 20 times that of the monomer, respectively, which is qualitatively consistent with the experimental result. The discrepancy in the magnitude may result from the effect of AgNR arrays on the AuNP clusters.
. This result shows that the AuNP cluster on the AgNR substrate not only provides a way to achieve multiplex detection, but also significantly enhances the SERS signal. In fact, recalling the FDTD results from Fig. 2D, for dp = 1 nm, the |E/E0|4 values of the dimer and trimer are 13 and 20 times that of the monomer, respectively, which is qualitatively consistent with the experimental result. The discrepancy in the magnitude may result from the effect of AgNR arrays on the AuNP clusters.
Conclusions
In summary, a FMTDN-modified AgNR array substrate is demonstrated for the multiplex detection of three lung cancer-related biomarkers, i.e., miR-21, miR-486 and CEA protein, with enhanced SERS signals. The SERS signals of a particular SERS tag are not only enhanced due to AgNRs, but also the hybridization induced clustering of AuNPs gives an additional enhancement. The sensor is highly specific to the three biomarkers. The high sensitivity and good specificity for the FMTDN-based SERS sensor are attributed to the following advantages: (1) The FMTDNs provide a template to align the probe molecules upright on the AgNR surfaces; the MTDNs with high rigidity allow themselves to immobilize on the Ag surface with an upright orientation. This could avoid possible steric crowding and electrostatic interactions often encountered by soft ssDNA probes,40 thus resulting in high hybridization efficiency of target molecules. (2) The assembled FMTDNs on the surface provide an appropriate space among adjacent probe molecules for high efficacy hybridization: the FMTDN, with a triangular pyramid structure plus a six-carbon spacer, theoretically forms a biomolecule layer (∼8 nm thick) on the AgNRs.41 Such a structure can provide a liquid-phase-like environment to enhance the hybridization efficiency with target molecules, resulting in high capture efficiency of target molecules. (3) The self-assembled AuNP clusters on the FMTDNs can provide extra hot spots on the AgNR surface for SERS enhancement: the spatial arrangement of AuNPs based on FMTDNs achieved a small interparticle distance, <2 nm, in the presence of biomarkers, facilitating an effective plasmonic coupling and, in turn, generated a high density of hot spots for SERS enhancement. (4) Large surface area of the AgNRs: the AgNR array substrates with large surface areas and rough surface topographies provide more space for assembling more FMTDNs so as to enhance the ability to capture SERS tags. With these advantages, the proposed DNA nanostructure functionalized AgNR SERS sensor shows great potential for screening and clinical diagnosis of cancer in the early stage.Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Chunyuan Song, Jingjing Zhang, Jie Chao, and Lianhui Wang were supported by the National Key Research and Development Program of China (2017YFA0205300), the National Natural Science Foundation of China (61871236) and the Qinglan Project of Jiangsu Province of China. Yanjun Yang, Hoang Mai Luong, and Yiping Zhao were partially supported by a National Science Foundation grant under Grant No. ECCS-1808271. We would like to thank Mr. Yoong Sheng Phang and Mr. James Haverstick for proofreading this manuscript.References
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161–171 CrossRef CAS PubMed
.
- Y. Shen, Y. Zhang, Z. F. Gao, Y. Ye, Q. Wu, H.-Y. Chen and J.-J. Xu, Nano Today, 2021, 38, 101121 CrossRef CAS
.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667 CrossRef CAS
.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed
.
- M. D. Porter, R. J. Lipert, L. M. Siperko, G. Wang and R. Narayanan, Chem. Soc. Rev., 2008, 37, 1001–1011 RSC
.
- S. P. Mulvaney, M. D. Musick, C. D. Keating and M. J. Natan, Langmuir, 2003, 19, 4784–4790 CrossRef CAS
.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, Nanomedicine, 2010, 6, 214–226 CrossRef CAS PubMed
.
- Y. Pang, C. Wang, J. Wang, Z. Sun, R. Xiao and S. Wang, Biosens. Bioelectron., 2016, 79, 574–580 CrossRef CAS PubMed
.
- J. Fang, S. Du, S. Lebedkin, Z. Li, R. Kruk, M. Kappes and H. Hahn, Nano Lett., 2010, 10, 5006–5013 CrossRef CAS PubMed
.
- K. Zhang, J. Zhao, H. Xu, Y. Li, J. Ji and B. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 16767–16774 CrossRef CAS PubMed
.
- N. A. Abu Hatab, J. M. Oran and M. J. Sepaniak, ACS Nano, 2008, 2, 377–385 CrossRef CAS PubMed
.
- T. Y. Jeon, S.-G. Park, D.-H. Kim and S.-H. Kim, Adv. Funct. Mater., 2015, 25, 4681–4688 CrossRef CAS
.
- X. Zhao, J. Wen, M. Zhang, D. Wang, Y. Wang, L. Chen, Y. Zhang, J. Yang and Y. Du, ACS Appl. Mater. Interfaces, 2017, 9, 7710–7716 CrossRef CAS PubMed
.
- T. Gao, Z. Xu, F. Fang, W. Gao, Q. Zhang and X. Xu, Nanoscale Res. Lett., 2012, 7, 399 CrossRef PubMed
.
- C. Song, N. Zhou, B. Yang, Y. Yang and L. Wang, Nanoscale, 2015, 7, 17004–17011 RSC
.
- J. Sun, R. Liu, J. Tang, Z. Zhang, X. Zhou and J. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 16730–16737 CrossRef CAS PubMed
.
- J. D. Driskell, S. Shanmukh, Y. Liu, S. B. Chaney, X. J. Tang, Y. P. Zhao and R. A. Dluhy, J. Phys. Chem. C, 2008, 112, 895–901 CrossRef CAS
.
- Y. J. Liu and Y. P. Zhao, Phys. Rev. B: Condens. Matter Mater. Phys., 2008, 78, 075436 CrossRef
.
- Y.-J. Liu, Z.-Y. Zhang, Q. Zhao, R. Dluhy and Y.-P. Zhao, J. Phys. Chem. C, 2009, 113, 9664–9669 CrossRef CAS
.
- C. Song, J. Chen, Y. Zhao and L. Wang, J. Mater. Chem. B, 2014, 2, 7488–7494 RSC
.
- C. Song, Y. Yang, B. Yang, Y. Sun, Y. P. Zhao and L.-H. Wang, Nanoscale, 2016, 8, 17365–17373 RSC
.
- R. Guo, F. Yin, Y. Sun, L. Mi, L. Shi, Z. Tian and T. Li, ACS Appl. Mater. Interfaces, 2018, 10, 25770–25778 CrossRef CAS PubMed
.
- H. Pei, X. Zuo, D. Zhu, Q. Huang and C. Fan, Acc. Chem. Res., 2014, 47, 550–559 CrossRef CAS PubMed
.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi and X. Zuo, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed
.
- L. Xu, W. Yan, W. Ma, H. Kuang, X. Wu, L. Liu, Y. Zhao, L. Wang and C. Xu, Adv. Mater., 2015, 27, 1706–1711 CrossRef CAS PubMed
.
- B. Liu, C. Song, D. Zhu, X. Wang, M. Zhao, Y. Yang, Y. Zhang, S. Su, J. Shi, J. Chao, H. Liu, Y. Zhao, C. Fan and L. Wang, Small, 2017, 13, 1603991 CrossRef PubMed
.
-
W. Choyke and E. D. Palik, Handbook of optical constants of solids, Academic Press, Inc, 1985, vol. 587 Search PubMed
.
- P. B. Johnson and R.-W. Christy, Phys. Rev. B: Solid State, 1972, 6, 4370 CrossRef CAS
.
- X. Wang, S.-C. Huang, S. Hu, S. Yan and B. Ren, Nat. Rev. Phys., 2020, 2, 253–271 CrossRef
.
- T.-H. Park and M. Galperin, Phys. Rev. B: Condens. Matter Mater. Phys., 2011, 84, 075447 CrossRef
.
- Y. P. Zhao, S. B. Chaney and Z. Y. Zhang, J. Appl. Phys., 2006, 100, 063527 CrossRef
.
- J. Shen, B. Luan, H. Pei, Z. Yang, X. Zuo, G. Liu, J. Shi, L. Wang, R. Zhou, W. Cheng and C. Fan, Adv. Mater., 2017, 29, 1606796 CrossRef PubMed
.
- G. R. H. Tabar and C. L. Smith, World Appl. Sci. J., 2010, 8, 16–21 Search PubMed
.
-
C. L. Smith, Patent Application Publication, US 2010/0254901 A1, 2010 Search PubMed
.
- L. Guerrini, Ž. Krpetić, D. van Lierop, R. A. Alvarez-Puebla and D. Graham, Angew. Chem., 2015, 127, 1160–1164 CrossRef
.
- J. L. Abell, J. M. Garren, J. D. Driskell, R. A. Tripp and Y. Zhao, J. Am. Chem. Soc., 2012, 134, 12889–12892 CrossRef CAS PubMed
.
- H. He, S. Yan, D. Lyu, M. Xu, R. Ye, P. Zheng, X. Lu, L. Wang and B. Ren, Anal. Chem., 2021, 93, 3653–3665 CrossRef CAS PubMed
.
- N. M. Ralbovsky and I. K. Lednev, Chem. Soc. Rev., 2020, 49, 7428–7453 RSC
.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS
.
- M. H. Lin, J. J. Wang, G. B. Zhou, J. B. Wang, N. Wu, J. X. Lu, J. M. Gao, X. Q. Chen, J. Y. Shi, X. L. Zuo and C. H. Fan, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed
.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu, H. Yan and C. Fan, Adv. Mater., 2010, 22, 4754–4758 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2nr00133k |
| This journal is © The Royal Society of Chemistry 2022 |