Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
TCF-based fluorescent probe for monitoring superoxide anion produced in bacteria under chloramphenicol- and heat-induced stress†
Kai-Cheng
Yan
 ac,
Bethany L.
Patenall
ac,
Bethany L.
Patenall
 a,
Jordan E.
Gardiner
a,
Jordan E.
Gardiner
 a,
Rachel A.
Heylen
a,
Rachel A.
Heylen
 a,
Naing
Thet
a,
Naing
Thet
 a,
Xiao-Peng
He
a,
Xiao-Peng
He
 c,
Adam C.
Sedgwick
c,
Adam C.
Sedgwick
 *b,
Tony D.
James
*b,
Tony D.
James
 *ad and
A. Toby A.
Jenkins
*ad and
A. Toby A.
Jenkins
 *a
*a
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk; a.t.a.jenkins@bath.ac.uk
bChemistry Research Laboratory, University of Oxford, Mansfield Road, OX1 3TA, UK
cKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai, 200237, China
dSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, 453007, China
First published on 7th November 2022
Abstract
We report on a superoxide anion (O2˙−) responsive fluorescent probe called TCF-OTf. TCF-OTf is able to monitor O2˙− production when the bacterial species Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, and Enterococcus faecalis are exposed to chloramphenicol and heat shock at 50 and 58 °C.
Reactive oxygen species (ROS) are products of intracellular mitochondrial respiration and electron transport.1,2 High levels of intracellular ROS can result in intracellular oxidative stress, generate cell aberrations, and thus result in cell dysfunction.3,4 Prokaryotes have simple and fragile cytoplasmic structures, and as such ROS are toxic to bacteria, where even a low concentration can lead to the oxidation of biomolecules and the denaturation of proteins.5 As a type of highly-reactive ROS, the formation and fate of O2˙− in eukaryotic cells is associated with diverse physiological and pathological processes, including innate immunity and metabolic homeostasis.6 However, its role in prokaryotic microorganisms has been much less investigated. Albesa et al. initially found that O2˙− is produced by bacteria under antibiotic treatment, especially chloramphenicol,7 yet this area of research has remained relatively unexplored. Furthermore, the production of unspecified ROS by Escherichia coli under heat stress has also been reported.8 However, the fluorescence-based detection of both ROS-related and heat-related oxidative stress remain underexplored.9,10
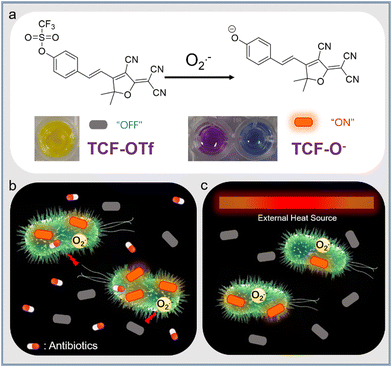
Herein, we report on a fluorescent probe, TCF-OTf, for monitoring O2˙− generated during antibiotic treatment and from heat-stressed bacteria. TCF-OTf was readily prepared, and cleavage of the trifluoromethanesulfonate group by O2˙− resulted in the formation of the fluorescent anion TCF-O−, which was confirmed via mass spectroscopic (MS) analysis (Fig. S1 and S2, ESI†). Reaction with O2˙− resulted in a fluorescence “turn-on” and colorimetric response from yellow to purple (aqueous phase) or blue (organic phase) (Scheme 1a). We then evaluated the probe using Gram-positive Staphylococcus aureus (NCTC 10788) and Enterococcus faecalis (NCTC 29212), and Gram-negative Pseudomonas aeruginosa (PAO1) and Escherichia coli (BW 25113), which are of significant clinical concern and exhibit high infection rates.11,12 Significantly, we detected O2˙− stress under both antibiotic and heat treatment (Scheme 1b and c, respectively). Compared with available commercially available sensors, our probe contains a near-infrared (NIR) fluorophore to enable minimal biological damage and ensure biocompatibility, effective penetration through tissues, excellent stability of the measurement signals, and minimal interference from background fluorescence, resulting in improved clinical expectations. Moreover, our probe, unlike current commercial systems, enables the real-time and in situ mapping of O2˙− production within a range of complex biological environments.
From our previous research we determined that the 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF-OH) fluorophore was an attractive platform for the study of enzymes and ROS in cancer cell-based applications.13–16 Here, we incorporated a trifluoromethanesulfonate ester, as a known selectively cleaved O2˙− responsive group, to study the specific ROS (O2˙−) production from a range of bacterial species that has not previously been reported. The TCF core was prepared using 3-hydroxy-3-methyl-2-butanone, malononitrile and NaOEt in EtOH. Subsequently, TCF, and 4-hydroxybenzaldehyde in EtOH with piperidine (cat.) afforded TCF-OH in 75% yield. Trifluoromethanesulfonate anhydride was then linked to TCF-OH (see ESI†). TCF-OTf was then evaluated during the addition of 0–50 equiv. of O2˙− in pH = 7.40 PBS buffer (10% DMSO). The fluorescence of TCF-OTf gradually increased at 606 nm, suggesting the formation of TCF-OH, while no response was observed in a pure organic phase (Fig. S3, ESI†). The UV-vis absorption changes for TCF-OTf upon the addition of O2˙− resulted in a colour change (Fig. S4, ESI†), which was different in the two phases. Based on the fluorescence observation, purple is believed to be the colour of the TCF-O− aggregates, while the dispersed form was blue (Fig. S3 and S4, ESI†). We then determined the sensitivity and response time, resulting in an acceptable sensing performance and full response within 30 min (Fig. S5, ESI†).
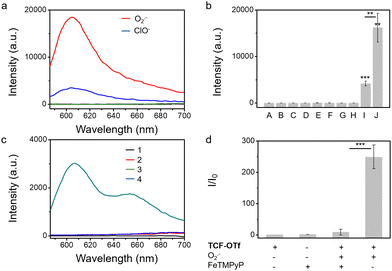
We also evaluated the effect of pH on the probes. We observed that TCF-OTf is only fluorescent at pH values above 7 (neutral to alkaline conditions) (Fig. S6, ESI†), which may indicate that the fluorescence originates from TCF-O− rather than TCF-OH. Fortunately, the conditions for bacterial growth are neutral or weakly alkaline, and under those conditions the probe functions well. As mentioned above, chemo-selectivity is important for the study of ROS in biological systems. Therefore, we challenged TCF-OTf with other ROS and other common species found in bacteria, such as glutathione,17 and Fe(III).18TCF-OTf was found to be selective for O2˙− (Fig. 1a and b). However, TCF-OTf also responds somewhat to hypochlorite (ClO−), but the ClO− concentration used was 10-fold greater than O2˙−, and ClO− is unlikely to be found in bacteria under normal conditions.19 Using a common O2˙− decomposition catalyst FeTMPyP,20 we confirmed that the O2˙− signal can be specifically quenched, which makes it an important tool for us to differentiate O2˙− signals from ClO− (Fig. 1c and d).
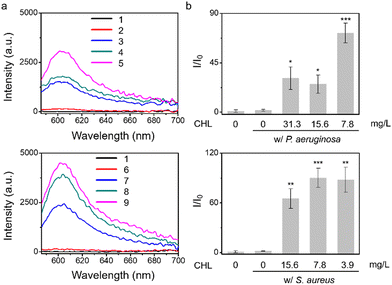
Given that treatment of S. aureus with chloramphenicol was known to increase O2˙− production,7 we evaluated chloramphenicol as a model antibiotic with S. aureus, P. aeruginosa, E. coli, and E. faecalis. To determine the efficiency of O2˙− production, we used standard minimum inhibitory concentration (MIC) assays, which determine the minimum concentration of a compound required to completely inhibit bacterial growth (Fig. S7 and S8, ESI†), then the sub-MIC wells (with antibiotic concentrations just below the full-inhibitory concentration) were selected and incubated with TCF-OTf (Fig. 2 and Fig. S9, ESI†). It was found that S. aureus produced the most O2˙− (further confirmed using an FeTMPyP inhibition assay (Fig. S10, ESI†) that removes the possibility of interference from ClO−). Clear fluorescence signal changes from the baseline were also detected when using P. aeruginosa. However, for E. coli and E. faecalis, no O2˙− was detected, even though clear therapeutic effects were seen via the MIC.
After assessing O2˙− in relation to chloramphenicol treatment, we screened an additional four antibiotics, including streptomycin, amikacin, tetracycline, and ciprofloxacin. Initially, we mixed TCF-OTf with the antibiotics in solution to evaluate the stability of the probes (Fig. S11, ESI†). MIC assays were performed for the four species (Fig. S12–S15, ESI†), where the sub-MIC wells with each antibiotic towards each bacterium with TCF-OTf were tested (Fig. S16, ESI†). Whilst modest levels of O2˙− were reported for ciprofloxacin-treated E. coli and E. faecalis,7 no signal was observed for ciprofloxacin or tetracycline towards all four species P. aeruginosa, S. aureus, E. coli, and E. faecalis. The absence of signals with ciprofloxacin may be because ciprofloxacin is a strong antibiotic and the MICs of ciprofloxacin (drug concentrations) towards the four species are very low (generally below 1 mg L−1). However, amikacin and streptomycin with P. aeruginosa and S. aureus exhibit some fluorescence signal, although it was still insufficient to observe the characteristic peak of TCF-OH. By contrast, E. coli and E. faecalis exhibited no fluorescence. Similarly, the screening was also confirmed using an FeTMPyP assay (Fig. S17, ESI†) which ensures that O2˙− was quenched. From these results, chloramphenicol is the most significant O2˙−-inducing antibiotic for P. aeruginosa and S. aureus.
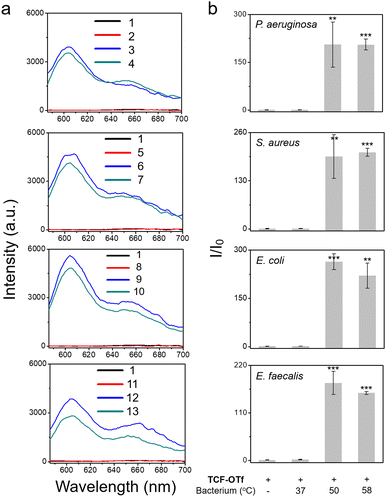
After determining that the probe was stable at higher temperatures (Fig. S18, ESI†) we explored the effect of heating on bacterial stress and the production of O2˙−. Marcen et al. reported that E. coli (BW 25113) was a bacterial strain that generates ROS under heat-stress.8 Hence, this strain served as a model system among three other additional strains, including P. aeruginosa, S. aureus, and E. faecalis, where ROS generation under heating was unknown. Gratifyingly, signals were observed amongst all the strains using TCF-OTf (Fig. 3), which was in accordance with bacterial mortality induced by heat from 37 °C to 50 °C and 58 °C (Fig. S19, ESI†), and the production of O2˙− was further confirmed using an FeTMPyP assay (Fig. S20, ESI†)−. Furthermore, time-dependent detection confirmed that the signals gradually increase with time (Fig. S21, ESI†) and correlated with bacterial cell death (Fig. S22, ESI†).
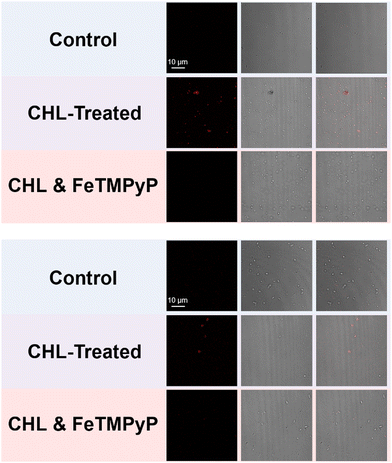
Confocal laser-scanning microscopy (CLSM) was subsequently used to examine the bioimaging feasibility of TCF-OTf among living bacterial species with antibiotic/heat treatments. TCF-OTf-stained chloramphenicol sub-MIC S. aureus exhibited clear and bright fluorescent images, but the three other species also exhibit some “turn-on” fluorescence, which could be quenched by FeTMPyP (Fig. 4, Fig. S23, and S24, ESI†). Significantly, we found that compared with P. aeruginosa and S. aureus, the E. coli and E. faecalis systems, which fail to display any spectroscopic output (Fig. S9, ESI†), exhibited some signal when imaged (Fig. S24, ESI†). TCF-OTf-stained and heated bacterial species, on the other hand, were modestly activated under imaging conditions and then quenched by FeTMPyP (Fig. S25–S27, ESI†).
In summary, a new trifluoromethanesulfonate-functionalized TCF-based long-wavelength fluorescent probe TCF-OTf for O2˙− was developed. The TCF-OTf probe was used to monitor O2˙− production by clinically relevant bacteria under chloramphenicol treatment. In addition, we monitored the production of O2˙− during the heat-treatment of bacteria. Pointedly, long-wavelength TCF-OTf enabled fluorescence cell staining/imaging. We are currently working to develop probes with improved selectivity in order to avoid interference by other highly-reactive ROS such as ClO−. However, in the meantime we anticipate that TCF-OTf could be used to assist the understanding of O2˙−-related pathological/therapeutic processes and the development of new treatments (antibiotics) for life-threatening bacterial pathogens.
A. T. A. J. wishes to thank the EPSRC for supporting this research through EP/V00462X/1 and EP/R003939/1. B. L. P. would like to thank The James Tudor Foundation for funding. R. A. H. thanks the Annette Trust and EPSRC for funding. T. D. J. wishes to thank the Royal Society for a Wolfson Research Merit Award and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University for support (2020ZD01). A. C. S. would like to thank the Glasstone Research fellowship (University of Oxford) and the major research grant from Jesus College Oxford for financial support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. W. Hall and T. F. Mah, FEMS Microbiol. Rev., 2017, 41, 276 CrossRef CAS PubMed.
- L. J. Stephens, M. V. Werrett, A. C. Sedgwick, S. D. Bull and P. C. Andrews, Future Med. Chem., 2020, 12, 2035 CrossRef CAS PubMed.
- C. H. Koh, M. Whiteman, Q. X. Li, B. Halliwell, A. M. Jenner, B. S. Wong, K. M. Laughton, M. Wenk, C. L. Masters, P. M. Beart, O. Bernard and N. S. Cheung, J. Neurochem., 2006, 98, 1278 CrossRef CAS PubMed.
- R. A. Cairns, I. S. Harris and T. W. Mak, Nat. Rev. Cancer, 2011, 11, 85 CrossRef CAS PubMed.
- C. Zhang, D.-T. Shi, K.-C. Yan, A. C. Sedgwick, G.-R. Chen, X.-P. He, T. D. James, B. Ye, X.-L. Hu and D. Chen, Nanoscale, 2020, 12, 23234 RSC.
- J. J. Hu, N.-K. Wong, S. Ye, X. Chen, M.-Y. Lu, A. Q. Zhao, Y. Guo, A. C.-H. Ma, A. Y.-H. Leung, J. Shen and D. Yang, J. Am. Chem. Soc., 2015, 137, 6837 CrossRef CAS PubMed.
- I. Albesa, M. C. Becerra, P. C. Battan and P. L. Paez, Biochem. Biophys. Res. Commun., 2004, 317, 605 CrossRef CAS PubMed.
- M. Marcen, V. Ruiz, J. Serrano, S. Condon and P. Manas, Int. J. Food Microbiol., 2017, 241, 198 CrossRef CAS PubMed.
- K.-C. Yan, A. C. Sedgwick, Y. Zang, G.-R. Chen, X.-P. He, J. Li, J. Yoon and T. D. James, Small Methods, 2019, 3, 1900013 CrossRef.
- J. Klockgether, A. Munder, J. Neugebauer, C. F. Davenport, F. Stanke, K. D. Larbig, S. Heeb, U. Schock, T. M. Pohl, L. Wiehlmann and B. Tummler, J. Bacteriol., 2010, 192, 1113 CrossRef CAS PubMed.
- B. W. Holloway, J. Gen. Microbiol., 1955, 13, 572 CAS.
- C. Shaw, J. M. Stitt and S. T. Cowan, J. Gen. Microbiol., 1951, 5, 1010 CrossRef CAS PubMed.
- A. C. Sedgwick, H.-H. Han, J. E. Gardiner, S. D. Bull, X.-P. He and T. D. James, Chem. Commun., 2017, 53, 12822 RSC.
- L. Gwynne, A. C. Sedgwick, J. E. Gardiner, G. T. Williams, G. Kim, J. P. Lowe, J.-Y. Maillard, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, J. Yoon and T. D. James, Front. Chem., 2019, 7, 255 CrossRef CAS PubMed.
- L. Gwynne, G. T. Williams, K.-C. Yan, J. E. Gardiner, K. L. F. Hilton, B. L. Patenall, J. R. Hiscock, J.-Y. Maillard, X.-P. He, T. D. James, A. C. Sedgwick and A. T. A. Jenkins, Isr. J. Chem., 2021, 61, 234 CrossRef CAS.
- C. Chen, Z. Song, X. Zheng, Z. He, B. Liu, X. Huang, D. Kong, D. Ding and B. Z. Tang, Chem. Sci., 2017, 8, 2191 RSC.
- L. Gwynne, G. T. Williams, K.-C. Yan, B. L. Patenall, J. E. Gardiner, X.-P. He, J.-Y. Maillard, T. D. James, A. C. Sedgwick and A. T. A. Jenkins, Biomater. Sci., 2021, 9, 4433 RSC.
- W. Chen and L. Zhang, J. Microbiol., 2004, 1, 46 Search PubMed.
- L. Wu, L. Liu, H.-H. Han, X. Tian, M. L. Odyniec, L. Feng, A. C. Sedgwick, X.-P. He, S. D. Bull and T. D. James, New J. Chem., 2019, 43, 2875 RSC.
- X. Liu and A. Shi, Chin. J. Clin. Pharmacol., 2002, 4, 302 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc04662h |
| This journal is © The Royal Society of Chemistry 2022 |