Open Access Article
Open Access ArticleMolecular tweezers – a new class of potent broad-spectrum antivirals against enveloped viruses
My-Hue
Le
 ,
Estelle S.
Taghuo K.
,
Estelle S.
Taghuo K.
 and
Thomas
Schrader
and
Thomas
Schrader
 *
*
University of Duisburg-Essen, Faculty of Chemistry, Universitätsstr. 7, 45117 Essen, Germany. E-mail: thomas.schrader@uni-due.de
First published on 4th February 2022
Abstract
A new supramolecular approach to broad spectrum antivirals utilizes host guest chemistry between molecular tweezers and lysine/arginine as well as choline. Basic amino acids in amyloid-forming SEVI peptides (semen-derived enhancers of viral infection) are included inside the tweezer cavity leading to disaggregation and neutralization of the fibrils, which lose their ability to enhance HIV-1/HIV-2 infection. Lipid head groups contain the trimethylammonium cation of choline; this is likewise bound by molecular tweezers, which dock onto viral membranes and thus greatly enhance their surface tension. Disruption of the envelope in turn leads to total loss of infectiosity (ZIKA, Ebola, Influenza). This complexation event also seems to be the structural basis for an effective inihibition of cell-to-cell spread in Herpes viruses. The article describes the discovery of novel molecular recognition motifs and the development of powerful antiviral agents based on these host guest systems. It explains the general underlying mechanisms of antiviral action and points to future optimization and application as therapeutic agents.
1. Introduction
Sexually transmitted viruses (HIV-1, HIV-2) rely on specific peptidic semen components which form amyloid fibrils, transport the oppositely charged virions to the host cell membrane and facilitate membrane fusion.1,2 This effect promotes HIV infection in vitro by several orders of magnitude, and likely is the cause for the large number of HIV infections worldwide.3 Small molecules which inhibit fibril formation, counteract virion attachment and remodel exisiting fibrils into benign unstructured material would be promising antiviral agents because they could abrogate the infectiosity increase mediated by these peptides. Unfortunately, available microbicides that target viral components have proven largely ineffective in preventing sexual virus transmission.Classical therapeutic strategies are mainly directed against viral replication; for each virus a specific protease or polymerase inhibitor is required.4 However, there is constant threat of zoonoses, such as, e.g., the newly emerged coronavirus, SARS-CoV-2. Broad-spectrum antivirals constitute a new powerful strategy against emerging viruses of unknown mechanism and a total lack of therapeutic agents. The underlying idea is external interference with the membrane fusion process which is essential for all enveloped viruses.5 This can in principle be achieved by inhibition of fusion proteins6 or modulation of membrane properties.7
Cell-to-cell spread is a common mechanism of fast viral dissemination/distribution especially for herpes viruses, poorly understood and very hard to prevent. In cell culture, HCMV as well as HSV-1 and HSV-2 resist even neutralizing antibodies.8,9
In recent years, molecular tweezers have been discovered to be active in all these above-mentioned aspects and therefore open new opportunities for powerful and mild intervention with viral infections. The article summarizes the development of these supramolecular host molecules and explains how their unique way to recognize various biologic cations is the key to understand their multiple and synergistic modes of action: a success story for Supramolecular Chemistry which offers new weapons against dangerous viruses, where no specific therapy exists.
2. Molecular tweezers and clips for supramolecular entrapment
2.1. Molecular tweezer CLR01
Molecular tweezers (MTs) consist of alternating benzene and norbornadiene rings which form a torus-shaped structure with an unpolar, electron-rich interior cavity open to receive cationic guests. They possess a unique binding mechanism and invite the side chains of arginine and lysine to be threaded through their cavity – endowing them with superior affinity for basic amino acids under physiological conditions.10 The parent molecule CLR01 gains high water-solubility by two phosphate groups attached to the central hydroquinone ring; these add to the overall binding energy, because they form salt bridges with the cationic head group of both basic amino acids. Through the combination of dispersive and electrostatic interactions with the hydrophobic effect, host–guest complexation occurs in a kinetically fast and reversible process in an equilibrium state. Tweezers bearing other anionic substituents like phosphonate and sulfate groups have similar properties, but display a certain degree of cytotoxicity, so that only the phosphate tweezer CLR01 evolved as the lead tweezer compound for potential medicinal applications. Furthermore, in buffered aqueous solution, CLR01 is essentially monomeric, with a very low dimerization constant (KDim = 60 M−1) which does not restrain guest binding.112.2. Specific lysine and arginine binding by CLR01
The selective threading of lysine and arginine side chains through the tweezer cavity is confirmed both experimentally as well as by calculation. Combined quantum mechanics and molecular mechanics (QM/MM) calculations, Monte Carlo (MC) and molecular dynamics (MD) simulations demonstrate this unique binding mechanism. Fluorescence and NMR titrations as well as isothermal titration calorimetry (ITC) measurements all produce affinities in the low micromolar regime for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes with these cationic guests in buffered aqueous solution (Table 1). Moreover, insertion of the amino acid side chain into the cavity is accompanied with efficient quenching of the tweezer fluorescence as well as with massive upfield-shifts of its methylene signals.10,11
1 complexes with these cationic guests in buffered aqueous solution (Table 1). Moreover, insertion of the amino acid side chain into the cavity is accompanied with efficient quenching of the tweezer fluorescence as well as with massive upfield-shifts of its methylene signals.10,11
2.3. Prototype molecules CLR03, CLR05 and PC
Besides the diphosphate tweezer CLR01, several related prototypes sharing structural similarity were also designed in order to study the influence of different anionic head groups, linkers and side walls. Among these,14 CLR03 and CLR05 as well as the molecular phosphate clip PC adopt a prominent position and will be considered in this article. Their structures are shown in Scheme 1. All prototypes CLR01, CLR03, CLR05 and PC display an identical central benzene spacer but differ in their designed sidewalls which dictate their particular binding behaviour.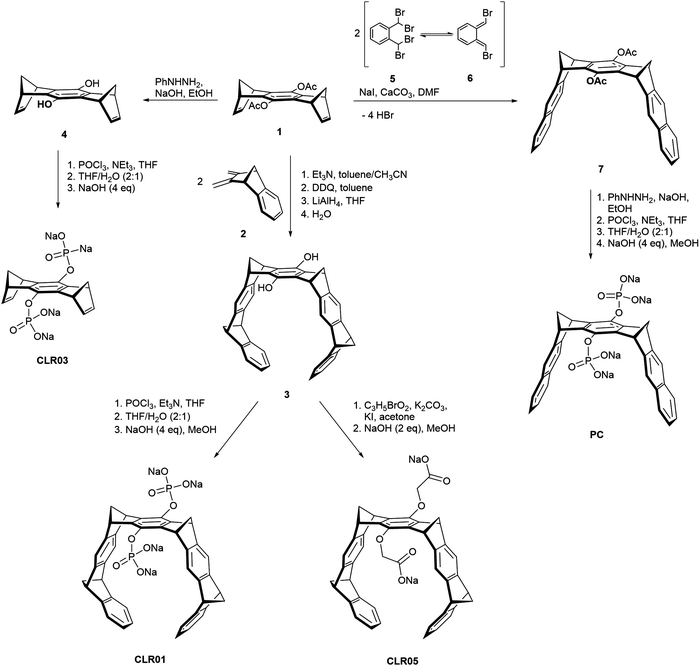 | ||
| Scheme 1 Synthesis of tweezers CLR01 and CLR05, phosphate clip PC and negative control CLR03 starting from acetylated hydroquinone center piece 1. | ||
The smallest derivate CLR03 contains only the doubly phosphorylated hydroquinone center piece but lacks the lateral side walls. It is therefore not able to include lysine and arginine side chains and serves as a negative control for the supramolecular mechanism of action.14,15 CLR05 has the same structural framework as CLR01 but both phosphate groups are exchanged with methylene carboxylate substituents. These carry the terminal carboxylate one carbon atom further away from their hydroquinone oxygen atom and possess only very weak affinity towards lysine and arginine. It can be explained by a competing external guest interaction outside tweezer cavity which is less efficient than guest inclusion. This second binding mode was revealed by calculations showing low energy structures with the cationic guest being chelated between both anionic tweezer groups above the central spacer unit.11 Finally, the molecular diphosphate clip PC carries two planar naphthalene sidewalls with an overall widened cavity; it does not form inclusion complexes with lysine and arginine at all. By contrast, it embraces planar cationic guest molecules through π–stacking and π–cation interactions with its flat aromatic side walls. Therefore, PC binds to biological cofactors such as nicotinamide adenine dinucleotide (NAD+), thiamine diphosphate (TPP) and S-adenosylmethionine (SAM) with high affinity and thus finds application in enzymatic processes, e.g., involving dehydrogenases.12,17,18
2.4. Synthesis of the tweezer and clip skeleton
All parent molecules can be synthesized starting from the hydroquinone spacer 1 according to published procedures. Their total syntheses are summarized in Scheme 1. In the key step, the belt-shaped tweezer cavity is constructed from two neutral Diels–Alder cycloadditions19 between 1 equivalent of spacer 1 as bisdienophile (1,4,5,8-bismethanotetrahydroanthracene) and 2 equivalents of diene 2 (2,3-bismethylene-5,6-benzonorbornene). Subsequent oxidative dehydrogenation with 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) and deprotection of the acetoxy groups with lithium aluminium hydride (LiAlH4) provide the free hydroquinone tweezer 3. Proceeding from this dihydroxy tweezer, the desired phosphate tweezer CLR01 can be obtained after double phosphorylation with phosphorus oxychloride (POCl3) and final neutralization using 4 equivalents of sodium hydroxide (NaOH).18,20,21Similarly, the dicarboxylate tweezer CLR05 becomes accessible after nucleophilic substitution of tweezer 3 with methyl bromoacetate in the presence of potassium carbonate (K2CO3) and potassium iodide (KI) followed by base hydrolysis of the methyl esters with aqueous NaOH.11
For the shortened control CLR03 without sidewalls, the acetylated hydroquinone spacer 1 is treated with phenylhydrazine (PhNHNH2)/NaOH and the resulting deprotected dihydroxy spacer 4 is functionalized with two phosphate groups again using POCl3.22
The synthesis of the phosphate clip is somewhat divergent from the tweezer route. It is based on a general method to annulate naphthalene to norbornadiene systems23–25 and was further optimized in the Klärner group.26,27 The clip skeleton is constructed in the key reaction step between bisdienophile 1 and two equivalents of α,α,α′,α′-tetrabromo-o-xylene (5). It involves a Finkelstein reaction of 5 with sodium iodide leading to o-quinodimethane 6 as a reactive preorganized intermediate which again undergoes a double Diels–Alder reaction with bisdienophile 1. After a four-fold elimination of HBr, molecular clip 7 is obtained with two attached acetoxy substituents. Treatment with PhNHNH2/NaOH cleaves both acetoxy groups, followed by phosphorylation of the resulting hydroquinone clip with POCl3. Neutralization of the free phosphoric acid with 4 equivalents of NaOH furnishes the phosphate-functionalized clip PC as the water-soluble sodium salt.
3. Molecular tweezer CLR01 prevents protein aggregation and amyloidosis
3.1. CLR01 obstructs protein aggregation
Due to its unique binding mode, CLR01 is able to complex well-accessible lysine and arginine residues both in peptides and proteins in a reversible recognition process with moderate affinity (Kd = 10–30 μM).11 In unstructured flexible peptides, almost each basic amino acid may be targeted as evidenced from ESI-ECD-ICR MS as well as NMR experiments of amyloid-β-42 and IAPP. In proteins roughly one third of all basic amino acids are bound by CLR01, again depending strongly on their steric accessibility for the threading mode. Since electrostatic and dispersive interactions between lysines/arginines and oppositely charged as well as aromatic residues are instrumental for pathologic protein aggregation, capping with CLR01 has a strong inhibiting influence on these processes. As a potent lead compound, CLR01 counteracts misfolding and pathologic aggregation of a significant number of critical peptides and proteins. In most cases, fibrils and well-ordered β-sheet-rich assemblies are not dissolved into monomers or even to toxic oligomers, but redirected to form benign amorphous solids.14,28,29 Since lysines play an important role in many of these aggregation processes, CLR01 is indeed able to prevent and revert pathologic aggregation of prominent amyloidogenic proteins such as Amyloid-β (Alzheimer's disease),30,31 α-synuclein (Parkinson's disease)32–34 and IAPP (Diabetes mellitus).35 Due to the entirely reversible threading process with fast on and off rates normal protein functions are not compromised at therapeutic doses, rendering the tweezers essentially nontoxic in mouse experiments.143.2. Anti-amyloid effect of CLR01 in human seminal fluid
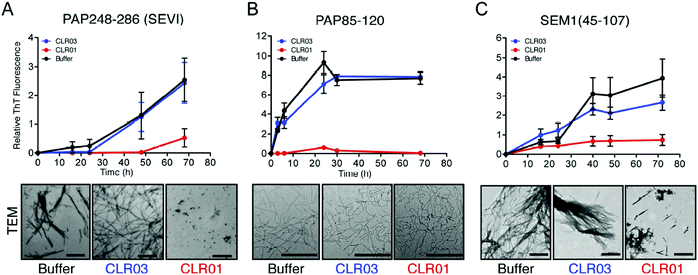
Human semen contains lysine- and arginine-rich polypeptides which likewise form fibrils, bind to virions and transport them to the human host cell, where they attach to the oppositely charged membrane. This process greatly enhances the infectiosity of human immunodeficiency viruses and trigger HIV infection.3 The study by Münch et al. demonstrates that only 1 to 3 viral particles are sufficient to initiate a productive infection in the presence of fibrils, while 1000 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 virions are required in the absence of fibrils.36 These fibrils are formed from fragments of prostatic acid phosphatase (PAP) which generate the peptide SEVI (Semen-derived Enhancer of Viral Infection),37,38 or from fragments of semenogelins (SEM1 and SEM2) which generate several peptides that stimulate infection in vitro.39 Intriguingly, the molecular tweezer CLR01 was found to efficiently block formation of these dangerous amyloid fibrils.40 By specifically targeting lysine and arginine residues in seminal PAP and SEM peptides, CLR01 prevents spontaneous fibril assembly of PAP248–286, PAP85–120 and SEM1(45–107) (Fig. 1), particularly effective against SEVI fibrils formed by PAP248–286.41 Remarkably, CLR01 also swiftly remodels already preformed PAP85–120 fibrils. The underlying cause is most likely a powerful neutralization of prevailing positive peptide surface charges found on PAP and SEM peptides when the triply negatively charged tweezers dock onto the cations. Without these positive charges the fibrils lose their viral infection promoting property.40,42 However, when the tweezers are treated with an excess of free lysine, their anti-amyloid activity against SEVI is strongly reduced, indicating that their mechanism of action requires a direct complexation of SEVI lysines and arginines. Taken together, the tweezers prevent formation of SEVI fibrils as well as neutralize existing fibrils and thus effectively abolish their HIV infectiosity increase.14
000 virions are required in the absence of fibrils.36 These fibrils are formed from fragments of prostatic acid phosphatase (PAP) which generate the peptide SEVI (Semen-derived Enhancer of Viral Infection),37,38 or from fragments of semenogelins (SEM1 and SEM2) which generate several peptides that stimulate infection in vitro.39 Intriguingly, the molecular tweezer CLR01 was found to efficiently block formation of these dangerous amyloid fibrils.40 By specifically targeting lysine and arginine residues in seminal PAP and SEM peptides, CLR01 prevents spontaneous fibril assembly of PAP248–286, PAP85–120 and SEM1(45–107) (Fig. 1), particularly effective against SEVI fibrils formed by PAP248–286.41 Remarkably, CLR01 also swiftly remodels already preformed PAP85–120 fibrils. The underlying cause is most likely a powerful neutralization of prevailing positive peptide surface charges found on PAP and SEM peptides when the triply negatively charged tweezers dock onto the cations. Without these positive charges the fibrils lose their viral infection promoting property.40,42 However, when the tweezers are treated with an excess of free lysine, their anti-amyloid activity against SEVI is strongly reduced, indicating that their mechanism of action requires a direct complexation of SEVI lysines and arginines. Taken together, the tweezers prevent formation of SEVI fibrils as well as neutralize existing fibrils and thus effectively abolish their HIV infectiosity increase.14
3.3. CLR05 and PC possess no anti-amyloid activity
It is interesting, that the structurally related carboxylate-functionalized tweezer CLR05 as well as the diphosphate clip PC both display a markedly reduced affinity for lysine and arginine. We asked if they were still active against the HIV promoting PAP and SEM peptides. Experimental evidence was obtained for the fact that both CLR05 and PC cannot hinder spontaneous amyloid fibrillization and hold no fibril remodeling property. However, while the clip PC has no effect on HIV infection, the tweezers CLR05 mildly suppress HIV infection.13 This has nothing to do with the SEVI proteins but is a direct antiviral effect which was discovered later. CLR03 finally served as negative control and does not interfere with SEVI amyloid aggregation nor with virus infection.154. Antiviral activity of molecular tweezer CLR01
4.1. CLR01 blocks infection of enveloped viruses
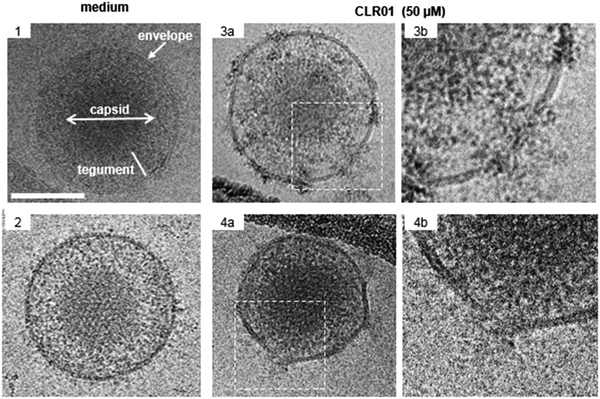
The viral envelope is a crucial factor for the stability of the virus against external influences and the ability of the virus to infect the cell.43 Resistance to external factors allows enveloped viruses to counteract or adapt to the host's immune defense more easily. The viral envelope is also the main instrument responsible for infection and determines the level of virulence or pathogenicity of the virus.44,45 All emerging viruses in humans that may cause pandemics are enveloped viruses such as HIV, SARS-CoV-2, influenza virus, Ebola virus, Zika.46It was therefore a great surprise when control experiments with HIV-1 virus and CLR01 revealed a direct destabilizing effect of the tweezer on the viral envelope. This eventually led to the discovery that CLR01 and even more so advanced molecular tweezers of the second generation represent broad-spectrum antiviral agents, which inhibit the viral action of enveloped viruses by disturbing their membrane. Several experiments have meanwhile unanimously proven that the interaction of CLR01 with the viral particle of enveloped viruses like HCMV leads to physical disruption of the membrane (Fig. 2). In sharp contrast, CLR01 has no effect against non-enveloped viruses such as, e.g., the human adenovirus. From the beginning it became clear that this must be a supramolecular effect, since the related molecule CLR03, which lacks the hydrophobic side arms, has no destabilizing influence on the viral membrane and hence no antiviral activity. Obviously, cation inclusion on the membrane was the key to understand the unknown mechanism of CLR01, and CLR03 therefore has served as a reliable negative control. It is also very important that normal healthy cells are not destabilized by the tweezers, and cell viability of many cell types treated with therapeutic doses of CLR01 remains ∼ 100% in numerous control experiments.
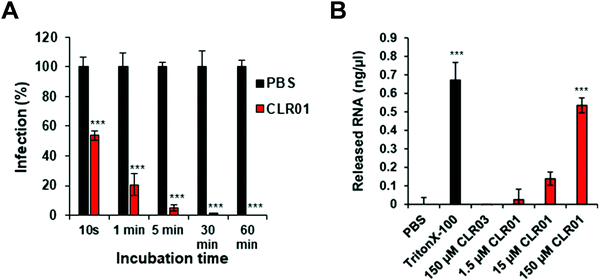
Further details were obtained from experiments with the ZIKA virus, well known to cause microcephaly, i.e. abnormally small heads of new born babies, sadly documented especially 2015/2016 in Brazil.47 To test the direct activity of CLR01 against the Zika virion, a kinetic analysis was performed, in which ZIKV was exposed to 10 μM CLR01 for 10 to 3600 s prior to infection. After 10 s of exposure, a reduction in infection of approximately 43% could be observed. As the incubation time increased, the infection decreased until nearly healthy cells were reached after 1800 s of incubation (Fig. 3A).
As described previously for the HIV-1 virus, CLR01 also causes a final disruption of the viral membrane in ZIKV. An experiment was performed measuring the amount of viral RNA released into solution, when CLR01, CLR03, PBS and Triton-X 100 were administered to ZIKV particles for 30 min at 37 °C. Triton-X 100 detergent served as a positive control, and its administration to ZIKV particles resulted in the detection of massive viral RNA, demonstrating that the detergent destroyed the viral particle. Similarly, incubation of ZIKV with CLR01 led to a release of high amounts of RNA, indicating physical destruction of the virus. In contrast, no viral RNA was released by incubating the ZIKV with CLR03 or PBS (Fig. 3B).48
4.2. CLR01 abrogates the formation of the ZIKV-induced cytopathic effect
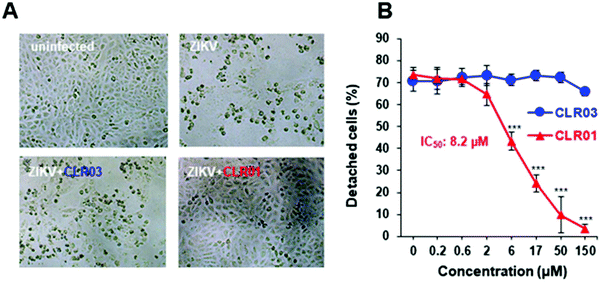
The ZIKV is responsible for cytopathic effects (CPE) characterized by morphological changes such as rounding or clumping of adherent cells. The ZIKV-mediated CPE also induce cell hypertrophy, growth restriction, cell-cycle dysregulation, and cell death or apoptosis, underlying the diseases associated with ZIKV.49–51 Since CLR01 leads to the destruction of the virus, it can also prevent the cytopathic effect caused by the ZIKV, as was observed in an experiment where VERO E6 cells were infected with the ZIKV. After 4 days of infection, in the absence of compounds or the presence of CLR03, an apparent CPE was evident by the formation of large plaques resulting from cell death. In the presence of 150 μM CLR01, no CPE effect was observed. A colorimetric MTT-assay showed that CLR01 suppressed the CPE formation with an IC50 of 8.2 μM (Fig. 4A and B).484.3. CLR01 suppresses viral infection of cells derived from the anogenital region and the brain
Some enveloped viruses such as ZIKV are neurotropic and affect cervical cells. CLR01 administered at a concentration of 150 μM has been shown to abrogate the infection of neuronal cells without causing any toxic effect at this concentration.CLR01 also suppresses ZIKV infectivity in cells of the anogenital region, such as cells derived from the cervix, colon, and primary foreskin cells, with IC50 values in the μM range. In general, CLR01 abrogated the infectivity of HIV, HCMV, ZIKV, and other sexually transmitted viruses in body fluids related to virus transmissions via semen, urine, saliva, and cerebrospinal fluid. It may thus bear a considerable potential as a therapeutic agent to reduce the transmission of sexually transmitted viral diseases. The molecular tweezer could be administered locally or topically. By contrast, a systematic application of CLR01 as a drug seems unlikely since the tweezer loses its antiviral activity in the presence of human serum, most likely by direct binding to human serum albumin.
4.4. CLR01 inhibits viral cell-to-cell spread of HCMV and Herpesvirus
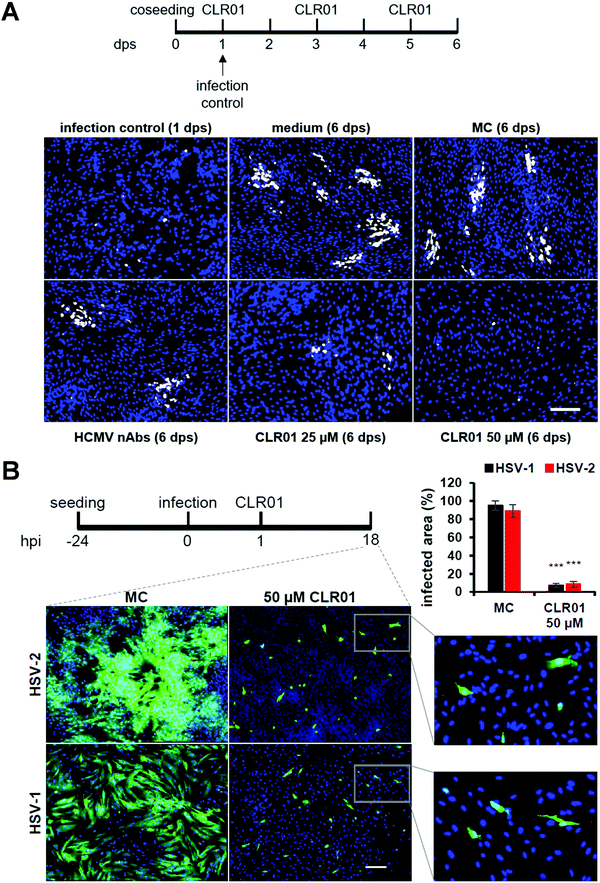
Viruses typically spread between host cells by releasing viral particles and diffusion of these particles, followed by attachment and penetration into new permissive cells. However, many pathogenic viruses, including HIV, HCMV, herpes simplex, can spread through direct cell-to-cell contact, and these viruses have developed diverse and “ingenious” mechanisms to achieve this. The transfer of viral particles across cell-to-cell contacts promotes immune evasion and thus greatly accelerates the infection of target cells. Understanding this mode of transmission plays an important role in the search of novel targets for drug design.52–54In a recent investigation, another highly desired effect was unraveled for CLR01: the molecular tweezer not only shows a rapid direct antiviral effect on viral particles, but it also blocks the cell-to-cell spread of HCMV and HSV-1 &-2. Focus expansion assays were performed using human fibroblasts seeded with cells infected with a clinical isolate of HCMV propagating only by direct cell-to-cell transmission. Infection control was monitored 24 hours after seeding, showing single infected cells before different concentrations of CLR01, nAbs, methylcellulose overlay medium, and medium only were added. Verification of cell-to-cell propagation was performed six days after seeding by detecting HCMV IE antigen-positive cells. When the virus was grown in medium only, i.e., in the presence of MC-medium and nAbs, the propagation of HCMV could not be inhibited, as shown in the experiment: foci in the presence of nAbs, MC-medium, and medium only contained an average of ten IE-positive cells. However, in the presence of 25 μM of CLR01, partial inhibition of the propagation could be observed, and at a concentration of 50 μM, CLR01 was able to prevent cell-to-cell propagation of HCMV completely as only single IE-positive cells were detected after six days (Fig. 5A).
The same experiment was performed with HSV-1 &-2 in a shortened time since herperviruses have a short replication cycle of about 6 hours, and the cell-to-cell propagation was verified 18 hours after infection (Fig. 5B). Thus, CLR01 effectively blocks the direct cell-to-cell propagation of HSV-1 and HSV-2 in human fibroblasts. It is important to note that the molecular tweezer CLR01 does not affect the viability of the cells when administered for an extended period.55
5. Mechanism of viral envelope destabilization
5.1. Approach to elucidate antiviral action mechanistically
It appears remarkable that CLR01 exerts a broad antiviral effect against enveloped viruses caused by the disruption of the lipid bilayer of viral membranes while leaving cellular membranes unimpaired. For several years it remained unknown how CLR01 (and to a smaller extent also CLCR05) executes this selective membrane destabilization after docking onto the lipid bilayer and how it eventually annihilated all tested viruses irreversibly. Hints came from the altered membrane composition of viral envelopes as opposed to their host cells, from which they originate: in most cases, viral membranes are highly enriched in lipid rafts, which constitute up to 30% of all membrane lipids, e.g. in HIV-1. These microdomains contain tightly packed cholesterol and sphingolipid molecules. Such a high content of rigid lipids in a viral membrane leads to increased surface tension, which makes them vulnerable for additional effects in the same direction. Indeed, experiments with liposomes with and without lipid rafts later confirmed our hypothesis, that the tweezers might work by raising the surface tension inside lipid rafts beyond the threshold value for membrane rupture.A second important discovery was made when the tweezers were titrated to isolated lipid molecules such as sphingomyelin and DOPC and a direct inclusion of cholin head groups inside the tweezer cavity was revealed by massive upfield shifts of their trimethylammonium moiety. Consequently, diverse computational studies, comparative NMR titrations and biological and virological/liposome experiments were conducted to probe this putative novel supramolecular antiviral mechanism. To compare the influence of different architectures, studies were carried out not only with the phosphate tweezer CLR01 and the carboxylate tweezer CLR05, but also with the so-called phosphate clip PC, which carries a different cavity with almost planar aromatic side walls.
5.2. Computational studies show altered orientation of lipid heads
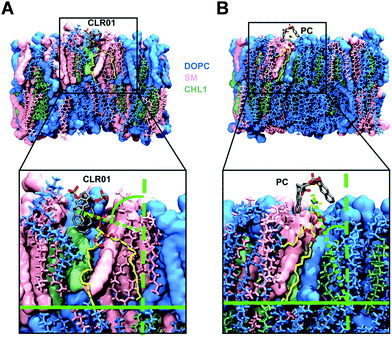
For the computational modeling a distinct amount of dipalmitoylphosphatidylcholine (DOPC), sphingomyelin (SM) and cholesterol (Chol) was chosen to build a simplified bilayer consisting of only DOPC and a mixed one made of all three lipids. Then the protocompounds CLR01, CLR05 and PC were placed 4 Å above the model membrane surface before running molecular dynamics simulations. The outcome hints at the engagement of all three ligands encapsulating the trialkylammonium group of DOPC and SM in both model bilayers. More specifically, the lipid heads were observed to approach the upright orientated tweezers from the side causing a ∼90° angle tilt of their choline groups (Fig. 6A), which in turn promotes surface tension and induces membrane disturbance. This alignment change was not observed for PC since the lipid heads can easily enter the widened clip cavity from below (Fig. 6B) and therefore do not lead to additional strain in the membrane – in good agreement with the absence of an antiviral effect. Moreover, quantum mechanics/molecular mechanics (QM/MM) calculations show how CLR01 forms thermodynamically more stable complexes with SM compared to DOPC not only inside the membrane but also in the aqueous environment. This is likely caused by a decreased cost for desolvation of the hydrated lipid heads in SM, originating from an additional intramolecular hydrogen bond of the phosphate group with the neighboring hydroxyl group.135.3. NMR titrations demonstrate inclusion of choline heads inside tweezers and clip
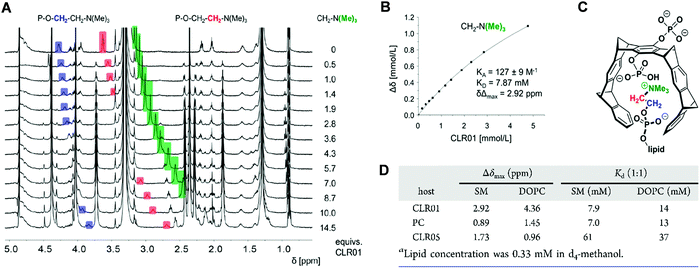
Comparative NMR titrations were carried out in methanol-d4 to characterize and quantify DOPC and SM lipid head group encapsulation by tweezers CLR01, CLR05 and clip PC. All three host molecules were added stepwise in increasing amounts to a defined lipid solution reaching a final ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9–14. These NMR studies unveiled strong chemical upfield shifts for the choline head protons, especially for the trimethylammonium moiety, accompanying complex formation within the host cavities (Fig. 7A–C). Absolute affinities are modest under these conditions – and Kd values from nonlinear regression all remain in the low millimolar range. Most likely, the close proximity between the choline phosphate ester anion and the anionic substituents of the tweezers or clip severely limits lipid affinities. While CLR01 and PC form somewhat tighter complexes (Kd values: SM ∼ 7 mM; DOPC ∼ 14 mM), much weaker affinities are determined for CLR05 (Kd value ∼40–60 mM) (Fig. 7D). The latter may be explained by additional external chelate formation above the carboxylated tweezer cavity, similar to lysine complexation.11 Since SM is especially enriched in viral membranes, its superior affinity towards CLR01 may point to a selective destabilization of viral membranes by lipid inclusion.13
9–14. These NMR studies unveiled strong chemical upfield shifts for the choline head protons, especially for the trimethylammonium moiety, accompanying complex formation within the host cavities (Fig. 7A–C). Absolute affinities are modest under these conditions – and Kd values from nonlinear regression all remain in the low millimolar range. Most likely, the close proximity between the choline phosphate ester anion and the anionic substituents of the tweezers or clip severely limits lipid affinities. While CLR01 and PC form somewhat tighter complexes (Kd values: SM ∼ 7 mM; DOPC ∼ 14 mM), much weaker affinities are determined for CLR05 (Kd value ∼40–60 mM) (Fig. 7D). The latter may be explained by additional external chelate formation above the carboxylated tweezer cavity, similar to lysine complexation.11 Since SM is especially enriched in viral membranes, its superior affinity towards CLR01 may point to a selective destabilization of viral membranes by lipid inclusion.13
5.4. Disruption of viral membranes rich in lipid rafts by tweezers and clip
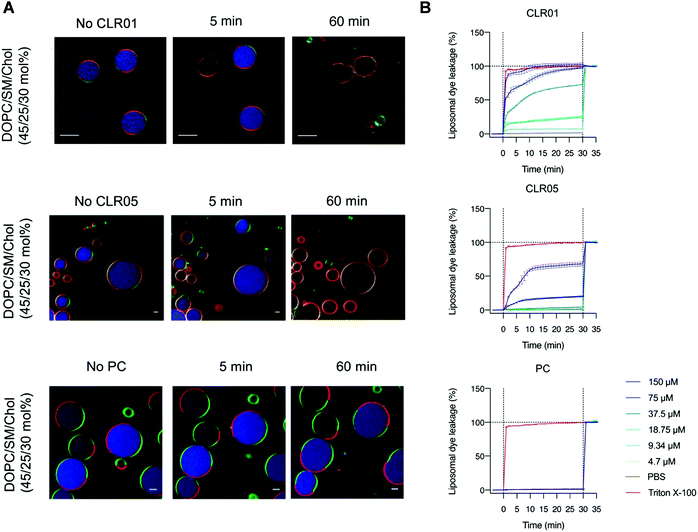
To examine the effect of lipid rafts on membrane disruption, we conducted experiments with simple model membranes of different lipid composition. Liposomes were prepared with a high content of lipid rafts found, e.g., in HIV-1 (DOPC/SM/Chol, 45/25/30 mol%) and were compared to liposomes containing only pure DOPC. Two different liposome studies were conducted monitoring with confocal fluorescence microscopy and dye leakage. The first experiment used two different giant unilamellar vesicles (GUVs) as model envelopes assembled from the lipid compositions mentioned above. GUVs were filled with a liposome dye (ATTO 647) and also contained different fluorescent labels on their liquid disordered (ld) and liquid ordered (lo) phase, so that they could be visualized microscopically after incubation with 150 μM tweezer or clip (Fig. 8A). For the second experiment, liposomes composed of the same lipid mixture (DOPC/SM/Chol, 45/25/30 mol%) were only filled with a self-quenched fluorescent dye (carboxyfluorescein). After addition of tweezers or clip, dye leakage caused by membrane disruption was then detected from the dose-dependent fluorescence intensity increase (Fig. 8B). Exposure of the pure DOPC liposomes with tweezers or clip had no effect and no dye was released. Instead, 78% dye leakage was observed for liposomes containing the lipid raft mixture when exposed to 150 μM CLR05 after 30 min incubation while vesicle integrity was partially maintained. More impressively, the same concentration of CLR01 led to complete liposome lysis after 5 min with a total dye release.16 By contrast, both phosphate clip PC and negative control CLR03, left all lipid vesicles intact even after one hour incubation time, regardless of the applied doses.13 These effects are all in very good agreement with our proposed mechanism: Only DOPC membranes with a high content in lipid rafts, mimicking viral envelopes, can be disrupted by tweezer complexation, with CLR01 being superior to CLR05. PC with its wide open cavity and the negative control CLR03 have no destabilizing effect because they do not increase surface tension.6. Advanced tweezers with additional aliphatic lipid anchors
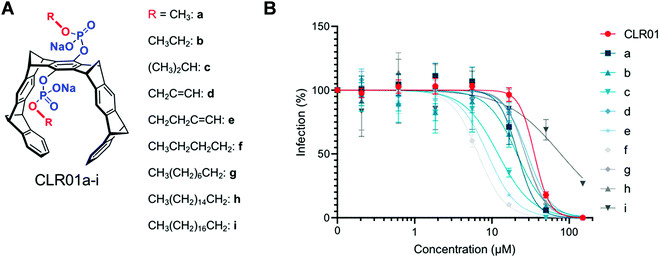
If the membrane targeting mechanism of CLR01 is correct and elevated surface tension leads to facilitated membrane rupture, the introduction of additional lipid anchors into the tweezer molecules should improve their antiviral activity. This straightforward optimization was tested with a series of novel tweezers of the second generation, developed to enhance anti-viral activity. To this end, the advanced tweezers were symmetrically equipped with diverse alkyl ester arms of various length and degree of branching (Fig. 9A). Specifically, alkyl esters were prepared between C1 and C16 chain length by monoesterification of both tweezer phosphates through activation with trichloroacetontrile (TCA) in pyridine.56 The inhibitory effect of all two-armed tweezers on HIV-1 infection was quantified in cell culture as before for CLR01. With all diarmed tweezers infection rates were decreased much more effectively than with CLR01, indicating a superior antiviral activity (Fig. 9B). Of all modified tweezer compounds, those with an attached C4 alkyl arm were the best inhibitors and retained very low toxicity. Likewise, liposome dye-leakage assays confirmed a highly efficient disruption of membrane models with lipid rafts (DOPC/SM/Chol). We conclude that the newly attached ester arms most likely operate as additional hydrophobic anchors which boost membrane surface tension and disruption by direct insertion of one or both arms into the lipid bilayer.13 These modified variants provide a promising starting point for further tweezer optimization.7. Conclusion and outlook
From the beginning, it was not anticipated that the same supramolecular host would offer several points of attack for antiviral action. The starting point was a novel approach to treat neurodegenerative diseases with molecular tweezers by complexation of critical arginines and lysines in the respective aggregating proteins, so that these cannot form small toxic oligomers and fibrils anymore. This strategy was transferred to a similar newly discovered feature of amyloid fibrils which greatly increase the infectiosity of the human immune deficiency virus. Thus, the inhibition of SEVI proteins by disaggregation, neutralisation and remodeling of their toxic amyloid fibrils was the initial focus, which effectively eliminates the infectivity increase of sexually transmitted viruses (HIV-1/HIV-2).Even more surprising was the general direct disruption of viral envelopes by another supramolecular host guest complexation: The trimethylammonium cation which constitutes the natural lipid head group can likewise be accommodated inside the tweezer cavity and leads to a significant increase in surface tension in viral membranes, which eventually causes their disruption (ZIKA, Ebola, Influenza). Mechanistic elucidation of this unique process triggered further optimization of supramolecular membrane destabilizers by introduction of additional lipid arms. It seems that their powerful synergistic influence on the viral membrane is also responsible for the recently discovered inhibition of cell-to-cell-spread which operates in herpes viruses.
Meanwhile, cell culture experiments have been conducted with a large number of viruses and the 2nd generation tweezers with their two additional lipid anchor arms appeared to be superior to all of the original prototype tweezer molecules from the first generation. There are also strong hints for a correlation between the type of lipid anchor and a molecular tweezer and its preferred virus – most likely a match with its specific lipid composition.
Preliminary animal experiments in mice following intranasal application of virus and tweezers already indicated full control over virus multiplication and spreading without adverse side effects. For a systemic application, the intranasal application must be improved by nebulization to carry the antivirals deep into the animal lungs. In addition, pharmacokinetics and histopathology studies are underway. Thus, a supramolecular host molecule which was invented for supramolecular cation encapsulation, currently turns out to provide entirely new opportunities for combatting viral infections.
Author contributions
M.-H. Le: data curation, investigation, methodology, writing-review & editing. E. S. Taghuo K.: data curation, investigation, methodology, writing – review & editing. T. Schrader: conceptualization, supervision, writing-review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge financial support by the European Union in the frame of Horizon 2020 (Fight-nCov, GA number 101003555) and by the DFG through the CRC 1093 “Supramolecular Chemistry on Proteins”.Notes and references
- K. A. Kim, M. Yolamanova, O. Zirafi, N. R. Roan, L. Staendker, W. G. Forssmann, A. Burgener, N. Dejucq-Rainsford, B. H. Hahn, G. M. Shaw, W. C. Greene, F. Kirchhoff and J. Münch, Retrovirology, 2010, 7, 55 CrossRef PubMed.
- S. Usmani, O. Zirafi, J. A. Müller, L. N. Sandi-Monroy, J. K. Yadav, C. Meier, T. Weil, N. R. Roan, W. C. Greene, P. Walther, K. Peter, R. Nilsson, P. Hammarström, R. Wetzel, C. D. Pilcher, F. Gagsteiger, M. Fändrich, F. Kirchhoff and J. Münch, Nat. Commun., 2014, 5, 3508 CrossRef PubMed.
- J. Münch, E. Rücker, L. Ständker, K. Adermann, C. Goffinet, M. Schindler, S. Wildum, R. Chinnadurai, D. Rajan, A. Specht, G. Giménez-Gallego, P. C. Sánchez, D. M. Fowler, A. Koulov, J. W. Kelly, W. Mothes, J.-C. Grivel, L. Margolis, O. T. Keppler, W.-G. Forssmann and F. Kirchhoff, Cell, 2007, 131, 1059–1071 CrossRef PubMed.
- N. Kumar, S. Sharma, R. Kumar, B. N. Tripathi, S. Barua, H. Ly and B. T. Rouse, Clin. Microbiol. Rev., 2020, 33, 1–36 CrossRef PubMed.
- F. Vigant, N. Santos and B. Lee, Nat. Rev. Microbiol., 2015, 13, 426–437 CrossRef CAS PubMed.
- L. Maofu and M. Kielian, J. Cell Biol., 2005, 171, 111–120 CrossRef PubMed.
- M. R. S. Vincent, C. C. Colpitts, A. V. Ustinov, M. Muqadas, M. A. Joyce, N. L. Barsby, R. F. Epand, R. M. Epand, S. A. Khramyshev, O. A. Valueva, V. A. Korshun, D. L. J. Tyrrell and L. M. Schang, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17339–17344 CrossRef PubMed.
- Q. Sattentau, Nat. Rev. Microbiol., 2008, 6, 815–826 CrossRef CAS PubMed.
- W. Mothes, N. M. Sherer, J. Jin and P. Zhong, J. Virol., 2010, 84, 8360–8368 CrossRef CAS PubMed.
- M. Fokkens, T. Schrader and F. G. Klärner, J. Am. Chem. Soc., 2005, 127, 14415–14421 CrossRef CAS PubMed.
- S. Dutt, C. Wilch, T. Gersthagen, P. Talbiersky, K. Rodriguez, M. Hanni, E. Sánchez-García, C. Ochsenfeld, F. G. Klärner and T. Schrader, J. Org. Chem., 2013, 78, 6721–6734 CrossRef CAS PubMed.
- P. Talbiersky, F. Bastkowski, F.-G. Klärner and T. Schrader, J. Am. Chem. Soc., 2008, 130, 9824–9828 CrossRef CAS PubMed.
- T. Weil, R. Groß, A. Röcker, K. Bravo-Rodriguez, C. Heid, A. Sowislok, M.-H. Le, N. Erwin, M. Dwivedi, S. M. Bart, P. Bates, L. Wettstein, J. A. Müller, M. Harms, K. Sparrer, Y. B. Ruiz-Blanco, C. M. Stürzel, J. Von Einem, S. Lippold, C. Read, P. Walther, M. Hebel, F. Kreppel, F.-G. Klärner, G. Bitan, M. Ehrmann, T. Weil, R. Winter, T. Schrader, J. Shorter, E. Sanchez-Garcia and J. Münch, J. Am. Chem. Soc., 2020, 142, 17024–17038 CrossRef CAS PubMed.
- T. Schrader, G. Bitan and F. G. Klärner, Chem. Commun., 2016, 52, 11318–11334 RSC.
- E. Lump, L. M. Castellano, C. Meier, J. Seeliger, N. Erwin, B. Sperlich, C. M. Stürzel, S. Usmani, R. M. Hammond, J. Von Einem, G. Gerold, F. Kreppel, K. Bravo-Rodriguez, T. Pietschmann, V. M. Holmes, D. Palesch, O. Zirafi, D. Weissman, A. Sowislok, B. Wettig, C. Heid, F. Kirchhoff, T. Weil, F. G. Klärner, T. Schrader, G. Bitan, E. Sanchez-Garcia, R. Winter, J. Shorter and J. A. Munch, eLife, 2015, 4, 1–33 CrossRef PubMed.
- T. Schrader, M. Fokkens, F.-G. Klärner, J. Polkowska and F. Bastkowski, J. Org. Chem., 2005, 70, 10227–10237 CrossRef CAS PubMed.
- F.-G. Klärner and T. Schrader, Acc. Chem. Res., 2013, 46, 967–978 CrossRef PubMed.
- M. Fokkens, C. Jasper, T. Schrader, F. Koziol, C. Ochsenfeld, J. Polkowska, M. Lobert, B. Kahlert and F.-G. Klärner, Chem. – Eur. J., 2005, 11, 477–494 CrossRef CAS PubMed.
- J. Benkhoff, R. Boese and F.-G. Klärner, Liebigs Ann./Recl., 1997, 501–516 CrossRef CAS.
- F.-G. Klärner, U. Burkert, M. Kamieth, R. Boese and J. Benet-Buchholz, Chem. – Eur. J., 1999, 5, 1700–1707 CrossRef.
- F.-G. Klärner and B. Kahlert, Acc. Chem. Res., 2003, 36, 919–932 CrossRef PubMed.
- I. Hadrovic, P. Rebmann, F. G. Klärner, G. Bitan and T. Schrader, Front. Chem., 2019, 7, 1–9 CrossRef PubMed.
- M. P. Cava and D. R. Napier, J. Am. Chem. Soc., 1957, 79, 1701–1705 CrossRef CAS.
- M. P. Cava and R. L. Shirley, J. Am. Chem. Soc., 1960, 82, 654–656 CrossRef CAS.
- M. N. Paddon-Row and H. K. Patney, Synthesis, 1986, 328–330 CrossRef CAS.
- F. G. Klärner, J. Panitzky, D. Bläser and R. Boese, Tetrahedron, 2001, 57, 3673–3687 CrossRef.
- F. G. Klärner, B. Kahlert, R. Boese, D. Bläser, A. Juris and F. Marchioni, Chem. – Eur. J., 2005, 11, 3363–3374 CrossRef PubMed.
- S. Sinha, D. H. Lopes, Z. Du, E. S. Pang, A. Shanmugam, A. Lomakin, P. Talbiersky, A. Tennstaedt, K. McDaniel, R. Bakshi, P. Y. Kuo, M. Ehrmann, G. B. Benedek, J. A. Loo, F. G. Klärner, T. Schrader, C. Wang and G. Bitan, J. Am. Chem. Soc., 2011, 133, 16958–16969 CrossRef CAS PubMed.
- S. Sinha, Z. Du, P. Maiti, F. G. Klärner, T. Schrader, C. Wang and G. Bitan, ACS Chem. Neurosci., 2012, 3, 451–458 CrossRef CAS PubMed.
- C. A. Lane, J. Hardy and J. M. Schott, Eur. J. Neurol., 2018, 25, 59–70 CrossRef CAS PubMed.
- X. Zheng, D. Liu, F. G. Klärner, T. Schrader, G. Bitan and M. T. Bowers, J. Phys. Chem. B, 2015, 119, 4831–4841 CrossRef CAS PubMed.
- S. Prabhudesai, S. Sinha, A. Attar, A. Kotagiri, A. G. Fitzmaurice, R. Lakshmanan, M. I. Ivanova, J. A. Loo, F. G. Klärner, T. Schrader, M. Stahl, G. Bitan and J. M. Bronstein, Neurotherapeutics, 2012, 9, 464–76 CrossRef CAS PubMed.
- S. Acharya, B. M. Safaie, P. Wongkongkathep, M. I. Ivanova, A. Attar, F. G. Klärner, T. Schrader, J. A. Loo, G. Bitan and L. J. Lapidus, J. Biol. Chem., 2014, 289, 10727–10737 CrossRef CAS PubMed.
- A. Lulla, L. Barnhill, G. Bitan, M. I. Ivanova, B. Nguyen, K. O'Donnell, M. C. Stahl, C. Yamashiro, F. G. Klärner, T. Schrader, A. Sagasti and J. M. Bronstein, Environ. Health Perspect., 2016, 124, 1766–1775 CrossRef CAS PubMed.
- D. H. J. Lopes, A. Attar, G. Nair, E. Y. Hayden, Z. Du, K. McDaniel, S. Dutt, K. Bravo-Rodriguez, S. Mittal, F.-G. Klärner, C. Wang, E. Sánchez-García, T. Schrader and G. Bitan, ACS Chem. Biol., 2015, 10, 1555–1569 CrossRef CAS PubMed.
- J. Münch, E. Rücker, L. Ständker, K. Adermann, C. Goffinet, M. Schindler, S. Wildum, R. Chinnadurai, D. Rajan, A. Specht, G. Giménez-Gallego, P. C. Sánchez, D. M. Fowler, A. Koulov, J. W. Kelly, W. Mothes, J.-C. Grivel, L. Margolis and F. Kirchhoff, Cell, 2007, 131, 1059–1071 CrossRef PubMed.
- R. Ren, S. Yin, B. Lai, L. Ma, J. Wen, X. Zhang, F. Lai and S. Liu, Retrovirology, 2018, 15, 49 CrossRef PubMed.
- K. Elias, D. Scanlon, I. F. Musgrave and J. A. Carver, Biochim. Biophys. Acta, Proteins Proteomics, 2014, 1844, 1591–1598 CrossRef PubMed.
- N. R. Roan, H. Liu, S. M. Usmani, J. Neidleman, J. A. Müller, A. Avila-Herrera, A. Gawanbacht, O. Zirafi, S. Chu, M. Dong, S. T. Kumar, J. S. Smith, K. S. Pollard, M. Fändrich, F. Kirchhoff, J. Münch, H. E. Witkowska and W. C. Greene, J. Virol., 2014, 88, 7221–7234 CrossRef PubMed.
- N. R. Roan, J. Münch, N. Arhel, W. Mothes, J. Neidleman, A. Kobayashi, K. Smith-McCune, F. Kirchhoff and W. C. Greene, J. Virol., 2009, 83, 73–80 CrossRef CAS PubMed.
- J. S. Olsen, J. T. M. DiMaio, T. M. Doran, C. Brown, B. L. Nilsson and S. Dewhurst, J. Biol. Chem., 2012, 287(15), 11842–11849 CrossRef CAS PubMed.
- D. Easterhoff, F. Ontiveros, L. R. Brooks, Y. Kim, B. Ross, J. N. Silva, J. S. Olsen, C. Feng, D. J. Hardy, P. M. Dunman and S. Dewhurst, Antimicrob. Agents Chemother., 2013, 57, 2443–2450 CrossRef CAS PubMed.
- D. S. Dimitrov, Nat. Rev., 2004, 2, 109 CrossRef PubMed.
- S. Firquet, S. Beaujard, P. Lobert, F. Sané, D. Caloone, D. Izard and D. Hober, Microbes Environ., 2015, 30, 140–144 CrossRef PubMed.
- J. Klasse, R. Bron and M. Marsh, Adv. Drug Delivery Rev., 1998, 34, 65–91 CrossRef PubMed.
- F. A. Rey and S. Lok, Cell, 2018, 172, 1319–1334 CrossRef CAS PubMed.
- W. K. De Oliveira, G. V. A. de França, E. H. Carmo, B. B. Duncan, R. de Souza Kuchenbecker and M. I. Schmidt, Lancet, 2017, 390, 861–870 CrossRef.
- A. E. Röcker, J. A. Müller, E. Dietzel, M. Harms, F. Krüger, C. Heid, A. Sowislok, C. F. Riber, A. Kupke, S. Lippold, J. von Einem, J. Beer, B. Knöll, S. Becker, J. Schmidt-Chanasit, M. Otto, O. Vapalahti, A. N. Zelikin, G. Bitan, T. Schrader and J. Münch, Antiviral Res., 2018, 152, 26–35 CrossRef PubMed.
- F. R. Cugola, I. R. Fernandes, F. B. Russo, B. C. Freitas, J. L. M. Dias, K. P. Guimarães, C. Benazzato, N. Almeida, G. C. Pignatari, S. Romero, C. M. Polonio, I. Cunha, C. L. Freitas, W. N. Brandão, C. Rossato, D. G. Andrade, D. de, P. Faria, A. T. Garcez, C. A. Buchpigel, C. T. Braconi, E. Mendes, A. A. Sall, P. M. de, A. Zanotto, J. P. S. Peron, A. R. Muotri and P. C. B. Beltrão-Braga, Nature, 2016, 534, 267–271 CrossRef CAS PubMed.
- C. Li, D. Xu, Q. Ye, S. Hong, Y. Jiang, X. Liu, N. Zhang, L. Shi, C.-F. Qin and Z. Xu, Cell Stem Cell, 2016, 19, 120–126 CrossRef CAS PubMed.
- G. Li, M. Poulsen, C. Fenyvuesvolgyi, Y. Yashiroda, M. Yoshida, J. M. Simard, R. C. Gallo and R. Y. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E376–E385 CrossRef CAS PubMed.
- C. L. Jacob, L. Lamorte, E. Sepulveda, I. C. Lorenz, A. Gauthier and M. Franti, Virology, 2013, 444, 140–147 CrossRef CAS PubMed.
- C. Sinzger, K. Schmidt, J. Knapp, M. Kahl, R. Beck, J. Waldman, H. Hebart, H. Einsele and G. Jahn, J. Gen. Virol., 1999, 80, 2867–2877 CrossRef CAS PubMed.
- J. J. Falk, M. Winkelmann, K. L. Sampaio, C. Paal, H. Schrezenmeier, M. Alt, R. Stanton, A. Krawczyk, R. Lotfi and C. Sinzger, Viruses, 2018, 10, 500 CrossRef PubMed.
- S. Brenner, B. Braun, C. Read, T. Weil, P. Walther, T. Schrader, J. Münch and J. von Einem, Viruses, 2021, 13, 1685 CrossRef CAS PubMed.
- C. Heid, A. Sowislok, T. Schaller, F. Niemeyer, F.-G. Klärner and T. Schrader, Chem. – Eur. J., 2018, 24, 11332 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |