Open Access Article
Open Access ArticleA high-throughput 3D bioprinted cancer cell migration and invasion model with versatile and broad biological applicability†
MoonSun
Jung
 abc,
Joanna N.
Skhinas
ab,
Eric Y.
Du
bd,
M. A. Kristine
Tolentino
abc,
Joanna N.
Skhinas
ab,
Eric Y.
Du
bd,
M. A. Kristine
Tolentino
 bd,
Robert H.
Utama
e,
Martin
Engel
e,
Alexander
Volkerling
e,
Andrew
Sexton
e,
Aidan P.
O'Mahony
e,
Julio C. C.
Ribeiro
e,
J. Justin
Gooding
bd,
Robert H.
Utama
e,
Martin
Engel
e,
Alexander
Volkerling
e,
Andrew
Sexton
e,
Aidan P.
O'Mahony
e,
Julio C. C.
Ribeiro
e,
J. Justin
Gooding
 *bd and
Maria
Kavallaris
*bd and
Maria
Kavallaris
 *abc
*abc
aChildren’s Cancer Institute, Lowy Cancer Research Centre, UNSW Sydney, Sydney, NSW, Australia. E-mail: m.kavallaris@ccia.unsw.edu.au
bAustralian Center for NanoMedicine, UNSW Sydney, Sydney, NSW, Australia
cSchool of Clinical Medicine, UNSW Medicine & Health, UNSW Sydney, Sydney, NSW, Australia
dSchool of Chemistry, UNSW Sydney, Sydney, NSW, Australia
eInventia Life Science Pty Ltd, Sydney, NSW, Australia
First published on 23rd September 2022
Abstract
Understanding the underlying mechanisms of migration and metastasis is a key focus of cancer research. There is an urgent need to develop in vitro 3D tumor models that can mimic physiological cell–cell and cell–extracellular matrix interactions, with high reproducibility and that are suitable for high throughput (HTP) drug screening. Here, we developed a HTP 3D bioprinted migration model using a bespoke drop-on-demand bioprinting platform. This HTP platform coupled with tunable hydrogel systems enables (i) the rapid encapsulation of cancer cells within in vivo tumor mimicking matrices, (ii) in situ and real-time measurement of cell movement, (iii) detailed molecular analysis for the study of mechanisms underlying cell migration and invasion, and (iv) the identification of novel therapeutic options. This work demonstrates that this HTP 3D bioprinted cell migration platform has broad applications across quantitative cell and cancer biology as well as drug screening.
Introduction
Metastasis is the leading cause of cancer-related death and thus an understanding of the underlying molecular and cellular processes continues to be a key focus of cancer research.1 Tumor dissemination is a multistep process. It involves cancer cell migration and invasion through the extracellular matrix (ECM) to allow entry into adjacent tissues, blood, and lymphatic vessels.2 The migration and invasion of cancer cells is a dynamic activity of cells, regulated by integrins, matrix-degrading enzymes, cell–cell adhesion molecules and cell–cell communication in living tissues.3 However, current understanding of the molecular mechanisms underlying cell migration and invasion remains heavily dependent on cancer cells grown on flat 2-dimensional (2D) plastic. These 2D model systems are frequently used in high throughput (HTP) drug discovery, although they do not always accurately predict drug response.4,5 A scarcity of appropriate screening platforms that can be directly translatable from in vitro to in vivo is a major contributor that hinders the development of drugs specifically targeting cancer metastasis. As such, there is a growing consensus regarding the use of 3D cell models to better mimic physiological cell–cell and cell–ECM interactions as more suitable approaches for identifying novel inhibitors disrupting cell migration than 2D cell culture.The classic in vitro 3D cell migration and invasion models are the Boyden chamber transwell-based assays. In the transwell assays, cells migrate through a physical barrier, including membrane pores and ECM-mimicking materials, toward a chemo-attractant gradient.6,7 The simplicity and low-cost for set-up make these methods an excellent tool in cancer research. The transwell assays however are often labor-intensive, low throughput and give only an endpoint readout. Recently, a microfluidic platform has been developed to be an advance over the conventional transwell models.8 The platform is a miniaturized device that can process or manipulate small amounts of fluid flow and cells using microchannel networks, which can mimic the dynamic events that constitute the metastatic processes.9,10 Polydimethylsiloxane (PDMS) is one of the most widely used polymers in manufacturing microfluidic devices, due to its biocompatibility, low-cost and ease of fabrication into shape using photolithography.11 However, PDMS is hydrophobic and requires surface treatment, such as coating with ECM-like hydrogels, to promote cell adhesion and proliferation.12,13 Therefore, additional advances could come from improvements in the tunability of the ECM mimic as this is an important component of any 3D cell migration and invasion assay, including the transwell assays and microfluidic models. Most 3D cell model systems currently use naturally derived hydrogels, such as collagen and Matrigel. Of those, Matrigel, a basement membrane matrix composed of a complex mixture of various proteins has been widely used with a variety of cell types.14 It is recognized as a “gold standard” scaffold for numerous in vitro 3D cell culture applications. Despite Matrigel being a powerful resource as an ECM mimic for the study of cell biology for decades, batch-to-batch variability and challenges in tuning the chemical and physical properties of Matrigel lead to a lack of reproducibility and hampers unambiguous determination of interactions between cell, physical, and biochemical ECM factors, including stiffness and integrin-binding proteins.15,16 Thus, it is important to develop matrix scaffolds that are more reproducible and tunable for their mechanical, chemical, physical, and biological properties for 3D cell models. Accordingly, an automated 3D model fabrication combined with tunable matrix materials would increase the reliability of in vitro cell migration and invasion experiments.
Numerous biomaterial-based fabrication technologies, including 3D bioprinting, have recently been developed for the recreation of 3D constructs using ECM mimics that have more tunable properties than Matrigel.17 3D bioprinting provides a new approach to fabricate cell-laden matrices in a well-controlled manner over the positioning of cells and synthetic hydrogels.18 Thus, it has emerged as a promising tool to engineer complex 3D tumor-microenvironments in vitro to study various biological processes including cell migration and invasion. 3D bioprinting can also be combined with microfluidics to engineer living tissues, which has gained popularity over the conventional transwell assays for studying chemotaxis due to its ability to capture the tumor microenvironment features.19,20 Despite recent advances in 3D bioprinting and numerous reports on its potential applications in cancer research, there are few bioprinting platforms that are simple to use, capable of HTP and suitable for multiple biological assays with high cell viability.21–26
Recently we developed an enabling technology consisting of a bespoke drop-on-demand 3D bioprinter, which employs a fly-by printing logic that allows for the ejection of ink droplets at a constant and rapid speed in multi-well plates.27,28 In our recent work, we demonstrated that the hydrogel system of a 4-arm poly(ethylene glycol) maleimide (PEG-4MAL) bioink and a bis-thiol activator that instantly cross-links the polymer can be used as the 3D ECM-like hydrogels, due to the highly tunable mechanical and biofunctional properties as well as their printability.27,29 The crosslinking efficiency of maleimide cross-linked in bioactive PEG hydrogel has also been reported to be close to 100%.30–33 In the present study, we leveraged the 3D bioprinting technology and tunable hydrogel system to develop a versatile HTP 3D bioprinting platform for studying cell migration and invasion. Herein, as model cell systems, we selected a combination of cancer cell lines with non-invasive and invasive phenotypes, which are known to express distinct epithelial and mesenchymal markers.34–36 We demonstrated that the platform can be utilized to identify cell-type specific ECM-like hydrogels by tuning the matrix stiffness and biochemical molecules, and capabilities for downstream phenotypic and molecular analysis in situ using non-invasive and invasive cancer cell models. Moreover, cell movement within the 3D constructs can be monitored, tracked and measured in real-time using the platform. The proposed bioprinted platform could serve as a powerful tool to understand the mechanisms of cancer cell migration and invasion and be used in HTP drug testing to discover drugs effective against cancer metastasis.
Experimental
Cell lines
MCF7 and H1299 cells were purchased from ATCC. MDA-MB-231 cells were kindly provided by Dr. Rose Boutros (Kids Research, Sydney, NSW, Australia). HEY was a generous gift from Georgia Chenevix-Trench (QIMR Berghofer, Brisbane, Australia). All cells were cultured in either DMEM (MCF7 and MDA-MB-231) or RPMI1640 (HEY and H1299) media supplemented with 10% FCS. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C and were mycoplasma free. All cell lines were authenticated using short tandem repeat profiling at Cell Bank Australia and Kinghorn Centre for Clinical Genomics, Australia within the last two years.Bioprinting of 3D cell laden models
Bioinks and activators for 4 hydrogel combinations were purchased from Inventia Life Science, Sydney, Australia (Cat. no. Px01.00, Px01.03P, Px02.00 and Px02.03P for 0.7 kPa, 0.7 kPa + RGD, 1.1 kPa and 1.1 kPa + RGD hydrogels respectively). 3D cell models were printed using the Rastrum 3D bioprinter (Inventia Life Science) as previously described.27 Briefly, the structure design and printing protocol were first created using RASTRUM Cloud (Inventia Life Science). Cells were primed (1 × 106 cells for MCF7, MDA-MB-231 and H1299 with a final seeding density of ∼500 cells per well and 0.5 × 106 cells for HEY with a final seeding density of ∼250 cells per well) prior to being printed within Small Plug or Imaging Model. Cells were also bioprinted using Large Plug for molecular downstream analysis (qPCR and western blotting) available through RASTRUM Cloud. All printing was conducted in flat bottom 96-well plates (Corning). In parallel, Matrigel-encapsulated cells were manually prepared in a 96 well plate as a control. 500 cells (MCF7, MDA-MB-231 and H1299) or 250 cells (HEY) were mixed in 2 μl of growth-factor reduced Matrigel solution (In Vitro Technologies), which was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diluted with cell culture media.
1 diluted with cell culture media.
Live and dead cell staining
Cell viability analysis was performed using live/dead viability/cytotoxicity kit, for mammalian cells (Invitrogen, Cat. no. L3224) according to the manufacturer's instructions. Briefly, cells were bioprinted in multi-well plates and cultured for 7 days post-printing. At day 7, cells were rinsed with DPBS and stained with 100 μL of live/dead stock solutions (10 μM Ethidium Homodimer-1 (EthD1) and 5 μM Calcein AM in DPBS) and incubated for 30 min. Images were taken at 5× magnification using green fluorescence channel for live cells and red fluorescence channel for dead cells using Celldiscoverer 7 (Zeiss). Images were analysed and visualized using Arivis 4D software.Alamar Blue cell proliferation assay
Cells were bioprinted with each hydrogel condition in a 96 well plate and cultured for up to 7 days. Cells were incubated with resazurin-based reagent (Sigma-Aldrich) at 10% of media volume, for 16 h at time-points of day 1, 2, 3 and 7 (12 wells per time point in a plate). The assay was read with Benchmark Plus plate reader (BIO-RAD) at 570–595 nm and percent viability normalized to ethanol treated cells (negative control).Immunofluorescent staining
Cells were bioprinted in a 96 well plate and cultured for up to 7 days. At day 7, cells were fixed and permeabilized in 4% paraformaldehyde/0.1% Triton-X100/DPBS solution in the plate for 2 h at room temperature followed by blocking with 5% BSA/TBST overnight at 4 °C. After removing the blocking solution, cells were incubated with primary antibodies including anti-E-Cadherin (Cat. no. 14472, Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), anti-vimentin (Cat. no. 5741, Cell Signaling Technology, 1
500 dilution), anti-vimentin (Cat. no. 5741, Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), anti-MMP2 (Cat. no. ab37150, Abcam, 1
500 dilution), anti-MMP2 (Cat. no. ab37150, Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) and anti-MMP9 (Cat. no. ab38898, Abcam, 1
200 dilution) and anti-MMP9 (Cat. no. ab38898, Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) overnight at 4 °C. All antibodies were diluted with 5% BSA/TBST. Each well was rinsed twice with TBST then cells were incubated with Alexa Fluor 488 labelled secondary antibodies (anti-rabbit IgG (Cat. no. A11034, Invitrogen, 1
200 dilution) overnight at 4 °C. All antibodies were diluted with 5% BSA/TBST. Each well was rinsed twice with TBST then cells were incubated with Alexa Fluor 488 labelled secondary antibodies (anti-rabbit IgG (Cat. no. A11034, Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution) or anti-mouse IgG (Cat. no. A32723, Invitrogen, 1
500 dilution) or anti-mouse IgG (Cat. no. A32723, Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution)) overnight at 4 °C. For F-actin staining, bioprinted cells were incubated with phalloidin conjugated to Alexa Fluor® 568 (Cat. no. A12380, Life Technologies, 1
500 dilution)) overnight at 4 °C. For F-actin staining, bioprinted cells were incubated with phalloidin conjugated to Alexa Fluor® 568 (Cat. no. A12380, Life Technologies, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) on a shaker 2 h at room temperature. After washing with TBST twice, nuclei were counterstained with 0.2 mM Hoechst 33342 (Cat. no. 62249, ThermoFisher Scientific, stock concentration 20 mM) in DPBS for 10 minutes at room temperature. The 3D bioprinted cells were then imaged using the Leica TCS SP8 DLS (Digital LightSheet) confocal microscope. Images were taken using 10× objective, argon laser (458 and 488 nm) and DPSS 561 Laser (561 nm) (inset zoomed-in images: 2× or 4× zoom). Z-Stacks were defined from the bottom to the top of the 3D hydrogels and an image stack on the z-axis was taken per well.
200 dilution) on a shaker 2 h at room temperature. After washing with TBST twice, nuclei were counterstained with 0.2 mM Hoechst 33342 (Cat. no. 62249, ThermoFisher Scientific, stock concentration 20 mM) in DPBS for 10 minutes at room temperature. The 3D bioprinted cells were then imaged using the Leica TCS SP8 DLS (Digital LightSheet) confocal microscope. Images were taken using 10× objective, argon laser (458 and 488 nm) and DPSS 561 Laser (561 nm) (inset zoomed-in images: 2× or 4× zoom). Z-Stacks were defined from the bottom to the top of the 3D hydrogels and an image stack on the z-axis was taken per well.
Cell recovery from 3D bioprinted matrices
RASTRUM Cell Retrieval Solution was purchased from Inventia Life Sciences (Cat. no. F235). Cells were bioprinted with each hydrogel condition in a 96 well plate and cultured for 7 days. Cells were retrieved and pellets were collected according to manufacturer's instructions. Briefly, cells were rinsed with DPBS and incubated with 75 μl Cell Retrieval Solution for 20 min at 37 °C/5% CO2. Solution was collected after pipetting up and down in each well several times, then wells were washed with DPBS and contents were combined with collected cell suspension. Cells were pelleted at 1200 rpm and pellets rinsed twice with cold DPBS. Final cell pellets were snap frozen on dry ice and stored at −80 °C for further analysis.Real-time qPCR (qPCR)
Cells were bioprinted with each hydrogel condition in a 96 well plate and cultured for 7 days before cells were extracted using method described above. RNA was extracted from cell pellets using QIAGEN RNeasy Mini Kit (Cat. no. 74104) according to manufacturer's instructions. RNA concentration was determined using ThermoScientific Nanodrop 2000 Spectrophotometer quantification. RNA was reverse transcribed using MMLV reverse transcriptase (Life Technologies) and qPCR was performed with the 7900HT Fast Real-Time PCR System (Life Technologies) as previously described.37 Briefly, qPCR was set up using 2 μg cDNA and KAPA Probe Fast master mix (Roche, Cat. no. KK4705). For target genes, TaqMan™ Gene Expression Assay probes for CDH1 (Hs01023895_m1), VIM (Hs00185584_m1), MMP2 (Hs01548727_m1) and MMP9 (Hs00957562_m1) were used to measure gene expression levels and normalized against GAPDH housekeeping gene (Hs02786624_g1). For each target gene, expression level was quantified in relation to the expression of a control gene using the ΔΔCt method to provide relative quantification. Gene expression values normalized to each control gene were calculated as the average of the expression value for each target gene.Western blot analysis
Western blot was performed as previous described.38 Briefly, lysates were made from cell pellets using RIPA buffer containing 1 mM EDTA and 10% Protease Inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured using Pierce™ BCA Protein assay (ThermoFisher Scientific, Cat. no. 23225) as per manufacturer's instructions. 20 μg protein was run on 4–15% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad, Cat. no. 4568085) before being transferred to nitrocellulose membranes. Antibodies include anti-E-cadherin (Cat. no. 14472, Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) and anti-vimentin (Cat. no. 5741, Cell Signaling Technology, 1
1000 dilution) and anti-vimentin (Cat. no. 5741, Cell Signaling Technology, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), anti-MMP2 (Cat. no. ab37150, Abcam, 1
1000 dilution), anti-MMP2 (Cat. no. ab37150, Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), anti-MMP9 (Cat. no. ab38898, Abcam, 1
1000 dilution), anti-MMP9 (Cat. no. ab38898, Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), GAPDH (Cat. no. ab8245, Abcam, 1
1000 dilution), GAPDH (Cat. no. ab8245, Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution) primary antibodies and anti-rabbit-HRP (Cat. no. P0448, DAKO, 1
000 dilution) primary antibodies and anti-rabbit-HRP (Cat. no. P0448, DAKO, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution) and anti-mouse (Cat. no. P0447. DAKO, 1
5000 dilution) and anti-mouse (Cat. no. P0447. DAKO, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution) secondary antibodies. All antibodies were diluted in 5% skim milk/TBST. Signals were detected using Clarity ECL Western Blotting Substrate (BIO-RAD).
5000 dilution) secondary antibodies. All antibodies were diluted in 5% skim milk/TBST. Signals were detected using Clarity ECL Western Blotting Substrate (BIO-RAD).
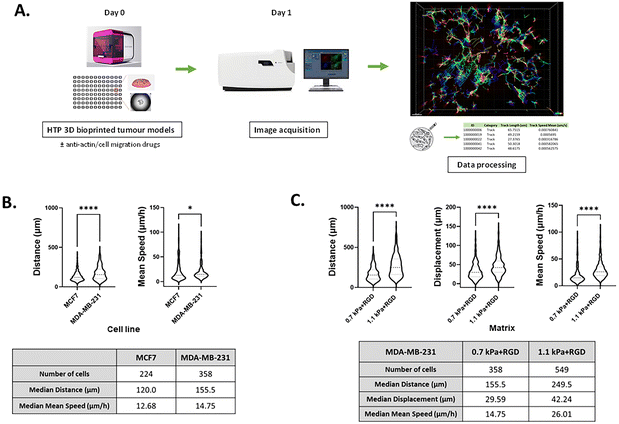
Live cell tracking
Cells were bioprinted with each hydrogel condition in a glass-bottom 96 well plate. The plates were incubated at 5% CO2 and 37 °C for 3 h prior to adding a ROCK inhibitor, Y-27632 (Cat. no. 72302, StemCell Technologies) and global myosin inhibitor, blebbistatin (Cat. no. B0560, Sigma-Aldrich), which were made up to 10 mM in DMSO. The drugged plates were then incubated for 24 h at 37 °C/5% CO2. Incucyte® Nuclight Rapid Red Dye (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in culture media, Sartorius, Cat. no. 4717) was added with fresh drugged media 1 h prior to cell tracking. Control samples for track analysis were treated with 80% ethanol for cell death to correct for non-cell movement. Z-Stack images were taken every 15 min for 24 h with a height of 300 μm using Zeiss CellDiscoverer 7 microscope (bright field and red fluorescence channel at 10× objective). The microscope incubator was set to 37 °C and 5% CO2. Cell tracking analyses were quantified in Imaris software (Imaris, RRID:SCR_007370, version 9.5.1, Bitplane AG, Zurich, Switzerland). The cells at each time point were detected from the fluorescence channel. Cells with diameter equal or greater than 10 μm were detected with 60 μm point-spread function modelled along z-axis. The detected cells were then tracked using autoregressive motion algorithm. Imaris calculated distance, displacement, and mean speed measured between first and last positions of the tracked cells. A minimum of 95 cell tracks per well were included in the analysis. The distance and mean speed values of live cells below 75% of the control samples for track analysis were considered to be artifacts and excluded in the analysis. The details of calculated values from Imaris software are described below.
1000 in culture media, Sartorius, Cat. no. 4717) was added with fresh drugged media 1 h prior to cell tracking. Control samples for track analysis were treated with 80% ethanol for cell death to correct for non-cell movement. Z-Stack images were taken every 15 min for 24 h with a height of 300 μm using Zeiss CellDiscoverer 7 microscope (bright field and red fluorescence channel at 10× objective). The microscope incubator was set to 37 °C and 5% CO2. Cell tracking analyses were quantified in Imaris software (Imaris, RRID:SCR_007370, version 9.5.1, Bitplane AG, Zurich, Switzerland). The cells at each time point were detected from the fluorescence channel. Cells with diameter equal or greater than 10 μm were detected with 60 μm point-spread function modelled along z-axis. The detected cells were then tracked using autoregressive motion algorithm. Imaris calculated distance, displacement, and mean speed measured between first and last positions of the tracked cells. A minimum of 95 cell tracks per well were included in the analysis. The distance and mean speed values of live cells below 75% of the control samples for track analysis were considered to be artifacts and excluded in the analysis. The details of calculated values from Imaris software are described below.
Distance refers to the total length of movement of a cell along a track and was calculated using eqn (1):
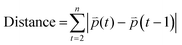 | (1) |
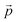 refers to the three-dimensional position coordinates x, y, and z, and t refers to time point. Displacement refers to the length between the first and the last position of the cell along the same track (eqn (2)).
refers to the three-dimensional position coordinates x, y, and z, and t refers to time point. Displacement refers to the length between the first and the last position of the cell along the same track (eqn (2)).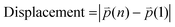 | (2) |
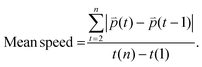 | (3) |
The difference between three-dimensional position coordinates is calculated using eqn (4).
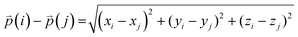 | (4) |
Statistical analysis
Statistical analyses were performed using the GraphPad Prism v9.0 software (GraphPad Software). For the cell movement analysis, unpaired, two-tailed Mann–Whitney tests were used to determine statistical differences between two different groups (cell types or gel conditions). For comparison of multiple samples, Kruskal–Wallis one-way analysis with a post hoc Dunn test was used.Results & discussion
Design of HTP bioprinting platform with tunable hydrogels
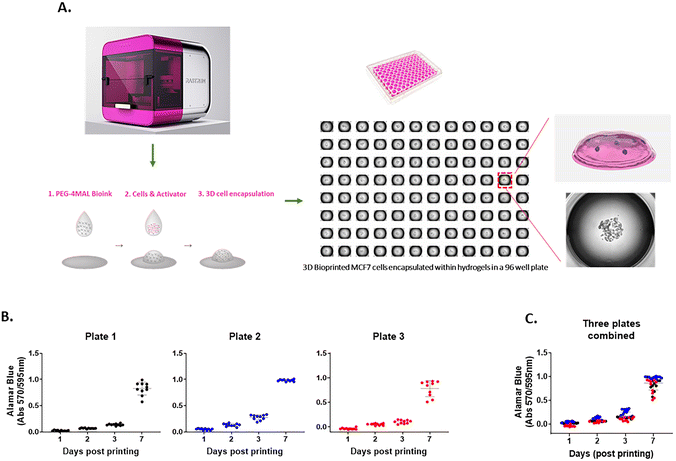
The 3D cell-laden hydrogel constructs were printed in multi-well plates via a two-droplet bioprinting process in which the first droplets containing PEG-4MAL bioink were printed, followed by printing of the second droplets of the bis-thiol activator mixed with cancer cells onto the bioink droplet for instant hydrogel formation as previously described.27 The process facilitated the generation of simple and highly reproducible 3D cancer models to study cell movement in well-defined ECM environments in a HTP manner. We demonstrated using the MCF7 breast cancer cell line that the PEG-4MAL hydrogel could successfully encapsulate cells as part of the bioprinting process and provide a platform for a stable 3D culture (Fig. 1A). To assess the accuracy of the printing platform, we repeatedly printed MCF7 cells in 96 well plates and measured the cell growth for up to 7 days using the Alamar Blue metabolic assay. For each time point, there was 10 replicates per plate and similar absorbance values for each replicate were obtained, confirming the high repeatability in one print run (Fig. 1B). Three independent print runs of the MCF7 cells were performed. We found that the absorbance readings of cell metabolic activity for technical replicates were very similar (Fig. 1C), indicating that the HTP bioprinting platform can print and encapsulate cells in the 3D hydrogels with high accuracy and reproducibility. Furthermore, by taking advantage of the HTP 3D bioprinting technology, the platform enables one print run to generate up to 3 different tumor models (i.e., different cell types and/or different hydrogel combinations) in the same multi-well plate with many replicates, enabling the testing of several cell types and hydrogels concurrently (ESI Fig. 1†). Altogether, these suggest that the bioprinting platform can produce various cell-laden ECM structures in multi-well plates in a rapid, highly reproducible and less-labor intensive way, offering a better alternative to traditional manual pipetting.Cytocompatibility of 3D bioprinted hydrogels
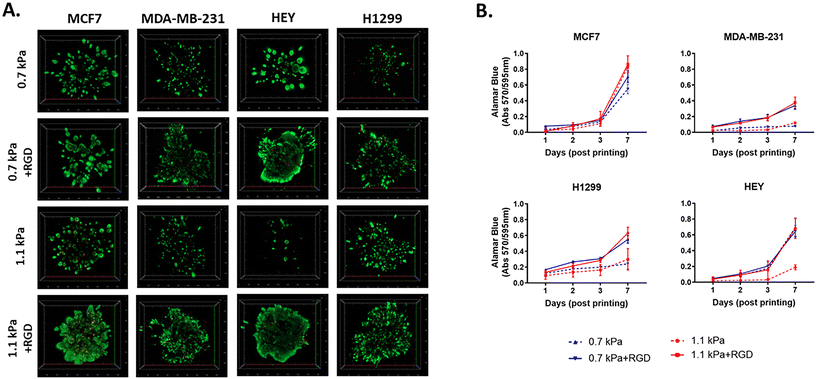
Next, to investigate the biocompatibility of the bioprinted hydrogels, we selected 4 different PEG-4MAL hydrogel combinations, which exhibited two different levels of stiffness, 0.7 kPa and 1.1 kPa. Each PEG-4MAL bioink was decorated with or without RGD (arginine–glycine–aspartic acid) cell adhesion peptides39,40 and crosslinked with MMP (matrix metalloproteinase)-cleavable activator: 0.7 kPa, 0.7 kPa + RGD, 1.1 kPa, and 1.1 kPa + RGD hydrogels. The stiffness values of 0.7 and 1.1 kPa rheology storage modulus values are equivalent to Young's modulus values (E) of approximately 1.5 and 3.2 kPa respectively as calculated using the formula previously reported.41 While the PEG-4MAL based hydrogel system can be expanded to higher mechanical stiffness by increasing concentration and/or reducing the polymer molecular weight as previously reported,42 we selected 0.7 kPa and 1.1 kPa as they are close to the stiffness levels of malignant breast tumors (0.2–2.5 kPa(E)43,44) and ovarian cancers (∼3 ± 2.5 kPa(E)45), indicating that the hydrogels were tuned in a biomimetic way. The addition of MMP-sensitive sites in the hydrogel systems enables cells to migrate and invade through the hydrogels. These four hydrogel combinations were thus chosen to demonstrate the effect of mechanical (stiffness) and biological molecules (the cell adhesion peptides) of a tumor-like microenvironment on cell responses (growth and migration). As cell models, we chose four epithelial cancer cell lines that are known to have differing migratory and invasive properties, including two variants of breast cancer cell lines, MCF7 (basal-like; non-invasive) and MDA-MB-231 (triple-negative; invasive), and two other invasive cell lines, HEY (high-grade serous ovarian cancer) and H1299 (metastatic lung cancer).To determine cell viability, each cancer cell line was bioprinted and encapsulated within the hydrogel systems in 96 well plates. The initial effect of the bioprinting on cell viability was tested using MCF7 cells. Post-bioprinting, cells were incubated at 37 °C for 2 h and the majority of MCF7 cells were viable, confirming that the bioprinting process has little effect on cell survival (ESI Fig. 2†). When cultured for 7 days post-printing, all four cell lines remained highly viable in those four hydrogel conditions (Fig. 2A). While MCF7 non-invasive breast cancer cells appeared to grow similarly in all 4 hydrogel conditions, MDA-MB-231, an invasive variant of breast cancer, was shown to be highly proliferative only in the presence of the cell adhesion peptides, RGD, irrespective of the different stiffness of the hydrogels (Fig. 2B). Similar to MDA-MB-231, H1299, an invasive lung cancer cell displayed high metabolic activity in the hydrogels incorporated with RGD, 0.7 kPa + RGD or 1.1 kPa + RGD hydrogels. In contrast, the growth of HEY cells, derived from aggressive high-grade serous ovarian cancer, was observed in both 0.7 kPa hydrogels with or without RGD, while in the 1.1 kPa hydrogels, the presence of RGD seemed to be vital for their growth (Fig. 2B). Altogether, these data suggest that while bioprinted PEG-4MAL hydrogel systems are highly cytocompatible, and each cancer cell type requires distinct matrix components for their growth.
Morphology of cancer cells encapsulated in ECM-like hydrogels
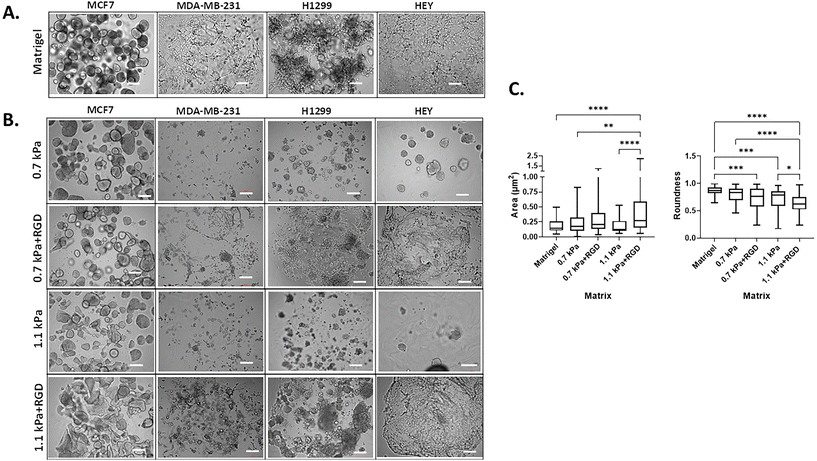
3D cell morphology can be used to define cellular behavior and function and predict the malignant potential of cells.46,47 Thus, we next determined the impact of the matrix conditions on cell morphology by modulating hydrogel stiffness and/or adhesion peptides. Cells were bioprinted and encapsulated within the hydrogels, which were optimal for their growth as determined in Fig. 2. In parallel, cells were encapsulated within Matrigel, a widely used biomaterial, as a reference control of their 3D morphology. As expected, the two variants of breast cancer cells showed distinct morphology when cultured within Matrigel. MCF7 cells were found to form multiple spheroids in Matrigel, whereas the MDA-MB-231 cells invaded through the Matrigel and exhibited stellate and protrusion morphology at day 7 (Fig. 3A, Movies 1 and 2†), in line with previous observations.46 When MCF7 cells were bioprinted in any of the four hydrogels, cells formed spherical structures and were predominantly proliferative from a single colony rather than migratory (Fig. 3B, Movie 3†). Interestingly, we found that the spheroid morphology appeared to vary between the hydrogel conditions, in terms of their size and roundness, a measure of how close the shape of the 2D spheroid image approaches a circle.48 In the presence of ECM mimics that have conjugated RGD peptides (0.7 kPa + RGD and 1.1 kPa + RGD hydrogels), the generated MCF7 3D models displayed spherical structures that were larger in size, irregular, and had less-round shapes (Fig. 3B). In contrast, the mean size and roundness of 3D bioprinted MCF7 models in the absence of RGD peptides were similar to those obtained with Matrigel (size: 0.18 ± 0.10 μm2; roundness 0.86 ± 0.089), especially the spheroids generated with 0.7 kPa hydrogel (size: 0.23 ± 0.18 μm2; roundness: 0.80 ± 0.12) (Fig. 3C). This suggests that despite the similar proliferation rate of MCF7 cells in all hydrogel conditions, the mechanical and biological characteristics of the matrix can affect cell morphology. MDA-MB-231 cells bioprinted in 0.7 kPa + RGD or 1.1 kPa + RGD hydrogels where the cells were highly proliferative (Fig. 2B) also showed the protrusion and network forming morphology and appeared to migrate through the hydrogels in a similar way to that observed in Matrigel (Fig. 3A and B, Movies 2 and 4†). Moreover, the morphology seemed not to be affected by the hydrogel stiffness. In addition to the metastatic breast cancer cells, a similar morphologic pattern was observed in metastatic lung cancer (H1299) and ovarian cancer (HEY) cell lines (Fig. 3B). The bioprinted 3D models for both H1299 and HEY cells had similar morphology in 0.7 kPa + RGD and 1.1 kPa + RGD hydrogels as they did in Matrigel. Yet, when HEY cells were bioprinted in 0.7 kPa hydrogels without RGD peptide, the cells displayed spheroid-like morphology (Fig. 3B). Hence, these indicate the matrix components, such as cell adhesion molecules can alter morphology of cells grown on the hydrogels.Expression of metastasis relevant genes and proteins in 3D bioprinted cancer cells
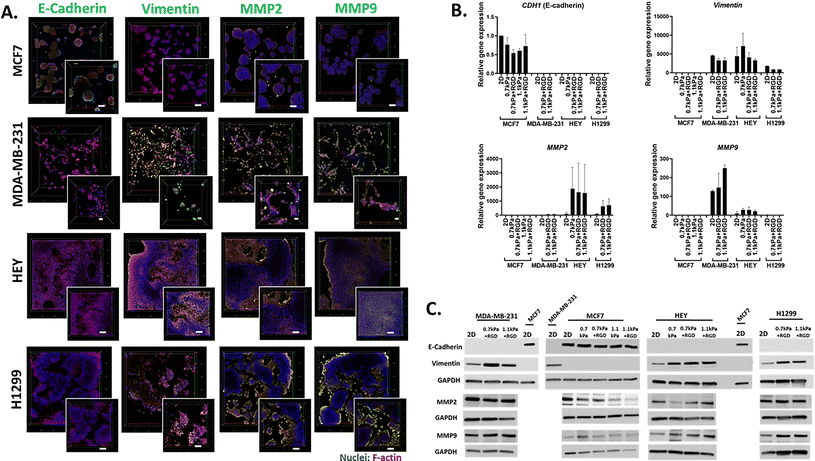
We next demonstrated the suitability of this platform to be used for the analysis of phenotypic markers relevant to cell migration and invasion in 3D cancer models. Epithelial-to-mesenchymal transition (EMT) has been implicated in cancer invasion and progression, which is associated with a loss of epithelial markers such as E-cadherin and a gain of mesenchymal markers such as vimentin.35,49,50 Bioprinted 3D models of each cell line in their optimized hydrogel conditions were cultured for 7 days, and then subjected to in situ immunofluorescent staining for the simultaneous detection of several proteins involved in cell migration including EMT process (E-cadherin and vimentin) as well as ECM-remodeling (MMP2 and MMP9). We confirmed that E-cadherin was predominantly expressed in MCF7 cells, while positive vimentin expression was detected in the invasive cancer cell lines, MDA-MB-231, H1299 and HEY, showing that the EMT phenotypic markers were retained in the 3D bioprinted cell models (Fig. 4A). Interestingly, while positive expression of MMP2 and MMP9 was found in all 3D bioprinted models, the expression of both MMP2 and MMP9 appeared to be more prominent in the invasive cell lines, MDA-MB-231, H1299 and HEY (Fig. 4A). This in situ analysis demonstrated that the MMP2 and MMP9 proteases were localized on the surface of invasive cancer cells, which may facilitate their proteolytic activation that in turn affects various cell functions, such as proliferation and migration.51,52 Therefore, increased production of MMPs may allow cells to remodel the PEG-MMP synthetic environment, migrate, and ultimately deposit their own ECM much like they do in vivo.53,54Having shown the qualitative expression of migration and invasion associated proteins in the hydrogel embedded cells, we next sought to investigate the quantitative expression of the relevant genes and proteins. Due to the presence of MMP-sensitive peptides within the PEG-4MAL hydrogel system, cells can be readily retrieved from the hydrogels via proteolytic degradation. Using this hydrogel feature, we next determined the versatility of the bioprinting platform for detailed molecular and protein analysis. Cells bioprinted in 96 well plates were recovered from the hydrogels through enzymatic approaches after 7 days in culture. Expression levels of metastasis relevant genes and proteins in the cells recovered from hydrogels was determined using qPCR and western blotting, respectively. MCF7 cells recovered from all 4 hydrogel conditions exhibited similar levels of E-cadherin (CDH1 gene) gene and protein expression to their 2D culture counterpart (Fig. 4B and C). As expected, vimentin was not expressed at the gene or protein levels in the MCF7 cells (Fig. 4B and C). On the contrary, all the invasive breast, lung and ovarian cancer cells (MDA-MB-231, H1299 and HEY respectively) showed expression of vimentin but no E-cadherin expression in any hydrogel conditions (Fig. 4B and C). Additionally, consistent with the in situ immunofluorescence image analysis (Fig. 4A), gene expression of the ECM-remodeling proteases was prominently detected in the invasive cell lines, MDA-MB-231, HEY cells and H1299 while neither MMP2 nor MMP9 mRNA expression was detectable in MCF7 cells (Fig. 4B). Despite the positive protein staining of MMP2 and MMP9 (Fig. 4A), only MMP2 mRNA expression was detected in H1299 cells (Fig. 4B). However, both MMP2 and MMP9 proteins were found to be positively expressed in all four bioprinted cell lines (Fig. 4C), suggesting that mRNA levels are not always proportional to protein levels.55 Altogether, these show that this 3D bioprinting platform employing PEG-4MAL hydrogel system can be a simple and versatile approach to be routinely used for molecular and in situ analysis to study cancer cell migration and invasion.
3D bioprinted platform for cell movement tracking in real-time
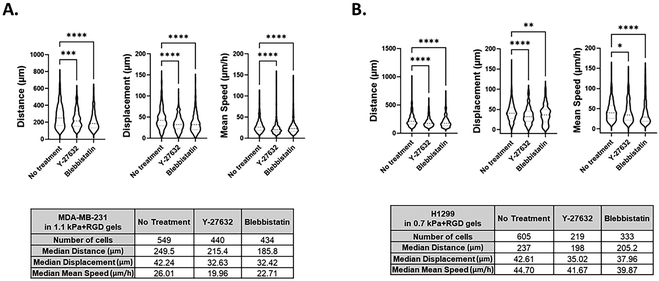
Although several methods have been developed for visualizing and analyzing cell migration in a 2D setting, no simple and straightforward cell tracking approach to quantify 3D cell movement has been established. Having demonstrated that invasive cancer cells retained their mesenchymal characteristics in the bioprinted any of the four hydrogels, we next sought to validate the potential of our bioprinting platform to be used as a methodology for studying cancer cell migration and invasion. To measure the dynamic movement of single cells within the bioprinted hydrogels, nuclei-labelled live cells were monitored for a period of 24 h on day 1 of post-printing using an automated widefield microscopic imaging system and then analyzed using object tracking. We quantified migratory properties including track length, displacement and mean speed of cell movement (Fig. 5A). Interestingly, while for the invasive cancer cell lines the effect of the matrix stiffness on cell growth and morphology was trivial, we observed that the migration of those cells was promoted in the stiffer bioprinted matrix. When comparing cell movement between the two breast cancer cell variants, MDA-MB-231 exhibited greater migratory behavior with a longer track length (P < 0.0001) and faster movement (P < 0.05) than MCF7 cells within the same hydrogel condition, 0.7 kPa + RGD (Fig. 5B). In MDA-MB-231 cells, the hydrogel stiffness impacted migration behavior. Greater migratory behavior of MDA-MB-231 cells was observed within the stiffer hydrogels (1.1 kPa + RGD) compared to the softer hydrogel system, 0.7 kPa + RGD (Fig. 5C). These data are in line with previous studies which reported that increasing matrix stiffness has been shown to induce malignant phenotypes.43,56 These findings demonstrate that the mechanical property of the hydrogels can affect cell migration behaviors.3D bioprinted cells as a screening platform for pharmacological inhibitors of migration and invasion
We further determined the feasibility of our 3D bioprinting platform as a preclinical model for anti-metastatic drug testing. Cancer cells can sense matrix stiffening through integrins that regulate cell migration via the activation of various signaling pathways, including the Rho/ROCK pathway.56,57 Here, we tested the effect of two known pharmacological inhibitors of cell migration, Y-27632 (a ROCK inhibitor) and blebbistatin (a global myosin inhibitor) on cell migratory behaviors within one of the hydrogels, 1.1 kPa + RGD. Once the 3D tumor models were bioprinted in 96 well plates, cells in each well were treated with either of the drugs for 48 h and tracked from 24 h to 48 h after printing. Using MDA-MB-231 cells, we successfully monitored 3D movement of cells treated with different drugs simultaneously and quantitate their track distance, displacement and mean speed of cell movement. As shown in Fig. 6A and Movies 5–7,† 3D migration of MDA-MB-231 cells was significantly impeded by the ROCK inhibitor (track distance P < 0.001; displacement P < 0.0001; mean speed P < 0.0001) or blebbistatin (P < 0.0001 for all parameters) compared to no drug-treated cells in 1.1 kPa + RGD hydrogel. The significant inhibitory effects on MDA-MB-231 cell movement were also observed in the softer gels (0.7 kPa + RGD; ESI Fig. 3†). For further validation, another invasive cancer cell line, H1299 bioprinted with 0.7 kPa + RGD hydrogels, was treated with either of the inhibitors. As observed in MDA-MB-231 cells, suppression of H1299 cell movement upon drug treatment could be monitored and quantitated using the platform (Fig. 6B and Movies 8–10†). Overall, the 3D bioprinted platform we propose here cannot only accurately measure cell migratory behaviors within the 3D matrices but also has application as a HTP preclinical anti-metastasis drug testing platform. A recent study has developed a HTP bioprinting platform using GelMA hydrogels and a digital light processing-based system.24 Similar to the bioprinting platform presented in this study, their platform was capable of printing for HTP in situ fabrication of up to 96 samples per batch. While application of the platform was demonstrated to be a drug-response assay in their study, we further advanced the biological applicability of the HTP bioprinted platform beyond drug screening, including in situ and detailed molecular analysis and real-time cell migration tracking. Further by taking advantage of the use of proteolytic degradable hydrogel systems in this platform, the downstream molecular analysis using retrieved cells can be performed to better understand the mechanisms of actions for novel drugs or inhibitors.Conclusions
3D bioprinting provides a unique approach to the fabrication of complex tissue constructs in vitro, yet the use of 3D bioprinted cell models for biological applications has not been fully explored. This 3D bioprinting platform presented herein exhibits several notable advantages over existing approaches for use in versatile biological applications; (1) the 3D bioprinting platform automates multi-well plate printing, thereby facilitating the generation of 3D cancer cell models in a simple, reproducible, viable and HTP manner. (2) The HTP 3D platform allows for the visualization of metastasis markers in situ and also for the simultaneous examination of cell movement and the effects of inhibitors. (3) The fine tunability of the hydrogel systems used in the bioprinting platform helps identify cell type specific matrix conditions. (4) Finally, the design of bioprinted hydrogels with sites for proteolytic breakdown enables cells to be retrieved from the hydrogels for downstream molecular analysis post-bioprinting. Therefore, we believe that this approach of combining the rapid HTP bioprinting platform and its biological applications offers the significant potential for better understanding cell migration and invasion processes and for the identification of novel anti-metastasis drugs.Author contributions
Conceptualisation: M. K., J. J. G., and M. J.; methodology: M. K., J. J. G., M. J., M. E., R. H. U., J. C. C. R., A. P. O. M., A. S, and A. V.; software: A. S. and A. P. O. M.; investigation: M. J., J. N. S., M. A. K. T., E. D.; writing – original draft: M. J.; writing – review & editing: M. K., J. J. G., and M. J.; visualisation: M. J. and J. N. S.; resources: J. C. C. R., M. E., R. H. U., A. V., A. P. O. M. and A. S.; funding acquisition: M. K., J. J. G. and J. C. C. R.Conflicts of interest
R. H. U., M. E., A. V., A. S., A. P. O. M. and J. C. C. R. are employees, shareholders, and/or optionees of Inventia Life Science Pty. Ltd. Inventia has an interest in commercializing the 3D bioprinting technology.Acknowledgements
This work was supported by the Children's Cancer Institute, which is affiliated with the University of New South Wales (UNSW Sydney), and the Sydney Children's Hospital Network and by grants from the Australian Research Council (ARC) Linkage Grant (LP170100623 to J. J. G., M. K., and J. C. C. R.); the National Health and Medical Research Council (NHMRC) Program Grant (APP1091261 to M. K. and J. J. G.); the NHMRC Principal Research Fellowship (APP1119152 to M. K.), the NHMRC Investigator (APP1196648 to J. J. G.), and grant support from the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology (CE140100036 to J. J. G. and M. K.). The authors would like to thank the Katharina Gaus Light Microscopy Unit, at the Mark Wainwright Analytical Center at UNSW Sydney for their support and resources involved in this work.References
- P. S. Steeg, Targeting metastasis, Nat. Rev. Cancer, 2016, 16(4), 201–218 CrossRef CAS PubMed.
- N. M. Novikov, S. Y. Zolotaryova, A. M. Gautreau and E. V. Denisov, Mutational drivers of cancer cell migration and invasion, Br. J. Cancer, 2021, 124(1), 102–114 CrossRef PubMed.
- P. Friedl and K. Wolf, Tumour-cell invasion and migration: diversity and escape mechanisms, Nat. Rev. Cancer, 2003, 3(5), 362–374 CrossRef CAS.
- M. Dhandapani and A. Goldman, Preclinical Cancer Models and Biomarkers for Drug Development: New Technologies and Emerging Tools, J. Mol. Biomarkers Diagn., 2017, 8(5), 356 Search PubMed.
- J. A. Hickman, R. Graeser, R. de Hoogt, S. Vidic, C. Brito and M. Gutekunst, et al., Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo, Biotechnol. J., 2014, 9(9), 1115–1128 CrossRef CAS PubMed.
- C. R. Justus, N. Leffler, M. Ruiz-Echevarria and L. V. Yang, In vitro cell migration and invasion assays, J. Visualized Exp., 2014,(88), 51046–51040 Search PubMed.
- S. Boyden, The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes, J. Exp. Med., 1962, 115(3), 453–466 CrossRef CAS.
- C. Zhang, M. P. Barrios, R. M. Alani, M. Cabodi and J. Y. Wong, A microfluidic Transwell to study chemotaxis, Exp. Cell Res., 2016, 342(2), 159–165 CrossRef CAS PubMed.
- G. M. Whitesides, The origins and the future of microfluidics, Nature, 2006, 442(7101), 368–373 CrossRef CAS PubMed.
- B. S. Wong, S. R. Shah, C. L. Yankaskas, V. K. Bajpai, P.-H. Wu and D. Chin, et al., A microfluidic cell-migration assay for the prediction of progression-free survival and recurrence time of patients with glioblastoma, Nat. Biomed. Eng., 2021, 5(1), 26–40 CrossRef CAS PubMed.
- T. Trantidou, Y. Elani, E. Parsons and O. Ces, Hydrophilic surface modification of PDMS for droplet microfluidics using a simple, quick, and robust method via PVA deposition, Microsyst. Nanoeng., 2017, 3(1), 16091 CrossRef CAS PubMed.
- K. C. Chaw, M. Manimaran, F. E. H. Tay and S. Swaminathan, Matrigel coated polydimethylsiloxane based microfluidic devices for studying metastatic and non-metastatic cancer cell invasion and migration, Biomed. Microdevices, 2007, 9(4), 597–602 CrossRef CAS PubMed.
- A. Morss Clyne, S. Swaminathan and A. Díaz Lantada, Biofabrication strategies for creating microvascular complexity, Biofabrication, 2019, 11(3), 032001 CrossRef PubMed.
- G. Benton, I. Arnaoutova, J. George, H. K. Kleinman and J. Koblinski, Matrigel: From discovery and ECM mimicry to assays and models for cancer research, Adv. Drug Delivery Rev., 2014, 79–80, 3–18 CrossRef CAS PubMed.
- M. J. Kratochvil, A. J. Seymour, T. L. Li, S. P. Paşca, C. J. Kuo and S. C. Heilshorn, Engineered materials for organoid systems, Nat. Rev. Mater., 2019, 4(9), 606–622 CrossRef CAS PubMed.
- Y. Fang and R. M. Eglen, Three-Dimensional Cell Cultures in Drug Discovery and Development, SLAS Discovery, 2017, 22(5), 456–472 CrossRef CAS.
- N. Peela, D. Truong, H. Saini, H. Chu, S. Mashaghi and S. L. Ham, et al., Advanced biomaterials and microengineering technologies to recapitulate the stepwise process of cancer metastasis, Biomaterials, 2017, 133, 176–207 CrossRef CAS.
- L. Moroni, J. A. Burdick, C. Highley, S. J. Lee, Y. Morimoto and S. Takeuchi, et al., Biofabrication strategies for 3D in vitro models and regenerative medicine, Nat. Rev. Mater., 2018, 3(5), 21–37 CrossRef CAS PubMed.
- C. Colosi, S. R. Shin, V. Manoharan, S. Massa, M. Costantini and A. Barbetta, et al., Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs Using Low-Viscosity Bioink, Adv. Mater., 2016, 28(4), 677–684 CrossRef CAS PubMed.
- H.-G. Yi, Y. H. Jeong, Y. Kim, Y.-J. Choi, H. E. Moon and S. H. Park, et al., A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy, Nat. Biomed. Eng., 2019, 3(7), 509–519 CrossRef CAS PubMed.
- H. Ding, N. P. Illsley and R. C. Chang, 3D Bioprinted GelMA Based Models for the Study of Trophoblast Cell Invasion, Sci. Rep., 2019, 9(1), 18854 CrossRef CAS.
- F. Meng, C. M. Meyer, D. Joung, D. A. Vallera, M. C. McAlpine and A. Panoskaltsis-Mortari, 3D Bioprinted In Vitro Metastatic Models via Reconstruction of Tumor Microenvironments, Adv. Mater., 2019, 31(10), 1806899 CrossRef PubMed.
- X. Zhou, W. Zhu, M. Nowicki, S. Miao, H. Cui and B. Holmes, et al., 3D Bioprinting a Cell-Laden Bone Matrix for Breast Cancer Metastasis Study, ACS Appl. Mater. Interfaces, 2016, 8(44), 30017–30026 CrossRef CAS PubMed.
- H. H. Hwang, S. You, X. Ma, L. Kwe, G. Victorine and N. Lawrence, et al., High throughput direct 3D bioprinting in multiwell plates, Biofabrication, 2021, 13(2), 025007 CrossRef CAS.
- S. Swaminathan, Q. Hamid, W. Sun and A. M. Clyne, Bioprinting of 3D breast epithelial spheroids for human cancer models, Biofabrication, 2019, 11(2), 025003 CrossRef CAS PubMed.
- M. A. Heinrich, R. Bansal, T. Lammers, Y. S. Zhang, R. Michel Schiffelers and J. Prakash, 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics, Adv. Mater., 2019, 31(14), 1806590 CrossRef.
- R. H. Utama, V. T. G. Tan, K. C. Tjandra, A. Sexton, D. H. T. Nguyen and A. P. O'Mahony, et al., A Covalently Crosslinked Ink for Multimaterials Drop-on-Demand 3D Bioprinting of 3D Cell Cultures, Macromol. Biosci., 2021, 2100125 CrossRef CAS.
- R. H. Utama, L. Atapattu, A. P. O'Mahony, C. M. Fife, J. Baek and T. Allard, et al., A 3D Bioprinter Specifically Designed for the High-Throughput Production of Matrix-Embedded Multicellular Spheroids, iScience, 2020, 23(10), 101621 CrossRef CAS PubMed.
- M. P. Lutolf, J. L. Lauer-Fields, H. G. Schmoekel, A. T. Metters, F. E. Weber and G. B. Fields, et al., Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(9), 5413–5418 CrossRef CAS PubMed.
- E. A. Phelps, N. O. Enemchukwu, V. F. Fiore, J. C. Sy, N. Murthy and T. A. Sulchek, et al., Maleimide Cross-Linked Bioactive PEG Hydrogel Exhibits Improved Reaction Kinetics and Cross-Linking for Cell Encapsulation and In Situ Delivery, Adv. Mater., 2012, 24(1), 64–70 CrossRef CAS PubMed.
- R. Cruz-Acuña, M. Quirós, S. Huang, D. Siuda, J. R. Spence and A. Nusrat, et al., PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery, Nat. Protoc., 2018, 13(9), 2102–2119 CrossRef.
- S. Ravi, V. R. Krishnamurthy, J. M. Caves, C. A. Haller and E. L. Chaikof, Maleimide–thiol coupling of a bioactive peptide to an elastin-like protein polymer, Acta Biomater., 2012, 8(2), 627–635 CrossRef CAS PubMed.
- M. Moser, T. Behnke, C. Hamers-Allin, K. Klein-Hartwig, J. Falkenhagen and U. Resch-Genger, Quantification of PEG-Maleimide Ligands and Coupling Efficiencies on Nanoparticles with Ellman's Reagent, Anal. Chem., 2015, 87(18), 9376–9383 CrossRef CAS.
- C.-Y. Liu, H.-H. Lin, M.-J. Tang and Y.-K. Wang, Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation, Oncotarget, 2015, 6(18), 15966–15983 CrossRef PubMed.
- L. S. Havel, E. R. Kline, A. M. Salgueiro and A. I. Marcus, Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity, Oncogene, 2015, 34(15), 1979–1990 CrossRef CAS PubMed.
- V. Swaminathan, K. Mythreye, E. T. Brien, A. Berchuck, G. C. Blobe and R. Superfine, Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines, Cancer Res., 2011, 71(15), 5075 CrossRef CAS PubMed.
- M. Jung, A. J. Russell, B. Liu, J. George, P. Y. Liu and T. Liu, et al., A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers, Cancer Res., 2017, 77(4), 971 CrossRef CAS PubMed.
- J. Gao, M. Jung, C. Mayoh, P. Venkat, K. M. Hannan and J. I. Fletcher, et al., Suppression of ABCE1-Mediated mRNA Translation Limits N-MYC–Driven Cancer Progression, Cancer Res., 2020, 80(17), 3706 CrossRef CAS PubMed.
- S. P. Singh, M. P. Schwartz, J. Y. Lee, B. D. Fairbanks and K. S. Anseth, A peptide functionalized poly(ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration, Biomater. Sci., 2014, 2(7), 1024–1034 RSC.
- M. D. Pierschbacher and E. Ruoslahti, Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule, Nature, 1984, 309(5963), 30–33 CrossRef CAS PubMed.
- S. R. Polio, A. N. Kundu, C. E. Dougan, N. P. Birch, D. E. Aurian-Blajeni and J. D. Schiffman, et al., Cross-platform mechanical characterization of lung tissue, PLoS One, 2018, 13(10), e0204765 CrossRef.
- S. Lee, X. Tong and F. Yang, The effects of varying poly(ethylene glycol) hydrogel crosslinking density and the crosslinking mechanism on protein accumulation in three-dimensional hydrogels, Acta Biomater., 2014, 10(10), 4167–4174 CrossRef CAS PubMed.
- I. Acerbi, L. Cassereau, I. Dean, Q. Shi, A. Au and C. Park, et al., Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration, Integr. Biol., 2015, 7(10), 1120–1134 CrossRef CAS PubMed.
- K. R. Levental, H. Yu, L. Kass, J. N. Lakins, M. Egeblad and J. T. Erler, et al., Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling, Cell, 2009, 139(5), 891–906 CrossRef CAS PubMed.
- T. I. R. Hopkins, V. L. Bemmer, S. Franks, C. Dunlop, K. Hardy and I. E. Dunlop, Micromechanical mapping of the intact ovary interior reveals contrasting mechanical roles for follicles and stroma, Biomaterials, 2021, 277, 121099 CrossRef CAS PubMed.
- G. Benton, H. K. Kleinman, J. George and I. Arnaoutova, Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells, Int. J. Cancer, 2011, 128(8), 1751–1757 CrossRef CAS PubMed.
- B. Weigelt and M. J. Bissell, Unraveling the microenvironmental influences on the normal mammary gland and breast cancer, Semin. Cancer Biol., 2008, 18(5), 311–321 CrossRef CAS PubMed.
- R. L. F. Amaral, M. Miranda, P. D. Marcato and K. Swiech, Comparative Analysis of 3D Bladder Tumor Spheroids Obtained by Forced Floating and Hanging Drop Methods for Drug Screening, Front. Physiol., 2017, 8, 605 CrossRef PubMed.
- J. Roche, The Epithelial-to-Mesenchymal Transition in Cancer, Cancers, 2018, 10(2), 52 CrossRef.
- T. Brabletz, R. Kalluri, M. A. Nieto and R. A. Weinberg, EMT in cancer, Nat. Rev. Cancer, 2018, 18(2), 128–134 CrossRef CAS PubMed.
- P. C. Brooks, S. Strömblad, L. C. Sanders, T. L. von Schalscha, R. T. Aimes and W. G. Stetler-Stevenson, et al., Localization of Matrix Metalloproteinase MMP-2 to the Surface of Invasive Cells by Interaction with Integrin αvβ3, Cell, 1996, 85(5), 683–693 CrossRef CAS.
- G. Murphy and H. Nagase, Localizing matrix metalloproteinase activities in the pericellular environment, FEBS J., 2011, 278(1), 2–15 CrossRef CAS.
- C. Bonnans, J. Chou and Z. Werb, Remodelling the extracellular matrix in development and disease, Nat. Rev. Mol. Cell Biol., 2014, 15(12), 786–801 CrossRef CAS PubMed.
- A. Malakpour-Permlid, I. Buzzi, C. Hegardt, F. Johansson and S. Oredsson, Identification of extracellular matrix proteins secreted by human dermal fibroblasts cultured in 3D electrospun scaffolds, Sci. Rep., 2021, 11(1), 6655 CrossRef CAS PubMed.
- A. Mehra, K. H. Lee and V. Hatzimanikatis, Insights into the relation between mRNA and protein expression patterns: I. theoretical considerations, Biotechnol. Bioeng., 2003, 84(7), 822–833 CrossRef CAS PubMed.
- F. Kai, H. Laklai and V. M. Weaver, Force Matters: Biomechanical Regulation of Cell Invasion and Migration in Disease, Trends Cell Biol., 2016, 26(7), 486–497 CrossRef CAS PubMed.
- P. Costa, T. M. E. Scales, J. Ivaska and M. Parsons, Integrin-Specific Control of Focal Adhesion Kinase and RhoA Regulates Membrane Protrusion and Invasion, PLoS One, 2013, 8(9), e74659 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm00651k |
| This journal is © The Royal Society of Chemistry 2022 |