Open Access Article
Open Access ArticleArticulation inspired by nature: a review of biomimetic and biologically active 3D printed scaffolds for cartilage tissue engineering
Donagh G.
O'Shea
 ac,
Caroline M.
Curtin
ac,
Caroline M.
Curtin
 abc and
Fergal J.
O'Brien
abc and
Fergal J.
O'Brien
 *abc
*abc
aTissue Engineering Research Group, Department of Anatomy and Regenerative Medicine, RCSI University of Medicine and Health Sciences, Dublin, Ireland. E-mail: fjobrien@rcsi.ie
bTrinity Centre for Biomedical Engineering, Trinity College Dublin, Ireland
cAdvanced Materials and Bioengineering Research Centre (AMBER), RCSI and TCD, Dublin, Ireland
First published on 22nd March 2022
Abstract
In the human body, articular cartilage facilitates the frictionless movement of synovial joints. However, due to its avascular and aneural nature, it has a limited ability to self-repair when damaged due to injury or wear and tear over time. Current surgical treatment options for cartilage defects often lead to the formation of fibrous, non-durable tissue and thus a new solution is required. Nature is the best innovator and so recent advances in the field of tissue engineering have aimed to recreate the microenvironment of native articular cartilage using biomaterial scaffolds. However, the inability to mirror the complexity of native tissue has hindered the clinical translation of many products thus far. Fortunately, the advent of 3D printing has provided a potential solution. 3D printed scaffolds, fabricated using biomimetic biomaterials, can be designed to mimic the complex zonal architecture and composition of articular cartilage. The bioinks used to fabricate these scaffolds can also be further functionalised with cells and/or bioactive factors or gene therapeutics to mirror the cellular composition of the native tissue. Thus, this review investigates how the architecture and composition of native articular cartilage is inspiring the design of biomimetic bioinks for 3D printing of scaffolds for cartilage repair. Subsequently, we discuss how these 3D printed scaffolds can be further functionalised with cells and bioactive factors, as well as looking at future prospects in this field.
1. Introduction
“Innovation inspired by nature” – this is how biomimicry is described by author and scientist Janine M. Benyus in her 1997 book of the same title and although a relatively new term, the concept of biomimicry is evident throughout the history of human innovation.1,2 From the study of birds to inspire the design of aeroplanes to the study of human anatomy to inspire next generation medical devices, mimicking nature has allowed us to create innovative new technologies and improve our own quality of life.But how does biomimicry apply to tissue engineering (TE)? TE, also known as regenerative medicine, is an emerging multi-disciplinary field of modern medicine which uses a combination of biomaterials, cells and bioactive factors or gene therapeutics, sometimes referred to as the ‘TE triad’, to bioengineer living tissues for a range of applications.3 These diverse applications range from disease modelling4 and organ-on-a-chip development,5 to the regeneration of a variety of tissue types including cardiac tissue,6 musculoskeletal tissue7 and skin,8 amongst others. In essence, the field of TE relies on the fabrication of biomaterial implants called ‘scaffolds’, which can support cell growth and whose microenvironment, architecture and functionality mimic that of native human tissue. Therefore, it could be argued that biomimicry forms the foundations of the field of TE.
In recent years, TE scaffolds have grown in popularity as a potential treatment option for a range of conditions. In the field of orthopaedic medicine, for example, these scaffolds have emerged as a promising treatment option for chondral defects (CDs), which are localised areas of damage to the articular cartilage of a synovial joint. In severe cases, the defect can penetrate into the underlying subchondral bone leading to the formation of an osteochondral defect.9 Once the friction-reducing articular cartilage tissue has worn away, the underlying bone surfaces can rub against one another causing significant stiffness and pain, thus hindering joint mobility.10 CDs can be caused by traumatic injury or wear and tear over time and can lead to the development of osteoarthritis, a degenerative joint disease which affects 9.6% of men and 18% of women over the age of 60 years worldwide.11 Unfortunately, due to the avascular and aneural nature of articular cartilage, these defects will not heal on their own.
Current treatment options for CDs include surgical procedures such as microfracture, autograft or allograft procedures, or cell-based techniques.12 Microfracture is a procedure whereby tiny fractures are made in the subchondral bone allowing the release of bone marrow stem cells into the defect which ultimately develop into fibrocartilage.13 However unlike articular cartilage, fibrocartilage is rich in collagen type I and thus possesses inferior mechanical properties. Autograft procedures, such as osteochondral autograft transfer systems (OATS) and mosaicplasty, involve transplanting osteochondral tissue from low load bearing regions of the knee into defects located in high load bearing regions.14,15 However the procedure is only suitable for smaller defects due to limited tissue availability at the donor site and can also be associated with donor site morbidity due to infection. Allografts are articular cartilage transplants taken from another donor and thus are not constrained by tissue availability at the donor site. However the availability of donors and the risk of the patient's immune system rejecting the graft limits the use of this procedure.16,17 Cell-based procedures such as autologous chondrocyte implantation (ACI) have shown promise and involve removing cells from healthy articular cartilage, expanding them in culture and then implanting the expanded chondrocytes into the chondral defect under a collagen membrane. However dedifferentiation of the chondrocytes during in vitro cell expansion can occur leading to a reduced capacity of the cells to lay down new ECM when implanted back into the defect.18 Therefore, despite providing much needed symptomatic relief to patients, current surgical procedures are not without limitations, often resulting in a variable healing response and the formation of non-durable tissue.16,19 This issue becomes even more prominent following surgical treatment of larger defects.20
Without successful intervention, the osteoarthritic joint can deteriorate to a point where total joint replacement is the only remaining option to relieve pain and discomfort. Total joint replacement involves complete removal of the arthritic joint and insertion of a prosthesis in its place. These prostheses are typically made from a metal such as a titanium alloy, polymers, ceramics or a composite.21 Although total joint replacement can result in dramatic improvements in patient quality of life following initial post-operative rehabilitation, revision surgery can be required over time due to implant failure. While the lifetime risk of requiring revision surgery is relatively low in patients aged over 70 years (1–6%), this risk is significantly higher in younger patients with 1 in 3 patients aged 50 to 55 years likely to require revision surgery. More than half of these revision surgeries are needed within 6 years of the initial joint replacement surgery.22 Therefore the benefits of this procedure need to be weighed against the potential risk of future surgeries and poor health outcomes, particularly for younger patients.
Several fabrication techniques have been investigated to engineer scaffolds which could be surgically implanted into these defects in order to support regrowth of cartilage and/or bone tissue. Examples of these techniques including electrospinning,23–29 solvent casting/melt moulding and particulate leaching,30–33 freeze drying34–36 and gas foaming techniques,37–42 amongst others. Scaffolds for bone and cartilage repair fabricated using the freeze drying method have been the subject of extensive research here in the RCSI Tissue Engineering Research Group (TERG). Using a controlled freeze drying cycle, our lab fabricates highly porous scaffolds from slurries of biomaterials native to the human body such as collagen type I, hyaluronic acid (HyA) and chondroitin sulfate (CS).43–47 The composition and stiffness of these scaffolds can be tailored to promote cartilage or bone regeneration as required.44
While significant progress in the field of TE for cartilage repair has been made using scaffolds fabricated via the techniques outlined above, the inability to mimic the complexity of native tissue has hindered the clinical translation of many products thus far.48,49 Fortunately, the development of 3D printed scaffolds has provided a potential solution. These scaffolds can be designed to mirror the complex zonal architecture of articular cartilage and can be reinforced with polymers to improve their mechanical strength. The biomaterial inks used to print these scaffolds can also be functionalised with cells such as mesenchymal stem cells (MSCs) or mature chondrocytes, bioactive factors such as growth factors, or gene therapeutics such as plasmid DNA (pDNA) or microRNA (miR) to promote cartilage growth.50–53 These functionalised biomaterial inks are also called ‘bioinks’.54 There are a number of 3D printing techniques used in this field including droplet-based,54–56 laser-based57,58 and extrusion-based methods.5 However, due to its versatility and compatibility with cells and a wide range of biomaterials, extrusion-based 3D printing is one of the most popular 3D printing methods in the field of TE for cartilage repair.59,60 Thus, this review will focus on the development of bioinks for extrusion-based 3D printing only.
In the field of 3D printing for cartilage repair, there is an ever growing emphasis placed on the importance of designing advanced biomimetic bioinks whose matrix composition reflects that of native articular cartilage. This biomimetic approach helps in the regeneration of functional tissue and minimises adverse reactions when the 3D printed scaffold is implanted in vivo. Therefore, the scope of this review will focus on how the architecture and composition of native articular cartilage is inspiring the design of biomimetic bioinks for extrusion-based 3D printing of scaffolds for cartilage repair.
2. Architecture and composition of articular cartilage
Articular cartilage performs two crucial functions in human synovial joints – the first is to facilitate frictionless movement of the joint, and the second is to withstand repeated compressive loading. Both the architecture and the composition of the tissue play a significant role in facilitating these functions. Therefore, a thorough understanding of the structure and both the biomaterial and cellular composition of native articular cartilage is crucial when designing biomimetic bioinks for cartilage repair.2.1. Architecture of articular cartilage
The extracellular matrix (ECM) of articular cartilage is largely composed of collagen (primarily type II), proteoglycans and water.61 This ECM is synthesised and maintained by highly specialised cells known as chondrocytes whose morphology, density and organisation varies with cartilage depth leading to the formation of three distinct zones within the articular cartilage.62 The uppermost superficial zone (10–20% articular cartilage thickness) is characterised by the presence of high density flattened chondrocytes embedded in an ECM with collagen fibres aligned parallel to the joint surface and the presence of the lubricant proteoglycan 4 (PRG4 or lubricin). The middle zone (40–60% articular cartilage thickness) consists of rounded chondrocytes within a more disorganised collagen matrix, while the deep zone (30–40% articular cartilage thickness) consists of large chondrocytes surrounded by a pericellular matrix consisting of collagen type VI, and thicker collagen fibres organised perpendicular to the joint surface.63,64 The deep zone is then underpinned by calcified cartilage and the subchondral bone. The structure of articular cartilage is outlined in Fig. 1 below.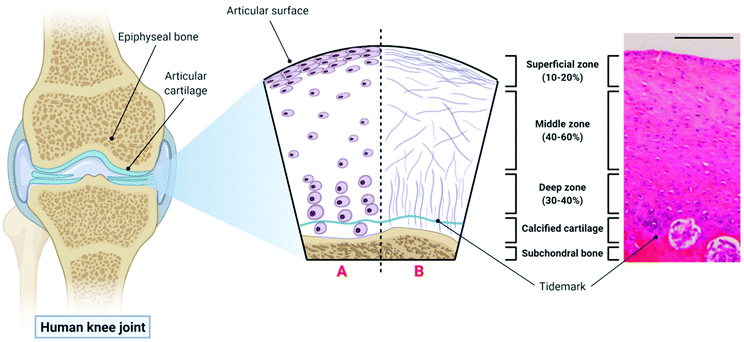 | ||
| Fig. 1 The structure of articular cartilage – the diagram on the left shows a synovial joint with a cross-sectional schematic depicting healthy articular cartilage: A – morphology and organisation of chondrocytes in the superficial, middle and deep zones respectively; B – orientation of collagen fibres in the superficial, middle and deep zones respectively. The histological image (haemotoxylin and eosin (H&E) staining) on the right is taken from the femoral condyle of a rabbit knee joint and demonstrates the zonal distribution of chondrocytes within articular cartilage (scale bar = 100 µm). Histological image reproduced and adapted from Matsiko et al.62 with permission from MDPI (Copyright © 2013, MDPI). Figure created with Biorender.com. | ||
2.2. Composition of articular cartilage
Collagen is a protein consisting of a triple helix of three polypeptide chains called α-chains, and is the primary macromolecule component found in the ECM of articular cartilage.65 There are many different types of collagen present in the human body however fibril-forming collagens, such as collagen types I and II, are by far the most abundant.66 Collagen type I, which is the main collagen type found in fibrocartilage, consists of two α1(I) chains and one α2(I) chain. In contrast, collagen type II, which is the primary collagen type found in articular cartilage, consists of three identical α1(II) chains which contain a higher proportion of hydroxylysine, glucosyl and galactosyl residues. These residues mediate interactions with surrounding proteoglycans in the ECM of articular cartilage.65 The tightly packed collagen type II and IX fibres found aligned parallel to the articulating surface in the superficial zone of articular cartilage are also largely responsible for its tensile properties.64Proteoglycans are the second largest macromolecule component of articular cartilage and consist of a protein core with glycosaminoglycan (GAG) side chains. GAGs are negatively charged polysaccharides and can be non-sulfated, such as HyA, or sulfated, such as CS or keratin sulfate. Aggrecan is the most abundant proteoglycan found in articular cartilage and interacts with HyA to form large negatively charged aggregates within the matrix of collagen type II fibrils.67 Water comprises 60–80% of the wet weight of cartilage and in this aqueous environment, aggrecan molecules become hydrated and swell. This swelling is resisted by the surrounding collagen fibrils, leading to the formation of an equilibrium between the swelling forces of aggrecan and the tensile forces of the collagen matrix.68,69 This is the mechanism by which articular cartilage withstands repeated compressive loading so effectively.
2.3. Chondrocytes – the resident cells of articular cartilage
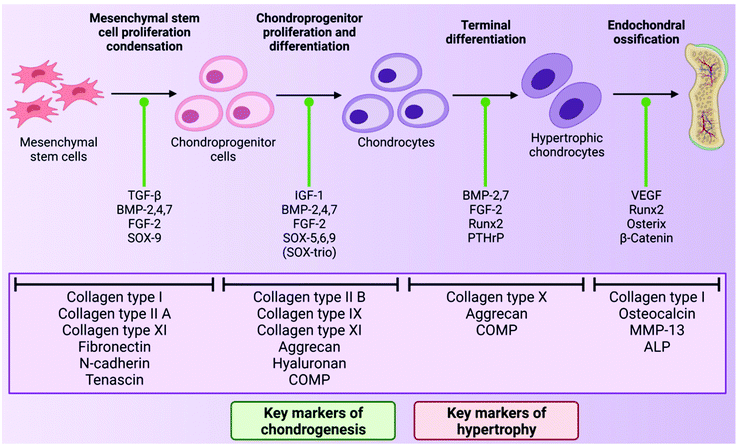
As mentioned above, chondrocytes are the primary cell type found in articular cartilage and play an integral role in the synthesis and maintenance of articular cartilage ECM. In the superficial and middle zones, chondrocytes primarily synthesise collagen types II, IX and XI and aggrecan, all of which are critical to the compressive and tensile properties of the tissue. However, in the deep zone, terminally differentiated chondrocytes mainly produce collagen type X and aggrecan.70 However, despite the importance of these cells, chondrocytes make up just 2% of the total cartilage tissue volume in a healthy adult and so are present in quite a small quantity.71 This sparse distribution means that very little cell–cell contact occurs within the tissue, therefore chondrocyte activity is largely dictated by interactions with the surrounding ECM.72During development, chondrocytes are formed following exposure of mesenchymal stem cells (MSCs) to certain biological cues in a process known as chondrogenesis. MSCs are a type of stem cell which are capable of differentiating into several connective tissue cell types including those from an adipogenic, osteogenic and chondrogenic lineage, depending on exposure to defined conditions.73,74 Chondrogenesis begins with proliferation and condensation of MSCs, leading to the initiation of cell–ECM and cell–cell interactions via gap junctions, and ultimately the formation of chondroprogenitor cells.75 Following this, chondroprogenitor cells differentiate into chondrocytes and this stage is characterised by the deposition of the cartilage ECM components collagen types II, IX and XI, and aggrecan.76 Production of these ECM components is a direct result of the expression of the genes Col2a1, Col9A1, Col11a1 and ACAN, and thus these genes are often considered to be markers of chondrogenesis in the field of TE for cartilage repair. This process is driven by a number of growth factors including bone morphogenetic proteins (BMPs), transforming growth factor-β1 and 3 (TGF-β1 and 3) and insulin-like growth factor-1 (IGF-1).77 The transcription factors, SOX-5, 6 and 9 (known as the SOX-trio), also play a key role in promoting chondrogenesis with SOX-9, in particular, directly influencing up-regulation of pro-chondrogenic genes such as Col2a1 and ACAN.78
In the formation of articular cartilage, the chondrogenic pathway stops here. However, chondrocytes can undergo terminal differentiation to form hypertrophic chondrocytes, which eventually leads to ossification of the tissue.79 This process is known as endochondral ossification and is the way in which long bones are formed during foetal development. These hypertrophic chondrocytes synthesise collagen type X (regulated by the gene Col10a1), and also express the enzymes matrix metalloproteinase-13 (MMP-13) and alkaline phosphatase (ALP), which are involved in the degradation of cartilage ECM and regulation of bone mineralisation respectively.76,80,81 Thus, the expression of Col10a1, MMP-13 and ALP are often considered markers of hypertrophy in cartilage TE.82 The transcription factor Runx2 is also known to promote chondrocyte hypertrophy, and the growth factor, vascular endothelial growth factor (VEGF), plays a key role in vascularisation of the tissue during endochondral ossification.83,84 In healthy articular cartilage, hypertrophic chondrocytes are found in the deep zone, near the border with the subchondral bone.64 The chondrogenic pathway is outlined in Fig. 2 below.
3. Biomimetic 3D printed scaffolds for cartilage repair
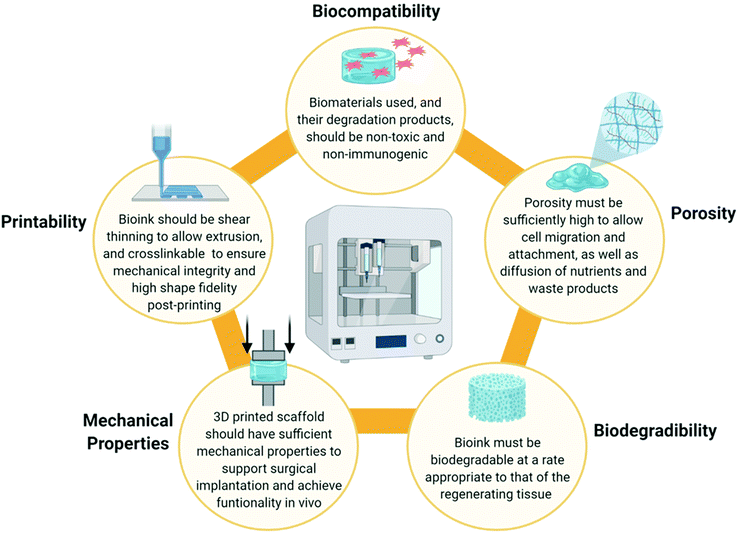
When designing a biomimetic 3D printed scaffold for cartilage repair, there are several key considerations which should be taken into account based on the biomaterial and cellular composition of native articular cartilage. Firstly, a highly hydrated environment is crucial not only to cell survival, but also to the biofunctionality of the tissue. From a 3D printing perspective, hydrogels are well placed to provide such an environment. Hydrogels are hydrophilic polymer networks which are extensively water swollen and are already widely employed as bioink components throughout the field of TE.85 Secondly, several biomaterials such as collagen, HyA and CS play a synergistic role in the unique mechanical and biological functions of articular cartilage and thus are desirable as bioink components. Thirdly, chondrocytes are required for tissue regeneration and thus the bioink could be functionalised with MSCs, chondroprogenitor cells or mature chondrocytes to accelerate healing. However, these cells must also be provided with appropriate biological cues in order for them to differentiate into, or maintain, a chondrogenic lineage and prevent hypertrophy.The physiochemical and rheological properties of the bioink used to develop the scaffold are also a key consideration. An ideal bioink should exhibit a decrease in viscosity during the printing process to allow extrusion (i.e. possess shear thinning properties), but also be capable of crosslinking immediately post-printing to retain its shape and ensure mechanical stability of the 3D printed construct.86,87 In addition to this, the 3D printed scaffold must also be sufficiently porous to facilitate the attachment, proliferation and differentiation of cells, as well as allowing diffusion of solutes and nutrients and deposition of ECM.54,88 This is often influenced by the polymer concentration or the crosslink density of the bioink. The bioink, and thus the 3D printed scaffold, must also be biodegradable at a rate appropriate to that of the regenerating tissue to allow for gradual replacement of the scaffold with deposited ECM from native cells.89 It is also critical that both the bioink and its degradation products are non-toxic and non-immunogenic.3 The ideal properties of a bioink are summarised in Fig. 3 below.
3.1. Mimicking the composition of articular cartilage when developing 3D printed scaffolds
Strategies for fabricating biomimetic 3D printed scaffolds for cartilage repair strive to maintain the synergistic biofunctionality of native biomaterials by using biomimetic bioink formulations with desirable physiochemical and rheological properties. This can be challenging as many biomaterials native to human articular cartilage do not innately possess good 3D printing properties. The following sections will look at how native biomaterials are being adapted to formulate biomimetic bioinks for cartilage repair with favourable 3D printing properties.One such approach involves controlling the physical crosslinking of COL-I hydrogels via the manipulation of hydrogel concentration, pH and temperature. Traditionally, COL-I hydrogels were formulated at lower concentrations of <10 mg mL−1 in acetic acid; however, these hydrogels were not cell friendly and they possessed very poor mechanical properties.93 However, more recent studies have been successful in formulating high concentration (up to 20 mg mL−1), neutralised COL-I hydrogels by carefully adjusting and buffering the pH of the formulation to physiological pH and salt concentration.94 Very high concentration COL-I hydrogels are also now available as commercial bioink formulations such as Lifeink® 200 (Advanced BioMatrix, CA, USA) which contains 35 mg mL−1 COL-I, or Viscoll (Imtek Ltd, Russia) which contains up to 80 mg mL−1 COL-I.95,96 These high concentration COL-I bioinks possess better 3D printing properties and mechanical properties than lower concentration COL-I formulations, resulting in greater shape fidelity of the printed construct and they can also support 3D bioprinting of cells.97,98 However, temperature control when 3D printing with COL-I bioinks is critical – the print head should be maintained at 4–10 °C to facilitate printing of the bioink, but the print bed should be maintained at 37 °C to facilitate thermal crosslinking of the deposited bioink filaments.94,99 COL-I prints can also be further crosslinked post-printing using cell friendly chemical crosslinkers such as genipin.100,101
Another approach to improving the mechanical properties of COL-I bioinks involves chemically modifying the COL-I hydrogel with crosslinkable chemical groups. The COL-I molecule may be chemically modified at the primary amine site on the lysine residue with photocrosslinkable methacrylate groups to form methacrylated collagen (ColMA).102–105 This allows the COL-I bioink to be crosslinked in the presence of a photoinitiator, such as Irgacure 2959 (I2959) or lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), and UV light during or immediately following the printing process. The addition of methacrylate groups has also been shown to confer thermoreversible gelation properties to the hydrogel without affecting the fibrillation process, biodegradability or bioactivity of COL-I.103,106 Similarly, COL-I has also been functionalised with norbornene groups to form a thiol–ene photocrosslinkable hydrogel which was shown to have improved 3D printing properties without a loss of bioactivity.107
Sacrificial support baths, as employed in Freeform Reversible Embedding of Suspended Hydrogels (FRESH), can also be used to facilitate 3D printing of COL-I bioinks.108 The FRESH method of 3D printing involves 3D printing a bioink into a thermoreversible gelatin slurry support bath which exhibits yield-stress rheological behaviour, similar to that exhibited by Bingham plastic fluids. This rheological phenomenon facilitates seamless movement of the print needle through the support bath while extruding the bioink, while also embedding the printed filament into the support bath to maintain the shape fidelity of the 3D printed construct109 (Fig. 4). The support bath can also be supplemented with specific crosslinking agents to crosslink the construct as it prints and once the print is completed, the support bath can be melted away at 37 °C. When 3D printing with a COL-I based bioink, the FRESH support bath is supplemented with a neutral pH buffer such as phosphate buffered saline (PBS) or 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer to allow fibrillation to occur. The fibrillation process is then completed upon incubation of the prints at 37 °C for at least 30 minutes and the melted gelatin slurry can then be discarded. This method has been used to 3D print complex biological structures using both low concentration,108 and high concentration110,111 COL-I bioink formulations.
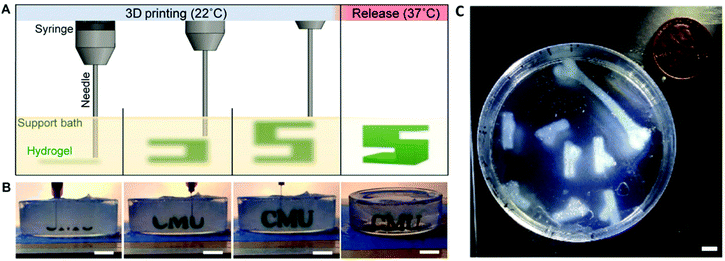 | ||
| Fig. 4 FRESH 3D printing is conducted by extruding a bioink into a thermoreversible gelatin slurry support bath which can subsequently be melted and removed by heating to 37 °C (A and B). Heating to 37 °C also allows thermal fibrillation of collagen-based bioinks to occur. The FRESH gelatin slurry provides mechanical support to the 3D printed bioink filaments and facilitates 3D printing of complex anatomical structures using collagen-based bioinks (C). A, B and C are re-printed and adapted with from Hinton et al.108 with permission from the American Association for the Advancement of Science (AAAS) (Copyright © 2015, The Authors). | ||
Collagen can also be partially hydrolysed to form gelatin, which is often used to formulate bioinks for cartilage repair. Gelatin has superior solubility and rheological properties when compared to native collagen and it also retains the RGD cell binding motif which is present in native collagen.112 Thus, it is a popular biomimetic collagen substitute for 3D printing purposes. Gelatin is also biocompatible and biodegradable, and already has approval from the United States Food and Drug Administration (US FDA) for use in the food and biomedical industry, thus it does not present the same regulatory hurdles as other biomaterials.113 However, gelatin in its unmodified state is not suitable for TE applications due to its low viscosity at body temperature.114 Therefore, gelatin is often incorporated into composite bioink formulations with other biomaterials such as silk fibroin115–117 or alginate,118 or else chemically modified to confer crosslinking functionality to the molecule. The most common example of this is the chemical modification of gelatin with methacrylate groups to form gelatin methacrylate (GelMA).119,120 Similar to ColMA, GelMA is photocrosslinkable in the presence of a photoiniator, such as I2959 or LAP, and UV light, and both GelMA-only bioinks and GelMA-containing composite bioinks have been shown to be biocompatible and promote regeneration of cartilage tissue.121–126 Lim et al. also further built on the functionality of GelMA by chemically modifying the molecule with tyrosine residues which aid integration of the 3D printed scaffold into the chondral defect.127
Due to their innate biocompatibility, chondrogenic potential and shear thinning properties, HyA hydrogels are popular as bioink formulations for cartilage repair.141 However, the main drawback with these hydrogels is that they are not readily crosslinkable for 3D printing applications and so have poor shape retention post-printing.142 Therefore, when used to formulate bioinks, HyA hydrogels are often blended with other biomaterials or chemically modified to confer crosslinking functionality to the bioink. Biomaterials commonly blended with HyA to formulate bioinks for cartilage repair include hydrogels such as alginate, which crosslinks in the presence of divalent cations,143–146 or GelMA,147 which is photocrosslinkable, amongst others. As alginate is also a hydrophilic linear polysaccharide, it is sometimes employed as a crosslinkable GAG mimetic, and may even be sulfated to further mimic naturally occurring sulfated GAGs such as CS.148 However, as alginate hydrogels do not possess any naturally occurring cell-binding moieties,149 blending with HyA can facilitate improved cell adhesion to the bioink.
HyA is also quite a chemically versatile molecule and can be modified at the carboxyl, hydroxyl and amide groups respectively to confer a wide range of crosslinking functionality.150 Similar to ColMA and GelMA which are discussed above, HyA can also be chemically modified with photocrosslinkable methacrylate groups to form methacrylated HyA or HyA methacrylate (often abbreviated to ‘MeHA’ or ‘HAMA’) which crosslinks in the presence of UV light and a photoiniator.123,151–156 Modification of HyA with other chemical groups such as norbornene groups,157 thiols,158,159 and amino acid derivatives,160,161 amongst others, also allow for the formation of crosslinkable hydrogels. Furthermore, the HyA molecule can be modified to allow for protein–ligand binding (e.g. biotin–avidin binding system) or enzymatic crosslinking functionality which has been shown to improve the printability of the hydrogels, and promote subsequent cell adhesion and proliferation.162,163 Some studies also report blending modified HyA and other biomaterials, such as alginate, to further improve the 3D printing properties and chondrogenic potential of the bioink,162 or co-printing with thermoplastics such as PLA or PCL to improve the mechanical properties of the 3D printed scaffold.143,158,159
However, when chemically modifying HyA hydrogels, it is important to consider the effects of the modification on the physiochemical properties and overall biofunctionality of the HyA molecule. For example, the carboxylic acid group on the glucuronic acid subunit is deprotonated at physiological pH and thus confers a negative charge on the molecule in vivo.164 This negative charge largely contributes to the swelling properties of HyA as the HyA hydrogel becomes hydrated and swells in response to electrostatic repulsion between adjacent deprotonated carboxylic acid groups.165 Therefore, chemical modification of the HyA molecule at the carboxylic acid group can affect the overall charge of the molecule and thus the swelling properties of the hydrogel. Similarly, chemical modification of HyA can also affect cell interactions with the hydrogel via CD44 binding. Kwon et al. demonstrated that high degrees of HyA chemical modification (approximately 40%) can negatively affect CD44-hydrogel interactions and subsequently have negative effects on MSC chondrogenesis in cell-laden hydrogels. However, HyA hydrogels with a lower degree of modification (approximately 10–20%) still exhibit improved CD44 binding and chondrogenic potential when compared to inert hydrogels.154 Therefore, the degree of HyA modification is an important consideration when formulating biomimetic bioinks for cartilage repair using modified HyA hydrogels.
Although CS hydrogels are more popular in the formulation of injectable, gradient and photocrosslinkable hydrogels for cartilage repair,173–180 they have also been used to formulate a number of biomimetic bioinks. When used in the formulation of bioinks, CS is typically included as part of a composite bioink and is often chemically modified with methacrylate (often referred to as CSMA)181,182 or catechol183,184 groups to improve bioink printability or adhesion to the cartilage defect in vivo. For example, Costantini et al. developed a cartilage ECM mimetic, crosslinkable bioink by blending alginate, GelMA and CS aminoethyl methacrylate which promoted chondrogenic differentiation of bone marrow-derived MSCs.181 Abbadessa et al. also developed a thermo-sensitive and photocrosslinkable CSMA and poly(N-(2-hydroxypropyl) methacrylamide-mono/dilactate)-polyethylene glycol triblock copolymer bioink with suitable 3D printing properties, and which also facilitated the fabrication of scaffolds with tailorable porosity that promoted chondrogenic cell proliferation.185
Cartilage tissue for formulating dECM-based bioinks is typically derived from porcine,189–192 caprine193 or bovine194 sources. Following the removal of cellular material and sterilisation of the dECM to remove any pathogenic compounds, the dECM must then be further processed into a hydrogel formulation before being used as a bioink. This is usually achieved by freeze-drying the dECM, then blending or grinding the resulting lyophilate into small particles which can then be solubilised under acidic conditions using the enzyme pepsin.195 Adjustment of the pH and salt concentration to physiological conditions then neutralises the pepsin and allows the formation of a dECM hydrogel which, similar to COL-I hydrogels, undergoes thermal crosslinking at 37 °C.189,195,196 Therefore, similar to COL-I hydrogels, complete gelation can take up to 30 mins which poses problems for the mechanical stability of the 3D printed construct post-printing.
To overcome this issue, dECM hydrogels are often blended with other biomaterials when formulating bioinks for cartilage repair, which also has the added benefit of adding additional functionality to the bioink. Blending dECM with synthetic biomaterials such as polyvinyl alcohol (PVA) or PEG allows for the formulation of biomimetic bioinks with the bioactivity of native cartilage ECM, but with the enhanced printability and crosslinking control of a synthetically made bioink.193,197 dECM has also been blended with silk fibroin to greatly improve its printability and fabricate structures which mimic the native architecture of cartilage tissue, and which can subsequently be crosslinked using 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS).191 Blending dECM hydrogels with a versatile biomaterial, such as alginate, allows formulation of bioinks with tuneable stiffness and good printability.192 dECM only or dECM composite bioinks can also be co-printed with polymers such as PCL to bring the mechanical properties of the 3D printed scaffold in line with those of native articular cartilage.191,192 In the case of dECM only bioinks, co-printing with a polymer also provides mechanical scaffolding for the bioink during the 3D printing and crosslinking processes.191
Similar to other biomimetic biomaterials, dECM can also be chemically modified with methacrylate groups to confer crosslinking functionality and thus improve mechanical stability of the printed construct. For example, Visscher et al. formulated a methacrylated dECM hydrogel for auricular cartilage regeneration, which was blended with gelatin, HyA and glycerol to improve bioink printability and initial mechanical stability of the 3D printed construct. This bioink was shown to be more stiff and maintain a higher level of chondrocyte viability and proliferation than GelMA controls.190 Overall, the inclusion of dECM in bioinks for cartilage repair has been shown to enhance the biocompatibility of the bioink and chondrogenesis of MSCs.189,191,197 These bioinks can also be further funtionalised to facilitate controlled release of bioactive factors, such as TGF-β, to further increase expression of pro-chondrogenic genes and subsequent deposition of cartilage ECM components.192,193
IPN bioinks offer several advantages over single polymer network bioinks or non-IPN composite bioinks in the field of cartilage TE. One such advantage is that IPN bioinks have enhanced compressive stiffness and toughness when compared to the individual biomaterial components that make up the bioink.201 For example, an alginate and polyacrylamide IPN hydrogel formulated by Liao et al. was found to have a compressive modulus approximately four times greater than its individual biomaterial counterparts.202 Similarly, Li et al. found that the equilibrium modulus of a gellan gum and PEG diacrylate (PEGDA) double network IPN was approximately 10 times higher than that of the respective constituent hydrogels.203 Thus, IPN bioinks can be tailored to mimic the mechanical stiffness of articular cartilage ECM, without significantly increasing the polymer concentration or crosslink density. This is beneficial for TE applications as high polymer concentrations or crosslink densities can hinder cell migration and attachment, as well as diffusion of nutrients and waste products.
Another advantage of IPN bioinks is the ability to tune material properties, such as viscoelasticity or stiffness, in an independent manner in order to influence cell behaviour.200 This fine control of the bioink microenvironment is desirable as it has been shown that the mechanical and rheological properties of a cell's environment have a significant influence on cell spreading, proliferation and differentiation.204–207 For example, Lee et al. demonstrated that chondrocytes embedded in fast relaxing viscoelastic alginate hydrogels undergo higher levels of proliferation and produce a more extensive and interconnected ECM than those embedded in slow relaxing hydrogels.208 Park et al. also demonstrated that MSCs cultured on stiffer substrates were more likely to differentiate into smooth muscle cells, while MSCs cultured on softer substrates tended towards a chondrogenic or adipogenic lineage (dependent on the presence or absence of TGF-β).205 This alludes to the critical role that bioink properties play in regulating chondrocyte behaviour and thus the ability to control these parameters is highly desirable in the formulation of bioinks for cartilage repair.
A number of research groups have employed IPNs of natural and synthetic biomaterials to formulate bioinks for cartilage repair with enhanced mechanical properties, biocompatibility and chondrogenic potential. Wang et al. and Schipani et al. both developed alginate and GelMA-based IPN bioinks, which were shown to possess superior mechanical properties to their constituent hydrogel components and promoted chondrogensis of MSCs and subsequent deposition of articular cartilage ECM components.124,148 Wu et al. designed a double network IPN bioink composed of gellan gum and PEGDA which had desirable 3D printing properties and could undergo non-covalent and covalent crosslinking post-printing respectively to fabricate scaffolds with versatile mechanical properties.209 Ni et al. optimised a double network bioink consisting of silk fibroin and hydroxyl propyl methyl cellulose methacrylate (HPMC-MA), whereby silk fibroin β-sheets formed the more brittle primary network and photocrosslinked HPMC-MA formed the ductile secondary network. Formation of the double network improved the mechanical properties of the bioink and the bioink was also shown to facilitate proliferation and chondrogenesis of MSCs.210 Thus, this demonstrates that a wide range of biomaterial properties can be synergistically combined through the fabrication of these biomimetic networks.
The main advantages and disadvantages of each bioink type outlined above are summarised in Table 1 below.
| Category | Biomaterial | Concentrations used | Crosslinking method | Key Advantages | Key Disadvantages | Ref. |
|---|---|---|---|---|---|---|
| Collagen | Collagen type I | 0.5–5% | Thermal crosslinking | Good cell viability (>80%) post-printing and contains RGD cell-binding motif | Hydrogels can be mechanically weak and thermal crosslinking process is difficult to control. Must be printed at low temperatures (2–8 °C) | 99, 108 and 211–213 |
| Methacrylated collagen | 0.3% | Thermal and photo-crosslinking | Good cell viability (>80%) post-printing and contains RGD cell-binding motif. Improved mechanical properties versus unmodified collagen type I bioinks | Must be printed at low temperatures (2–8 °C). Use of UV light during photo-crosslinking may have negative effects on cell viability | 104 | |
| Gelatin | 4.5–10% (as part of a composite bioink) | Not readily crosslinkable | Good cell viability (>90%) post-printing and contains RGD cell-binding motif. Already has US FDA approval for use in the biomedical industry | Not readily crosslinkable for 3D printing purposes in its unmodified state. Low viscosity at body temperature | 115–117, 214 and 215 | |
| Methacrylated gelatin | 5–30% | Photo-crosslinking | Good cell viability (>80%) post-printing and contains RGD cell-binding motif. Improved mechanical properties versus unmodified gelatin bioinks | Use of UV light during photo-crosslinking may have negative effects on cell viability | 119–126 and 216 | |
| Glycosaminoglycan | Hyaluronic acid | 1–30% (as part of a composite bioink) | Not readily crosslinkable | Good cell viability (>80%) post-printing and contains CD44 cell-binding domain. Plays important role in the biological and mechanical properties of articular cartilage | Not readily crosslinkable for 3D printing purposes in its unmodified state. Poor print resolution unless blended with other biomaterials or chemically modified | 143–147 |
| Methacrylated hyaluronic acid | 0.5–4% | Photo-crosslinking | Good cell viability (>80%) post-printing and contains CD44 cell-binding domain. Improved mechanical properties versus unmodified hyaluronic acid hydrogels | Higher degrees of functionalisation can negatively affect CD44 cell-binding and chondrogenesis | 123, 142 and 151–156 | |
| Chondroitin sulfate | 1–10% (as part of a composite bioink) | Not readily crosslinkable | Good cell viability (>90%) post-printing. Plays important role in the biological and mechanical properties of articular cartilage | Not readily crosslinkable for 3D printing purposes in its unmodified state. Poor print resolution unless blended with other biomaterials or chemically modified | 217 | |
| Chondroitin sulfate methacrylate | 2–4% (as part of a composite bioink) | Photo-crosslinking | Good cell viability (>80%) post-printing. Improved mechanical properties versus unmodified chondroitin sulfate hydrogels | Often required to be formulated as part of a composite bioink to improve printability | 181, 182 and 185 | |
| Composite | Decellularised ECM | 0.2–20% | Thermal crosslinking | Good cell viability (>70%) post-printing. Possesses bioactivity of several biomaterials native to articular cartilage. Can be chemically modified to confer crosslinking functionality and improve printability | Thermal crosslinking of hydrogels can take up to 1 h at 37 °C and is a difficult process to control. Poor print resolution unless blended with other biomaterials, chemically modified or printed using a support bath | 190–193 and 197 |
| Interpenetrating networks | — | — | Have enhanced compressive stiffness and toughness when compared to the individual biomaterial components that make up the bioink. Ability to tune material properties, such as viscoelasticity or stiffness, in an independent manner in order to influence cell behaviour. Can possess bioactivity of several biomaterials native to articular cartilage | Disadvantages dependent on biomaterial selection | 124, 148, 202, 203, 209 and 210 |
3.2. Functionalising 3D printed scaffolds for cartilage repair with cells
Complete regeneration of functional cartilage tissue requires mature chondrocytes which are capable of producing a collagen type II and GAG-rich ECM to gradually replace the implanted TE scaffold. In the field of TE for cartilage repair, there are two ways in which cells may be incorporated into the scaffold. The first method involves implanting a cell-free scaffold with sufficient porosity into the cartilage defect and allowing the patient's own cells to infiltrate the scaffold and lay down their own ECM.43 The second method involves functionalising a bioink with cells which have been harvested from the patient and expanded in vitro, and subsequently 3D printing a scaffold embedded with the patient's expanded cells in a process known as ‘3D bioprinting’.5 However, 3D bioprinting scaffolds for cartilage repair is not without its challenges. Thus, the following sections will discuss the considerations that should be taken into account when formulating cell-laden bioinks for 3D bioprinting scaffolds for cartilage repair.MSCs are one of the most popular cell types used in cartilage TE due to their proliferative and differentiation abilities, and are usually obtained from bone marrow or adipose tissue.218,219 In fact, MSCs are currently employed clinically to promote cartilage regeneration as part of the microfracture surgical procedure, during which tiny fractures are made in the subchondral bone allowing the release of bone marrow MSCs into the defect which ultimately develop into fibrocartilage.13 There are extensive reports in the literature that show that MSCs from human,104,123,192,215,220 rat,221,222 bovine,157 rabbit223,224 and porcine124,192 sources can be successfully directed towards a chondrogenic lineage when encapsulated within a suitably formulated biomimetic bioink. For example, Rathan et al. reported that MSCs encapsulated in a dECM-functionalised alginate bioink expressed higher levels of the pro-chondrogenic genes Col2a1 and ACAN than those encapsulated in the non-functionalised alginate control. However, gene expression of Col1a1 and RUNX2 was also elevated, suggesting that a proportion of MSCs had also followed an endochondral or osteogenic pathway.192 This highlights the importance of bioink design when attempting to direct the differentiation of MSCs.
Articular chondroprogenitor cells (ACPCs) can also be used to functionalise bioinks for cartilage repair, and similar to MSCs, have been shown to produce a collagen type II and GAG rich matrix when encapsulated within biomimetic bioinks.214,225,226 ACPCs can be more advantageous than MSCs in the field of TE for cartilage repair as they have already started on a chondrogenic pathway and express high levels of SOX-9, also known as the master regulator of chondrogenesis.227,228 Similar to MSCs, ACPCs also have a good proliferative capacity, are capable of migration and express similar cell surface markers.229
Mature chondrocytes from human, porcine, rabbit, bovine and equine sources have also been successfully shown to produce articular cartilage-like ECM components in biomimetic 3D bioprinted scaffolds for cartilage repair.99,124,143,212,230–232 However, the main drawback with using mature articular chondrocytes is that availability of tissue from which to isolate these cells is limited and the tissue that can be harvested contains quite a low density of chondrocytes.233 Mature chondrocytes also have a tendency to undergo dedifferentiation when cultured in vitro, as is often seen following the autologous chondrocyte implantation procedure (ACI).234 However, similar to MSCs, this risk can be reduced by providing the cells with the correct biological and physical cues through appropriate selection of biomaterials and/or inclusion of bioactive factors, in order to maintain an articular cartilage phenotype.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mm3 in the superficial zone, to 10
000 cells per mm3 in the superficial zone, to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mm3 in the middle zone, to 7000 cells per mm3 in the deep zone.235 In bioprinting terms, this is equivalent to approximately 24M cells per mL in the superficial zone, 10M cells per mL in the middle zone and 7M cells per mL in the deep zone. Therefore, the majority of bioinks for cartilage repair are functionalised with MSCs,123,157,210,221–224 ACPCs,225,226 mature articular chondrocytes,212,230,236 or co-cultures of MSCs and chondrocytes124,232 at cell densities of 5M to 20M cells per mL to mimic the native cellular composition of human articular cartilage.
000 cells per mm3 in the middle zone, to 7000 cells per mm3 in the deep zone.235 In bioprinting terms, this is equivalent to approximately 24M cells per mL in the superficial zone, 10M cells per mL in the middle zone and 7M cells per mL in the deep zone. Therefore, the majority of bioinks for cartilage repair are functionalised with MSCs,123,157,210,221–224 ACPCs,225,226 mature articular chondrocytes,212,230,236 or co-cultures of MSCs and chondrocytes124,232 at cell densities of 5M to 20M cells per mL to mimic the native cellular composition of human articular cartilage.
However, studies have also been conducted to investigate the effects of low cell densities, in the range of 1 to 2M cells per mL, on chondrogenesis in 3D bioprinted scaffolds. Henrionnet et al. fabricated scaffolds using an alginate, gelatin and fibrinogen bioink, functionalised with MSCs at two respective cell densities – 1M cells per mL and 2M cells per mL. Interestingly, following 28 days culture in TGF-β1 supplemented media, scaffolds containing the lower cell density showed more enhanced expression of the chondrogenic markers Col2a1, ACAN and SOX-9.215 Similarly, Koo et al. 3D bioprinted a collagen-based scaffold containing rabbit articular chondrocytes at a density of 1M cells per mL which successfully facilitated neo-cartilage formation in osteochondral defects of the rabbit knee.99 Thus, when provided with a suitable micro-environment, low cell densities can also be used to promote cartilage tissue regeneration.
Due to the pivotal role that GFs play in promoting proliferation and chondrogenic differentiation of MSCs during long bone formation, they are the most common bioactive factor used to functionalise 3D printed scaffolds for cartilage repair. A number of growth factor families have attracted interest in the field of TE for cartilage repair. The TGF-β family are known promoters of chondrogenesis, however TGF-β1 and 3 in particular has been shown to elicit a strong chondrogenic response during in vitro and in vivo studies.247–250 Thus, these TGF-β isoforms are particularly popular in the field of 3D printing for cartilage repair, both as a supplement for chondrogenic cell media and as a bioink component to enhance chondrogenesis of MSCs,148,192,193,223,251–253 The BMP family are a subgroup of the TGF-β superfamily that have been shown to regulate almost every step of the chondrogenic pathway. Of particular relevance to cartilage repair, BMP-2, 4 and 7 promote MSC condensation at the beginning of the chondrogenic pathway, as well as promoting chondrogenic differentiation of chondroprogenitor cells by maintain the expression of the pro-chondrogenic SOX transcription factors.76,254 Similar to TGF-β, BMPs can also be added to bioinks for osteochondral repair.255
Platelet-rich plasma (PRP) is also used as a source of endogenous GFs for cartilage TE. PRP is obtained by centrifuging a sample of peripheral blood to create a concentrated pellet of platelets, which release proteins and GFs following activation.256 GFs obtained from platelets include IGF-1, PDGF, TGF-β1 and basic fibroblast growth factor (bFGF), amongst others, and intra-articular injections of PRP have been shown to promote the repair of smaller cartilage defects in clinical studies.257–259 Thus, a number of recent studies have attempted to harness the chondrogenic potential of PRP by incorporating it into 3D printed scaffolds for cartilage repair.221,260–262 For example, Irmak et al. developed a patient-specific photo-activated PRP and GelMA bioink which facilitated the controlled release of the GFs PDGF, TGF-β1 and bFGF, and enhanced the expression of pro-chondrogenic genes and deposition of articular cartilage matrix components.260 The platelets adhered to the GelMA-based bioink via integrin receptors and were activated upon exposure to near-infrared light, allowing for the controlled release of GFs. Similarly, Luo et al. incorporated freshly activated PRP into a 3D bioprinted MSC-laden GelMA scaffold and implanted it intramuscularly into a mouse. They found that addition of the PRP promoted chondrogenic differentiation of the embedded MSCs and deposition of articular cartilage-specific ECM components.221
3.3. Gradient 3D printed scaffolds for cartilage repair
In recent years, there has been a growing acceptance within the field of TE for cartilage repair that homogenous biomaterial scaffolds with a uniform architecture and composition are not well positioned to mirror the complexity and functionality of the native tissue. This is where 3D printing has really come into its own, allowing researchers to fabricate gradient scaffolds whose structure and composition mimic the zonal architecture of articular cartilage. Gradients within 3D printed scaffolds can be achieved by creating zonal differences in parameters such as biomaterial composition, pore size, cell density or presence of bioactive factors to promote deposition of zone-specific matrix components. Some examples of 3D printed gradient scaffolds for cartilage repair are outlined below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio). They found that ECM production was positively correlated with cell density and that there was zonal differences in ECM properties based on cell density, similar to those seen in the native tissue.236 Similarly, Dimaraki et al. 3D bioprinted PCL-reinforced alginate-based zonal scaffolds containing human chondrocytes at a density of 20M cells per mL in the superficial zone, 10M cells per mL in the middle zone and 5M cells per mL in the deep zone. They found that distinct zonal cell densities could be partially maintained over a 25 day culture period and that, similar to Ren et al., creating a cell density gradient resulted in gradient deposition of cartilage ECM components.269
1 ratio). They found that ECM production was positively correlated with cell density and that there was zonal differences in ECM properties based on cell density, similar to those seen in the native tissue.236 Similarly, Dimaraki et al. 3D bioprinted PCL-reinforced alginate-based zonal scaffolds containing human chondrocytes at a density of 20M cells per mL in the superficial zone, 10M cells per mL in the middle zone and 5M cells per mL in the deep zone. They found that distinct zonal cell densities could be partially maintained over a 25 day culture period and that, similar to Ren et al., creating a cell density gradient resulted in gradient deposition of cartilage ECM components.269
4. Bioink formulation – what works best?
Thus far in this review we have discussed how a variety of different biomimetic biomaterials, cell types and densities, and bioactive factors can be used to formulate bioinks for cartilage repair. In doing so, we have highlighted a number of biomimetic bioink formulations, each of which possesses desirable 3D printing properties, biocompatibility and chondrogenic potential. However, clear evidence is yet to accrue on which bioink formulation works best. Although it is a relatively simple and logical question to pose, differences in experimental procedures make it difficult to draw direct comparisons between studies.Despite this fact, several formulation trends can still be observed across studies. With regards biomaterial composition of bioinks, the majority of studies use composite bioinks consisting of two or more biomaterial components. In these studies, one of these components is often a GAG-derived (e.g. a HyA-derived biomaterial) or a GAG-mimetic (e.g. alginate) biomaterial, while the other is a collagen-derived (e.g. gelatin or GelMA) biomaterial. At least one of these biomaterial components also possesses crosslinking functionality. With regards the cellular composition of these bioinks, the use of MSC and/or chondrocyte-laden bioinks is more popular than ACPC-laden bioinks. However, regardless of the cell type used, higher cell densities (in the range of 10 to 20M cells per mL) are more commonly employed when a single cell density is present throughout the entire 3D bioprinted scaffold. Where a cell density gradient is employed in a 3D bioprinted scaffold, it generally mirrors that found in native articular cartilage, i.e. approximately 20M mL−1 in the superficial zone, 10M mL−1 in the middle zone and 5M mL−1 in the deep zone.
Investigation of the composition of 3D printed scaffolds used in successful in vivo studies also indicates which bioink formulations achieve functional regeneration of articular cartilage in a living, moving organism. These scaffolds are usually 3D printed ex vivo and surgically implanted into the cartilage defect, however in some instances the scaffold may be 3D printed in situ directly into the defect (Fig. 5C).270,271 Similar to in vitro studies, it is difficult to draw direct comparisons between the results of in vivo studies due to differences in animal models used and experimental design.272 However, similar to above, there are formulation trends present across studies. Firstly, in a large number of successful in vivo studies, the bioink (either composite or single component) is co-printed with a polymer to improve the mechanical properties of the 3D printed scaffold99,125,223,267,270,273,274 (Fig. 5B). These polymers are usually thermoplastics with relatively low melting points, such as PCL. Secondly, gradient 3D printed scaffolds are frequently used to promote chondral or osteochondral zonal differentiation of cells and deposition of ECM components (e.g. pore size gradient, as shown in Fig. 5A).267
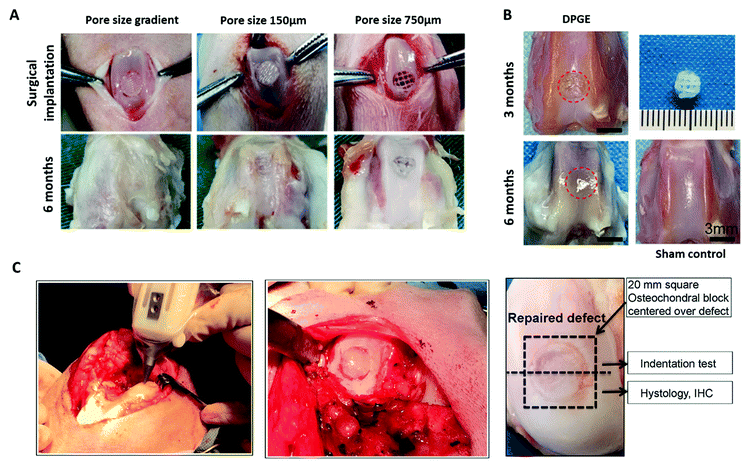 | ||
| Fig. 5 3D bioprinted biomimetic scaffolds for cartilage repair have been shown to facilitate regeneration of articular cartilage in vivo. (A) Surgical implantation of a PCL-reinforced gelatin, fibrinogen, HA and glycerol with a pore size gradient (ranging from 150 µm in superficial zone to 750 µm in the deep zone). Following 6 months implantation, the gradient scaffolds displayed better tissue repair than the non-gradient scaffolds in chondral defects of the rabbit knee. (B) Repair of articular cartilage 3 months and 6 months post-implantation of a 3D bioprinted PCL-reinforced aptamer-functionalised GelMA and dECM scaffold. (C) In situ 3D bioprinting of a scaffold for cartilage repair using a GelMA and MeHA MSC-laden bioink in a chondral defect of the sheep knee, and subsequent repair of the defect following implantation for 2 months. A is re-printed and adapted from Sun et al.267 with permission from Elsevier (Copyright © 2021, Elsevier B.V.), B is re-used and adapted with permission from Yang et al.270 (Copyright © 2021, American Chemical Society), and C is re-used and adapted with permission from Di Bella et al.271 (Copyright © 2017, John Wiley & Sons, Ltd). | ||
5. Future perspectives
From its inception with the development of the first TE biomaterial-based scaffolds for skin repair in the 1970s and 1980s, TE has positioned itself as having immense potential in the field of modern medicine.275 The advent of 3D printed scaffolds has further advanced this field, allowing for the fabrication of complex biomimetic biomaterial structures with defined architecture, which can be functionalised with cells and bioactive factors in a manner that mimics the native tissue. However, the question still remains – how can the gap between bench and bedside, which currently hinders the clinical use of 3D printed scaffolds for the treatment of cartilage defects, finally be bridged? The answer to this question may lie in the field of gene therapy.Gene therapy involves the introduction of a specific genetic sequence into a cell via complexation with a targeted delivery vector in order to introduce a new gene, or promote or silence expression of a pre-existing gene. This, in turn, leads to an increase or decrease in the expression of a particular protein of interest. At the beginning the century, the field of gene therapy suffered a severe setback following reports of severe side-effects and deaths in clinical trials for gene therapeutic treatments of X-linked severe combined immunodeficiency (SCID) and ornithine transcarbamylase deficiency respectively.276 However, in recent years a range of gene therapeutics have been granted marketing authorisations worldwide for the treatment of a number of orphan diseases and cancers, amongst other conditions.277 In the last year, in particular, the field of gene therapy has been catapulted into the limelight following the successful development and regulatory approval of two messenger RNA (mRNA)-based vaccines against SARS-CoV-2 (also known as Covid-19); the mRNA-1273 SARS-CoV-2 vaccine,278 and the BNT162b2 mRNA Covid-19 vaccine279 (colloquially known as the ‘Moderna’ and ‘Pfizer/BioNTech’ vaccines respectively). Both of these vaccines contain mRNA encoding for the SARS-CoV-2 spike protein which is encapsulated in lipid nanoparticles to enable uptake by cells, subsequently leading to expression of the spike protein, followed by the desired immune response.280
Aside from mRNA, other examples of genetic sequences which can be introduced into cells include pDNA, miRNA and silencing RNA (siRNA), amongst others.281 Delivery vectors used to transport these genetic sequences into the cell can be viral or non-viral in nature. Lentiviral and adeno-associated viral vectors are the most commonly used viral vectors in the field of gene therapy and although they exhibit high transfection efficiency, safety concerns such as insertional mutagenesis have traditionally hindered their clinical use.276,282,283 Therefore, non-viral delivery vectors including layered double hydroxides (LDHs),284 lipid-based vectors such as Lipofectamine,285 polymers such as polyethylenimine (PEI),286 inorganic nanoparticles such as nanohydroxyapatite (nHA)240 and cell-penetrating peptides (CPPs) such as the RALA amphipathic peptide287 and glycosaminoglycan binding enhanced transduction (GET) peptides288 are becoming increasingly popular in this field. Our own research group has been pioneering the concept of gene-activated TE scaffolds for a myriad of applications including cartilage repair,245 bone repair,241,286,289 skin repair290–292 and peripheral nerve repair.293
Incorporating these gene therapeutic nanoparticles into bioinks for cartilage repair would allow for the fabrication of multi-layered scaffolds with distinct zonal biological cues. While this approach has been taken with the use of GFs, their release from 3D printed scaffolds can be difficult to control and can result in off-target side-effects.276 Thus, gene therapy can be used to promote the expression of pro-chondrogenic factors in vivo without the risk of adverse effects. The concept of gene-activation of bioinks was pioneered by members of our group, Gonzalez-Fernandez et al., who developed a MSC-laden, pore-forming alginate-based bioink which facilitated the controlled release of RALA and pDNA gene therapeutic nanoparticles. This bioink was used to 3D print a PCL-reinforced gradient scaffold for osteochondral repair which contained nanoparticles consisting of RALA and pDNA for TGF-β3, BMP-2 and SOX-9 respectively in the cartilage layer, and nanoparticles consisting of nHA and pDNA for BMP-2 in the bone layer. In an in vivo mouse model, these bi-layered gene-activated scaffolds facilitated the production of a bone layer, overlaid with a collagen type II and GAG-rich articular cartilage layer.50
Therefore, the development of these novel bioinks is building on recent successes in the fields of TE and gene therapy to deliver next generation 3D printed gene-activated scaffolds. These gene-activated scaffolds could potentially bridge the aforementioned gap between the bench and bedside, leading to a novel clinical treatment for a myriad of conditions including chondral and osteochondral defects, skin wounds and spinal cord injury, to name a few. Thus, this field has the potential to greatly improve quality of life for patients worldwide.
6. Conclusion
To conclude, the development of TE scaffolds, which are 3D printed using biomimetic bioinks functionalised with cells and bioactive factors or gene therapeutics, may finally offer the opportunity to develop scaffolds which fully recapitulate the complex zonal architecture of native articular cartilage and thus increase the likelihood of an effective clinical treatments for cartilage repair. The ability to restore functionality to the articular joints of patients would have a profound effect on the quality of life of millions of patients worldwide.Author contributions
DOS, CC and FOB devised the structure of the manuscript, and DOS drafted and wrote the manuscript. DOS created figures used unless otherwise stated. DOS, CC, and FOB revised the manuscript critically and suggested references. CC and FOB supervised the project and FOB provided funding for the project. All authors have read and approved the final submitted manuscript.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The authors acknowledge funding from the European Research Council under the European Community's Horizon 2020 research and innovation programme under ERC Advanced Grant agreement n°788753 (ReCaP). Dr. Curtin acknowledges funding from the Health Research Board (HRB) in Ireland under grant agreement ILP-POR-2019-023. All original figures were created with Biorender.com.References
- J. F. V. Vincent, O. A. Bogatyreva, N. R. Bogatyrev, A. Bowyer and A.-K. Pahl, J. R. Soc., Interface, 2006, 3, 471–482 CrossRef PubMed.
- J. M. Benyus, Biomimicry: Innovation Inspired by Nature, William Morrow & Company, 1997 Search PubMed.
- F. J. O’Brien, Mater. Today, 2011, 14(3), 88–95 CrossRef.
- C. M. Curtin, J. C. Nolan, R. Conlan, L. Deneweth, C. Gallagher, Y. J. Tan, B. L. Cavanagh, A. Z. Asraf, H. Harvey, S. Miller-Delaney, J. Shohet, I. Bray, F. J. O'Brien, R. L. Stallings and O. Piskareva, Acta Biomater., 2018, 70, 84–97 CrossRef CAS PubMed.
- W. Sun, B. Starly, A. C. Daly, J. A. Burdick, J. Groll, G. Skeldon, W. Shu, Y. Sakai, M. Shinohara, M. Nishikawa, J. Jang, D.-W. Cho, M. Nie, S. Takeuchi, S. Ostrovidov, A. Khademhosseini, R. D. Kamm, V. Mironov, L. Moroni and I. T. Ozbolat, Biofabrication, 2020, 12, 022002 CrossRef CAS PubMed.
- R. Chaudhuri, M. Ramachandran, P. Moharil, M. Harumalani and A. K. Jaiswal, Mater. Sci. Eng., C, 2017, 79, 950–957 CrossRef CAS PubMed.
- M. Lemoine, S. M. Casey, J. M. O’Byrne, D. J. Kelly and F. J. O’Brien, Biochem. Soc. Trans., 2020, 48(4), 1443–1445 CrossRef PubMed.
- S. MacNeil, Mater. Today, 2008, 11(5), 26–35 CrossRef CAS.
- J. F. Mano and R. L. Reis, J. Tissue Eng. Regener. Med., 2007, 1, 261–273 CrossRef CAS PubMed.
- R. Wittenauer, L. Smith and K. Aden, Background Paper 6.12 Osteoarthritis, 2013. Available from: https://www.who.int/medicines/areas/priority_medicines/BP6_12Osteo.pdf [Accessed on 18/02/21] Search PubMed.
- WHO Department of Chronic Diseases and Health Promotion, Chronic rheumatic conditions, 2021. Available from: https://www.who.int/chp/topics/rheumatic/en/ [Accessed on 18/02/21] Search PubMed.
- B. S. Dunkin and C. Lattermann, Operative Techniques in Sports Medicine, 2013, vol. 21, pp. 100–107 Search PubMed.
- J. R. Steadman, B. S. Miller, S. G. Karas, T. F. Schlegel, K. K. Briggs and R. J. Hawkins, J. Knee Surg., 2003, 16, 83–86 Search PubMed.
- H.-L. Ma, S.-C. Hung, S.-T. Wang, M.-C. Chang and T.-H. Chen, Injury, 2004, 35, 1286–1292 CrossRef PubMed.
- H. Robert, Orthop. Traumatol.: Surg. Res., 2011, 97, 418–429 CAS.
- H. Kwon, W. E. Brown, C. A. Lee, D. Wang, N. Paschos, J. C. Hu and Kyriacos A. Athanasiou, Nat. Rev. Rheumatol., 2019, 15, 550–570 CrossRef PubMed.
- F. J. O'Brien, Mater. Today, 2011, 14, 88–95 CrossRef.
- K. Pelttari, H. Lorenz, S. Boeuf, M. F. Templin, O. Bischel, K. Goetzke, H.-Y. Hsu, E. Steck and W. Richter, Arthritis Rheum., 2008, 58, 467–474 CrossRef CAS PubMed.
- E. B. Hunziker, K. Lippuner, M. J. B. Keel and N. Shintani, Osteoarthritis Cartilage, 2015, 23, 334–350 CrossRef CAS PubMed.
- W. J. Choi, K. K. Park, B. S. Kim and J. W. Lee, Am. J. Sports Med., 2009, 37, 1974–1980 CrossRef PubMed.
- K. S. Katti, Colloids Surf., B, 2004, 39, 133–142 CrossRef CAS PubMed.
- L. E. Bayliss, D. Culliford, A. P. Monk, S. Glyn-Jones, D. Prieto-Alhambra, A. Judge, C. Cooper, A. J. Carr, N. K. Arden, D. J. Beard and A. J. Price, Lancet, 2017, 387, 1424–1430 CrossRef.
- D. I. Braghirolli, D. Steffens and P. Pranke, Drug Discovery Today, 2014, 19, 743–753 CrossRef CAS PubMed.
- N. Munir, A. McDonald and A. Callanan, Bioprinting, 2019, 16, e00056 CrossRef.
- M. Rafiei, E. Jooybar, M. J. Abdekhodaie and M. Alvi, Mater. Sci. Eng., C, 2020, 113, 110913 CrossRef CAS PubMed.
- N. Toyokawa, H. Fujioka, T. Kokubu, I. Nagura, A. Inui, R. Sakata, M. Satake, H. Kaneko and M. Kurosaka, Arthroscopy, 2010, 26, 375–383 CrossRef PubMed.
- T. Nascimento da Silva, R. P. Gonçalves, C. L. Rocha, B. S. Archanjo, C. A. G. Barboza, M. B. R. Pierre, F. Reynaud and P. H. de Souza Picciani, Mater. Sci. Eng., C, 2019, 97, 602–612 CrossRef CAS PubMed.
- J. A. Matthews, E. D. Boland, G. E. Wnek, D. G. Simpson and G. L. Bowlin, J. Bioact. Compat. Polym., 2003, 18, 125 CrossRef CAS.
- K. J. Shields, M. J. Beckman, G. L. Bowlin and J. S. Wayne, Tissue Eng., 2004, 10(9–10), 1510–1517 CrossRef CAS PubMed.
- J. Reignier and M. A. Huneault, Polymer, 2006, 47, 4703–4717 CrossRef CAS.
- C.-J. Liao, C. F. Chen, J. H. Chen, S.-F. Chiang, Y.-J. Lin and K. Y. Chang, J. Biomed. Mater. Res., 2002, 59, 676–681 CrossRef CAS PubMed.
- D. C. Sin, X. Miao, G. Liu, F. Wei, G. Chadwick, C. Yan and T. Friis, Mater. Sci. Eng., C, 2010, 30, 78–85 CrossRef CAS.
- N. Thadavirul, P. Pavasant and P. Supaphol, J. Biomed. Mater. Res., 2014, 102, 3379–3392 CrossRef.
- V. Vishwanath, K. Pramanik and A. Biswas, J. Biomater. Sci., Polym. Ed., 2016, 27, 657–674 CrossRef CAS PubMed.
- Q. Zhang, H. Lu, N. Kawazoe and G. Chen, Acta Biomater., 2014, 10, 2005–2013 CrossRef CAS PubMed.
- W. Dai, N. Kawazoe, X. Lin, J. Dong and G. Chen, Biomaterials, 2010, 31, 2141–2152 CrossRef CAS PubMed.
- S. A. Poursamar, A. N. Lehner, M. Azami, S. Ebrahimi-Barough, A. Samadikuchaksaraei and A. P. M. Antunes, Mater. Sci. Eng., C, 2016, 63, 1–9 CrossRef CAS PubMed.
- W. L. Murphy, R. G. Dennis, J. L. Kileny and D. J. Mooney, Tissue Eng., 2002, 8, 43–52 CrossRef CAS PubMed.
- D. J. Mooney, D. F. Baldwin, N. P. Suh, J. P. Vacanti and R. Langer, Biomaterials, 1996, 17, 1417–1422 CrossRef CAS PubMed.
- L. D. Harris, B.-S. Kim and D. J. Mooney, J. Biomed. Mater. Res., 1998, 42, 396–402 CrossRef CAS PubMed.
- A. Salerno, E. Di Maio, S. Iannace and P. A. Netti, J. Porous Mater., 2012, 19, 181–188 CrossRef CAS.
- I. Manavitehrani, T. Y. L. Le, S. Daly, Y. Wang, P. K. Maitz, A. Schindeler and F. Dehghani, Mater. Sci. Eng., C, 2019, 96, 824–830 CrossRef CAS PubMed.
- C. M. Murphy, M. G. Haugh and F. J. O'Brien, Biomaterials, 2010, 31, 461–466 CrossRef CAS PubMed.
- C. M. Murphy, A. Matsiko, M. G. Haugh, J. P. Gleeson and F. J. O'Brien, J. Mech. Behav. Biomed. Mater., 2012, 11, 53–62 CrossRef CAS PubMed.
- F. J. O'Brien, B. A. Harley, I. V. Yannas and L. J. Gibson, Biomaterials, 2005, 26, 433–441 CrossRef PubMed.
- T. J. Levingstone, A. Matsiko, G. R. Dickson, F. J. O'Brien and J. P. Gleeson, Acta Biomater., 2014, 10, 1996–2004 CrossRef CAS PubMed.
- C. M. Tierney, M. G. Haugh, J. Liedl, F. Mulcahy, B. Hayes and F. J. O'Brien, J. Mech. Behav. Biomed. Mater., 2009, 2, 202–209 CrossRef PubMed.
- L. Moroni, J. A. Burdick, C. B. Highley, S. J. Lee, Y. Morimoto and S. Takeuchi, Nat. Rev. Mater., 2018, 3, 21–37 CrossRef CAS PubMed.
- R. M. Raftery, D. P. Walsh, L. B. Ferreras, I. M. Castaño, G. Chen, M. Lemoine, G. Osman, K. M. Shakesheff, J. E. Dixon and F. J. O’Brien, Biomaterials, 2019, 216, 119277 CrossRef CAS PubMed.
- T. Gonzalez-Fernandez, S. Rathan, C. Hobbs, P. Pitacco, F. E. Freeman, G. M. Cunniffe, N. J. Dunne, H. O. McCarthy, V. Nicolosi, F. J. O'Brien and D. J. Kelly, J. Controlled Release, 2019, 301, 13–27 CrossRef CAS PubMed.
- X. Hu, Y. Man, W. Li, L. Li, J. Xu, R. Parungao, Y. Wang, S. Zheng, Y. Nie, T. Liu and K. Song, Polymers, 2019, 11, 10 Search PubMed.
- A. C. Daly, F. E. Freeman, T. Gonzalez-Fernandez, S. E. Critchley, J. Nulty and D. J. Kelly, Adv. Healthcare Mater., 2017, 6, 22 Search PubMed.
- M. E. Prendergast and J. A. Burdick, Adv. Mater., 2020, 32, 13 CrossRef PubMed.
- J. Malda, J. Visser, F. P. Melchels, T. Jüngst, W. E. Hennink, W. J. A. Dhert, J. Groll and D. W. Hutmacher, Adv. Mater., 2013, 25, 5011–5028 CrossRef CAS PubMed.
- M. Singh, H. M. Haverinen, P. Dhagat and G. E. Jabbour, Adv. Mater., 2010, 22, 673–685 CrossRef CAS PubMed.
- A. Negro, T. Cherbuin and M. P. Lutolf, Sci. Rep., 2018, 8, 17099 CrossRef PubMed.
- Dassault Systèmes, 3D Printing – Additive, (accessed 01/04/2020).
- L. Koch, M. Gruene, C. Unger and B. Chichkov, Curr. Pharm. Biotechnol., 2013, 14, 91–97 CAS.
- N. Paxton, W. Smolan, T. Böck, F. P. Melchels, J. Groll and T. Jüngst, Biofabrication, 2017, 9, 4 CrossRef PubMed.
- J. Groll, J. A. Burdick, D.-W. Cho, B. Derby, M. Gelinksy, S. C. Heilshorn, T. Jüngst, J. Malda, V. A. Mironov, K. Nakayama, A. Ovsianikov, W. Sun, S. Takeuchi, J. J. Yoo and T. B. F. Woodfield, Biofabrication, 2019, 11, 1 Search PubMed.
- A. M. Bhosale and J. B. Richardson, Br. Med. Bull., 2008, 87, 77–95 CrossRef PubMed.
- A. Matsiko, T. J. Levingstone and F. J. O'Brien, Materials, 2013, 6, 637–668 CrossRef CAS PubMed.
- A. R. Armiento, M. J. Stoddart, M. Alini and D. Eglin, Acta Biomater., 2018, 65, 1–20 CrossRef CAS PubMed.
- A. J. S. Fox, A. Bedi and S. A. Rodeo, Sports Health, 2009, 1, 461–468 CrossRef PubMed.
- K. Gelse, E. Poschl and T. Aigner, Adv. Drug Delivery Rev., 2003, 55, 1531–1546 CrossRef CAS PubMed.
- K. E. Kadler, A. Hill and E. G. Canty-Laird, Curr. Opin. Cell Biol., 2008, 20, 495–501 CrossRef CAS PubMed.
- J. Casale and J. S. Crane, Biochemistry, Glycosaminoglycans, StatPearls Publishing LLC, 2021, https://www.ncbi.nlm.nih.gov/books/NBK544295/ [Accessed on 18/05/21] Search PubMed.
- P. J. Roughley and J. S. Mort, J. Exp. Orthop., 2014, 1, 8 CrossRef PubMed.
- A. R. Poole, T. Kojima, T. Yasuda, F. Mwale, M. Kobayashi and S. Laverty, Clin. Orthop. Relat. Res., 2001, 391S, S26–S33 CrossRef PubMed.
- H. Akkiraju and A. Nohe, J. Dev. Biol., 2015, 3, 177–192 CrossRef PubMed.
- J. W. Alford and Brian J. Cole, Am. J. Sports Med., 2005, 33(2), 295–306 CrossRef PubMed.
- J. A. Buckwalter and H. J. Mankin, Instr. Course Lect., 1998, 47, 477–486 CAS.
- D. Baksh, L. Song and R. S. Tuan, J. Cell. Mol. Med., 2004, 8, 301–316 CrossRef CAS PubMed.
- M. Dominici, K. Le Blanc, I. Mueller, I. Slaper-Cortenbach, F. C. Marini, D. S. Krause, R. J. Deans, A. Keating, D. J. Prockop and E. M. Horwitz, Cryotherapy, 2006, 8, 315–317 CrossRef CAS PubMed.
- C. N. D. Coelho and R. A. Kosher, Dev. Biol., 1991, 144, 47–53 CrossRef CAS PubMed.
- M. B. Goldring, K. Tsuchimochi and K. Ijiri, J. Cell. Biochem., 2006, 97, 33–44 CrossRef CAS PubMed.
- L. Shum and G. Nuckolls, Arthritis Res., 2002, 4, 94–106 CrossRef CAS PubMed.
- J. D. Green, V. Tollemar, M. Dougherty, Z. Yan, L. Yin, J. Ye, Z. Collier, M. K. Mohammed, R. C. Haydon, H. H. Luu, R. Kang, M. J. Lee, S. H. Ho, T.-C. He, L. L. Shi and A. Athiviraham, Genes Dis., 2015, 2, 307–327 CrossRef PubMed.
- L. Yang, K. Y. Tsang, H. C. Tang, D. Chan and K. S. E. Cheah, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12097–12102 CrossRef CAS PubMed.
- M. Wang, E. R. Sampson, H. Jin, J. Li, Q. H. Ke, H.-J. Im and D. Chen, Arthritis Res. Ther., 2013, 15, 1 Search PubMed.
- U. Sharma, D. Pal and R. Prasad, Indian J. Clin. Biochem., 2014, 29, 269–278 CrossRef CAS PubMed.
- R. Dreier, Arthritis Res. Ther., 2010, 12, 216 CrossRef PubMed.
- M. Ding, Y. Lu, S. Abbassi, F. Li, X. Li, Y. Song, V. Geoffroy, H.-J. Im and Q. Zheng, J. Cell. Physiol., 2012, 227, 3446–3456 CrossRef CAS PubMed.
- Y.-Q. Yang, Y.-Y. Tan, R. Wong, A. Wenden, L.-K. Zhang and A. B. M. Rabie, Int. J. Oral Sci., 2012, 4, 64–68 CrossRef CAS PubMed.
- E. M. Ahmed, J. Adv. Res., 2015, 6, 105–121 CrossRef CAS PubMed.
- S. Uman, A. P. Dhand and J. A. Burdick, J. Appl. Polym. Sci., 2019, 137, 25 Search PubMed.
- M. Guvendiren, H. D. Lu and J. A. Burdick, Soft Matter, 2012, 2, 260–272 RSC.
- F. L. C. Morgan, L. Moroni and M. B. Baker, Adv. Healthcare Mater., 2020, 9, 15 Search PubMed.
- S. Ji and M. Guvendiren, Front. Bioeng. Biotechnol., 2017, 5, 23 Search PubMed.
- A. Panwar and L. P. Tan, Molecules, 2016, 21, 685 CrossRef PubMed.
- E. O. Osidak, V. I. Kozhukhov, M. S. Osidak and S. P. Domogatsky, Int. J. Bioprint., 2020, 6, 270 CAS.
- M. Hospodiuk, M. Dey, D. Sosnoski and I. T. Ozbolat, Biotechnol. Adv., 2017, 35, 217–239 CrossRef CAS PubMed.
- V. L. Cross, Y. Zheng, N. W. Choi, S. S. Verbridge, B. A. Sutermaster, L. J. Bonassar, C. Fischback and A. D. Stroock, Biomaterials, 2010, 31, 8596–8607 CrossRef CAS PubMed.
- S. Rhee, J. L. Puetzer, B. N. Mason, C. A. Reinhart-King and L. J. Bonassar, ACS Biomater. Sci. Eng., 2016, 2(10), 1800–1805 CrossRef CAS PubMed.
- Advanced BioMatrix, Lifeink 200 – Neutralized Type I Collagen Bioink, 3 5 mg mL−1 (accessed 27/05/21).
- Imtek Ltd, Viscoll Collagen Solution – Pig collagen type I, sterile solution (accessed 27/05/21).
- E. O. Osidak, P. A. Karalkin, M. S. Osidak, V. A. Parfenov, D. E. Sivogrivov, F. D. A. S. Pereira, A. A. Gryadunova, E. V. Koudan, Y. D. Khesuani, V. A. Kasyanov, S. I. Belousov, S. V. Krasheninnikov, T. E. Grigoriev, S. N. Chvalun, E. A. Bulanova, V. A. Mironov and S. P. Domogatsky, J. Mater. Sci.: Mater. Med., 2019, 30(31) CAS.
- J. M. Lee, S. K. Q. Suen, W. L. Ng, W. C. Ma and W. Y. Yeong, Macromol. Biosci., 2021, 21, 1 CrossRef PubMed.
- Y. W. Koo, E.-J. Choi, J. Y. Lee, H.-J. Kim, G. H. Kim and S. H. Do, J. Ind. Eng. Chem., 2018, 66, 343–355 CrossRef CAS.
- G. Montalbano, G. Borciani, G. Cerqueni, C. Licini, F. Banche-Niclot, D. Janner, S. Sola, S. Fiorilli, M. Mattioli-Belmonte, G. Ciapetti and C. Vitale-Brovarone, Nanomaterials, 2020, 10, 1681 CrossRef CAS PubMed.
- Y. B. Kim, H. Lee and G. H. Kim, ACS Appl. Mater. Interfaces, 2016, 8, 32230–32240 CrossRef CAS PubMed.
- T.-U. Nguyen, K. E. Watkins and V. Kishore, J. Biomed. Mater. Res., Part A, 2019, 107, 1541–1550 CrossRef CAS PubMed.
- K. E. Drzewiecki, A. S. Parmar, I. D. Gaudet, J. R. Branch, D. H. Pike, V. Nanda and D. I. Shreiber, Langmuir, 2014, 30, 11204–11211 CrossRef CAS PubMed.
- N. S. Kajave, T. Schmitt, T.-U. Nguyen and V. Kishore, Mater. Sci. Eng., C, 2020, 107, 110290 CrossRef CAS PubMed.
- N. S. Kajave, T. Schmitt, T.-U. Nguyen, A. K. Gaharwar and V. Kishore, Biomed. Mater., 2021, 16, 3 Search PubMed.
- I. D. Gaudet and D. I. Shreiber, Biointerphases, 2012, 7, 25 CrossRef CAS PubMed.
- K. Guo, H. Wang, S. Li, H. Zhang, S. Li, H. Zhu, Z. Yang, L. Zhang, P. Chang and X. Zheng, ACS Appl. Mater. Interfaces, 2021, 13, 7037–7050 CrossRef CAS PubMed.
- T. J. Hinton, Q. Jallerat, R. N. Palchesko, J. H. Park, M. S. Grodzicki, H.-J. Shue, M. H. Ramadan, A. R. Hudson and A. W. Feinberg, Sci. Adv., 2015, 1, e1500758 CrossRef PubMed.
- D. J. Shiwarski, A. R. Hudson, J. W. Tashman and A. W. Feinberg, APL Bioeng., 2021, 5, 010904 CrossRef PubMed.
- A. Lee, A. R. Hudson, D. J. Shirwarski, J. W. Tashman, T. J. Hinton, S. Yerneni, J. M. Bliley, P. G. Campbell and A. W. Feinberg, Science, 2019, 365, 482–487 CrossRef CAS PubMed.
- T. Sousa, N. S. Kajave, P. Dong, L. Gu, S. Florczyk and V. Kishore, 3D Printing and Additive Manufacturing, 2021, Ahead of print Search PubMed.
- M. Santoro, A. M. Tatara and A. G. Mikos, J. Controlled Release, 2014, 190, 210–218 CrossRef CAS PubMed.
- M. B. Łabowska, K. Cierluk, A. M. Jankowska, J. Kulbacka, J. Detyna and I. Michalak, Materials, 2021, 14, 858 CrossRef PubMed.
- W. Schuurman, P. A. Levett, M. W. Pot, P. R. van Weeren, W. J. A. Dhert, D. W. Hutmacher, F. P. W. Melchels, T. J. Klein and J. Malda, Macromol. Biosci., 2013, 13, 551–561 CrossRef CAS PubMed.
- Y. P. Singh, A. Bandyopadhyay and B. B. Mandal, ACS Appl. Mater. Interfaces, 2019, 11, 33684–33696 CrossRef CAS PubMed.
- S. Chawla, A. Kumar, P. Admane, A. Bandyopadhyay and S. Ghosha, Bioprinting, 2017, 7, 1–13 CrossRef.
- S. Chawla, G. Desando, E. Gabusi, A. Sharma, D. Trucco, J. Chakraborty, C. Manferdini, M. Petretta, G. Lisignoli and S. Ghosh, J. Mater. Res., 2021, 36, 4051–4067 CrossRef CAS.
- S. Schwarza, S. Kuthab, T. Distler, C. Gögele, K. Stölzel, R. Detsch, A. R. Boccaccini and G. Schulze-Tanzil, Mater. Sci. Eng., C, 2020, 116, 111189 CrossRef PubMed.
- J. Visser, B. Peters, T. J. Burger, J. Boomstra, W. J. A. Dhert, F. P. W. Melchels and J. Malda, Biofabrication, 2013, 5, 3 CrossRef PubMed.
- Q. Gao, X. Niu, L. Shao, L. Zhou, Z. Lin, A. Sun, J. Fu, Z. Chen, J. Hu and Y. Liu, Biofabrication, 2019, 11, 3 CrossRef PubMed.
- J. Liu, L. Li, H. Suo, M. Yan, J. Yin and J. Fu, Mater. Des., 2019, 171, 107708 CrossRef CAS.
- V. H. M. Mouser, R. Levato, A. Mensinga, W. J. A. Dhert, D. Gawlitta and J. Malda, Connect. Tissue Res., 2018, 61, 137–151 CrossRef PubMed.
- J. S. Lee, H. S. Park, H. Jung, H. Lee, H. Hong, Y. J. Lee, Y. J. Suh, O. J. Lee, S. H. Kim and C. H. Park, Addit. Manuf., 2020, 33, 101136 CAS.
- R. Schipani, S. Scheurer, R. Florentin, S. E. Critchley and D. J. Kelly, Biofabrication, 2020, 12, 3 CrossRef PubMed.
- F. Gao, Z. Xu, Q. Liang, H. Li, L. Peng, M. Wu, X. Zhao, X. Cui, C. Ruan and W. Liu, Adv. Sci., 2019, 6, 15 Search PubMed.
- S. Duchi, C. Onofrillo, C. D. O’Connell, R. Blanchard, C. Augustine, A. F. Quigley, R. M. I. Kapsa, P. Pivonka, G. G. Wallace, C. Di Bella and P. F. M. Choong, Sci. Rep., 2017, 7, 5387 CrossRef PubMed.
- K. S. Lim, F. Abinzano, P. N. Bernal, A. A. Sanchez, P. Atienza-Roca, I. A. Otto, Q. C. Peiffer, M. Matsusaki, T. B. F. Woodfield, J. Malda and R. Levato, Adv. Healthcare Mater., 2020, 9, 1901792 CrossRef CAS PubMed.
- J. A. Burdick and G. D. Prestwich, Adv. Mater., 2011, 23, H41–H56 CrossRef CAS PubMed.
- E. D. Bonnevie, D. Galesso, C. Secchieri, I. Cohen and L. J. Bonassar, PLoS One, 2015, 10, e0143415 CrossRef PubMed.
- C. B. Knudson, Birth Defects Res., Part C, 2003, 69, 174–196 CrossRef CAS PubMed.
- C. Chung and J. A. Burdick, Tissue Eng., Part A, 2008, 15, 2 Search PubMed.
- W. S. Toh, T. C. Lim, M. Kurisawa and M. Spector, Biomaterials, 2012, 33, 3835–3845 CrossRef CAS PubMed.
- I. L. Kim, S. Khetan, B. M. Baker, C. S. Chen and J. A. Burdick, Biomaterials, 2013, 34, 5571–5580 CrossRef CAS PubMed.
- I. E. Erickson, A. H. Huang, S. Sengupta, S. Kestle, J. A. Burdick and R. L. Mauck, Osteoarthritis Cartilage, 2009, 17, 1639–1648 CrossRef CAS PubMed.
- A. A. Hegewald, J. Ringe, J. Bartel, I. Krüger, M. Notter, D. Barnewitz, C. Kaps and M. Sittinger, Tissue Cell, 2004, 36, 431–438 CrossRef CAS PubMed.
- R. B. Jakobsen, A. Shahdadfar, F. P. Reinholt and J. E. Brinchmann, Knee Surg. Sports Traumatol. Arthrosc., 2010, 18, 1407–1416 CrossRef PubMed.
- A. Matsiko, T. J. Levingstone, F. J. O'Brien and J. P. Gleeson, J. Mech. Behav. Biomed. Mater., 2012, 11, 41–52 CrossRef CAS PubMed.
- I. L. Kim, R. L. Mauck and J. A. Burdick, Biomaterials, 2011, 32, 8771–8782 CrossRef CAS PubMed.
- L. A. Solchaga, J. E. Dennis, V. M. Goldberg and A. I. Caplan, J. Orthop. Res., 1999, 17, 205–213 CrossRef CAS PubMed.
- Y. Jin, R. H. Koh, S.-H. Kim, K. M. Kim, G. K. Park and N. S. Hwang, Mater. Sci. Eng., C, 2020, 115, 111096 CrossRef CAS PubMed.
- L. Valot, J. Martinez, A. Mehdi and G. Subra, Chem. Soc. Rev., 2019, 48, 3999–4340 RSC.
- D. Petta, U. D’Amora, L. Ambrosio, D. W. Grijpma, D. Eglin and M. D’Este, Biofabrication, 2020, 12, 3 CrossRef PubMed.
- C. Antich, J. de Vicente, G. Jiménez, C. Chocarro, E. Carrillo, E. Montañez, P. Gálvez-Martín and J. A. Marchal, Acta Biomater., 2020, 106, 114–123 CrossRef CAS PubMed.
- M. Lafuente-Merchan, S. Ruiz-Alonso, A. Espona-Noguera, P. Galvez-Martin, E. López-Ruiz, J. A. Marchal, M. L. López-Donaire, A. Zabala, J. Ciriza, L. Saenz-del-Burgo and J. L. Pedraz, Mater. Sci. Eng., C, 2021, 126, 112160 CrossRef CAS PubMed.
- S. J. Lee, J. M. Seok, J. H. Lee, J. Lee, W. D. Kim and S. A. Park, Polymers, 2021, 13, 794 CrossRef CAS PubMed.
- C. C. Piras and D. K. Smith, J. Mater. Chem. B, 2020, 8, 8171–8188 RSC.
- I. Noh, N. Kim, H. N. Tran, J. Lee and C. Lee, Biomater. Res., 2019, 23, 3 CrossRef PubMed.
- B. Wang, P. J. Díaz-Payno, D. C. Browe, F. E. Freeman, J. Nulty, R. Burdis and D. J. Kelly, Acta Biomater., 2021, 128, 130–142 CrossRef CAS PubMed.
- K. Y. Lee and D. J. Mooney, Prog. Polym. Sci., 2012, 37, 106–126 CrossRef CAS PubMed.
- C. B. Highley, G. D. Prestwich and J. A. Burdick, Curr. Opin. Biotechnol., 2016, 40, 35–40 CrossRef CAS PubMed.
- B. S. Spearman, N. K. Agrawal, A. Rubiano, C. S. Simmons, S. Mobini and C. E. Schmidt, J. Biomed. Mater. Res., 2020, 108A, 279–291 CrossRef PubMed.
- M. T. Poldervaart, B. Goversen, M. de Ruijter, A. Abbadessa, F. P. W. Melchels, F. C. Öner, W. J. A. Dhert, T. Vermonden and J. Alblas, PLoS One, 2017, 12, 6 CrossRef PubMed.
- J. A. Burdick, C. Chung, X. Jia, M. A. Randolph and R. Langer, Biomacromolecules, 2005, 6, 386–391 CrossRef CAS PubMed.
- M. Y. Kwon, C. Wang, J. H. Galarraga, E. Puré, L. Han and J. A. Burdick, Biomaterials, 2019, 222, 119451 CrossRef CAS PubMed.
- Q. Mei, J. Rao, H. P. Bei, Y. Liu and X. Zhao, Int. J. Bioprint., 2021, 7, 367 CrossRef PubMed.
- M. Kesti, M. Müller, J. Becher, M. Schnabelrauch, M. D'Este, D. Eglin and M. Zenobi-Wong, Acta Biomater., 2015, 11, 162–172 CrossRef CAS PubMed.
- J. H. Galarraga, M. Y. Kwon and J. A. Burdick, Sci. Rep., 2019, 9, 19987 CrossRef CAS PubMed.
- S. Stichler, T. Böck, N. Paxton, S. Bertlein, R. Levato, V. Schill, W. Smolan, J. Malda, J. Teßmar, T. Blunk and J. Groll, Biofabrication, 2017, 9, 4 CrossRef PubMed.
- J. Hauptstein, T. Böck, M. Bartolf-Kopp, L. Forster, P. Stahlhut, A. Nadernezhad, G. Blahetek, A. Zernecke-Madsen, R. Detsch, T. Jüngst, J. Groll, J. Teßmar and T. Blunk, Adv. Healthcare Mater., 2020, 9(15) CAS.
- D. Nguyen, D. A. Hägg, A. Forsman, J. Ekholm, P. Nimkingratana, C. Brantsing, T. Kalogeropoulos, S. Zaunz, S. Concaro, M. Brittberg, A. Lindahl, P. Gatenholm, A. Enejder and S. Simonsson, Sci. Rep., 2017, 7, 658 CrossRef PubMed.
- A. K.-X. Lee, Y.-H. Lin, C.-H. Tsai, W.-T. Chang, T.-L. Lin and M.-Y. Shie, Biomedicines, 2021, 9(7), 714 CrossRef CAS PubMed.
- S. Nedunchezian, P. Banerjee, C.-Y. Lee, S.-S. Lee, C.-W. Lin, C.-W. Wu, S.-C. Wu, J.-K. Chang and C.-K. Wang, Mater. Sci. Eng., C, 2021, 124, 112072 CrossRef CAS PubMed.
- P. L. Thi, J. Y. Son, Y. Lee, S. B. Ryu, K. M. Park and K. D. Park, Macromol. Res., 2020, 28, 400–406 CrossRef.
- K. J. Wolf and S. Kumar, ACS Biomater. Sci. Eng., 2019, 5, 3753–3765 CrossRef CAS PubMed.
- J.-T. Kim, D. Y. Lee, Y.-H. Kim, I.-K. Lee and Y.-S. Song, J. Sens. Sci. Technol., 2012, 21, 256–262 CrossRef.
- C. Kiani, L. Chen, Y. J. Wu, A. J. Yee and B. B. Yang, Cell Res., 2002, 12, 19–32 CrossRef PubMed.
- H. Watanabe, Y. Yamada and K. Kimata, J. Biochem., 1998, 124, 687–693 CrossRef CAS PubMed.
- J. J. Lim and J. S. Temenoff, Biomaterials, 2013, 34, 5007–5018 CrossRef CAS PubMed.
- J. Radhakrishnan, A. Subramanian, U. M. Krishnan and S. Sethuraman, Biomacromolecules, 2017, 18, 1–26 CrossRef CAS PubMed.
- S. Varghese, N. S. Hwang, A. C. Canver, P. Theprungsirikul, D. W. Lin and J. Elisseeff, Matrix Biol., 2008, 27, 12–21 CrossRef CAS PubMed.
- C. S. Moura, J. C. Silva, S. Faria, P. R. Fernandes, C. L. da Silva, J. M. S. Cabral, R. Linhardt, P. J. Bártolo and F. C. Ferreira, J. Biosci. Bioeng., 2020, 129, 756–764 CrossRef CAS PubMed.
- E. A. Aisenbrey and S. J. Bryant, Biomaterials, 2019, 190–191, 51–62 CrossRef CAS PubMed.
- T. Li, X. Song, C. Weng, X. Wang, L. Sun, X. Gong, L. Yang and C. Chen, Appl. Mater. Today, 2018, 10, 173–183 CrossRef.
- C. Tang, B. D. Holt, Z. M. Wright, A. M. Arnold, A. C. Moy and S. A. Sydlik, J. Mater. Chem. B, 2019, 7, 2442–2453 RSC.
- X. Li, Q. Xu, M. Johnson, X. Wang, J. Lyu, Y. Li, S. McMahon, U. Greiser, S. A and W. Wang, Biomater. Sci., 2021, 9, 4139–4148 RSC.
- S. Lee, J. Choi, J. Youn, Y. Lee, W. Kim, S. Choe, J. Song, R. L. Reis and G. Khang, Biomolecules, 2021, 11, 8 Search PubMed.
- M. Liu, X. Zeng, C. Ma, H. Yi, Z. Ali, X. Mou, S. Li, Y. Deng and N. He, Bone Res., 2017, 5, 17014 CrossRef CAS PubMed.
- P. A. Levett, F. P. W. Melchels, K. Schrobback, D. W. Hutmacher, J. Malda and T. J. Klein, Acta Biomater., 2014, 10, 214–223 CrossRef CAS PubMed.
- A. Khanlari, M. S. Detamore and S. H. Gehrke, Macromolecules, 2013, 46, 9609–9617 CrossRef CAS.
- X. Liu, S. Liu, R. Yang, P. Wang, W. Zhang, X. Tan, Y. Ren and B. Chi, Carbohydr. Polym., 2021, 270, 118330 CrossRef CAS PubMed.
- M. Costantini, J. Idaszek, K. Szöke, J. Jaroszewicz, M. Dentini, A. Barbetta, J. E. Brinchmann and W. Święszkowski, Biofabrication, 2016, 8(3) CrossRef PubMed.
- A. Abbadessa, V. H. M. Mouser, M. M. Blokzijl, D. Gawlitta, W. J. A. Dhert, W. E. Hennink, J. Malda and T. Vermonden, Biomacromolecules, 2016, 17, 2137–2147 CrossRef CAS PubMed.
- A. Scalzone, M. A. Bonifacio, S. Cometa, F. Cucinotta, E. De Giglio, A. M. Ferreira and P. Gentile, Front. Bioeng. Biotechnol., 2020, 8, 712 CrossRef PubMed.
- J. Shin, E. H. Kang, S. Choi, E. J. Jeon, J. H. Cho, D. Kang, H. Lee, I. S. Yun and S.-W. Cho, ACS Biomater. Sci. Eng., 2021, 7, 4230–4243 CrossRef CAS PubMed.
- A. Abbadessa, M. M. Blokzijl, V. H. M. Mouser, P. Marica, J. Malda, W. E. Hennink and T. Vermonden, Carbohydr. Polym., 2016, 149, 163–174 CrossRef CAS PubMed.
- A. J. Vernengo, S. Grad, D. Eglin, M. Alini and Z. Li, Adv. Funct. Mater., 2020, 30, 1909044 CrossRef CAS.
- T. W. Gilbert, T. L. Sellaro and S. F. Badylak, Biomaterials, 2006, 27, 3675–3683 CAS.
- P. M. Crapo, T. W. Gilbert and S. F. Badylak, Biomaterials, 2011, 32, 3233–3243 CrossRef CAS PubMed.
- F. Pati, J. Jang, D.-H. Ha, S. W. Kim, J.-W. Rhie, J.-H. Shim, D.-H. Kim and D.-W. Cho, Nat. Commun., 2014, 5, 3935 CrossRef CAS PubMed.
- D. O. Visscher, H. Lee, P. P. M. van Zuijlen, M. N. Helder, A. Atala, J. J. Yoo and S. J. Lee, Acta Biomater., 2021, 121, 193–203 CrossRef CAS PubMed.
- C. S. Jung, B. K. Kim, J. Lee, B.-H. Min and S.-H. Park, Tissue Eng. Regener. Med., 2018, 15, 155–162 CrossRef CAS PubMed.
- S. Rathan, L. Dejob, R. Schipani, B. Haffner, M. E. Möbius and D. J. Kelly, Adv. Healthcare Mater., 2019, 8, 7 Search PubMed.
- X. Zhang, Y. Liu, C. Luo, C. Zhai, Z. Li, Y. Zhang, T. Yuan, S. Dong, J. Zhang and W. Fan, Mater. Sci. Eng., C, 2021, 118, 111388 CrossRef CAS PubMed.
- W. Chen, Y. Xu, Y. Li, L. Jia, X. Mo, G. Jiang and G. Zhou, Chem. Eng. J., 2020, 382, 122986 CrossRef CAS.
- A. Abaci and M. Guvendiren, Adv. Healthcare Mater., 2020, 9, 2000734 CrossRef CAS PubMed.
- D. O. Freytes, J. Martin, S. S. Velankar, A. S. Lee and S. F. Badylak, Biomaterials, 2008, 29, 1630–1637 CrossRef CAS PubMed.
- M. Setayeshmehr, S. Hafeez, C. van Blitterswijk, L. Moroni, C. Mota and M. B. Baker, Int. J. Mol. Sci., 2021, 22, 8 Search PubMed.
- M. S. Silverstein, Polymer, 2020, 207, 122929 CrossRef CAS.
- S. L. Vega, M. Y. Kwon and J. A. Burdick, Eur. Cells Mater., 2017, 30, 59–75 CrossRef PubMed.
- A. P. Dhand, J. H. Galarraga and J. A. Burdick, Trends Biotechnol., 2021, 39, 519–538 CrossRef CAS PubMed.
- D. Chimene, R. Kaunas and A. K. Gaharwar, Adv. Mater., 2020, 32, 1 CrossRef PubMed.
- I.-C. Liao, F. T. Moutos, B. T. Estes, X. Zhao and F. Guilak, Adv. Funct. Mater., 2013, 23, 5833–5839 CrossRef CAS PubMed.
- W. Li, D. Wu, D. Hu, S. Zhu, C. Pan, Y. Jiao, L. Li, B. Luo, C. Zhou and L. Lu, Mater. Sci. Eng., C, 2020, 107, 110333 CrossRef CAS PubMed.
- O. Chaudhuri, J. Cooper-White, P. A. Janmey, D. J. Mooney and V. B. Shenoy, Nature, 2020, 584, 535–546 CrossRef CAS PubMed.
- J. S. Park, J. S. Chu, A. D. Tsou, R. Diop, Z. Tang, A. Wang and S. Li, Biomaterials, 2011, 32, 3921–3930 CrossRef CAS PubMed.
- A. S. Mao, J.-W. Shin and D. J. Mooney, Biomaterials, 2016, 98, 184–191 CrossRef CAS PubMed.
- A. J. Steward and D. J. Kelly, J. Anat., 2015, 227, 717–731 CrossRef PubMed.
- H.-p. Lee, L. Gu, D. J. Mooney, M. E. Levenston and O. Chaudhuri, Nat. Mater., 2017, 16, 1243–1251 CrossRef CAS PubMed.
- D. Wu, Y. Yu, J. Tan, L. Huang, B. Luo, L. Lu and C. Zhou, Mater. Des., 2018, 160, 486–495 CrossRef CAS.
- T. Ni, M. Liu, Y. Zhang, Y. Cao and R. Pei, Bioconjugate Chem., 2020, 31, 1938–1947 CrossRef CAS PubMed.
- X. Yang, Z. Lu, H. Wu, W. Li, Li Zheng and J. Zhao, Mater. Sci. Eng., C, 2018, 83, 195–201 CrossRef CAS PubMed.
- N. Diamantides, C. Dugopolski, E. Blahut, S. Kennedy and L. J. Bonassar, Biofabrication, 2019, 11, 045016 CrossRef CAS PubMed.
- A. Schwab, C. Hélary, R. G. Richards, M. Alini, D. Eglin and M. D’Este, Mater. Today Bio, 2020, 7, 100058 CrossRef CAS PubMed.
- Y. Zhou, R. Qin, T. Chen, K. Zhang and J. Gui, Mater. Des., 2021, 203, 109621 CrossRef CAS.
- C. Henrionnet, L. Pourchet, P. Neybecker, O. Messaoudi, P. Gillet, D. Loeuille, D. Mainard, C. Marquette and A. Pinzano, Stem Cells Int., 2020, 2020, 2487072 Search PubMed.
- I. Pepelanova, K. Kruppa, T. Scheper and A. Lavrentieva, Bioengineering, 2018, 5, 55 CrossRef CAS PubMed.
- M. Lafuente-Merchan, S. Ruiz-Alonso, A. Zabala, P. Gálvez-Martín, J. A. Marchal, B. Vázquez-Lasa, I. Gallego, L. Saenz-del-Burgo and J. L. Pedraz, Macromol. Biosci., 2022, 22(3), 2100435 CrossRef CAS PubMed.
- R. Contentin, M. Demoor, M. Concari, M. Desancé, F. Audigié, T. Branly and P. Galéra, Stem Cell Rev. Rep., 2020, 16(1), 126–143 CrossRef CAS PubMed.
- J. A. Semba, A. A. Mieloch and J. D. Rybk, Bioprinting, 2020, 18, e00070 CrossRef.
- O. Jeon, Y. B. Lee, T. J. Hinton, A. W. Feinberg and E. Alsberg, Mater. Today Chem., 2019, 12, 61–70 CrossRef CAS PubMed.
- C. Luo, R. Xie, J. Zhang, Y. Liu, Z. Li, Y. Zhang, X. Zhang, T. Yuan, Y. Chen and W. Fan, Tissue Eng., Part C, 2020, 26, 306–316 CrossRef CAS PubMed.
- J. Yin, M. Yan, Y. Wang, J. Fu and H. Suo, ACS Appl. Mater. Interfaces, 2018, 10, 6849–6857 CrossRef CAS PubMed.
- Y. Sun, Y. You, W. Jiang, B. Wang, Q. Wu and K. Dai, Sci. Adv., 2020, 6(37) Search PubMed.
- Y. Sun, Y. You, W. Jiang, Z. Zhai and K. Dai, Theranostics, 2019, 9(23), 6949–6961 CrossRef CAS PubMed.
- R. Levato, W. R. Webb, I. A. Otto, A. Mensinga, Y. Zhang, M. van Rijen, R. van Weeren, I. M. Khan and J. Malda, Acta Biomater., 2017, 1(61), 41–53 CrossRef PubMed.
- P. Diloksumpan, M. de Ruijter, M. Castilho, U. Gbureck, T. Vermonden, P. R. van Weeren, J. Malda and R. Levato, Biofabrication, 2020, 12(2) CrossRef CAS PubMed.
- E. Vinod, R. Parameswaran, B. Ramasamy and U. Kachroo, Cartilage, 2021, 12(2_suppl), 34S–52S CrossRef PubMed.
- I. M. Khan, J. C. Bishop, S. Gilbert and C. W. Archer, Osteoarthritis Cartilage, 2009, 17, 518–528 CrossRef CAS PubMed.
- C. T. Jayasuriya and Q. Chen, Connect. Tissue Res., 2015, 56(4), 265–271 CrossRef CAS PubMed.
- H. Chen, F. Fei, X. Li, Z. Nie, D. Zhou, L. Liu, J. Zhang, H. Zhang, Z. Fei and T. Xu, Bioact. Mater., 2021, 6(10), 3580–3595 CrossRef CAS PubMed.
- J. M. Baena, G. Jiménez, E. López-Ruiz, C. Antich, C. Griñán-Lisón, M. Perán, P. Gálvez-Martín and J. A. Marchal, Exp. Biol. Med., 2019, 244, 1 CrossRef PubMed.
- J. Idaszek, M. Costantini, T. A. Karlsen, J. Jaroszewicz, C. Colosi, S. Testa, E. Fornetti, S. Bernardini, M. Seta, K. Kasarełło, R. Wrzesień, S. Cannata, A. Barbetta, C. Gargioli, J. E. Brinchman and W. Święszkowski, Biofabrication, 2019, 11, 4 CrossRef PubMed.
- K. M. Hubka, R. L. Dahlin, V. V. Meretoja, F. K. Kasper and A. G. Mikos, Tissue Eng., Part B, 2014, 20(6), 641–654 CrossRef PubMed.
- K. Pelttari, H. Lorenz, S. Boeuf, M. F. Templin, O. Bischel, K. Goetzke, H.-Y. Hsu, E. Steck and W. Richter, Arthritis Rheum., 2008, 58(2), 467–474 CrossRef CAS PubMed.
- E. B. Hunziker, T. M. Quinn and H.-J. Häuselmann, Osteoarthritis Cartilage, 2002, 10, 564–572 CrossRef CAS PubMed.
- X. Ren, F. Wang, C. Chen, X. Gong, L. Yin and L. Yang, BMC Musculoskeletal Disord., 2016, 17, 301 CrossRef PubMed.
- L. Zhou, G. J. V. M. van Osch, J. Malda, M. J. Stoddart, Y. Lai, R. G. Richards, K. K.-w. Ho and L. Qin, Adv. Healthcare Mater., 2020, 9(23) Search PubMed.
- R. M. Raftery, I. M. Castaño, S. Sperger, G. Chen, B. Cavanagh, G. A. Feichtinger, H. Redl, A. Hacobian and F. J. O’Brien, J. Controlled Release, 2018, 283, 20–31 CrossRef CAS PubMed.
- C. M. Curtin, E. G. Tierney, K. McSorley, S.-A. Cryan, G. P. Duffy and F. J. O'Brien, Adv. Healthcare Mater., 2015, 4, 223–227 CrossRef CAS PubMed.
- C. M. Curtin, G. M. Cunniffe, F. G. Lyons, K. Bessho, G. R. Dickson, G. P. Duffy and F. J. O'Brien, Adv. Mater., 2012, 24, 749–754 CrossRef CAS PubMed.
- R. M. Raftery, I. M. Castaño, G. Chen, B. Cavanagh, B. Quinn, C. M. Curtin, S.-A. Cryan and F. J. O’Brien, Biomaterials, 2017, 149, 116–127 CrossRef CAS PubMed.
- Z. Yu, Y. Li, Y. Wang, Y. Chen, M. Wu, Z. Wang, M. Song, F. Lu, X. Lu and Z. Dong, Biosci. Rep., 2019, 39 Search PubMed.
- J. R. García, A. Y. Clark and A. J. García, J. Biomed. Mater. Res., Part A, 2016, 104, 889–900 CrossRef PubMed.
- A. I. Caplan and D. Correa, J. Orthop. Res., 2011, 29, 1795–1803 CrossRef CAS PubMed.
- R. M. Raftery, A. G. Gonzalez Vazquez, G. Chen and F. J. O’Brien, Adv. Healthcare Mater., 2020, 9, 1901827 CrossRef CAS PubMed.
- N. van Gastel, S. Stegen, G. Eelen, S. Schoors, A. Carlier, V. W. Daniëls, N. Baryawno, D. Przybylski, M. Depypere, P.-J. Stiers, D. Lambrechts, R. Van Looveren, S. Torrekens, A. Sharda, P. Agostinis, D. Lambrechts, F. Maes, J. V. Swinnen, L. Geris, H. Van Oosterwyck, B. Thienpont, P. Carmeliet, D. T. Scadden and G. Carmeliet, Nature, 2020, 579, 111–117 CrossRef CAS PubMed.
- I. Sekiya, J. T. Vuoristo, B. L. Larson and D. J. Prockop, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4397–4402 CrossRef CAS PubMed.
- B. Johnstone, T. M. Hering, A. I. Caplan, V. M. Goldberg and J. U. Yoo, Exp. Cell Res., 1998, 238, 265–272 CrossRef CAS PubMed.
- F. Barry, R. E. Boynton, B. Liu and J. M. Murphy, Exp. Cell Res., 2001, 268, 189–200 CrossRef CAS PubMed.
- M. E. Joyce, A. B. Roberts, M. B. Sporn and M. E. Bolander, J. Cell Biol., 1990, 110, 2195–2207 CrossRef CAS PubMed.
- L. Bian, D. Y. Zhai, E. Tous, R. Rai, R. L. Mauck and J. A. Burdick, Biomaterials, 2011, 32, 6425–6434 CrossRef CAS PubMed.
- X. Cui, K. Breitenkamp, M. G. Finn, M. Lotz and D. D. D'Lima, Tissue Eng., Part A, 2012, 18, 1304–1312 CrossRef CAS PubMed.
- J. Kundu, J.-H. Shim, J. Jang, S.-W. Kim and D.-W. Cho, J. Tissue Eng. Regener. Med., 2015, 9(11), 1286–1297 CrossRef CAS PubMed.
- B. S. Yoon and K. M. Lyons, J. Cell. Biochem., 2004, 93(1), 93–103 CrossRef CAS PubMed.
- F. E. Freeman, P. Pitacco, L. H. A. Van Dommelen, J. Nulty, D. C. Browe, J.-Y. Shin, E. Alsberg and D. J. Kelly, Sci. Adv., 2020, 6(33) CAS.
- Y. Zhu, M. Yuan, H. Y. Meng, A. Y. Wang, Q. Y. Guo, Y. Wang and J. Peng, Osteoarthritis Cartilage, 2013, 21(11), 1627–1637 CrossRef CAS PubMed.
- E. Kon, R. Buda, G. Filardo, A. Di Martino, A. Timoncini, A. Cenacchi, P. M. Fornasari, S. Giannini and M. Marcacci, Knee Surg. Sports Traumatol. Arthrosc., 2010, 18, 472–479 CrossRef PubMed.
- G. Filardo, E. Kon, R. Buda, A. Timoncini, A. Di Martino, A. Cenacchi, P. M. Fornasari, S. Giannini and M. Marcacci, Knee Surg. Sports Traumatol. Arthrosc., 2011, 19, 528–535 CrossRef PubMed.
- E. Kon, B. Mandelbaum, R. Buda, G. Filardo, M. Delcogliano, A. Timoncini, P. M. Fornasari, S. Giannini and M. Marcacci, Arthroscopy, 2011, 27, 1490–1501 CrossRef PubMed.
- G. Irmak and M. Gümüşderelioğlu, Biomed. Mater., 2020, 15, 6 Search PubMed.
- Z. Li, X. Zhang, T. Yuan, Y. Zhang, C. Luo, J. Zhang, Y. Liu and W. Fan, Tissue Eng., Part A, 2020, 26, 15–16 CrossRef PubMed.
- G. Irmak and M. Gümüşderelioğlu, Mater. Sci. Eng., C, 2021, 125, 112092 CrossRef CAS PubMed.
- R. Burdis and D. J. Kelly, Acta Biomater., 2021, 126, 1–14 CrossRef CAS PubMed.
- I. Martin, J. Malda and N. C. Rivron, Curr. Opin. Organ Transplant., 2019, 24, 562–567 CrossRef PubMed.
- B. S. Schon, G. J. Hooper and T. B. F. Woodfield, Ann. Biomed. Eng., 2017, 45, 100–114 CrossRef CAS PubMed.
- H. Zhang, H. Huang, G. Hao, Y. Zhang, H. Ding, Z. Fan and L. Sun, Adv. Funct. Mater., 2021, 31(1), 2006697 CrossRef CAS.
- Y. Sun, Q. Wu, Y. Zhang, K. Dai and Y. Wei, Nanomedicine, 2021, 37, 102426 CrossRef CAS PubMed.
- Y. Cao, P. Cheng, S. Sang, C. Xiang, Y. An, X. Wei, Z. Shen, Y. Zhang and P. Li, Regener. Biomater., 2021, 8, 3 Search PubMed.
- A. Dimaraki, P. J. Díaz-Payno, M. Minneboo, M. Nouri-Goushki, M. Hosseini, N. Kops, R. Narcisi, M. J. Mirzaali, G. J. V. M. van Osch, L. E. Fratila-Apachitei and A. A. Zadpoor, Appl. Sci., 2021, 11, 7821 CrossRef CAS.
- Z. Yang, T. Zhao, C. Gao, F. Cao, H. Li, Z. Liao, L. Fu, P. Li, W. Chen, Z. Sun, S. Jiang, Z. Tian, G. Tian, K. Zha, T. Pan, X. Li, X. Sui, Z. Yuan, S. Liu and Q. Guo, ACS Appl. Mater. Interfaces, 2021, 13, 23369–23383 CrossRef CAS PubMed.
- C. Di Bella, S. Duchi, C. D. O'Connell, R. Blanchard, C. Augustine, Z. Yue, F. Thompson, C. Richards, S. Beirne, C. Onofrillo, S. H. Bauquier, S. D. Ryan, P. Pivonka, G. G. Wallace and P. F. Choong, Tissue Eng. Regener. Med., 2018, 12, 611–621 CrossRef CAS PubMed.
- A. G. González Vázquez, L. A. Blokpoel Ferreras, K. E. Bennett, S. M. Casey, P. A. J. Brama and F. J. O’Brien, Adv. Healthcare Mater., 2021, 10(20), e2100878 CrossRef PubMed.
- M. Chen, Y. Li, S. Liu, Z. Feng, H. Wang, D. Yang, W. Guo, Z. Yuan, S. Gao, Y. Zhang, K. Zha, B. Huang, F. Wei, X. Sang, Q. Tian, X. Yang, X. Sui, Y. Zhou, Y. Zheng and Q. Guo, Bioact. Mater., 2021, 6, 1932–1944 CrossRef CAS PubMed.
- S. E. Critchley, E. J. Sheehy, G. M. Cunniffe, P. Diaz-Payno, S. F. Carroll, O. Jeon, E. Alsberg, P. A. J. Brama and D. J. Kelly, Acta Biomater., 2020, 113, 130–143 CrossRef CAS PubMed.
- F. Berthiaume, T. J. Maguire and M. L. Yarmush, Annu. Rev. Chem. Biomol. Eng., 2011, 2, 403–430 CrossRef PubMed.
- D. C. Kelly, R. M. Raftery, C. M. Curtin, C. M. O'Driscoll and F. J. O'Brien, J. Orthop. Res., 2019, 37, 1671–1680 CrossRef PubMed.
- C.-C. Ma, Z.-L. Wang, T. Xu, Z.-Y. He and Y.-Q. Wei, Biotechnol. Adv., 2020, 40, 107502 CrossRef CAS PubMed.
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller and T. Zaks, N. Engl. J. Med., 2021, 384, 403–416 CrossRef CAS PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck Jr., L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- L. Schoenmaker, D. Witzigmann, J. A. Kulkarni, R. Verbeke, G. Kersten, W. Jiskoot and D. J. A. Crommelin, Int. J. Pharm., 2021, 601, 120586 CrossRef CAS PubMed.
- T. Gonzalez-Fernandez, D. J. Kelly and F. J. O’Brien, Adv. Ther., 2018, 1(7), 1800038 CrossRef.
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359(6372), eaan4672 CrossRef PubMed.
- X. Guo and L. Huang, Acc. Chem. Res., 2012, 45, 971–979 CrossRef CAS PubMed.
- L. S. Costard, D. C. Kelly, R. N. Power, C. Hobbs, S. Jaskaniec, V. Nicolosi, B. L. Cavanagh, C. M. Curtin and F. J. O’Brien, Pharmaceutics, 2020, 12, 1219 CrossRef CAS PubMed.
- F. Cardarelli, L. Digiacomo, C. Marchini, A. Amici, F. Salomone, G. Fiume, A. Rossetta, E. Gratton, D. Pozzi and G. Caracciolo, Sci. Rep., 2016, 6, 25879 CrossRef CAS PubMed.
- E. G. Tierney, G. P. Duffy, A. J. Hibbitts, S.-A. Cryan and F. J. O'Brien, J. Controlled Release, 2012, 158, 304–311 CrossRef CAS PubMed.
- H. O. McCarthy, J. McCaffrey, C. M. McCrudden, A. Zholobenko, A. A. Ali, J. W. McBride, A. S. Massey, S. Pentlavalli, K.-H. Chen, G. Cole, S. P. Loughran, N. J. Dunne, R. F. Donnelly, V. L. Kett and T. Robson, J. Controlled Release, 2014, 189, 141–149 CrossRef CAS PubMed.
- J. E. Dixon, G. Osman, G. E. Morris, H. Markides, M. Rotherham, Z. Bayoussef, A. J. El Haj, C. Denning and K. M. Shakesheff, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E291–E299 CrossRef CAS PubMed.
- D. P. Walsh, R. M. Raftery, R. Murphy, G. Chen, A. Heise, F. J. O'Brien and S.-A. Cryan, Biomater. Sci., 2021, 9, 4984–4999 RSC.
- A. L. Laiva, R. M. Raftery, M. B. Keogh and F. J. O’Brien, Int. J. Pharm., 2018, 544(2), 372–379 CrossRef CAS PubMed.
- A. L. Laiva, F. J. O’Brien and M. B. Keogh, Biotechnol. Bioeng., 2021, 118(2), 725–736 CrossRef CAS PubMed.
- M. Suku, A. L. Laiva, F. J. O'Brien and M. B. Keogh, J. Pers. Med., 2021, 11, 4 CrossRef PubMed.
- W. A. Lackington, R. M. Raftery and F. J. O'Brien, Acta Biomater., 2018, 75, 115–128 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |