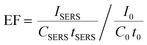Open Access Article
Open Access ArticleSurface-enhanced Raman scattering-based molecular encoding with gold nanostars for anticounterfeiting applications†
Yifeng
Huo
a,
Samantha
Curry
a,
Andrew
Trowbridge
ab,
Xurong
Xu
ac and
Chaoyang
Jiang
 *a
*a
aDepartment of Chemistry and Center for Fluorinated Functional Materials, University of South Dakota, Vermillion, South Dakota 57069, USA. E-mail: CY.Jiang@usd.edu
bDepartment of Chemistry, Concordia College, Moorhead, Minnesota 56562, USA
cQiushi Academy for Advanced Studies, Zhejiang University, Hangzhou, Zhejiang, 310027, P. R. China
First published on 7th June 2021
Abstract
Recently, surface-enhanced Raman scattering (SERS) joined other optical methods in making novel anticounterfeiting materials due to the fact that abundant molecular fingerprints in Raman spectra can be less susceptible to fraud. Using these molecular features, it is critical to make novel nanostructures with increased SERS enhancement and stability. Herein, we synthesized star-shaped gold nanoparticles as SERS substrates and applied various Raman probes with these gold nanostars to make SERS tags. The encoded molecular information was successfully decoded using principal component analysis (PCA). These colloidal tags can be further stabilized when embedded in a polymer matrix. We made a prototype ballpoint pen that can do simple writing with these secret SERS inks.
Introduction
The development of anticounterfeiting technology has received increasing attention due to ongoing global economic activities, as well as the protection of intellectual properties. In fighting counterfeiting activities, anticounterfeiting labels have been widely used to authenticate the genuineness of commercial goods, financial documents, and advanced pharmaceutical products. Various techniques and materials have been used in anticounterfeiting, such as watermarks,1 microscale patterns,2,3 fluorescent quantum dots,4 and upconversion nanomaterials.5,6 However, many of these well-developed anticounterfeiting techniques could be targeted by counterfeiters, thus being faked. It is thus essential to introduce new techniques into the field of anticounterfeiting, which will be less known to counterfeiters. In this regard, security labels based on a surface-enhanced Raman scattering (SERS) technique could be a very promising candidate due to their molecular fingerprint features, high sensitivity, and sufficient complexity. As we know, Raman spectroscopy is a noninvasive analytical method that can provide molecular spectra that contain narrow spectral bands associated with chemical structures.7,8 In the SERS study, Raman signals are dramatically increased with an enhancement factor of 106 or higher when Raman probes are placed at the vicinity of rough noble metals or other plasmonic materials.9,10 These enhanced signals allow the low-dose presence of molecular labels as well as the capability of using multiple SERS probes. Such probe-encoded Raman and SERS tags have actually been developed and applied in bioanalysis and bioimaging.11–16 It is worth noting that SERS labels are novel, complicated, and have a high storage capacity, all of which are critical for the development of new generation anticounterfeiting labels.Recently, there has been continuous interest in developing encoded SERS labels for anticounterfeiting applications.17–29 For example, Ling and co-workers fabricated plasmonic patterns of silver nanorods and studied their applications in anticounterfeiting with multiple Raman probes.17 In addition, there were also examples of combining SERS labels with bar codes, quick response (QR) codes, or physically unclonable features.27–31 In these studies, SERS-active substrates are a key component for the successful application of their materials in anticounterfeiting. While lithography methods were still widely used in making SERS-active plasmonic substrates, chemical syntheses of plasmonic nanomaterials have several advantages such as high tunability, easy processability, and low cost. For example, recently, Ye and co-workers reported the synthesis of gap-enhanced Raman tags and demonstrated their remarkable applications in anticounterfeiting.28 There is clearly an increasing demand in developing facile synthetic approaches to make novel SERS substrates for anticounterfeiting applications.
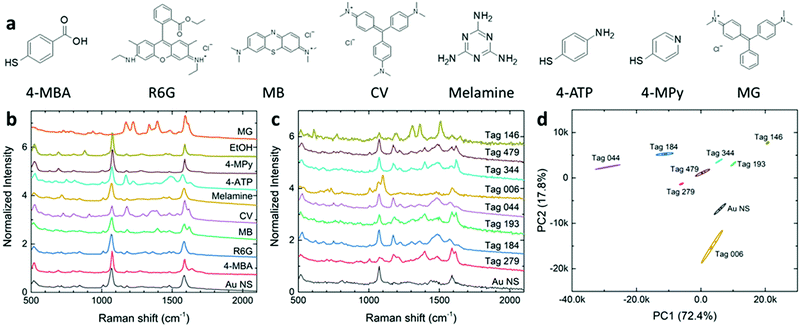
On the other hand, gold nanostars (AuNSs) have been synthesized through either seedless32,33 or seed-mediated34 methods. These nanostars have been widely utilized in sensing,35 catalysis,36 therapy,37 and SERS.38,39 Recently, we reported the synthesis of gold nanostars with sharp branches34 and studied their SERS performance.40 We found that AuNSs have a higher SERS enhancement as compared to normal gold nanoparticles.41 These AuNSs with the modification of a proper capping agent can be suspended in aqueous media and behave as colloidal SERS substrates.41,42 Recently, there were also reports on the use of encoded AuNSs for various applications such as cell discrimination.15,16 However, some of the preparation methods are rather complicated. Furthermore, using colloidal AuNS substrates in the field of anticounterfeiting applications has not been completely investigated.
In this paper, we present a new approach to design anticounterfeiting labels by using the SERS spectra of molecules on AuNSs to encrypt secret information. Colloidal AuNS solutions were mixed with multiple Raman probe molecules to form SERS tags. Their SERS spectra with unique features were then analysed, which are associated with the adsorption of Raman probes on the surface of nanostars. We also demonstrated a simple way to build a spectral database using a combination of binary coding and principal component analysis (PCA). These SERS tags can have long-term stablity after being embedded in the polymer matrix. Furthermore, a prototype ink containing the branched gold nanostars was formulated and the ink can be used to make anti-counterfeiting markings using a normal ballpoint pen.
Experimental
Chemicals and materials
Gold chloride solution (HAuCl4·3H2O, 30 wt%) and melamine were purchased from Sigma Aldrich. Trisodium citrate, ascorbic acid, and methylene blue (MB) were purchased from Alfa Aesar. Hydroquinone (HQ), rhodamine 6G (R6G), crystal violet (CV), 4-aminothiolphenol (4-ATP), 4-mercaptopyridine (4-MPy), poly(ethylene oxide) (PEO, Mw ∼ 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), and polyvinylpyrrolidone (PVP, Mw ∼ 55
000), and polyvinylpyrrolidone (PVP, Mw ∼ 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Acros Organics. 4-Mercaptobenzoic acid (4-MBA) was purchased from TCI America. Malachite green (MG) was purchased from Thermo Fisher Scientific. Anhydrous ethanol was purchased from Pharmco-AAPER. All the chemicals were of analytical grade and used without further purification.
000) were purchased from Acros Organics. 4-Mercaptobenzoic acid (4-MBA) was purchased from TCI America. Malachite green (MG) was purchased from Thermo Fisher Scientific. Anhydrous ethanol was purchased from Pharmco-AAPER. All the chemicals were of analytical grade and used without further purification.
Synthesis of colloidal labels
Colloidal AuNS solutions were synthesized using a known seed-mediated method that was recently developed in our laboratory.34 To make an anti-counterfeiting label, probe solutions were first prepared by dissolving one or more listed chemicals, including 4-MBA, R6G, MB, CV, melamine, 4-ATP, 4-MPy, ethanol, and MG. A typical concentration of each chemical in the probe stock solution was 100 μM. Then, 100 μL of probe solution was added to 900 μL of AuNS solution to produce a uniform solution as the colloidal anti-counterfeiting label.Production of AuNS-PEO thin-film labels
To make thin-film anti-counterfeiting labels, we first dissolved PEO powders into the colloidal labels with a PEO concentration of 10 wt.%. The AuNS-PEO thin films were prepared by spin coating 10 drops of the AuNS-PEO mixture on aluminum foils (3000 rpm). The coated aluminum foils were then dried in an oven (100 °C) to produce solid AuNS-PEO thin films.Production of writable AuNS-PVP inks
To produce writable labels, 25 mL of AuNS solutions were rinsed with a centrifugation–redispersion cycle (8000 rpm for 10 minutes) and then concentrated into 1 mL. Then 50 μL of the concentrated AuNS solution was mixed with 50 μL of probe solution and incubated for two hours. 0.4 mL of 2 wt% PVP solution was then further concentrated into 40 μL (anti-counterfeiting inks). Ballpoint pen cartridges were ultrasonically cleaned with water and ethanol. Finally, anti-counterfeiting inks were injected into the cartridges and the ballpoint pens were ready for writing.Characterizations
The UV–vis spectra of colloidal AuNS solutions in glass vials (diameter 15 mm) were measured using a GENESYS 30 Visible spectrophotometer. The AuNS morphologies were characterized using a field emission scanning electron microscope (FE-SEM) (Zeiss) and a transmission electron microscope (Tecnai Spirit G2 Twin, FIE Company) with a LaB6 filament operated at 120 kV. Electron micrographs were obtained by projection using a Gatan Orius SC200 CCD or Gatan Rio16 CMOS Digital Camera. Raman spectra of both colloidal solutions and thin-film samples were acquired using an i-Raman EX portable Raman spectrometer (B&W Tek). The 1064 nm laser with a power of 373 mW was used in the Raman measurements.Principal component analysis
The principal component analysis (PCA) was conducted using Origin Software with the PCA for the spectroscopy function when the SERS spectra were processed. Multiple SERS spectra (typically 5) were acquired from each sample and then normalized before the PCA process. The Origin Software used a mathematical process to differentiate all these spectra and quantify the difference in the spectra by plotting the graphs of multiple principal components (PCs). In the PCA plots, the deviations of each batch were calculated based on a 95% confidence level and shown as ellipses in the score plot. These ellipses provide clear conclusions on the differences in the samples.Results and discussion
SERS spectra on plasmonic gold nanostars
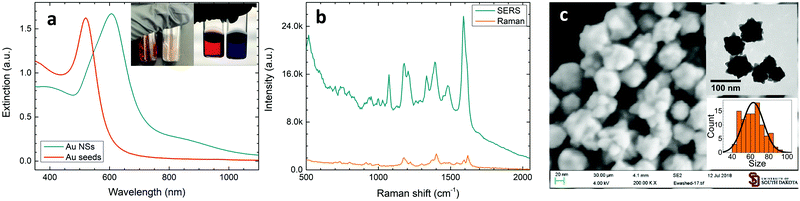
While the formation of sharp branches on AuNSs has been discussed in detail in our previous reports,34 here, we would like to mention that the strong extinction of AuNSs can be realized when observing a fast color change from light red to brownish blue during the chemical synthesis. The as-synthesized AuNS solutions are significantly different as compared to their gold seeds, in both optical images and UV–visible spectra (Fig. 1a). The AuNS solution has a strong extinction peak at 606 nm, which is red shifted from the peak of gold seeds at 520 nm, mainly due to the increase of the nanoparticle size.These AuNSs exhibited excellent SERS activity when Raman probe molecules were introduced into the colloidal solutions. Shown in Fig. 1b is the SERS spectrum of MG in the AuNS solution, in which characteristic MG Raman peaks can be easily observed. It is worth noting that this SERS spectrum was directly recorded using a colloidal solution in a glass vial, which will ensure great signal stability and data reproducibility. The Raman peak of MG at 1396 cm−1 associated with the N-phenyl stretching mode43 was then used to calculate the analytical enhancement factor (EF) for our AuNS colloidal solution. Using the equation:
Recently, there have been several reports on using PCA as a mathematic method in distinguishing the tiny difference in SERS spectra when a trace number of probe molecules was introduced.34,44–46 In this work, the Raman spectra of AuNSs with R6G at a concentration of 0.5 nM only showed increases of 1073 and 1590 cm−1 peaks compared to the spectra of bare AuNSs and there was no obvious R6G Raman peak (Fig. S2a, ESI†). These two peaks were assigned to the aromatic ring vibrational mode of 4-MBA, a stabilizing agent used in the AuNS synthesis.47 The intensity increase was temporally assigned to the disturbance of a trace amount of R6G, as we have discussed previously.41 From the PCA plot, we clearly observed the difference caused by R6G addition, as shown by the good separation of the two ellipses drawn at a confidence level of 95% (see Fig. S3b, ESI†). On the contrary, the PCA can hardly differentiate the spectral difference when quasi-spherical gold nanoparticles (Au NPs) were used (Fig. S2 and S3b, ESI†), which is consistent with that reported recently.41 In this work, the PCA processing will be used to identify Raman probes added to the colloidal AuNS solutions, as well as to develop anti-counterfeiting SERS labels.
SERS coding with various Raman probes
Knowing that various Raman probes can produce characteristic SERS spectra, here, we designed a unique approach to make anti-counterfeiting labels, where the spectroscopic information was encoded by mixing Raman probes with AuNSs and then decoded using PCA. The spectroscopic information was generated by measuring the SERS spectra on AuNSs with various chemicals. Here, nine chemicals were selected in this trial work, including 4-MBA, R6G, MB, CV, melamine, 4-ATP, 4-MPy, ethanol, and MG (Fig. 2a). Some of the selected chemicals contain a thiol group which could strongly bind to gold, and some chemicals were dyes with a relatively large Raman cross section. We also included ethanol in our list, which can likely cause a small change in the solvent environment. The SERS spectra of various AuNS stock solutions are shown in Fig. 2b, in which the individual chemicals were added. It is quite clear that some characteristic Raman peaks can be found in certain spectra. For example, the Raman spectra of AuNSs with R6G has a unique peak at 1365 cm−1, which can be associated with the aromatic C–C stretching of R6G molecules.48 Overall, we also found that there are some common Raman signals in these spectra, such as a peak at 1073 cm−1 that can be attributed to the residual 4-MBA used in the AuNS synthesis. It should be noted that the intensity of this peak varies in these spectra which could be due to some interference from other chemicals.It is anticipated that multiple chemicals mixed in the AuNS solution can result in complicated SERS spectra as a readout which can only be recognized with a known spectral database. Here, the combination of these nine SERS-active chemicals will be tried to produce unique spectra that will carry binary codes with a maximum of nine digits. To design the binary code, the presence of a chemical stands for “1” while the absence of the chemical stands for “0”. With this, the total number of combinations, P, can be calculated as follows:
| P = mn | (1) |
SERS spectra of SERS tags were recorded using simple direct measurements when these tags were placed in glass vials. As shown in Fig. 2c, it is not surprising that these SERS spectra contain complete and detailed Raman features due to the possible presence of nine Raman probes. Each SERS tag has its own unique SERS spectra with various peak positions, widths, and intensities. With multiple Raman probes in the tag, the exact peak assignments could be tricky and challenging. For example, both tag 184 and tag 044 contain CV and 4-ATP, and the peak of CV at 1480 cm−1 was overlapped with a peak at 1490 cm−1 from 4-ATP. Here, we would emphasize that the SERS spectrum of a SERS tag is NOT a simple addition of Raman spectra of individual chemicals. This is due to the possibility that various chemicals might have a competition when being adsorbed onto the AuNS surface. In this regard, a better understanding of the co-adsorption of multiple chemical ligands onto the AuNS surface would be necessary if someone wants to re-generate the “combined” spectra. It is therefore an additional challenge if the counterfeiter wants to duplicate these SERS tags. Fortunately, the public can simply use the combined SERS fingerprints in the authentication process.
The unique spectral information of these SERS tags can be presented in a PCA score plot. As shown in Fig. 2d, five data points of duplicate measurements on individual tags can be well clustered together. There are enough distances between any neighboring data sets. The well-separated 95% ellipses indicated the adequate capability of distinguishing any of these eight SERS tags. We designed an “unknown” tag and included its data in the PCA score plot (Fig. S4, ESI†). It is found that the location of the “unknown” tag in the PCA score plot is overlapped with the SERS tag 344, which is consistent with the record on the lab notebook. This example confirmed that one can identify the encrypted information by measuring the Raman spectra of the SERS tag and then compare it with a well-established database. Furthermore, this result also proved the excellent reproducibility of SERS-coded nanostars, which is an important issue in anticounterfeiting applications.
Additional experiments were conducted to confirm the reproducible performance of these SERS tags via preparing various sets of SERS tags and using various batches of the synthesized AuNSs. A typical result of this reproducibility study is shown in Fig. 3, in which four sets of SERS tags were examined. The corresponding SERS tag IDs are listed in Table S2 (ESI†). Sets I and II were prepared with one batch of AuNSs while sets III and IV were made with different AuNS batches. All sets include a common SERS tag of tag 330 for comparison. As shown in Fig. 3, the four ellipses associated with tag 330 overlap very well to each other. This overlap demonstrates the excellent reproducibility of these SERS tags in various batches. Similarly, the reproducibility from different batches of AuNSs can also be observed with tag 159 and tag 225. On the other hand, SERS tags with various combinations of chemical probes are well separated which, again, confirmed the powerful coding capacity using the approach of SERS labelling.
Optimization of the SERS coding
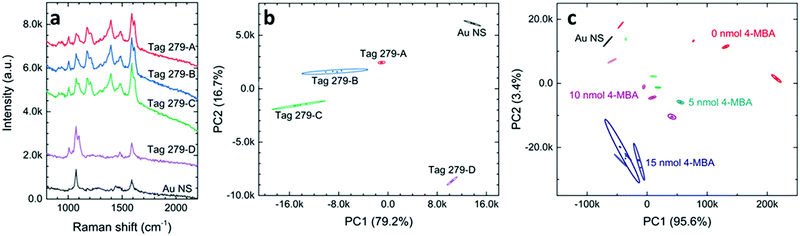
The capacity of SERS tags for storing the encrypted information is an important figure of merit (FOM) for an anticounterfeiting product. According to eqn (1), one approach to enlarge the information capacity is using additional types of Raman probe molecules (n) in the SERS tags. Another way is to increase the number of states for chemicals (m). This can probably be achieved by varying the chemical concentrations in the SERS tags in a discrete manner as far as their spectral differences, mainly in peak intensities, can be well distinguished in the PCA score plots. We first carefully applied four levels of probe concentrations on tag 279 with the MG concentrations of 100, 50, 25, and 0 μM, which are labeled tag 279-A, tag 279-B, tag 279-C, and tag 279-D (see Table S3 for detailed chemical combinations, ESI†). It is quite clear that Raman intensities varied for these SERS tags. In Fig. 4a, we can observe that the peak of MG, located at 1614 cm−1, increases with the increase of the MG concentration. Using the PCA score plot, we found that the ellipses of these four data clusters were separated (Fig. 4b). This result indicates that there is enough recognition of the encrypted information in these tags and, therefore, it is a practical approach to expand the tag capacity by varying the chemical concentration. This is quite exciting as we will be able to make 49 = 262,144 combinations using only nine chemicals if all these chemicals can be used with four different levels of concentrations.Now the question would be, how small an increment step can we use in the SERS tags? This question is rather challenging since the ability to distinguish various sets of SERS tags not only depends on the difference in SERS spectra, but also relies on the data process. Using the commercial mathematic software, Origin Pro, we have been able to separate some tags even when there is hardly an observable difference in their SERS spectra (Fig. S5, ESI†). Fig. 4c shows the PCA score plot of some SERS tags that contain both 4-MBA and MG, where the varying amounts of MG are as low as 1 nmol. The SERS tags with low concentrations of 4-MBA can be well distinguished with various concentrations of MG at micromolar regions. We also found that SERS tags with higher concentrations of 4-MBA can have different performances. Additional optimizations on the direction of changing the Raman probe concentration will be reported elsewhere. Nevertheless, we have demonstrated that changing the concentration of Raman probes can dramatically increase the information capacity of SERS tags, thus making forgery of these SERS tags more difficult.
Application of SERS tags as thin films and writable inks
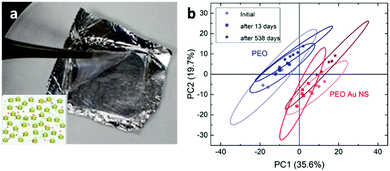
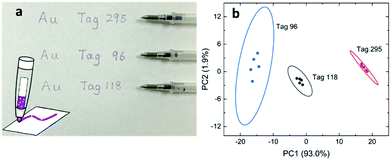
Stability and reproducibility are two major issues in the field of SERS study. For the application of SERS tags in anticounterfeiting, we also need to address these two issues. Compared to the SERS spectra from a solid state, the colloidal SERS has its intrinsic uniformity and reproducibility, as far as the problem of long-term colloidal stability can be solved. Here, we mixed the SERS tags in a PEO solution and prepared polymer thin films containing SERS tags using a spin-coating method (Fig. 5a). These polymeric nanocomposite thin films showed a pale pink color due to the existence of a low concentration of gold nanostars. The SERS spectra of these PEO composite thin films were recorded (Fig. S6, ESI†) and the PCA score plot (Fig. 5b) showed that the PEO film with SERS tags can be easily separated from the pure PEO film. Furthermore, the SERS data in the PCA score plot can be well overlapped for the sample measured at different times (immediately, 13 days, and 538 days). This result indicates that these SERS tags are very stable in the polymer matrix. With increased long-term stability, these SERS tag embedded polymer films are readily applied as plastic packing for good authentication and they might be further cooperated with other existing anti-counterfeiting techniques.Another way to use the SERS tags is to disperse them into polymer solutions (sols) which can then be applied as an ink for either printing or writing. There have been several reports on using gold nanoparticle calligraphy to produce SERS substrates.49,50 In our approach, we dispersed SERS tags into PVP solutions and made “special” inks for the possible anticounterfeiting applications. In this work, SERS inks with numbers 295, 96, and 118 were prepared (refer to Table S4 for the chemical combinations, ESI†). Fig. 6a shows the schematic of the SERS ink and the image of three ballpoint pens filled with these SERS inks in cartridges, respectively. The color of the SERS ink is decent pink due to the high concentration of AuNSs. The appearances of the letters and numbers are rather similar as shown in Fig. 6a. However, the SERS spectra of writing samples were quite different (Fig. S7, ESI†). From the PCA score plot (Fig. 6b), we can see that these three writing samples can be easily identified, given a known database of these SERS tags. The ellipse of data cluster for ink 96 is larger than the other two, which means that there are additional needs to improve the ink formulation. Our results showed that the PVP polymer in the ink does not leave any negative impact on identifying the SERS tags, a spectroscopic decoding process. As far as we know, this is the first report on using AuNSs in SERS ink for security applications. With further optimization, this kind of SERS ink can be used to produce encrypted security writing and printing, which will be an essential part in the mass production of security printing labels.
Overall, SERS-based molecular coding uses a unique cryptography method for anticounterfeiting applications. As we know, a typical cryptography method uses certain algorithms to generate public keys and private keys. In our approach, the private key was used to encrypt the information, and the encrypted information could be decoded by the public key without knowing the encryption method of the private key. Here, a chemical process, the adsorption of Raman probe molecules onto gold nanostars, was used as the “chemical algorithm” to generate keys. A private key is the combination of chemicals to produce a label from the label database. The public key is the relationship between Raman data positions and certain goods. In practical operation, a company could use the private key to encrypt certain type of goods by attaching a printed label, and the resellers or customers could measure the Raman spectra and use the public key to determine whether it is genuine or counterfeit. Compared to the other solid SERS substrates, the colloidal AuNS in this work demonstrated satisfactory SERS performance due to their branched nanostructures. With the enhanced Raman signals, SERS spectra of these encoded AuNSs can be well distinguished with a rather short acquisition time (typically a few seconds or less). The fast readout of these encoded SERS labels is very important for the purpose of anticounterfeiting applications. Furthermore, considering the complexity of surface adsorption, as well as the large pool of possible chemicals as Raman probes and nanomaterial morphology-dependent SERS characteristics, it is almost impossible for counterfeiters to forge the security labels. Using the potential of the encoded AuNSs for anticounterfeiting applications, SERS readout devices, cloud storage, spectra recognition, as well as approaches of Artificial Intelligence (machine learning) need to be developed and integrated.
Conclusions
In conclusion, we have demonstrated new colloidal SERS tags based on star-shaped gold nanoparticles that have potential in anti-counterfeiting. Various chemicals have been selected to mix with AuNSs, which produced unique SERS spectra. Using PCA analyses, we can measure the SERS spectra and recognize “unknown” tags. The capacity of the stored molecular information can be further increased by varying the concentration of chemical labels. In addition, we found that the polymeric packing could stabilize the SERS tag and extend the shelf lifetime. These AuNS-based SERS tags can be formulated into writable inks so that secret writings can be accomplished using a simple ballpoint pen. Further investigation on the impact of surface adsorption on the SERS spectra will provide a better understanding in promoting these anticounterfeiting labels.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Center for Security Printing & Anti-Counterfeiting Technology (SPACT) and the Center of Functional Fluorinated Materials (CFFM) from the state of South Dakota, Governor's Office of Economic Development, partially by four grants from the National Science Foundation (Award No. CHE-1337707, EEC-1560323, EEC-1852306, and CHE-2018422) and a seed grant from the NSF OIA-1849206 project.Notes and references
- S. Huang and J. K. Wu, IEEE Trans. Inf. Forensics Secur., 2007, 2, 164–173 Search PubMed.
- S. Han, H. J. Bae, J. Kim, S. Shin, S. E. Choi, S. H. Lee, S. Kwon and W. Park, Adv. Mater., 2012, 24, 5924–5929 CrossRef CAS PubMed.
- H. J. Bae, S. Bae, C. Park, S. Han, J. Kim, L. N. Kim, K. Kim, S. H. Song, W. Park and S. Kwon, Adv. Mater., 2015, 27, 2083–2089 CrossRef CAS PubMed.
- Y. Liu, F. Han, F. Li, Y. Zhao, M. Chen, Z. Xu, X. Zheng, H. Hu, J. Yao, T. Guo, W. Lin, Y. Zheng, B. You, P. Liu, Y. Li and L. Qian, Nat. Commun., 2019, 10, 2409 CrossRef PubMed.
- J. M. Meruga, W. M. Cross, P. S. May, Q. Luu, G. A. Crawford and J. J. Kellar, Nanotechnology, 2012, 23, 395201 CrossRef PubMed.
- M. You, J. Zhong, Y. Hong, Z. Duan, M. Lin and F. Xu, Nanoscale, 2015, 7, 4423–4431 RSC.
- C. V. Raman, Indian J. Phys., 1928, 2, 387–398 CAS.
- D. A. Long, Raman spectroscopy, New York, 1977, pp. 1–12 Search PubMed.
- N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS PubMed.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- J.-H. Kim, J.-S. Kim, H. Choi, S.-M. Lee, B.-H. Jun, K.-N. Yu, E. Kuk, Y.-K. Kim, D. H. Jeong and M.-H. Cho, Anal. Chem., 2006, 78, 6967–6973 CrossRef CAS PubMed.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516 CrossRef CAS PubMed.
- Y. Wang, B. Yan and L. Chen, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- H. Kang, S. Jeong, Y. Koh, M. Geun Cha, J.-K. Yang, S. Kyeong, J. Kim, S.-Y. Kwak, H.-J. Chang, H. Lee, C. Jeong, J.-H. Kim, B.-H. Jun, Y.-K. Kim, D. Hong Jeong and Y.-S. Lee, Sci. Rep., 2015, 5, 10144 CrossRef PubMed.
- D. Jimenez de Aberasturi, A. B. Serrano-Montes, J. Langer, M. Henriksen-Lacey, W. J. Parak and L. M. Liz-Marzán, Chem. Mater., 2016, 28, 6779–6790 CrossRef CAS.
- S. Rodal-Cedeira, A. Vázquez-Arias, G. Bodelón, A. Skorikov, S. Núñez-Sánchez, A. Laporta, L. Polavarapu, S. Bals, L. M. Liz-Marzán, J. Pérez-Juste and I. Pastoriza-Santos, ACS Nano, 2020, 14, 14655–14664 CrossRef PubMed.
- Y. Cui, R. S. Hegde, I. Y. Phang, H. K. Lee and X. Y. Ling, Nanoscale, 2014, 6, 282–288 RSC.
- Y. Cui, I. Y. Phang, Y. H. Lee, M. R. Lee, Q. Zhang and X. Y. Ling, Chem. Commun., 2015, 51, 5363–5366 RSC.
- T. Fukuoka, A. Yamaguchi, R. Hara, T. Matsumoto, Y. Utsumi and Y. Mori, 2015 International Conference on Electronics Packaging and iMAPS All Asia Conference (ICEP-IAAC), 2015 432-435.
- K. J. Si, D. Sikdar, L. W. Yap, J. K. K. Foo, P. Z. Guo, Q. Q. Shi, M. Premaratne and W. L. Cheng, Adv. Opt. Mater., 2015, 3, 1710–1717 CrossRef CAS.
- D. Li, L. Tang, J. Wang, X. Liu and Y. Ying, Adv. Opt. Mater., 2016, 4, 1475–1480 CrossRef CAS.
- R. Li, Y. Zhang, J. Tan, J. Wan, J. Guo and C. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 9384–9394 CrossRef CAS PubMed.
- X. Liu, J. Wang, L. Tang, L. Xie and Y. Ying, Adv. Funct. Mater., 2016, 26, 5515–5523 CrossRef CAS.
- Y. Liu, Y. H. Lee, Q. Zhang, Y. Cui and X. Y. Ling, J. Mater. Chem. C, 2016, 4, 4312–4319 RSC.
- L. Tian, K.-K. Liu, M. Fei, S. Tadepalli, S. Cao, J. A. Geldmeier, V. V. Tsukruk and S. Singamaneni, ACS Appl. Mater. Interfaces, 2016, 8, 4031–4041 CrossRef CAS PubMed.
- Y. Liu, Y. H. Lee, M. R. Lee, Y. Yang and X. Y. Ling, ACS Photonics, 2017, 4, 2529–2536 CrossRef CAS.
- C. L. Lay, C. S. L. Koh, J. Wang, Y. H. Lee, R. Jiang, Y. Yang, Z. Yang, I. Y. Phang and X. Y. Ling, Nanoscale, 2018, 10, 575–581 RSC.
- Y. Gu, C. He, Y. Zhang, L. Lin, B. D. Thackray and J. Ye, Nat. Commun., 2020, 11, 516 CrossRef CAS PubMed.
- Y. Zhou, G. Zhao, J. Bian, X. Tian, X. Cheng, H. Wang and H. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 28532–28538 CrossRef CAS PubMed.
- H. Cheng, Y. Lu, D. Zhu, L. Rosa, F. Han, M. Ma, W. Su, P. S. Francis and Y. Zheng, Nanoscale, 2020, 12, 9471–9480 RSC.
- S. Pekdemir, H. H. Ipekci, M. Serhatlioglu, C. Elbuken and M. S. Onses, J. Colloid Interface Sci., 2020, 584, 11–18 CrossRef PubMed.
- M. Yamamoto, Y. Kashiwagi, T. Sakata, H. Mori and M. Nakamoto, Chem. Mater., 2005, 17, 5391–5393 CrossRef CAS.
- J. Xie, J. Y. Lee and D. I. Wang, Chem. Mater., 2007, 19, 2823–2830 CrossRef CAS.
- X. Meng, A. Baride and C. Jiang, Langmuir, 2016, 32, 6674–6681 CrossRef CAS PubMed.
- M. S. Verma, P. Z. Chen, L. Jones and F. X. Gu, RSC Adv., 2014, 4, 10660–10668 RSC.
- P. Ndokoye, X. Li, Q. Zhao, T. Li, M. O. Tade and S. Liu, J. Colloid Interface Sci., 2016, 462, 341–350 CrossRef CAS PubMed.
- H. Chen, P. Kannan, L. Guo, H. Chen and D.-H. Kim, J. Mater. Chem., 2011, 21, 18271–18278 RSC.
- C. Hrelescu, T. K. Sau, A. L. Rogach, F. Jäckel and J. Feldmann, Appl. Phys. Lett., 2009, 94, 153113 CrossRef.
- A. D. S. Indrasekara, S. Meyers, S. Shubeita, L. C. Feldman, T. Gustafsson and L. Fabris, Nanoscale, 2014, 6, 8891–8899 RSC.
- O. M. Bakr, B. H. Wunsch and F. Stellacci, Chem. Mater., 2006, 18, 3297–3301 CrossRef CAS.
- X. Meng, J. Dyer, Y. Huo and C. Jiang, Langmuir, 2020, 36, 3558–3564 CrossRef CAS PubMed.
- W. Xi and A. J. Haes, J. Am. Chem. Soc., 2019, 141, 4034–4042 CrossRef CAS PubMed.
- L. He, N.-J. Kim, H. Li, Z. Hu and M. Lin, J. Agric. Food Chem., 2008, 56, 9843–9847 CrossRef CAS PubMed.
- I. Patel, W. Premasiri, D. Moir and L. Ziegler, J. Raman Spectrosc., 2008, 39, 1660–1672 CrossRef CAS PubMed.
- A. F. Chrimes, K. Khoshmanesh, S.-Y. Tang, B. R. Wood, P. R. Stoddart, S. S. Collins, A. Mitchell and K. Kalantar-Zadeh, Biosens. Bioelectron., 2013, 49, 536–541 CrossRef CAS PubMed.
- H. Yilmaz, S. H. Bae, S. Cao, Z. Wang, B. Raman and S. Singamaneni, ACS Appl. Nano Mater., 2019, 2, 3897–3905 CrossRef CAS.
- A. Michota and J. Bukowska, J. Raman Spectrosc., 2003, 34, 21–25 CrossRef CAS.
- P. Hildebrandt and M. Stockburger, J. Phys. Chem., 1984, 88, 5935–5944 CrossRef CAS.
- L. Tian, S. Tadepalli, M. E. Farrell, K.-K. Liu, N. Gandra, P. M. Pellegrino and S. Singamaneni, J. Mater. Chem. C, 2014, 2, 5438–5446 RSC.
- L. Polavarapu, A. L. Porta, S. M. Novikov, M. Coronado-Puchau and L. M. Liz-Marzán, Small, 2014, 10, 3065–3071 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: SEM graphs of AuNSs, gold nanoparticles, Raman spectra of R6G with AuNSs or Au NPs, zoom-in PCA score plot, Raman spectra of SERS tags with 4-MBA and MG, PEO films, AuNS-PEO film, and written tags, and tables of coding tags. See DOI: 10.1039/d1ma00348h |
| This journal is © The Royal Society of Chemistry 2021 |