Structurally screening calixarenes as peptide transport activators†
De-Yi
Zhang
a,
Zhe
Zheng
ab,
Hong
Zhao
a,
Huan-Yu
Wang
a,
Fei
Ding
a,
Hua-Bin
Li
a,
Yu-Chen
Pan
*a and
Dong-Sheng
Guo
 *a
*a
aCollege of Chemistry, Key Laboratory of Functional Polymer Materials (Ministry of Education), State Key Laboratory of Elemento-Organic Chemistry, Tianjin Key Laboratory of Biosensing and Molecular Recognition, Nankai University, Tianjin 300071, P. R. China. E-mail: panyuchen@mail.nankai.edu.cn; dshguo@nankai.edu.cn
bSchool of Chemical Engineering & Technology, China University of Mining and Technology, Xuzhou 221000, Jiangsu, P. R. China
First published on 8th November 2021
Abstract
Calixarenes are reportedly excellent activators that can remarkably improve the transport efficiencies of cell penetrating peptides. We employed eight calixarenes to systematically study the influence of structure on activation efficiency, which revealed that the scaffold, head group, and alkyl chain are all significant factors for activation efficiency by affecting affinities with the peptide and membrane.
Cellular membranes protect cells from harmful exogenous molecules while preventing some useful biologically active molecules from entering cells; hence transporting biologically active molecules into cells in a safe and effective manner remains a significant topic of interest.1,2 Cell penetrating peptides (CPPs) are short peptides rich in arginine and very membrane permeable.3–6 Benefiting from their low cytotoxicities, low immunogenicities, and high efficiencies,1,3,7,8 these peptides have been widely used to transport proteins, peptides, nucleotides, RNA, and DNA across membranes.1,4,8–12 Some counteranions (activators) improve CPP transport efficiency.13–18 Hence, designing and developing excellent activators have been scientific interests for many years; an ideal activator should recognize the CPP and interact with the membrane.
Calixarenes, which are third-generation supramolecular hosts, have pre-organized structures and tunable recognition and assembly abilities;19 hence, they are potentially excellent activators.20 As early as 2005, Matile et al. reported calixarene-based activators,21 and our group recently reported that p-sulfonatocalix[4]arene tetrapentyl ether acts as an extremely effective activator, with EC50 values more than three orders of magnitude smaller than those of classic activators.22 Our group also reported that amphiphilic sulfonatocalix[5]arene (sCx5-6C) activates lysine-rich peptide and protein transport, which cannot be achieved using other established counterion activators.17 While calixarene has a tunable scaffold that is easily modified, leading to diverse structures,20 the effect of calixarene structure on activation efficiency has not been explored. Investigating the calixarene structure–activity relationship is helpful for understanding their ultra-high activation efficiencies and for designing new activators.
In this work, we studied the effect of calixarene structure on activation efficiency by employing eight calixarenes (Fig. 2 and Fig. S1, ESI†) and dividing them into three groups: amphiphilic sulfonatocalix[4]arene (sCx4-6C), sCx5-6C, amphiphilic sulfonatocalix[6]arene (sCx6-6C), and amphiphilic sulfonatocalix[8]arene (sCx8-6C) were used to study the influence of the scaffold; sCx5-6C, amphiphilic carboxycalix[5]arene (cCx5-6C), and amphiphilic phosphinocalix[5]arene (pCx5-6C) were used to study the influence of the head group, while sulfonatocalix[4]arene (sCx4), sCx4-6C, and amphiphilic sulfonatocalix[4]arene (sCx4-12C) were used to study the influence of the alkyl chain length.
The carboxyfluorescein (CF) assay (Fig. 1) (see ESI† for details) was used to study the activation efficiencies of the chosen calixarenes,16,17,22,24,25 and nonaarginine (Arg9) as well as α-poly-L-lysine (polyLys) were used in this study (Fig. 2). The time-dependent fluorescence of CF-encapsulated large unilamellar egg-yolk phosphatidylcholine vesicles (EYPC-LUVs⊃CF) (Fig. S5, ESI†) was measured in response to varying concentrations of calixarene and appropriate concentrations of Arg9 or polyLys (0.350 μM for Arg9 or 0.125 μM for polyLys; Fig. S6 and S7, ESI†). No CF efflux was observed prior to the addition of Arg9 or polyLys, which indicates that the calixarenes do not cause transient pores or vesicle lysis (Fig. S6–S9, ESI†).
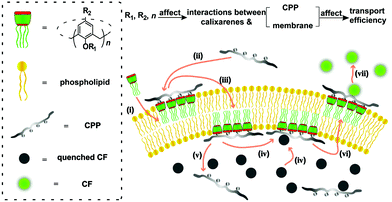 | ||
| Fig. 1 Illustrating how a calixarene activates CPP transport into an artificial membrane and the influence of calixarene structure on activation efficiency, with the CF assay as an example. (i) Interactions of calixarenes with the membrane; (ii) complexation of peptides with calixarenes; (iii) membrane translocation; (iv) complexation of CF; (v) release of peptides; (vi) membrane translocation; (vii) release of CF.17,22,23 | ||
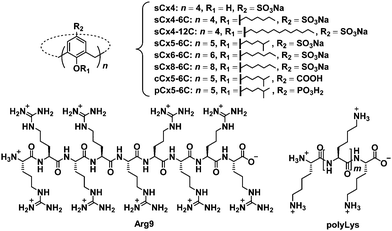 | ||
| Fig. 2 Chemical structures of the investigated calixarenes (sCx4, sCx4-6C, sCx4-12C, sCx5-6C, sCx6-6C, sCx8-6C, cCx5-6C, pCx5-6C), Arg9, and polyLys. | ||
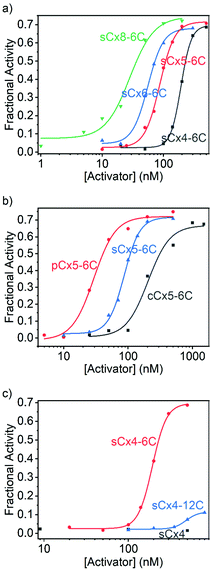
The data were fitted to the Hill equation (ESI,† eqn (S2)), which provided maximum transmembrane activity (Ymax), EC50 values and Hill coefficient (N). Transport efficiency (E), which is defined in eqn (S3) in the ESI,† was introduced to simultaneously reflect Ymax and EC50;21 a large E value corresponds to a high Ymax and low EC50 (i.e., high transport efficiency). Calixarenes with various scaffolds exhibited activation efficiencies for Arg9 transport that follow the order: sCx8-6C > sCx6-6C ≈ sCx5-6C > sCx4-6C (Fig. 3a and Table 1); the ordering for calixarenes with different head groups is: pCx5-6C > sCx5-6C > cCx5-6C (Fig. 3b and Table 1), while that for calixarenes with different alkyl chains is: sCx4-6C > sCx4-12C > sCx4 (Fig. 3c and Table 1). A Ymax value above 0.5, which is a meaningful criterion for effective transport activity, is considered to correspond to a positive hit.17,21,26Table 1 reveals that, with the exception of sCx4 and sCx4-12C, all calixarenes are robust activators for Arg9, with pCx5-6C the best among them.
| Activator | Y max | EC50 (nM) | N | E |
|---|---|---|---|---|
| a Indicates that the activity is too low to be detected. | ||||
| sCx4 | —a | — | — | — |
| sCx4-6C | 0.696 ± 0.009 | 193 ± 2 | 5.15 ± 0.30 | 12.5 ± 0.2 |
| sCx4-12C | 0.115 ± 0.010 | 483 ± 31 | 5.99 ± 2.39 | 1.85 ± 0.18 |
| sCx5-6C | 0.716 ± 0.016 | 86.9 ± 2.6 | 3.53 ± 0.32 | 14.1 ± 0.4 |
| sCx6-6C | 0.680 ± 0.012 | 55.7 ± 1.6 | 3.45 ± 0.37 | 14.0 ± 0.3 |
| sCx8-6C | 0.744 ± 0.048 | 28.2 ± 4.5 | 2.18 ± 0.61 | 16.4 ± 1.3 |
| cCx5-6C | 0.668 ± 0.044 | 204 ± 30 | 2.56 ± 1.01 | 12.0 ± 1.0 |
| pCx5-6C | 0.721 ± 0.025 | 29.4 ± 2.6 | 2.58 ± 0.56 | 15.9 ± 0.7 |
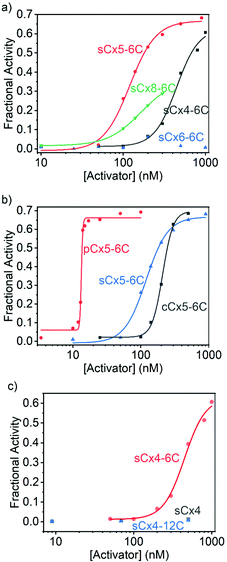
On the other hand, the activation efficiencies for polyLys transport using calixarenes with different scaffolds are ordered: sCx5-6C > sCx4-6C > sCx8-6C > sCx6-6C (Fig. 4a and Table 2), consistent with our previous results.17 In other words, the calix[5]arene scaffold is most suitable for transporting lysine-rich peptides. The activation efficiencies of calixarenes with different head groups are ordered: pCx5-6C > sCx5-6C > cCx5-6C (Fig. 4b and Table 2), while among calixarenes with different alkyl chains, only sCx4-6C activated polyLys transport, while neither sCx4 nor sCx4-12C were active (Fig. 4c and Table 2). Once again, pCx5-6C was the best activator among those examined.
| Activator | Y max | EC50 (nM) | N | E |
|---|---|---|---|---|
| sCx4 | — | — | — | — |
| sCx4-6C | 0.628 ± 0.049 | 446 ± 36 | 3.22 ± 0.70 | 10.2 ± 0.9 |
| sCx4-12C | — | — | — | — |
| sCx5-6C | 0.667 ± 0.018 | 120 ± 6 | 2.72 ± 0.35 | 12.7 ± 0.4 |
| sCx6-6C | — | — | — | — |
| sCx8-6C | 0.362 ± 0.005 | 164 ± 3 | 2.15 ± 0.05 | 6.65 ± 0.11 |
| cCx5-6C | 0.698 ± 0.018 | 210 ± 5 | 5.60 ± 0.67 | 12.5 ± 0.4 |
| pCx5-6C | 0.662 ± 0.015 | 13.3 ± 0.1 | 40.1 ± 8.0 | 15.7 ± 0.4 |
Calixarenes with different scaffolds show different effects when activating the transport of Arg9 and polyLys. The activation efficiencies for Arg9 are ordered: sCx8-6C > sCx6-6C ≈ sCx5-6C > sCx4-6C, from which we conclude that the charge and skeleton flexibility are important for Arg9 activation. As the negative charge increases, the interaction between calixarene and Arg9 will become stronger. In addition, a flexible skeleton can adapt well to the Arg9 sites and interact with more sites. Compared to arginine-rich peptides that exhibit “arginine magic”,21,27,28 lysine-rich peptides are more challenging to translocate29,30 because the ammonium group in lysine is kosmotropic, while the guanidinium in arginine is chaotropic.31 We conclude that the translocation of lysine-rich peptides requires special complexation rather than electrostatic interactions to provide the strong affinity required to promote the desolvation of the ammonium groups. To verify this, we performed the 1H NMR experiments. We previously reported that lysine was encapsulated into the cavity of sCx5-6C in a threading manner,17 and the NMR results obtained in this work show that the binding mode has not changed when lysine becomes part of the peptide (Ac-GKG-NH2). As shown in Fig. S11 and S12 (ESI†), the complexation-induced shifts become larger and larger from lysine protons α to ε, indicating increasing proximity to the cavity center, which is consistent with the threading mode.32,33 As control, the complexation-induced shifts were also observed in the case of Ac-GKG-NH2 with sCx4-6C, Ac-GRG-NH2 with sCx4-6C or sCx5-6C (Fig. S13–S15, ESI†), but not as remarkable as Ac-GKG-NH2 with sCx5-6C. This is because the cavity of calix[4]arene is too small to form the threading complex. The guanidinium residue is too large, regardless of whether for sCx4-6C or sCx5-6C, so it cannot form the threading complex either. Therefore, calix[5]arene, which has the most appropriate cavity size among the calix[n]arene (n = 4, 5, 6, 8) family for recognizing lysine, exhibits the highest activation efficiency.
Calixarenes with phosphonate groups exhibit the best activation efficiencies among the calix[5]arenes, regardless of whether for Arg9 or polyLys. There may be two factors contributing to the good efficiency of pCx5-6C. On one hand, it is well-established that phosphonates had strong affinities for guanidinium and ammonium.34–36 Moreover, phosphonate/guanidinium binds at vesicle surfaces reportedly stronger than in water.37–39 On the other hand, the solubility of pCx5-6C is much poorer than that of sCx5-6C, which indicates that pCx5-6C is less hydrophilic and may therefore be able to cross the phospholipid membrane more easily. It should be noted that the activator needs to bind moderately strongly (rather than strongly) to peptides, as an affinity that is too strong impedes peptide release, thereby reducing transport efficiency.
The sCx4-6C calixarene showed the best activation efficiency among sCx4, sCx4-6C, and sCx4-12C. The length of the alkyl chain appears to mainly affect interactions between the calixarene and the membrane. We used fluorescence polarization experiments to study the abilities of the calixarenes to embed in the membrane (see ESI† for details).40 Fig. S16 (ESI†) shows that the addition of sCx4-6C or sCx4-12C increases the fluorescence anisotropy of membrane-bound 1,6-diphenyl-1,3,5-hexatriene in EYPC-LUVs, while the addition of sCx4 resulted in little change, which indicates that sCx4-6C and sCx4-12C are embedded in the membrane, whereas sCx4 hardly interacts with the membrane. Moreover, the addition of sCx4-12C resulted in a stronger change in fluorescence anisotropy than the addition of sCx4-6C, which suggests that sCx4-12C binds more-strongly to the membrane (Fig. S16, ESI†). The above results suggest that the activator needs to be amphiphilic and interact with the membrane; however, excessive membrane interaction restricts the ability of the activator–peptide complex to move, which reduces activation efficiency.
According to above experiments, pCx5-6C has the best activation efficiency among calixarenes we used. In order to verify whether this conclusion is still valid for peptides/proteins other than Arg9 and polyLys, we further employed histones. Histones are the main components of chromatin, which are rich in both arginine and lysine.41 pCx5-6C still has the best activation efficiency among all calixarenes (Fig. S10, ESI†), consisting with the conclusion we got when using Arg9 and polyLys.
In summary, we studied and compared the activation efficiencies of eight calixarenes for the transmembrane transportation of arginine- and lysine-rich peptides, and discussed the influence of activator structure on activation efficiency. The results show that the influence of scaffold is related to the recognition mode. The negative charge and flexible scaffold are conducive to the translocation activation of arginine-rich peptides, whereas cavity size is more important for lysine-rich peptides. The head group facilitates calixarene/peptide complexation, thereby improving activation efficiency. Alkyl chains of appropriate length that provide moderate interactions with the membrane are also necessary; a weak affinity will result in activation failure, while an affinity that is too strong restricts the ability of the activator–peptide complex to move, which reduces activation efficiency. This study provides meaningful information for the design of new peptides and (even) protein transport activators.
This work was supported by NSFC (51873090 and 31961143004), Fundamental Research Funds for the Central Universities, and the NCC Fund (grant no. NCC2020FH04), which are gratefully acknowledged.
Conflicts of interest
The authors declare no conflicts of interest.Notes and references
- D. M. Copolovici, K. Langel, E. Eriste and Ü. Langel, ACS Nano, 2014, 8, 1972 CrossRef CAS PubMed.
- S. Chen, Y. Wang, T. Nie, C. Bao, C. Wang, T. Xu, Q. Lin, D.-H. Qu, X. Gong, Y. Yang, L. Zhu and H. Tian, J. Am. Chem. Soc., 2018, 140, 17992 CrossRef CAS PubMed.
- F. Heitz, M. C. Morris and G. Divita, Br. J. Pharmacol., 2009, 157, 195 CrossRef CAS PubMed.
- G. Guidotti, L. Brambilla and D. Rossi, Trends Pharmacol. Sci., 2017, 38, 406 CrossRef CAS PubMed.
- M. Kristensen, D. Birch and H. Mørck Nielsen, Int. J. Mol. Sci., 2016, 17, 185 CrossRef PubMed.
- T. Tashima, Bioorg. Med. Chem. Lett., 2017, 27, 121 CrossRef CAS PubMed.
- S. Pescina, C. Ostacolo, I. M. Gomez-Monterrey, M. Sala, A. Bertamino, F. Sonvico, C. Padula, P. Santi, A. Bianchera and S. Nicoli, J. Controlled Release, 2018, 284, 84 CrossRef CAS PubMed.
- H. Derakhshankhah and S. Jafari, Biomed. Pharmacother., 2018, 108, 1090 CrossRef CAS PubMed.
- P. P. Tripathi, H. Arami, I. Banga, J. Gupta and S. Gandhi, Oncotarget, 2018, 9, 37252 CrossRef PubMed.
- F. Wang, Y. Wang, X. Zhang, W. Zhang, S. Guo and F. Jin, J. Controlled Release, 2014, 174, 126 CrossRef CAS PubMed.
- W. L. L. Munyendo, H. Lv, H. Benza-Ingoula, L. D. Baraza and J. Zhou, Biomolecules, 2012, 2, 187 CrossRef CAS PubMed.
- H. Gao, Q. Zhang, Y. Yang, X. Jiang and Q. He, Int. J. Pharm., 2015, 478, 240 CrossRef CAS PubMed.
- S. Futaki, H. Hirose and I. Nakase, Curr. Pharm. Des., 2013, 19, 2863 CrossRef CAS PubMed.
- T. Takeuchi, M. Kosuge, A. Tadokoro, Y. Sugiura, M. Nishi, M. Kawata, N. Sakai, S. Matile and S. Futaki, ACS Chem. Biol., 2006, 1, 299 CrossRef CAS PubMed.
- J. B. Rothbard, T. C. Jessop, R. S. Lewis, B. A. Murray and P. A. Wender, J. Am. Chem. Soc., 2004, 126, 9506 CrossRef CAS PubMed.
- F. Perret, M. Nishihara, T. Takeuchi, S. Futaki, A. N. Lazar, A. W. Coleman, N. Sakai and S. Matile, J. Am. Chem. Soc., 2005, 127, 1114 CrossRef CAS PubMed.
- Y.-C. Pan, A. Barba-Bon, H.-W. Tian, F. Ding, A. Hennig, W. M. Nau and D.-S. Guo, Angew. Chem., Int. Ed., 2021, 60, 1875 CrossRef CAS PubMed.
- S. Katayama, I. Nakase, Y. Yano, T. Murayama, Y. Nakata, K. Matsuzaki and S. Futaki, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2134 CrossRef CAS PubMed.
- S. Biros and F. Hof, Supramol. Chem., 2012, 4, 1909 CAS.
- Y.-C. Pan, X.-Y. Hu and D.-S. Guo, Angew. Chem., Int. Ed., 2021, 60, 2768 CrossRef CAS PubMed.
- M. Nishihara, F. Perret, T. Takeuchi, S. Futaki, A. N. Lazar, A. W. Coleman, N. Sakai and S. Matile, Org. Biomol. Chem., 2005, 3, 1659 RSC.
- S. Peng, A. Barba-Bon, Y.-C. Pan, W. M. Nau, D.-S. Guo and A. Hennig, Angew. Chem., Int. Ed., 2017, 56, 15742 CrossRef CAS PubMed.
- T. Miyatake, M. Nishihara and S. Matile, J. Am. Chem. Soc., 2006, 128, 12420 CrossRef CAS PubMed.
- S. Matile, N. Sakai and A. Hennig, Supramol. Chem., 2012, 2, 473 CAS.
- T. Takeuchi, V. Bagnacani, F. Sansone and S. Matile, ChemBioChem, 2009, 10, 2793 CrossRef CAS PubMed.
- N. Chuard, K. Fujisawa, P. Morelli, J. Saarbach, N. Winssinger, P. Metrangolo, G. Resnati, N. Sakai and S. Matile, J. Am. Chem. Soc., 2016, 138, 11264 CrossRef CAS PubMed.
- M. Vazdar, E. Wernersson, M. Khabiri, L. Cwiklik, P. Jurkiewicz, M. Hof, E. Mann, S. Kolusheva, R. Jelinek and P. Jungwirth, J. Phys. Chem. B, 2013, 117, 11530 CrossRef CAS PubMed.
- N. Sakai and S. Matile, J. Am. Chem. Soc., 2003, 125, 14348 CrossRef CAS PubMed.
- H. L. Åmand, K. Fant, B. Nordén and E. K. Esbjörner, Biochem. Biophys. Res. Commun., 2008, 371, 621 CrossRef PubMed.
- A. Shaurya, G. A. E. Garnett, M. J. Starke, M. C. Grasdal, C. C. Dewar, A. K. Dheri, E. L. S. Thomson and F. Hof, ChemRxiv, 2020 DOI:10.26434/chemrxiv.11852529.v1.
- T. Hong, K. Iwashita and K. Shiraki, Curr. Protein Pept. Sci., 2018, 19, 746 CrossRef CAS PubMed.
- G. Gattuso, A. Notti, M. F. Parisi, I. Pisagatti, M. E. Amato, A. Pappalardo and S. Pappalardo, Chem. – Eur. J., 2010, 16, 2381 CrossRef CAS PubMed.
- Z. Zheng, W.-C. Geng, J. Gao, Y.-Y. Wang, H. Sun and D.-S. Guo, Chem. Sci., 2018, 9, 2087 RSC.
- P. Talbiersky, F. Bastkowski, F.-G. Klärner and T. Schrader, J. Am. Chem. Soc., 2008, 130, 9824 CrossRef CAS PubMed.
- R. Pinalli and E. Dalcanale, Acc. Chem. Res., 2013, 46, 399 CrossRef CAS PubMed.
- A. Späth and B. König, Beilstein J. Org. Chem., 2010, 6, 32 Search PubMed.
- R. Zadmard and T. Schrader, J. Am. Chem. Soc., 2005, 127, 904 CrossRef CAS PubMed.
- K. Ariga and T. Kunitake, Acc. Chem. Res., 1998, 31, 371 CrossRef CAS.
- Y.-C. Pan, H.-W. Tian, S. Peng, X.-Y. Hu and D.-S. Guo, Chin. Chem. Lett., 2017, 28, 787 CrossRef CAS.
- J. R. Lundblad, M. Laurance and R. H. Goodman, Mol. Endocrinol., 1996, 10, 607 CAS.
- R. D. Kornberg and Y. Lorch, Cell, 1999, 98, 285 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc05414g |
| This journal is © The Royal Society of Chemistry 2021 |