Characterization and application of fluidic properties of trinucleotide repeat sequences by wax-on-plastic microfluidics†
Ahmad Zaman
Qamar
a,
Narges
Asefifeyzabadi
a,
Motahareh
Taki
a,
Swati
Naphade
b,
Lisa M.
Ellerby
b and
Mohtashim Hassan
Shamsi
 *a
*a
aDepartment of Chemistry & Biochemistry, Southern Illinois University at Carbondale, 1245 Lincoln Dr, Carbondale, IL 62901, USA. E-mail: mshamsi@siu.edu
bThe Buck Institute for Research on Aging, 8001 Redwood Blvd, Novato, CA 94945, USA
First published on 19th December 2019
Abstract
Trinucleotide repeat (TNR) sequences introduce sequence-directed flexibility in the genomic makeup of all living species leading to unique non-canonical structure formation. In humans, the expansions of TNR sequences are responsible for almost 24 neurodegenerative and neuromuscular diseases because their unique structures disrupt cell functions. The biophysical studies of these sequences affect their electrophoretic mobility and spectroscopic signatures. Here, we demonstrate a novel strategy to characterize and discriminate the TNR sequences by monitoring their capillary flow in the absence of an external driving force using wax-on-plastic microchannels. The wax-on-plastic microfluidic system translates the sequence-directed flexibility of TNR into differential flow dynamics. Several variables were used to characterize sequences including concentration, single- vs. double-stranded samples, type of repeat sequence, length of the repeat sequence, presence of mismatches in duplex, and presence of metal ion. All these variables were found to influence the flow velocities of TNR sequences as these factors directly affect the structural flexibility of TNR at the molecular level. An overall trend was observed as the higher flexibility in the TNR structure leads to lower capillary flow. After testing samples derived from relevant cells harboring expanded TNR sequences, it is concluded that this approach may transform into a reagent-free and pump-free biosensing platform to detect microsatellite expansion diseases.
Introduction
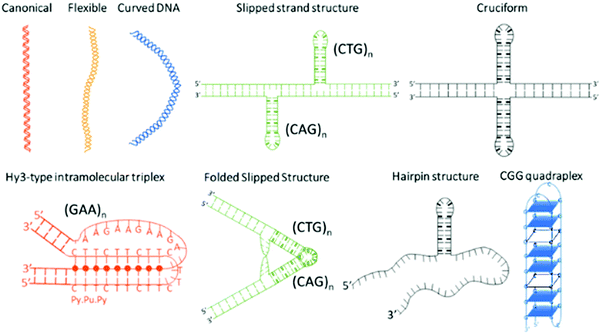
Microsatellite repeats expansions refer to expansion of short repetitive sequences (2–6 nucleotide combination) in a continuum or sporadic fashion in genomic makeup across living species.1 Repeat sequences comprise ∼3% of human genome and expansion of various trinucleotide repeat (TNR) sequences have been associated with more than 24 neurodegenerative and neuromuscular diseases.2,3 Disease relevant expansions include CAG repeat associated with Huntington's disease (HD), CGG with Fragile X, CTG with Myotonic Dystrophy, and GAA expansion with Friedreich's ataxia depending on location in the gene and length of the expansion.4 The sequence type and repetition of TNR cause formation of unique non-canonical conformations such as hairpins, bulges, cruciform, triplexes, quadruplexes etc. as depicted in Scheme 1.5–10 Some investigators have theorized that the non-canonical structure formation disrupts transcription/translation processes inside the cell that leads to these fatal diseases.7The biophysical properties of these sequences are sequence and length dependent; and normally characterized by electrophoresis,11,12 nuclear magnetic resonance,13,14 mass spectroscopy,15 circular dichroism,16 optical melting study and differential scanning calorimetry,17,18 and atomic force microscopy.7 However, detection methods that are currently used to diagnose repeat expansion diseases include two dimensional gel electrophoresis,19 repeat expansion detection method (RED),20,21 triplet repeat prime polymerase chain reaction (TP-PCR), and small pool PCR.22,23 These methods have their own strengths and limitations that are summarized in Table 1. Despite the advances, current methods of detection are prone to imprecise interpretation of repeat lengths leading to false positive or false negative results.24 Apart from sophisticated methods for precise repeat length determination,25–29 a simple and rapid test to distinguish between normal and pathogenic sequence is desirable for genetic testing. The cost of testing for over 20 distinct diseases poses a financial challenge to our health system and has been discussed as an issue for health care providers. Therefore, additional tools are still under exploration to address these challenges.
| Technologies | Advantages | Limitations |
|---|---|---|
| 2D gel electrophoresis | Migration of bands related to MW, high separation | Gel preparation, time consuming, high cost, poor band resolution, low reproducibility, require potential, low throughput.30 |
| Capillary electrophoresis | High throughput separation, automation | Pressure driven, require high electrical potential, high cost.31,32 |
| PCR | Inexpensive, easy to design, sensitive | Non-specific binding, increased risk false negative, time consuming.32,33 |
| RED | Reliable detection range of 40–150 repeats | Minimum detection limit of repeats is 24 units.34 |
| Wax-on-plastic microfluidics | No external force, low cost, time efficient, reagent free, label free | Separation has not been established on these platforms.35 |
In this study, we characterize and detect TNR sequences by measuring their unique pump-free capillary flow using wax-on-plastic microchannels. Wax-on-plastic microfluidic system is the fastest microfluidic system among wax-printed microanalytical devices that allowed us to perform a bioassay within 60 seconds.35 In the case of TNR, we expect a unique flow dynamics from these sequences because binding, folding, compression and extension occur during the flow of DNA in a microfluidic regime, and the extent of change depends on the nucleotide sequences in DNA.36–43 Employing the pump-free capillary flow dynamics, the TNR sequences were characterized based on their concentration, type of repeat sequence, length of repeat sequence, single and double stranded, presence of mismatches in a double-stranded sequence, and the effect of metal ion. Finally, we employed samples from patient cells to distinguish normal and pathological length of CAG repeats associated with Huntington's disease (HD). To best of our knowledge, this is the first study that translates the sequence-directed flexibility of neurodegenerative trinucleotide repeat sequences into distinct capillary flow and may have broad application to all microsatellite repeat expansions diseases.
Experimental section
Materials
Digital Microscope (Dino-Lite Edge 3.0) and UV excitation source (Dino-lite RB excitation source with long pass filter) were purchased from Electron Microscopy Sciences (USA) and was used to capture the capillary flow of the DNA and RNA samples. Fluorescent adaptor was purchased from Nightsea and coupled with digital microscope to monitor fluorescently tagged TNR sequence. Xerox colorQube 8580/DN was used to print wax microchannels on polyethylene terephthalate (PET), which was purchased from Novacentrix. Polyethylene sheets were locally obtained while an adjustable micropipette (Eppendorf research plus) with volume range of 0.5–10 μL was purchased from Fisher Scientific. Tween 20 (Sigma Aldrich) surfactant was used to adjust surface tension of TNR solutions. Fetal bovine serum was kindly provided by Prof. Keith Gagnon (SIUC) and used to test the matrix effect on the sensitivity of the TNR flow. For characterization, synthetic TNR oligonucleotides were procured from Integrated DNA Technologies (USA). A fluorophore 6-carboxyfluorescein (FAM) was covalently linked on 5′ position of single-stranded sequence to monitor flow by the portable fluorescence microscope. TNR solutions were made in 5× PBS buffer (pH = 7.4). Optical flow velocity of PBS buffer was obtained by addition of surfactant, Tween 20. Tween 20 was added in different concentrations ranging from 0% to 0.1%. 1 μL volume of all samples (10 μM) was used and run through 400 μm wide wax printed microfluidic system. The TNR sequences are listed in Table 2. The single-stranded CGG-8, CAG-8 and GAA-8 were prehybridized with their complementary strands, CCG-8, CTG-8 and TTC-8 respectively, by heating above their melting temperature followed by annealing at room temperature. To study the effect of K+ on the flow velocity of TNR, 1–200 mM concentration of K+ made in PBS buffer were added in 10 μM of CGG-8 and CCG-8 sequences. To study the cell-extracted samples, the RNA strands containing CAG repeats from normal and HD neural stem cells (NSCs) were generated from human HD induced pluripotent cells (iPSC)44 and isogenic control C116 using dual SMAD inhibition protocol.45 Plates (6 cm, Corning, Nunclon Delta Surface) were coated with Matrigel (1:60; BD Corning) for 1 h. NSCs were plated and cultured in Neural Proliferation Medium (NPM) in humidified incubator under 37 °C, 5% CO2. NPM was prepared using Neurobasal medium supplemented with 1X B-27 supplement (Life Technologies), 2 mM L-glutamine, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 10 ng mL−1 human leukemia inhibitory factor (LIF) (Peprotech, 300-05), and 25 ng mL−1 human basic fibroblast growth factor (bFGF) (Peprotech, 100-18B). Complete media changes were performed every 2–3 days. When confluent, total RNA was isolated from NSCs using ISOLATE II RNA Mini Kit (Bioline). The samples were obtained in the quantity of 10 μg mL−1 which was diluted with PBS buffer containing 0.1% Tween 20 to achieve 0.14 μM concentrations.| ID | TNR sequence | Repeats# | Flexibility |
|---|---|---|---|
| Based on the crystal structure database, flexibility of any TNR is due to the unique flexibility of each dinucleotide step present in that sequence by accumulation of their variances in roll (ρ) and tilt (τ) parameters. For e.g. (CTG)n repeats have CT, TG, and GC dinucleotide steps.46 | |||
| CGG-8 | 5′-CGG CGG CGG CGG CGG CGG CGG CGG-3′ | 8 | Highest (14.3°) |
| CCG-8 | 5′-CCG CCG CCG CCG CCG CCG CCG CCG-3′ | 8 | |
| CTG-8 | 5′-CTG CTG CTG CTG CTG CTG CTG CTG-3′ | 8 | Lower (14.0°) |
| CAG-8 | 5′-CAG CAG CAG CAG CAG CAG CAG CAG-3′ | 8 | |
| CAG-18 | 5′-CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG-3′ | 18 | |
| CAG-21 | 5′-CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG-3′ | 21 | |
| TTC-5 | 5′-TTC TTC TTC TTC TTC-3′ | 5 | Least (10.1°) |
| TTC-8 | 5′-TTC TTC TTC TTC TTC TTC TTC-3’ | 8 | |
| GAA-5 | 5′-GAA GAA GAA GAA GAA-3′ | 5 | |
| GAA-8 | 5′-GAA GAA GAA GAA GAA GAA GAA GAA-3′ | 8 | |
Device fabrication and TNR microfluidic characterization
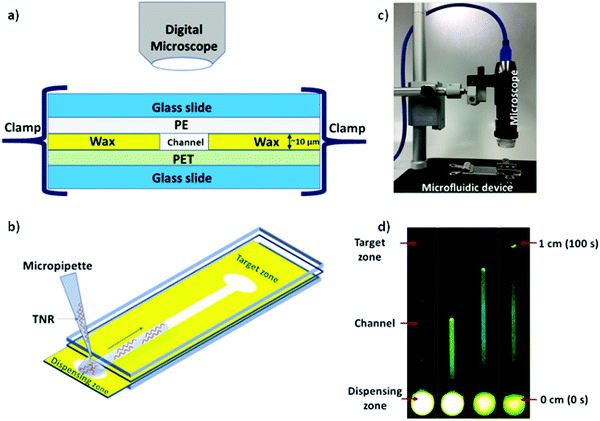
Microfluidic system was fabricated and assembled as we reported previously.35 In brief, rectangular channels of 10 μm × 400 μm × 1 cm dimensions were designed on vector based graphic software “inkscape 0.92” and printed on a polyethylene terephthalate (PET) substrate using a Xerox wax printer. Photo quality for printing was used, which provides channel height of 10 μm. On single 4” × 6” PET substrate, 36 microchannels were printed in a single batch. Then, rectangular shaped area (2 × 1 cm2) with a microchannel was cut from PET sheet and placed on a clean glass slide. Same size of a transparent polyethylene (PE) sheet was placed on top of wax printed microchannel. Then, the layers were closed with another glass slide cover using clamps while keeping the dispensing zone exposed in order to drop the liquid samples on the device as shown in Fig. 1. To observe the capillary flow of TNR sequences in the wax-on-plastic microchannels, the wax-on-plastic platform was covered by a styrofoam box which has two holes perpendicular to each other. The top hole is to introduce camera with fluorescence adaptor. The adaptor has emission bandpass filter of 500 to 560 nm. While the other hole allows us to introduce Nightsea excitation light source (SFA-DL-RB, 440–460 nm) with a long pass filter. The high-quality image at up to frame rate of 15 fps at 2560 × 1920 and 45 fps at 1280 × 960 resolution was recorded. In all experiments, 1 μL of sample liquids was dropped on the dispensing zone of the channels and allowed to flow in the channel to cover the distance of 1 cm.Results and discussion
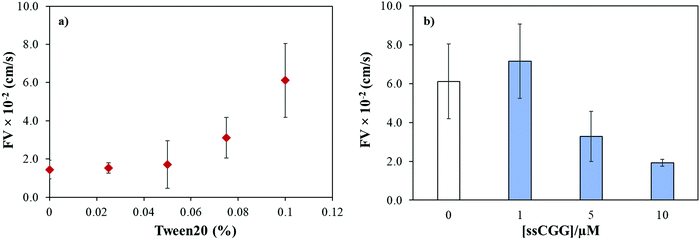
The capillary flow dynamics in a microfluidic channel depends on the interplay between surface energy of a capillary and rheological properties of the liquid under observation, i.e. surface tension and viscosity.47 In case of DNA, its rheological properties may be directly influenced by sequence-directed helical flexibilities and molecular conformations as shown in Scheme 1. To translate the conformational flexibility of TNR sequences into unique capillary flow without an external force, wax-on-plastic microfluidic platform was employed as described in Fig. 1. The enclosed rectangular-shaped microchannel (10 μm × 400 μm × 1 cm), Fig. 1a, is surrounded by PET substrate (bottom), wax (side walls) and PE (top cover) boundaries, sandwiched between glass slides, and clenched by clamps. The experiments were performed by simply dropping 1 μL of TNR sequences made in PBS buffer (pH = 7.4) into a dispensing zone exposed outside the glass coverslip and allowed to flow in the 1 cm long microchannel until reaching the target zone on the other end of the channel as shown in Fig. 1b. The capillary flow of fluorescently tagged TNR solutions were captured by a portable microscope equipped with camera (Fig. 1c). Fig. 1d shows the frames from a video recorded to capture flow of FAM tagged 1 μL volume of 10 μM double-stranded TTC-5 solution, ds-TTC (the real-time video with 5× speed is given in the ESI†). The capillary flow velocity (FV) of the TNR measured in this study is the ratio of total distance covered by TNR in cm and the time elapsed to cover the distance in seconds.We previously reported that the capillary flow of fluid in wax-on-plastic platforms is driven by difference in the surface energies of the channel boundaries where three boundaries are hydrophobic (wax and polyethylene layer) and the hydrophilic PET substrate.35 Moreover, linear correlation was found between the channel width and flow velocities from 100 to 400 μm wide channels.35 Therefore, 400 μm wide channels were used throughout this study to achieve optimum flow. To achieve high rate of capillary flow, the surface tension of the matrix (PBS buffer) was tuned by adding varying concentrations of a surfactant, Tween 20. In particular, the PBS buffer with varying concentrations of Tween 20 was dropped in clean and dry channels followed by observing and recording the meniscus of the flow under the microscope. The flow velocity (FV) was calculated by dividing the distance by time of the flow. Fig. 2a shows that the maximum FV was achieved at 0.1% concentration. However, further addition of the surfactant substantially reduces surface tension of the liquid which spills the liquid out of the channel boundaries. Later, all the TNR solutions used in this study were prepared in PBS buffer containing 0.1% surfactant. To understand how the number of TNR strands in the given volume affects the capillary flow, the concentration of the single-stranded CGG-8 sequence was varied 1–10 μM and allowed to flow in the microchannel, where the flow velocity decreased with the increase in TNR concentration as shown in Fig. 2b. This can be rationalized as the increase in number of strands in unit volume increases viscosity, which leads to higher friction between molecules that slows down the capillary flow.35 In comparison to negative control or PBS, maximum change in flow velocity was found for 10 μM CGG-8. For rest of the study, 10 μM concentrations of the synthetic TNR oligonucleotides were used.
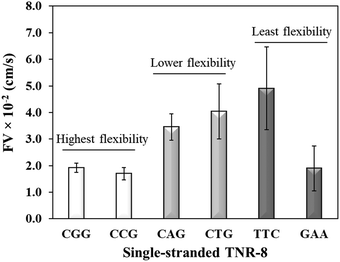
Sequence-directed flexibility of repeats is a unique characteristic, which is responsible for the non-canonical structures (Scheme 1).7 In literature, TNR sequences have been categorized into 12 groups based on their flexibility scale where the least flexible sequences have flexibility value of 9.8° and the most flexible sequences stand at 14.5°.46 Therefore, CGG and CCG are among the higher flexible TNRs in the scale, CAG and CTG are among the middle flexibility sequences, and GAA and TTC belong to least flexible sequences.46 The effect of the molecular flexibility on the flow velocity of TNR sequences was measured by capturing capillary flow of single-stranded TNRs with 8 repeats in the microchannels. Fig. 3 compares the capillary flow of two member sequences from three flexibility groups given in Table 1, i.e. highest flexibility (CGG and CCG), lower flexibility (CAG and CTG), and least flexibility (TTC and GAA). The overall trend of increase in flow rate with the decrease in flexibility of sequences was observed. Interestingly, the statistically notable difference was observed in flow velocities of the highest and lower flexibility groups despite having the difference of only 0.3° degree in the flexibility scale. Moreover, the members from same groups show similar flow velocity for the highest flexibility (CGG vs. CCG) and lower flexibility groups (CAG vs. CTG) with the exception of GAA which shows significantly lower flow velocity in contrast to its counterpart TTC. We attribute this unexpected behavior of GAA to its tendency to form Hy-3 type intramolecular triplex structure through non-Watson–Crick pairing (Scheme 1), which may lead to complex intra-strand entanglement thus lowering the capillary flow. Thus, these results demonstrate that the higher conformational flexibility or any other factor that promotes higher intra- and inter-strand entanglement may slow down the capillary flow.
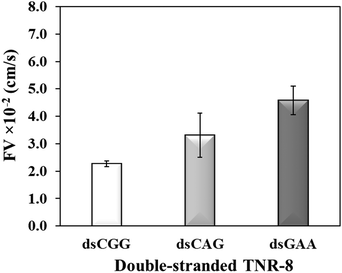
To further investigate whether higher flexibility of DNA sequence may resist capillary flow, we compared the double-stranded TNRs with 8 repeat units from three flexibility groups. Base-pairing in nucleic acids as a result of hybridization restricts molecular conformations and makes a rigid duplex structure, which significantly impacts various biophysical properties such as surface densities of molecular assemblies, charge transport properties, electrostatics, molecular mechanics, stability of surface-tethered states, interactions with surface and neighboring environment.48 To study the effect of hybridization, dsCGG, dsCAG, and dsGAA sequences were dispensed into the microchannel using same volume (1 μL), concentration (10 μM), length (8 repeats), and buffer as used to study the single-stranded counterparts in Fig. 3. It is evident from Fig. 4 that the correlation between the flexibility still holds same for ds-TNR as it was observed for ss-TNR, i.e. capillary flow decreases with the increase in structural flexibility. Interestingly, the flow of dsGAA repeats was enhanced 3-fold after hybridization because double helical conformation formed by complementary sequences is more stable and less flexible than the single-stranded conformations. Further, the extent of increase in flow after hybridization depends on the type of sequence and/or inherent flexibility. The additional advantage of this experiment is the significant discrimination between the neurodegenerative CGG, CTG, and GAA repeats in the double helical conformation, which may be useful to develop a simple genetic test to detect the associated diseases.
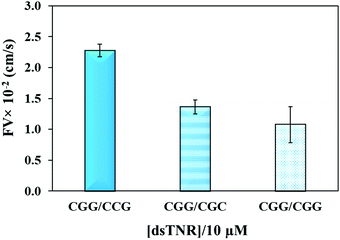
The flexibility of double helical conformations can be affected by the presence of base-pair mismatches, which introduce defects in the structure and eventually leads to various genetic diseases. Moreover, presence of a mismatch in a CAG over expansion may provoke left-handed Z-DNA conformation.49 To study the effect of mismatches, we compared the flow of CGG/CCG (completely matched) sequence with the mismatched sequences, i.e. CGG/CGC (2 mismatches per triplet = 16 mismatches per strand) and CGG/CGG (single mismatch per triplet = 8 mismatches per strand). The experiment was performed using the same experimental conditions (volume, concentration, repeat units, and buffer) as above. Theoretically, imperfection in the double helix due to presence of base-pair mismatch is expected to add flexibility and consequently impeding capillary flow. Secondly, presence of multiple mismatches in a strand significantly reduces hybridization efficiency and strands either partially clings to each other or remain unhybridized. If the unhybridized strands of a mismatched sequence have tendency of forming secondary structure, e.g. hairpin, then their overall flow is expected to be different from mixture of strands where one strand or no strand has tendency of making secondary structure. Fig. 5 confirms the assumption where both mismatched sequences flow at significantly lower rate than perfectly matched double-stranded CGG/CCG. Interestingly, the flow decelerates in case of single-mismatched CGG/CGG sequence more than the double-mismatched CGG/CGC. The lower flow of CGG/CGG can be attributed to the tendency of both strands of the complex to form stable hairpin structure,50 whereas only one strand in CGG/CGC complex has tendency to form stable hairpin. The result proves that the capillary flow velocity can distinguish base-pair mismatching as well as number of mismatches between sequences. However, one may expect a different extent of change or even trend if different types of mismatches are introduced. Although interesting, we have not yet studied this.
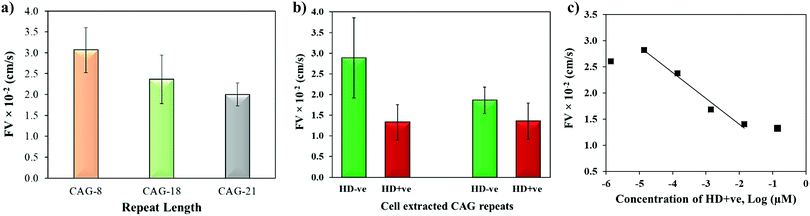
The biophysical properties of repeat sequences are also dependent on their length which has an important impact on human health as the expansion of certain TNR sequences beyond their threshold lengths lead to neurodegenerative diseases.51 For instance, CAG expands up to 35 repeats in a normal individual while pathogenicity begins when it expands beyond 38 repeats resulting in HD.52 To study the effect of repeat length on the capillary flow, varying lengths of single-stranded CAG repeats (8, 18, and 21 repeats) were dispensed in the microchannels. Fig. 6a shows that the flow velocity decreases with the increase in length of CAG repeats. The statistical T-test (p-value = 0.0038) confirms that there is a significant difference between CAG-8 and CAG-21 sequences, which implies that distinct capillary flow can be assumed for the difference in repeat lengths >10 repeat units. Practically, in the disease causing gene, the associated repeats expand hundreds to thousands in number.2 To study the effect of length on normal versus abnormal lengths of repeats, RNA samples with CAG repeats associated with HD were extracted from normal and diseased cells followed by evaluation of their flow in wax-on-plastic microchannels after diluting the samples to 0.14 μM in PBS buffer. The flow velocity of patient-derived DNA samples with CAG repeats (HD +ve with 72 repeats) was compared with a control (HD −ve with 21 repeats) extracted from iPSC cells in Fig. 6b. The flow of the HD +ve CAG repeats was found 2-fold slower than the control (HD −ve). The retardation in the flow of abnormal length is statistically significant, which proves the diagnostic value of the wax-on-plastic platforms as a tool for pump-free and reagent-free detection of neurodegenerative diseases. We assumed that due to 3-fold difference in the number of repeats it is easy to discriminate in aqueous medium of PBS buffer. However, it will be rationale to test the sensitivity in a complex medium such as fetal bovine serum. To test the matrix effect, cell-extracted samples of HD −ve and HD +ve of 0.14 μM concentrations were prepared in 10% fetal bovine serum (FBS) and allowed to flow in wax microchannel. The flow velocity of HD −ve was found to decrease in the viscous medium of 10% FBS while the response of HD +ve remains the same. Despite the change observed, their response still follows the trend as in a buffer medium. Moreover, the sensitivity of this method for sample concentrations was also determined by diluting the HD +ve samples in buffer and the flow velocity was obtained for up to six dilutions in the microfluidic channel. Fig. 6c shows that the flow velocity increases with the dilution, perhaps due to decrease in the viscosity, where the linear range was found in the range of 14 pM to 14 nM with the correlation coefficient R2 = 0.975. The method is simpler and the concentration range detected here is comparable to other sensitive methods that have been developed to replace PCR based methods. For instance, amperometric detection of CAG repeats involved molecular beacon labeled with ferrocene used to capture CAG repeats followed by enzyme-catalyzed reaction to produce electroactive product for signal detection with the concentration range of 1 pM to 100 nM.53
Sequence specific interaction between metal ions and DNA is a well-established phenomenon, which leads to unique structure formation and these interactions are useful for metal ion sensing.54,55 To understand the effect of metal ion (e.g. K+) on the flow velocity of these two sequences of distinct properties, we spiked these sequences with 1–200 mM K+ and allowed to run in the microchannels. Previously, it was reported that single-stranded CGG and CCG sequences form hairpin structures without metal mediated basepairing.16 However, the CGG hairpin structures were found more stable than CCG hairpin structures. In Fig. 7a and b, we observed the similar flow velocity of both sequences in absence of K+ (0 mM), which may be attributed to their tendency of making hairpin structures in absence of K+. However, it is known that CGG repeats form G-quadruplex structure in ≥155 mM K+,15,56 while no such conformational change has been observed for CCG sequence in presence of K+. It is evident from Fig. 7a and b that the addition of K+ changes the flow of both sequences confirming that K+ interferes with the interstrand and intrastrand basepairing (non-canonical structures) thus causing an increase in the flow. The interference at lower concentration of K+ is assumed to open the strands and form less flexible linear strands that allows the strands to flow faster. While, the systematic increase in K+ concentration gradually decreases the flow velocity of ssCGG. The flow behavior at 200 nM K+ was reached similar to flow at 0 mM K+. We attribute this behavior to the folding of CGG strands again and formation of interstrand and intrastrand G-quadruplexes through coordination with K+ ions. In contrast, the bimodal distribution of flow was observed for ssCCG with overall higher flow velocity at all concentrations of K+. We assume that this behavior was observed because the CCG strands do not fold back to higher order structure even without K+ coordination. Previously, TNRs were grouped into four different structural classes of (a) unstructured, (b) semi-stable hairpins, (c) fairly stable hairpins, and (d) very stable G-quadruplexes.57 The order of thermodynamic stability of the hairpin structure was found to be CAG > CGG > CUG > CCG in 100 mM KCl.50 Thus, we attribute the bimodal behavior of CCG repeats to its unstable hairpin structure in presence of K+ ion, which leads to mixed population of unstructured and linear forms at >100 mM K+.
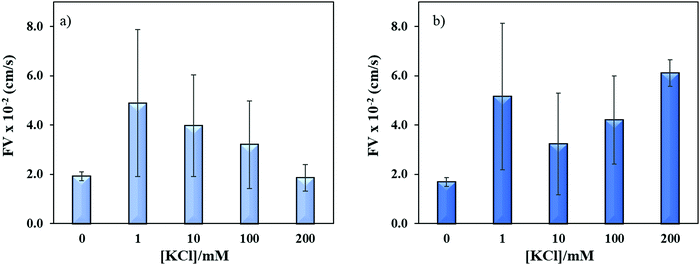 | ||
| Fig. 7 Effect of potassium ion concentrations on the flow velocity of (a) ssCGG-8 and (b) ssCCG-8. Error bar shows standard deviation for three replicates. | ||
Conclusion
In this study, we demonstrate that the biophysical properties of TNR sequences can affect their capillary flow, which is also the function of the TNR concentration, sequence type, repeat length, single-stranded vs. duplex structures, presence of mismatches in the duplexes, and K+ ions. The parameters studied here affect the conformational change or flexibility at molecular level that translates into change in the capillary flow. The general observation was any factor that leads to higher flexibility and inter- and intra-strands complex structures tend to resist the capillary flow. In future, this method can be used to characterize 64 possible combinations of TNRs. Moreover, the clear distinction between the flow of patient-derived normal and abnormal CAG repeats associated with HD proves that simple capillary flow on wax-on-plastic analytical devices can be used as a tool for research and diagnostics. Metal ion detection using repeat sequences can also be a venue of application on wax-on-plastic platforms.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MHS acknowledges NSF EAGER award #1940716 to fund the project and NIH R01 NS100529 and NS094422 to L. M. E.References
- H. Fan and J.-Y. Chu, A brief review of short tandem repeat mutation, Genomics, Proteomics Bioinf., 2007, 5(1), 7–14 CrossRef CAS.
- K. J. Rohilla and K. T. Gagnon, RNA biology of disease-associated microsatellite repeat expansions, Acta Neuropathol. Commun., 2017, 5(1), 63 CrossRef PubMed.
- W.-H. Tseng, C.-k. Chang, P.-C. Wu, N.-J. Hu, G.-H. Lee, C.-C. Tzeng, S. Neidle and M.-H. Hou, Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel–Chromomycin Complex Resulting in Large-Scale DNA Deformation, Angew. Chem., Int. Ed., 2017, 56(30), 8761–8765 CrossRef CAS PubMed.
- H. Paulson, Repeat expansion diseases, Handb. Clin. Neurol., 2018, 147, 105–123 Search PubMed.
- R. R. Iyer, A. Pluciennik, M. Napierala and R. D. Wells, DNA Triplet Repeat Expansion and Mismatch Repair, Annu. Rev. Biochem., 2015, 84(1), 199–226 CrossRef CAS PubMed.
- C.-W. Ni, Y.-J. Wei, Y.-I. Shen and I. R. Lee, Long-Range Hairpin Slippage Reconfiguration Dynamics in Trinucleotide Repeat Sequences, J. Phys. Chem. Lett., 2019, 3985–3990 CrossRef CAS PubMed.
- R. R. Sinden, V. N. Potaman, E. A. Oussatcheva, C. E. Pearson, Y. L. Lyubchenko and L. S. Shlyakhtenko, Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA, J. Biosci., 2002, 27(Suppl. 1), 53–65 CrossRef CAS PubMed.
- A. M. Gacy, G. M. Goellner, C. Spiro, X. Chen, G. Gupta, E. M. Bradbury, R. B. Dyer, M. J. Mikesell, J. Z. Yao, A. J. Johnson, A. Richter, S. B. Melancon and C. T. McMurray, GAA instability in Friedreich's Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases, Mol. Cell, 1998, 1(4), 583–593 CrossRef CAS PubMed.
- A. Halabi, K. T. B. Fuselier and E. Grabczyk, GAA·TTC repeat expansion in human cells is mediated by mismatch repair complex MutL gamma and depends upon the endonuclease domain in MLH3 isoform one, Nucleic Acids Res., 2018, 46(8), 4022–4032 CrossRef CAS PubMed.
- S. M. Mirkin, DNA structures, repeat expansions and human hereditary disorders, Curr. Opin. Struct. Biol., 2006, 16(3), 351–358 CrossRef CAS PubMed.
- P. D. Chastain 2nd, E. E. Eichler, S. Kang, D. L. Nelson, S. D. Levene and R. R. Sinden, Anomalous rapid electrophoretic mobility of DNA containing triplet repeats associated with human disease genes, Biochemistry, 1995, 34(49), 16125–16131 CrossRef CAS PubMed.
- M. Gomes-Pereira and D. G. Monckton, Ethidium Bromide Modifies The Agarose Electrophoretic Mobility of CAG·CTG Alternative DNA Structures Generated by PCR, Front. Cell. Neurosci., 2017, 11 Search PubMed.
- M. Zheng, X. Huang, G. K. Smith, X. Yang and X. Gao, Genetically Unstable CXG Repeats are Structurally Dynamic and Have a High Propensity for Folding. An NMR and UV Spectroscopic Study, J. Mol. Biol., 1996, 264(2), 323–336 CrossRef CAS PubMed.
- Structure, Motion, Interaction and Expression of Biological Macromolecules, ed. R. H. Sarma and M. H. Sarma, Adenine Press, Albany, vol. 2 Search PubMed.
- M. Malgowska, D. Gudanis, R. Kierzek, E. Wyszko, V. Gabelica and Z. Gdaniec, Distinctive structural motifs of RNA G-quadruplexes composed of AGG, CGG and UGG trinucleotide repeats, Nucleic Acids Res., 2014, 42(15), 10196–10207 CrossRef CAS PubMed.
- K. Sobczak, G. Michlewski, M. de Mezer, E. Kierzek, J. Krol, M. Olejniczak, R. Kierzek and W. J. Krzyzosiak, Structural Diversity of Triplet Repeat RNAs, J. Biol. Chem., 2010, 285(17), 12755–12764 CrossRef CAS PubMed.
- A. M. Paiva and R. D. Sheardy, Influence of Sequence Context and Length on the Structure and Stability of Triplet Repeat DNA Oligomers, Biochemistry, 2004, 43(44), 14218–14227 CrossRef CAS PubMed.
- J. Huang and S. Delaney, Unique Length-Dependent Biophysical Properties of Repetitive DNA, J. Phys. Chem. B, 2016, 120(18), 4195–4203 CrossRef CAS PubMed.
- M. M. Krasilnikova and S. M. Mirkin, Analysis of Triplet Repeat Replication by Two-Dimensional Gel Electrophoresis, in Trinucleotide Repeat Protocols, ed. Y. Kohwi, Humana Press, Totowa, NJ, 2004, pp. 19–28 Search PubMed.
- K. Lindblad, P.-O. Nylander, A. De bruyn, D. Sourey, C. Zander, C. Engström, G. Holmgren, T. Hudson, J. Chotai, J. Mendlewicz, C. Van Broeckhoven, M. Schalling and R. Adolfsson, Detection of expanded CAG repeats in Bipolar Affective Disorder using the repeat expansion detection (RED) method, Neurobiol. Dis., 1995, 2(1), 55–62 CrossRef CAS PubMed.
- Q.-P. Yuan and M. Schalling, Detection and Isolation of Trinucleotide Repeat Expansions Using the RED Method, in Trinucleotide Repeat Protocols, ed. Y. Kohwi, Humana Press, Totowa, NJ, 2004, pp. 47–59 Search PubMed.
- M. Gomes-Pereira, S. I. Bidichandani and D. G. Monckton, Analysis of Unstable Triplet Repeats Using Small-Pool Polymerase Chain Reaction in Trinucleotide Repeat Protocols, ed. Y. Kohwi, Humana Press, Totowa, NJ, 2004, pp. 61–76 Search PubMed.
- J. P. Warner, L. H. Barron, D. Goudie, K. Kelly, D. Dow, D. R. Fitzpatrick and D. J. Brock, A general method for the detection of large CAG repeat expansions by fluorescent PCR, J. Med. Genet., 1996, 33(12), 1022–1026 CrossRef CAS PubMed.
- A. Crook, A. McEwen, J. A. Fifita, K. Zhang, J. B. Kwok, G. Halliday, I. P. Blair and D. B. Rowe, The C9orf72 hexanucleotide repeat expansion presents a challenge for testing laboratories and genetic counseling, Amyotrophic Lateral Scler. Frontotemporal Degener., 2019, 20(5–6), 310–316 CrossRef CAS PubMed.
- E. Dolzhenko, J. J. F. A. van Vugt, R. J. Shaw, M. A. Bekritsky, M. van Blitterswijk, G. Narzisi, S. S. Ajay, V. Rajan, B. R. Lajoie, N. H. Johnson, Z. Kingsbury, S. J. Humphray, R. D. Schellevis, W. J. Brands, M. Baker, R. Rademakers, M. Kooyman, G. H. P. Tazelaar, M. A. van Es, R. McLaughlin, W. Sproviero, A. Shatunov, A. Jones, A. Al Khleifat, A. Pittman, S. Morgan, O. Hardiman, A. Al-Chalabi, C. Shaw, B. Smith, E. J. Neo, K. Morrison, P. J. Shaw, C. Reeves, L. Winterkorn, N. S. Wexler, U. S. V. C. R. Group, D. E. Housman, C. W. Ng, A. L. Li, R. J. Taft, L. H. van den Berg, D. R. Bentley, J. H. Veldink and M. A. Eberle, Detection of long repeat expansions from PCR-free whole-genome sequence data, Genome Res., 2017, 27(11), 1895–1903 CrossRef CAS PubMed.
- H. Tang, E. F. Kirkness, C. Lippert, W. H. Biggs, M. Fabani, E. Guzman, S. Ramakrishnan, V. Lavrenko, B. Kakaradov, C. Hou, B. Hicks, D. Heckerman, F. J. Och, C. T. Caskey, J. C. Venter and A. Telenti, Profiling of Short-Tandem-Repeat Disease Alleles in 12
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 632 Human Whole Genomes, Am. J. Hum. Genet., 2017, 101(5), 700–715 CrossRef CAS PubMed.
632 Human Whole Genomes, Am. J. Hum. Genet., 2017, 101(5), 700–715 CrossRef CAS PubMed. - S. P. Sadedin, H. Dashnow, P. A. James, M. Bahlo, D. C. Bauer, A. Lonie, S. Lunke, I. Macciocca, J. P. Ross, K. R. Siemering, Z. Stark, S. M. White, A. Melbourne Genomics Health, G. Taylor, C. Gaff, A. Oshlack and N. P. Thorne, Cpipe: a shared variant detection pipeline designed for diagnostic settings, Genome Med., 2015, 7(1), 68 CrossRef PubMed.
- M. Bahlo, M. F. Bennett, P. Degorski, R. M. Tankard, M. B. Delatycki and P. J. Lockhart, Recent advances in the detection of repeat expansions with short-read next-generation sequencing, F1000Res., 2018, 7, 736 Search PubMed.
- K. Hsieh, A. S. Patterson, B. S. Ferguson, K. W. Plaxco and H. T. Soh, Rapid, Sensitive, and Quantitative Detection of Pathogenic DNA at the Point of Care through Microfluidic Electrochemical Quantitative Loop-Mediated Isothermal Amplification, Angew. Chem., Int. Ed., 2012, 51(20), 4896–4900 CrossRef CAS PubMed.
- N. Bitarte, E. Bandrés, R. Zárate, N. Ramirez and J. Garcia-Foncillas, Moving forward in colorectal cancer research, what proteomics has to tell, World J. Gastroenterol., 2007, 13(44), 5813–5821 CrossRef CAS PubMed.
- K. Klepárník and P. Boček, DNA Diagnostics by Capillary Electrophoresis, Chem. Rev., 2007, 107(11), 5279–5317 CrossRef PubMed.
- E. Lyon, T. Laver, P. Yu, M. Jama, K. Young, M. Zoccoli and N. Marlowe, A simple, high-throughput assay for Fragile X expanded alleles using triple repeat primed PCR and capillary electrophoresis, J. Mol. Diagn., 2010, 12(4), 505–511 CrossRef CAS PubMed.
- R. J. Roberts, M. O. Carneiro and M. C. Schatz, The advantages of SMRT sequencing, Genome Biol., 2013, 14(7), 405 CrossRef PubMed.
- L. Martorell, M. A. Pujana, V. Volpini, A. Sanchez, J. Joven, E. Vilella and X. Estivill, The repeat expansion detection method in the analysis of diseases with CAG/CTG repeat expansion: Usefulness and limitations, Hum. Mutat., 1997, 10(6), 486–488 CrossRef CAS PubMed.
- A. Z. Qamar, G. Parker, G. R. Kinsel and M. H. Shamsi, Evolution of wax-on-plastic microfluidics for sub-microliter flow dynamics and its application in distance-based assay, Microfluid. Nanofluid., 2019, 23(6), 81 CrossRef.
- B. C. Durney, C. L. Crihfield and L. A. Holland, Capillary electrophoresis applied to DNA: determining and harnessing sequence and structure to advance bioanalyses (2009–2014), Anal. Bioanal. Chem., 2015, 407(23), 6923–6938 CrossRef CAS PubMed.
- P. LeDuc, C. Haber, G. Bao and D. Wirtz, Dynamics of individual flexible polymers in a shear flow, Nature, 1999, 399(6736), 564–566 CrossRef CAS PubMed.
- P. J. Shrewsbury, D. Liepmann and S. J. Muller, Concentration Effects of a Biopolymer in a Microfluidic Device, Biomed. Microdevices, 2002, 4(1), 17–26 CrossRef CAS.
- S. Gulati, S. J. Muller and D. Liepmann, Flow of DNA solutions in a microfluidic gradual contraction, Biomicrofluidics, 2015, 9(5), 054102 CrossRef PubMed.
- J. W. Larson, G. R. Yantz, Q. Zhong, R. Charnas, C. M. D’Antoni, M. V. Gallo, K. A. Gillis, L. A. Neely, K. M. Phillips, G. G. Wong, S. R. Gullans and R. Gilmanshin, Single DNA molecule stretching in sudden mixed shear and elongational microflows, Lab Chip, 2006, 6(9), 1187–1199 RSC.
- T. T. Perkins, S. R. Quake, D. E. Smith and S. Chu, Relaxation of a single DNA molecule observed by optical microscopy, Science, 1994, 264(5160), 822 CrossRef CAS PubMed.
- T. T. Perkins, D. E. Smith and S. Chu, Single Polymer Dynamics in an Elongational Flow, Science, 1997, 276(5321), 2016 CrossRef CAS PubMed.
- L. Rems, D. Kawale, L. J. Lee and P. E. Boukany, Flow of DNA in micro/nanofluidics: From fundamentals to applications, Biomicrofluidics, 2016, 10(4), 043403 CrossRef PubMed.
- M. C. An, N. Zhang, G. Scott, D. Montoro, T. Wittkop, S. Mooney, S. Melov and L. M. Ellerby, Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells, Cell Stem Cell, 2012, 11(2), 253–263 CrossRef CAS PubMed.
- S. M. Chambers, C. A. Fasano, E. P. Papapetrou, M. Tomishima, M. Sadelain and L. Studer, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling (vol. 27, p. 275, 2009), Nat. Biotechnol., 2009, 27(5), 485 CrossRef CAS.
- A. Bacolla, R. Gellibolian, M. Shimizu, S. Amirhaeri, S. Kang, K. Ohshima, J. E. Larson, S. C. Harvey, B. D. Stollar and R. D. Wells, Flexible DNA: Genetically unstable CTG·CAG and CGG·CCG from human hereditary neuromuscular disease genes, J. Biol. Chem., 1997, 272(27), 16783–16792 CrossRef CAS PubMed.
- A. Olanrewaju, M. Beaugrand, M. Yafia and D. Juncker, Capillary microfluidics in microchannels: from microfluidic networks to capillaric circuits, Lab Chip, 2018, 18(16), 2323 RSC.
- A. N. Rao and D. W. Grainger, Biophysical properties of nucleic acids at surfaces relevant to microarray performance, Biomater. Sci., 2014, 2(4), 436–471 RSC.
- N. Khan, N. Kolimi and T. Rathinavelan, Twisting right to left: A⋯A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation, PLoS Comput. Biol., 2015, 11(4), e1004162 CrossRef PubMed.
- M. Broda, E. Kierzek, Z. Gdaniec, T. Kulinski and R. Kierzek, Thermodynamic Stability of RNA Structures Formed by CNG Trinucleotide Repeats. Implication for Prediction of RNA Structure, Biochemistry, 2005, 44(32), 10873–10882 CrossRef CAS PubMed.
- H. Budworth and C. T. McMurray, A brief history of triplet repeat diseases, Methods Mol. Biol., 2013, 1010, 3–17 CrossRef CAS PubMed.
- N. Zhang, B. J. Bailus, K. L. Ring and L. M. Ellerby, iPSC-based drug screening for Huntington's disease, Brain Res., 2016, 1638(Pt A), 42–56 CrossRef CAS PubMed.
- J. Li, Y. L. Liu, X. Q. Zhu, G. Chang, H. P. He, X. H. Zhang and S. F. Wang, A Novel Electrochemical Biosensor Based on a Double-Signal Technique for d(CAG)(n) Trinucleotide Repeats, ACS Appl. Mater. Interfaces, 2017, 9(50), 44231–44240 CrossRef CAS PubMed.
- W. Zhou, R. Saran and J. Liu, Metal Sensing by DNA, Chem. Rev., 2017, 117(12), 8272–8325 CrossRef CAS PubMed.
- M. Shamsi and H. B. Kraatz, Interactions of Metal Ions with DNA and Some Applications, J. Inorg. Organomet. Polym., 2013, 23(1), 4–23 CrossRef CAS.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res., 2006, 34(19), 5402–5415 CrossRef CAS PubMed.
- A. Ciesiolka, M. Jazurek, K. Drazkowska and W. J. Krzyzosiak, Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions, Front. Cell. Neurosci., 2017, 11, 97 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9tb02208b |
| This journal is © The Royal Society of Chemistry 2020 |