Open Access Article
Open Access ArticleOpen microfluidics: droplet microarrays as next generation multiwell plates for high throughput screening
Robert
Strutt
,
Bijing
Xiong
,
Vanessa Fabienne
Abegg
and
Petra S.
Dittrich
 *
*
Department of Biosystems Science and Engineering, ETH Zürich, Schanzenstrasse 44, 4056 Basel, Switzerland. E-mail: petra.dittrich@bsse.ethz.ch
First published on 6th February 2024
Abstract
Multiwell plates are prominent in the biological and chemical sciences; however, they face limitations in terms of throughput and deployment in emerging bioengineering fields. Droplet microarrays, as an open microfluidic technology, organise tiny droplets typically in the order of thousands, on an accessible plate. In this perspective, we summarise current approaches for generating droplets, fluid handling on them, and analysis within droplet microarrays. By enabling unique plate engineering opportunities, demonstrating the necessary experimental procedures required for manipulating and interacting with biological cells, and integrating with label-free analytical techniques, droplet microarrays can be deployed across a more extensive experimental domain than what is currently covered by multiwell plates. Droplet microarrays thus offer a solution to the bottlenecks associated with multiwell plates, particularly in the areas of biological cultivation and high-throughput compound screening.
Introduction
Multiwell plates are the gold-standard platform in biological, pharmaceutical and chemical applications.1 These injection-moulded plastic plates which typically consist of 96, 384 or even 1536 wells are routinely employed in both industry and academia to facilitate analysis of discrete volumes in the range of a few to hundreds of microlitres. In operational terms, multiwell plates are optimised for situations where the same assay or analytical procedure is carried out repeatedly under consistent conditions, often with minor variations like dilution series of a single compound. Supplementing workflows with liquid pipetting robots, allows for filling and removal of liquids as well as use of instruments designed for workflow automation. Multiwell plates are therefore ideally suited for high-throughput applications and are flexible with respect to assays and analysis. However, workflows are limited when it comes to reducing the analyte volume and increasing the experimental throughput beyond the well count. These aspects are especially evident in specific applications such as single-cell analysis or in advanced 3D cell culture techniques. Moreover, multiwell plates suffer from drawbacks such as significant economic and environmental costs attributed to the use of single-use plastics and large experimental volumes.Microfluidics is a technology dedicated to precisely meter, guide and manipulate small volumes of liquids typically in the picolitre to microlitre range. For more than three decades, many demonstrations of this technology have redefined chemical, biological and clinically relevant settings such as point of care (PoC) testing,2 3D cell culture (“organs-on-chip”),3 and single-cell sequencing.4 Owing to the maturity of the field, microfluidic technologies now span the natural life sciences, with continuous invention of innovative architectural and functional chip features. Despite these widely disseminated achievements, microfluidic devices are sometimes sophisticated engineered tools that need trained individuals to manufacture and operate them. Perhaps considering this, an engineering trend of reducing chip complexity has begun to emerge, defining paper-based microfluidics,5 open microfluidics,6 fabrication through 3D printing and rapid prototyping.7 This simplification is crucial for transitioning microfluidic methods into the market and facilitating their adoption by industry beyond academic labs. Perhaps the most influential use of microfluidics as of this article are the rapid tests using lateral flow assays, which can be considered as paper-based microfluidics,8 where the end-user needs to do nothing else than position a sample in the defined introduction spot.
In a similar experimental space as multiwell plates, droplet microfluidics rapidly emerged as an elegant and versatile approach to compartmentalise experiments.9,10 The ease of droplet generation in microdevices or capillary systems led to a fast spread over many groups and disciplines. Droplet microfluidics has its strength particularly in high-throughput applications, where parallelisation is essential, but no further addition or exchange of fluids is necessary.11 The most successful example of droplet microfluidics is in single-cell sequencing, where single cells and barcoded beads are encapsulated together and after cell lysis and nucleic acid hybridisation, the emulsion is broken, and all further steps are done in bulk.12 Other procedures require more effort. Whilst addition of compounds (e.g. pico-injection), fusion, splitting, in-droplet separation, sorting and other ways of droplet manipulation have been shown, it remains challenging to integrate these various modules in series, on a single platform.
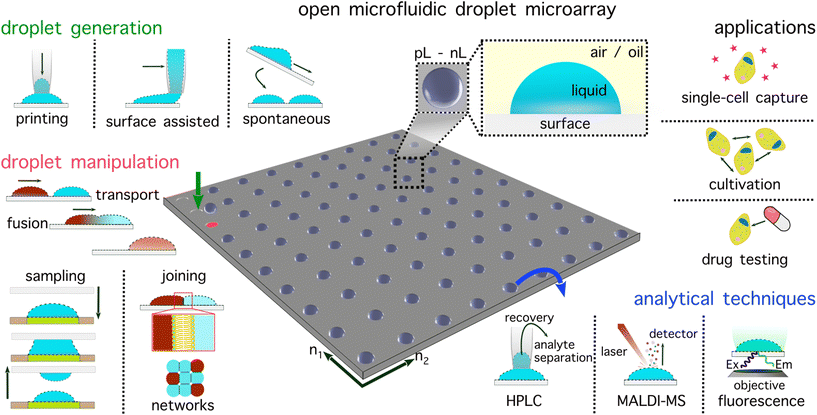
Droplet microarrays harness the strengths of two approaches: experiments are confined within remarkably small and discrete containers akin to droplet microfluidics, yet they also remain open, accessible, and indexable as in multiwell plates (Fig. 1). Droplet microarrays occupy a small footprint (typically glass slide dimensions, i.e., 75 × 25 mm) enabling fluid handling robots to be assembled around the open microfluidic plate. To date, droplet microarrays have been implemented with several thousand individual experiments (or droplets), enabling ultra-throughput and massively parallel experimentation. A key advantage of a droplet microarray is that each methodological component (experiment production, manipulation, and analysis) is isolated from the other, enabling huge analytical and operational customisability appropriate across a range of biological and chemical fields.
Droplet microarrays are capable of all functions of a multiwell plate and beyond; the wall-less design facilitates novel procedures, e.g. connecting droplets through control of their relative positions. Capitalising on these advantages, numerous research groups in recent years have showcased the immense potential of droplet microarrays for high-throughput applications, including examples of fully automated platforms with integrated feedback loop control. This perspective article explores open droplet microarray systems as the next generation of multiwell plates. This interdisciplinary technology emerged through unifying developments in droplet production, manipulation, and analysis. Perspective is therefore provided on each of these areas in the chronology of a droplet microarray workflow. Droplet microarrays are defined by their openness, accessibility and potential for automation, encompassing aspects of sessile droplets, open-droplet microfluidics, hanging droplet platforms and digital microfluidics. This perspective aims to organise and present diverse droplet microarray-based technologies highlighting their flexibility with respect to operation, analysis and applications.
Droplet generation and surface modification
A droplet microarray consists of droplets with typically a volume in the picolitre to nanolitre range. The lowest operational volume for a standard 1536 multiwell plate is around 3 μL.1 To address droplet evaporation, which is a well-known aspect when working with such small volumes, droplets can be generated under gas permeable oils such as hydrofluoroether (HFE) oil,16 Fluorinert™ FC-40,17 or mineral oil.18 For droplet microarrays formed in air, a humid environment can be used to reduce droplet evaporation.19 Depending on the application and droplet generation method, surfactant may be used to stabilise the water–oil interface. In droplet microarrays, droplets must be localised through contact with a solid substrate (Fig. 2a). Surface modification enhances the durability of droplet attachment. Central to this process is the generation of a surface pattern, delineating hydrophilic areas that serve as the sites for droplet hosting, on an otherwise hydrophobic surface. Defined surface patterns with different wettability can be obtained in various ways, e.g. by laser desorption of hydrophobic surfaces,20 by photopolymerisation through a photo mask,21 or thiol–yne based click chemistry using a nanoporous polymer layer atop a glass slide.22 Photolithography allows the generation of hydrophilic wells or hydrophilic pillar arrays.23 Droplet microarray spot geometry is not limited to two dimensions, for example hemispherical micro bowls have been achieved by manipulating the diffusion of polymerisation moieties and controlling the exposure.24,25 Finally, the fact that surface properties dictate the hosting sites enables the creation of droplets with varying diameters, shapes, distances, and even connections between droplets.26,27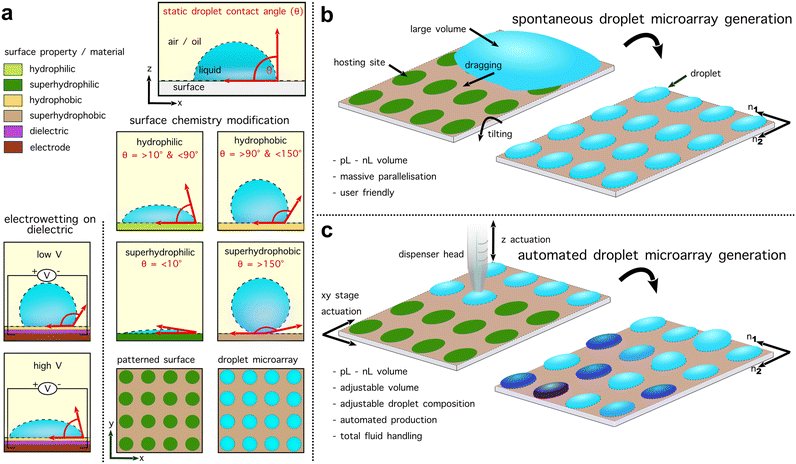 | ||
| Fig. 2 Droplet generation techniques for droplet microarrays. (a) A surface can be defined by the static water droplet contact angle (θw) as either hydrophilic (>10° and <90°)/hydrophobic (>90° and <150°) or superhydrophobic (>150°)/superhydrophilic (<10°).13,14 For untreated PDMS the θw is around 100°.15 Controlling wettability aligns droplets in droplet microarrays through patterns of droplet ‘hosting sites’. Additionally, the wettability and thus contact angle can be controlled via electrowetting on dielectric (EWOD). (b) Overview of spontaneous droplet microarray generation methods. By initially depositing a large volume on the plate, this can be fractionated into smaller droplets of the same content and volume to achieve an ultra-throughput, parallelised, low volume droplet microarray. (c) Overview of automated droplet microarray generation. Automated droplet microarrays typically utilise a programmable stage (for example, one on a commercial microscope) to control the plate position in two dimensions with a dispenser arranged perpendicular to the plate on an additional motorised stage. This allows an experiment to be localised at a defined position of the plate by arranging the dispenser above a hosting site. Automated droplet microarrays facilitate complex experimental operations and characteristics. | ||
In open microfluidic systems, passive droplet generation without pumps can be achieved through a balance of hydrostatic and capillary pressures emergent from channel geometry.28 In droplet microarrays, a crucial aspect that also defines the generation throughput is the way droplets are deposited or directly formed on the plate. On surfaces with large wettability differences, discontinuous wetting autonomously fractionates a volume through a periodic wettability pattern (Fig. 2b). The Levkin group, along with other research groups, has pioneered discontinuous wetting as an approach for forming droplet microarrays by dragging or displacing a volume across a hydrophilic–hydrophobic patterned surface.29–32 Surface modification-free approaches have also been shown where 2 × 2 pillar arrangements can be used to split a bulk volume and retain a fragment in the centre of the pillars.33
Alternatively, by embedding electrodes in the plate, in situ manipulation can be performed. Cole et al., utilised dielectrophoretic trapping in so called ‘nanowells’ via a polarizability difference between the droplet and a constant on-plate electric field.34 Electrodes can also enable EWOD – an approach for directly manipulating the droplet contact angle by controlling the plate wettability via an electric field between the droplet and the plate (Fig. 2a).35 This allows for the control of surface wettability at the edge of an initial volume, enabling fractionation and subsequent regulation of the spatial–temporal distribution of the resulting daughter droplets.36
Beyond this, more sophisticated generation procedures have been shown where the precision of a dispenser head is used to arrange a droplet microarray. As such, these approaches may not necessarily require surface hosting sites.37 Inkjet printing, for example utilises a dispenser head with a channel containing fluid that is deformed via a piezo element to dispense droplets.37 Mechanistically similar is echo acoustic droplet dispensing, where commercially available fluid handling robots have been deployed in droplet microarray contexts.38 Depending on the dispenser head, some droplet generation approaches may be unable to work under oil, perform sample removal from a surface and dispensation may be correlated with the fluid viscosity. To circumvent some of these issues, automated droplet microarrays are typically software-controlled custom-made platforms with advanced fluid handling (Fig. 2c).
Some examples of these technologies not only enable the deposition of distinct volumes but can also aspirate them, replicating a fundamental operational capability of multiwell plates. The Fang lab has defined such integrated systems as examples of sequential operation droplet arrays (SODA).18,39 Droplet printing or ‘spotting’, as pioneered by the Dittrich, Fang and Abate labs, utilises a closed microfluidic system to generate droplets prior to contact with the surface, thus focusing on oil immersed plates. Surface assisted droplet generation lies somewhere between automated and spontaneous approaches where a continuous flow dispenser is driven across a wettability-patterned surface. Droplets are formed as the dispenser passes over each hosting site, where a portion of the liquid is attached as dictated by the wettability pattern.23 Droplet microarrays have been formed with a wide variety of different methods and a comprehensive review is provided by Zhang et al.40
Experimental operations with droplets
Operations on droplets, whether during or after generation, allow for the manipulation of various properties, including the droplet chemical composition, size, and spatial distribution. The number of operations that can be done on droplet microarrays is informed by the generation method and thus directly decides the achievable complexity of experimental settings as well as the flexibility of analysis. Here we summarise and discuss the experimental operations that can change the droplet chemistry (i.e. reagent addition and dilution) and/or droplet functionality (i.e. spatially-structured yet communicating droplet networks).Reagent addition and dilution
Many chemical and biological experimental protocols require multistep and/or dynamic procedures. Hence, flexible and controlled reagent addition/dilution is a basic but important operation in creating complex experimental settings. In droplet microarrays, reagent addition/dilution can be achieved with sequential liquid deposition (Fig. 3a, akin to direct injection in multiwells)34,41,46,47 or via droplet transport & fusion (such as in digital microfluidics, Fig. 3b).42,48 Sequential liquid deposition is easily performed in automated droplet microarrays, where an integrated dispenser deposits a reagent (or a dilution) to pre-seeded droplets, rendering high-throughput and automated experimental procedures (Fig. 3a). A major technical challenge of sequential liquid deposition, however, is that the reagent (or the dilution liquid) needs to be deposited selectively, without generating cross-contamination issues. Using a contact-free liquid printing technique, where reagent or dilution liquid was ejected with high speed on top of an array and thereby without the need to be in contact with the oil or the pre-seeded droplets, Sun et al. report a method to automatically print secondary droplets with different compositions and volumes.49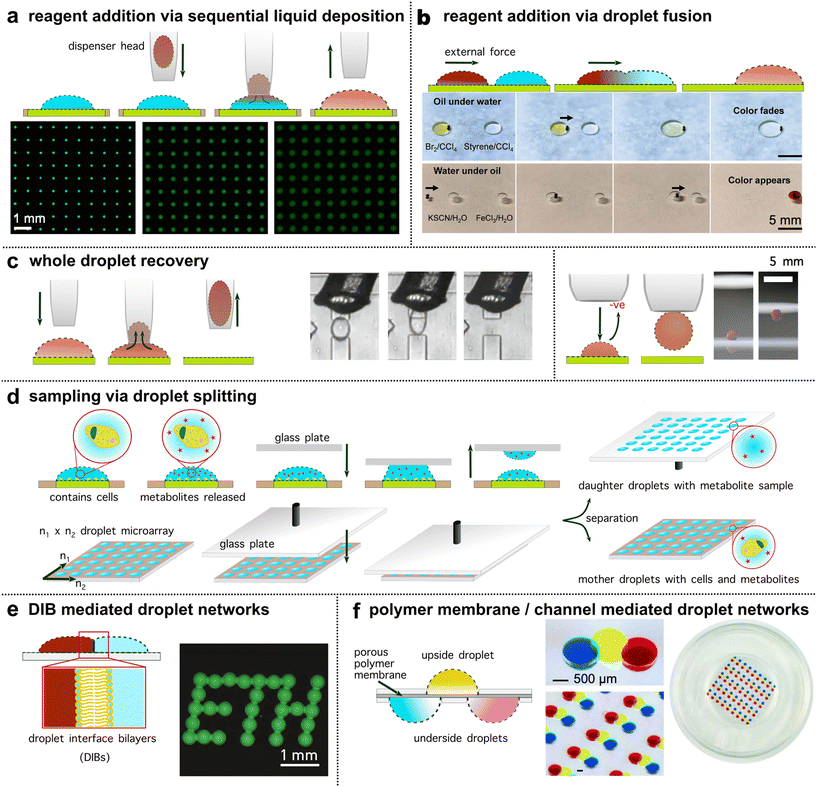 | ||
| Fig. 3 Operations on droplet microarrays. (a) Reagent addition/dilution via sequential liquid deposition as sketched in the top row. The micrographs show a droplet microarray with increasing volumes and a sequential decrease in the concentration of fluorescein (reproduced from Zhu et al.41 with permission from Nature Portfolio under a Creative Commons License CC BY 4.0.). (b) Reagent addition/dilution via droplet transport & fusion. The bottom micrographs show droplet transport and fusion actuated by magnetic beads (black dots indicated by arrow) for reagent addition (reproduced from Li et al.42 with permission from the American Association for the Advancement of Science under a Creative Commons License CC BY 4.0.). (c) Whole droplet recovery. Left panel: Droplet recovery using a capillary pipette connected to a negative pressure pump to aspirate a whole droplet (reproduced from Cole et al.34 with permission from the American Association for the Advancement of Science, copyright 2017). Right panel: A snapshot of seizing a blood droplet using a charged pipette (reproduced from Sun et al.43 with permission from Springer Nature Limited, copyright 2019). (d) Parallel droplet volume sampling via droplet splitting using the “sandwich approach” (reproduced from Haidas et al.44 with permission from the American Chemical Society, copyright 2020). (e) Droplet connections to form spatially structured yet communicating droplet networks via droplet interface bilayers (reproduced from Bachler et al.27 with permission from Nature Portfolio under a Creative Commons License CC BY 4.0.). (f) Droplet chain array connected by a porous polymer membrane. This conformation can render chemical gradients within the droplet chain to study chemoattractant-induced cell migrations (reproduced from Ma et al.45 with permission from the Royal Society of Chemistry, copyright 2016). | ||
In addition to sequential liquid deposition, reagent addition or dilution can also be achieved by the transport and fusion of droplets with explicit contents (i.e.Fig. 3b). To actuate the droplets, various external forces including electricity,50 magnets,42,48 acoustics,51 light52 and charged43 or wetting surfaces53 have been deployed. As with EWOD, in these contexts, consideration of surface properties and functionality of the plate is required. For example, by employing a magnetically functionalised polydimethylsiloxane (PDMS) membrane with micro-pillar arrays, Chen et al. report a platform with the potential to realise high-throughput and parallel on-plate droplet manipulation.48 In this study, parallel control of three droplets pairs was demonstrated with transport, fusion and mixing. Alternatively, the FlipDrop platform described by Zhang et al. merged droplets positioned on two microarrays (n = 36 (6 × 6) droplets per array) by flipping one chip on top of the other. This facilitated chemical combinations with orthogonal concentration gradients.54 Overall, both sequential liquid deposition and droplet–droplet fusion allow for (repetitive) addition of controlled quantities of reagents to pre-existing droplets to manipulate their chemical environment.
Droplet recovery and sampling
In evolutionary biotechnology or screening assays, volume sampling or recovery of specific experiments for additional rounds of culture or analysis is often required. Recovery of whole droplets from microarrays is usually accomplished by sucking/vacuuming the droplets with an aspiration probe from the substrate (i.e., Fig. 3c, left panel). By using a capillary pipette, Cole et al. described a method to selectively recover a specific droplet or to indiscriminately recover mass numbers of droplets from a printed array.34 In addition, using a pump-free, charged surface pipette, Sun et al. developed a method capable of seizing a sessile droplet (Fig. 3c, right panel) from the substrate and releasing it to an external location.43 Interestingly, during the entire transport process, droplets maintained their spherical shape and no mass loss was observed.In certain procedures, the objective is to preserve the overall volume while extracting a smaller sample for analysis. Here, the measurement resolution is defined by the number of fractions the bulk can be split into. Retrieving a small fraction from a tiny droplet can be challenging. For this task, droplet splitting offers a solution. By wetting the top of the microarray droplets onto another glass plate (the ‘sandwich approach’), the glass plate can then be removed to create ‘daughter’ droplets whilst leaving behind volume reduced droplets on the ‘mother’ droplet microarray (Fig. 3d). Using this plate-based droplet splitting method,44 Haidas et al. were able to split in parallel about 6000 droplets (initial droplet volume = 8 nL) within seconds. The ‘daughter’ droplets were interfaced with mass spectrometry to analyse a protein secreted by yeast cells which remained alive in the ‘mother’ droplets after the splitting, where their spatial resolution was maintained. Of note, even with the help of pipetting robots, this level of sampling (about 6000 droplets within seconds) is not yet feasible in multiwell-based systems.
Formation of droplet pairs and droplet networks
In contrast to multiwell-based systems, droplets deposited on open microarrays are wall-less and this fosters one of their most unique operations – the formation of spatially structured yet communicating droplet networks (Fig. 3e and f). These networks can mimic interactions among cells, interactions between cells and the environment, as well as the complex and cooperative structures and functions of living tissues or organs (referred to as “body-on-a-chip”).55 To construct such functional networks, droplets can be precisely arranged and interconnected via the formation of droplet interface bilayers (DIBs, Fig. 3e), a porous polymer membrane (Fig. 3f), or via microchannels, e.g. in hanging droplet arrays where cell assemblies and spheroids are connected.56DIBs are formed when two aqueous droplets in oil, each stabilised by a monolayer of phospholipids, are brought together. At the droplet–droplet interface a bilayer spontaneously forms.57 2D and 3D networks of droplets can thereby be assembled through controlled deposition and arrangement of lipid coated droplets.58,59 Membrane permeable compounds can pass passively through the bilayer by diffusion,26,60 or through integrated membrane pores.27 Krishna Kumar et al. report a hydrogel droplet-based method to arrange bacterial communities.61 Precision, inkjet-based printing arranged a 2D droplet microarray on quartz, using DIBs to initially stabilise droplet contact. DIBs were then removed following a gelation step, rendering inter-population communication across a patterned hydrogel droplet network with encapsulated bacterial populations.
Alternatively, a porous polymer membrane can separate two or more droplets to allow free diffusion of solutes between the droplets, conceptionally akin to transwell plates, where two compartments are separated by a permeable membrane/support (i.e. in Fig. 3f). Depending on the pore size, the passage of particles or cells can be controlled. Ma et al. deploy this strategy with upside and downside droplets separated by a polymer membrane for establishing a chemoattractant concentration gradient.45 Different configurations of droplets on either side of the membrane enabled a platform technology for the study of cell migration. Content exchange has also been demonstrated through microchannels embedded in the microarray plate. By hanging droplets on a patterned PDMS plate,56 Frey et al. enabled parallelised formation of different microtissues/organs in a physiological order, facilitating nutrient supply, substance dosage and inter-organ communication. Such environments (discrete compartmentalisation yet communication between the subunits) cannot easily be achieved in multiwell plates where each individual well is isolated with physical barriers. Droplet microarrays can thus serve as a platform for creating so called ‘minimal tissues’ where the resolution of biological patterning is defined by the droplet size and arrangement precision.62
Applications
Analytical methods beyond microscopy
Droplet microarrays excel in rapid and minimal sample chemical analyses. Due to the open platform, droplets are physically accessible, which unlocks alternative options for analysis beyond optical methods. Droplet microarrays are amenable to label-free techniques such as mass spectrometry, achieved on plate or high-performance liquid chromatography (HPLC), requiring droplet recovery.27 Evaporation of liquid content on plate, which is accelerated by the inherently reduced volumes, can thus be exploited to up-concentrate an analyte. For example, by letting droplets evaporate on a super-hydrophobic surface created by micropillars (Fig. 4a), De Angelis et al. concentrated analytes on the tips of the pillars and achieved high-sensitivity surface-enhanced Raman spectroscopy (SERS).63 With this approach, it was possible to detect a single lambda DNA molecule from a 3 μL droplet with an initial concentration of 1 aM, as well as 100 molecules of rhodamine with an initial concentration of 10 aM. Similarly, Song et al. report a high throughput, mini-pillar device for the detection of breast cancer markers.64 Of note, besides the decreasing volume, different physical phenomena also arise during droplet evaporation. The coffee-ring effect occurs during evaporation where particles and molecules of different properties explicitly deposit on the substrate surface at the hosting site perimeter. Devineau and colleagues manipulated the coffee-ring effect to create a distribution of analytes from the perimeter based on size (and other properties) and separated bovine serum albumin (BSA, <7 nm) from nanoparticles (<500 nm).65 Dynamic changes in the liquid chemistry, substrate surface or addition of electrode patterns can have additional benefits, e.g., assisted by dielectrophoresis, Jung et al. introduced a method to separate bacteria, mammalian cells and antibodies in an evaporating droplet (Fig. 4b).66 To suppress the coffee ring effect and coat the whole hosting site, Zargartalebi et al. tailored the wettability of the plate for a homogeneous coating of functionalised nanoparticles.67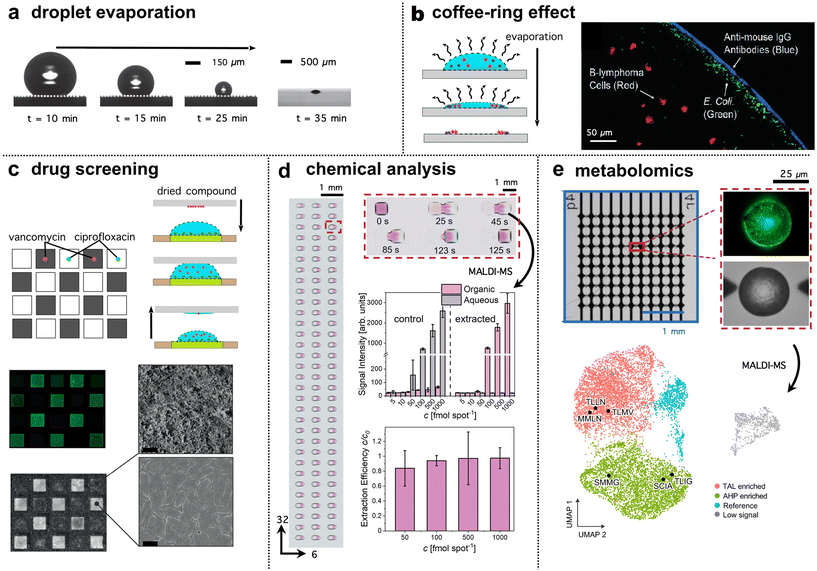 | ||
| Fig. 4 Droplet microarray applications and analysis. (a) Droplet evaporation shown by optical images (reproduced from De Angelis et al.,63 with permission from Springer Nature Limited, copyright 2011) on a super-hydrophobic surface. The droplet maintains its quasi-spherical shape to concentrate and deposit an analyte within an area of a few square micrometres for detection. (b) The separation of bacteria, mammalian cells and antibodies via the coffee-ring effect (reproduced from Jung et al.,66 and Garcia-Cordero et al.77 with permission from the American Chemical Society (copyright 2007) and the Royal Society of Chemistry (copyright 2017) respectively). Evaporation behaviour can thus depend on the plate surface. (c) Droplet microarrays are deployable across multiple research areas. An example of drug screening (cell viability) reproduced from Lei et al.29 with permission from Wiley under a Creative Commons License CC BY 4.0. (d) An example of chemical analysis, here with a miniaturised liquid–liquid extraction reproduced from Wiedmann et al.78 with permission from Wiley-VCH under a Creative Commons License CC BY 4.0. (e) An example of metabolic profiling reproduced from Xu et al.24 with permission from Nature Portfolio under a Creative Commons License CC BY 4.0. In both d and e, MALDI-MS was deployed. | ||
Droplet microarray platforms hyphenated with detection via mass spectrometry pose an exciting direction for label-free analysis of complex mixtures of chemicals and/or cells. Depending on the target, analytical instrument and method, sample preparation such as prior separation or cleavage steps may be necessary incurring additional fluid handling. Kelly and co-workers introduced a workflow for proteomic analysis for few or even single cells in nanowells, where fluids for cell lysis and protein digestion were delivered through reagent addition via a capillary dispenser.68,69 The platform successfully demonstrated the analysis of cellular protein markers to differentiate between cell types. Although currently realised for a relatively small number of wells (5 × 13), the method has great potential for further upscaling and automation. Alternatively, sample pre-treatment in the form of desalination is important for electrospray ionisation (ESI)-MS. Desalination was realised in a droplet microarray through addition of functionalised magnetic beads, which facilitated solid-phase extraction and implementation of washing steps, before the analytes were eluted and analysed by ESI-MS.70
Another ionisation method is matrix assisted laser desorption/ionisation mass spectrometry (MALDI-MS), where the sample is mixed and co-crystallised with a matrix compound that strongly absorbs laser light and supports desorption and ionisation of the analytes. Commercial MALDI-MS substrates are conductive steel plates in a 384-well multiwell format. The coupling of droplet microarrays to MALDI-MS is straightforward and has been shown for various applications including cell,71 protein,44 and chemical analyses.38 Most of these applications require the mixture with a matrix, but no further sample pre-treatment steps, however some care should be taken in the choice of oil and if required surfactant.72 To render the droplet microarray plate conductive, a coating of indium tin oxide is commonly used. At a defined point, the whole plate with seeded droplets can be directly transferred into a MALDI-MS instrument. This pipeline can be supplemented with the integration of commercial separation instruments. Küster et al. demonstrate fractionation of the eluate from nanoflow liquid chromatography (nano-LC) directly onto a droplet microarray.73 Notably, the incoming analyte band was spread over several droplets, fractionating at a resolution dictated by the band width and droplet size. This system enabled the analysis of post-translational protein modifications, such as phosphate groups and glycosylation patterns. Enzymatic cleavage steps were implemented in the droplets before downstream MS measurements.74
Finally, chemical analysis in droplets with nuclear magnetic resonance (NMR) has mostly been coupled to closed microfluidic systems and is underutilised in a droplet microarray context.75 By placing microfluidic devices within NMR detection chambers, additional analytical opportunities may be enabled as compared to capillary loading. An interesting platform was engineered by Swyer et al. for performing diffusion ordered spectroscopy (DOSY) in microliter droplets sandwiched between electrode layers with EWOD enabled sample manipulation.76 In this platform, droplets were shielded from evaporation and could be mixed on-demand without having to remove samples from the detection chamber. This utility may prove highly significant for automating NMR analytical pipelines.
Diverse high throughput applications
Droplet microarrays are powerful for applications where high throughput is required. Here, droplet microarray applications for cell cultivation and/or chemical analysis are highlighted. In principle, each droplet serves as a miniaturised culture chamber, i.e. a microcosm, where a determined chemical environment and nutrients are provided at the beginning of the experiment, together with cells. In such simple approaches, cell distribution follows the Poisson distribution, where not all droplets are occupied by the same cell number.79 As shown by Abate and co-workers, droplet sorting can be employed to select droplets within the distribution that contain single cells.34,80 After fractionation but before hosting site attachment, droplet contents can be screened and added to the microarray based on a droplet content readout.34 In this way, closed microfluidic droplet analysis systems act as the orchestrators of droplet printing. For example, droplets can be measured via in-line optics, enabling UV absorbance-based discrimination.81,82 Depending on the sorting technique, cells or populations of cells with a defined phenotype can be placed in selected positions on the droplet microarray. Since the operational process is repeatable, cells with selected properties can be paired deterministically, enabling controlled multicellular experiments in high throughput. The environment in each droplet can be further controlled. To mimic the extracellular matrix more closely, hydrogel droplets allow for cell culture and cell-based assays in 3D.83 Cells distributed in arrays can be subjected to relevant assays such as quantifying gene expression in individual cells by reverse transcription quantitative PCR.41Cells can produce valuable compounds of industrial interest, such as drugs or chemical building blocks. Droplet based cultivation retains such compounds in the aqueous phase,16 where they can be identified and further analysed e.g. for bioactivity. This is pertinent for protein engineering as it links genotype and phenotype in one compartment – a prerequisite for directed evolution. While the protection by oil enables the incubation of cells and hinders hydrophilic substances from leaving the compartment, it can be detrimental when considering hydrophobic compounds. Therefore, oil removal and use of humidified air supports the retention of compounds in each compartment, independent of their propensity to partition between phases.84 An elegant approach recently introduced by Wiedmann et al. brought together a water and organic droplet to miniaturise liquid–liquid extraction (Fig. 4d).78 This approach was scaled up to a droplet microarray format capable of performing high-throughput (192 droplets pairs per microarray), selective separation of single compounds from a complex solution of reagents. Assessed on-chip with MALDI-MS, miniaturisation conferred improved efficiency compared to the same extraction at the millilitre scale.
The maintenance of droplet composition is crucial in another notable application of droplet microarrays, specifically in high-throughput cell viability assays. This allows for not only the parallel testing of a larger number of samples but also a substantial reduction in volumes and, consequently, drug amounts. For example, Fang and co-workers report an automated droplet microarray to determine the effect of one or two drugs on cells in different concentrations and combinations.17 The system thereby enabled cultivation over three days by repeated addition of medium every 24 hours. Recently, Li et al. introduced a similar platform for antibiotic susceptibility testing (AST),85 where live cell imaging and real-time data analysis provided a fast (<4 h) sample-to-report turnaround time as required for AST. One ongoing technical challenge involves demonstrating numerous structurally diverse drugs simultaneously on a single plate. A solution was introduced by Lei et al.,29 in this work, a droplet array with dried drugs was arranged in contact with a bacteria-containing array. This plate with dried drugs on the surface mirrored the bacteria-containing droplets so that, once in contact, the drug dissolved into the aqueous droplets (Fig. 4c). Alternatively, droplet microarrays can be pre-seeded with a drug library and the cells added in a second step.19 To handle the multiple sample inputs from a drug library, a commercial fluid handling robot was implemented in both examples.
The transfection of cells allows manipulation of genetic content. In droplet microarray platforms, transfection was demonstrated by Zhou et al.,86 using a fluid handling approach that facilitated efficient exchange of medium with a liposome-based transfection mixture. More recently, transfection enhancers on three different cell types were screened at high throughput.19 In individual droplets (each 20 nL), the effect of 774 FDA-approved drugs on the transfection efficiency was tested, leading to 14 hit compounds that were further evaluated in traditional multiwell plate format. Through transfection of yeast cells with a plasmid library encoding enzymes, Xu et al. describe a new approach for characterising enzymatic activity.24 The platform consisted of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets generated in concave droplet hosting sites to concentrate material to the centre. The enzymes, synthesised within the yeast cells, influenced cell metabolism, and led to the formation of various products (Fig. 4e). This metabolic profile, as measured by MALDI-MS, corresponded to specific enzyme phenotypes which were further clustered based on product pattern similarity. This study demonstrated a complete screen of 9000 mutants on a single plate, elegantly utilising all the benefits of a droplet microarray for high throughput applications. Beyond these applications, we believe that further, currently semi-automated synthesis and screening applications such as multiplex DNA synthesis,87 DNA-encoded chemical libraries and their use in screening for binding partners,88 and related methods in combinatorial chemistry could benefit from ultra-high throughput by means of droplet microarrays.
000 droplets generated in concave droplet hosting sites to concentrate material to the centre. The enzymes, synthesised within the yeast cells, influenced cell metabolism, and led to the formation of various products (Fig. 4e). This metabolic profile, as measured by MALDI-MS, corresponded to specific enzyme phenotypes which were further clustered based on product pattern similarity. This study demonstrated a complete screen of 9000 mutants on a single plate, elegantly utilising all the benefits of a droplet microarray for high throughput applications. Beyond these applications, we believe that further, currently semi-automated synthesis and screening applications such as multiplex DNA synthesis,87 DNA-encoded chemical libraries and their use in screening for binding partners,88 and related methods in combinatorial chemistry could benefit from ultra-high throughput by means of droplet microarrays.
Conclusion
Microfluidic technologies enable cutting-edge approaches spanning from chemical synthesis to biological cultivation. With the rise of artificial intelligence, machine learning and automation, throughput is an essential factor in the development of new modalities. Droplet microarray technologies represent an approach to chemical and biological experimentation that can meet the escalating demand for massive data generation. Due to the flexibility in the choice of analytical methods, droplet microarrays facilitate multimodal data acquisition, pertinent to the field of biological analysis. Beyond typical multiwell plate approaches, droplet microarrays open new avenues for creating programmable networks of interconnecting wells facilitating a platform for the design of biologically inspired materials and experimentation. Future applications may encompass combinatorial chemical synthesis for generating compound libraries, along with large-scale drug screening, protein engineering, and high-throughput surface chemistry and binding assays. For custom systems, sharing operational principles and open-source code for imaging and data analysis could lead to even more rapid advancement and uptake.In the coming years, we therefore anticipate new applications and measurements will be enabled with droplet microarrays. This raises the question of how to strategically integrate these plates into pre-existing analytical machines and operational protocols. Scaling up droplet microarrays to the dimensions of multiwell plates is one potential solution. Improved uptake will also lead to further cost reductions in plate engineering and maintenance. Keeping this perspective in mind, it is crucial to examine the interplay between the droplet generation method, plate reusability, selected analytical technique, experimental throughput, and operational simplicity especially when compared to the same assay performed in a multiwell plate. The ongoing adoption of droplet microarrays will further illuminate and enhance an understanding of this multiparameter relationship from both an academic and industrial perspective. Compared to multiwell plates, droplet microarray-based assays can condense more experimental information into a smaller material space. By providing a sustainable alternative, droplet microarray technologies can embrace reduced plastic waste and exercise judicious usage of environmentally harmful substances. Taken together, there is therefore a high likelihood for a broad acceptance of droplet microarray technologies across the life sciences.
Author contributions
RS conceptualized the work, wrote the manuscript and produced and edited figs. BX, VFA wrote the manuscript and produced and edited figs. PD conceptualized the work, wrote the manuscript and supervised the project.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to thank Maximilian Breitfeld for assistance in producing the table of contents figure and for proof reading of the manuscript. The authors would also like to thank Simon F. Berlanda for proof reading of the manuscript. The authors acknowledge funding from the Swiss National Science Foundation (National Centre of Competence in Research (NCCR) Molecular Systems Engineering, Grant No. 51NF40-182895, and NCCR Antiresist, Grant No. 51NF40_180541). BX would like to thank the ETH Zürich Postdoctoral Fellowship, Grant No. 23-1 FEL-041.References
- D. S. Auld, P. A. Coassin, N. P. Coussens, P. Hensley, C. Klumpp-Thomas, S. Michael, G. S. Sittampalam, O. J. Trask, B. K. Wagner, J. R. Weidner, M. J. Wildey and J. L. Dahlin, Microplate Selection and Recommended Practices in High-throughput Screening and Quantitative Biology, 2004 Search PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Front. Bioeng. Biotechnol., 2021, 8, 602659 CrossRef PubMed.
- X. Li, A. V. Valadez, P. Zuo and Z. Nie, Bioanalysis, 2012, 4, 1509–1525 CrossRef CAS PubMed.
- B. Hwang, J. H. Lee and D. Bang, Exp. Mol. Med., 2018, 50, 96 CrossRef PubMed.
- S. Nishat, A. T. Jafry, A. W. Martinez and F. R. Awan, Sens. Actuators, B, 2021, 336, 129681 CrossRef CAS.
- Y. Zeng, J. W. Khor, T. L. Van Neel, W.-C. Tu, J. Berthier, S. Thongpang, E. Berthier and A. B. Theberge, Nat. Rev. Chem., 2023, 7, 439–455 CrossRef PubMed.
- P. Prabhakar, R. K. Sen, N. Dwivedi, R. Khan, P. R. Solanki, A. K. Srivastava and C. Dhand, Front. Nanotechnol., 2021, 3, 609355 CrossRef.
- C. Carrell, A. Kava, M. Nguyen, R. Menger, Z. Munshi, Z. Call, M. Nussbaum and C. Henry, Microelectron. Eng., 2019, 206, 45–54 CrossRef CAS.
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198 RSC.
- S. F. Berlanda, M. Breitfeld, C. L. Dietsche and P. S. Dittrich, Anal. Chem., 2021, 93, 311–331 CrossRef CAS PubMed.
- A. Stucki, J. Vallapurackal, T. R. Ward and P. S. Dittrich, Angew. Chem., Int. Ed., 2021, 60, 24368–24387 CrossRef CAS PubMed.
- A. M. Klein, L. Mazutis, I. Akartuna, N. Tallapragada, A. Veres, V. Li, L. Peshkin, D. A. Weitz and M. W. Kirschner, Cell, 2015, 161, 1187–1201 CrossRef CAS PubMed.
- A. Samanta, Q. Wang, S. K. Shaw and H. Ding, Mater. Des., 2020, 192, 108744 CrossRef CAS.
- K.-Y. Law, J. Phys. Chem. Lett., 2014, 5, 686–688 CrossRef CAS PubMed.
- T. Trantidou, Y. Elani, E. Parsons and O. Ces, Microsyst. Nanoeng., 2017, 3, 16091 CrossRef CAS PubMed.
- D. Haidas, S. Bachler, M. Köhler, L. M. Blank, R. Zenobi and P. S. Dittrich, Anal. Chem., 2019, 91, 2066–2073 CrossRef CAS PubMed.
- G.-S. Du, J.-Z. Pan, S.-P. Zhao, Y. Zhu, J. M. J. Den Toonder and Q. Fang, Anal. Chem., 2013, 85, 6747 Search PubMed.
- Y. Zhu, Y.-X. Zhang, L.-F. Cai and Q. Fang, Anal. Chem., 2013, 85, 6731 Search PubMed.
- Y. Liu, T. Tronser, R. Peravali, M. Reischl and P. A. Levkin, Adv. Biosyst., 2020, 4, e1900257 CrossRef PubMed.
- S. K. Küster, S. R. Fagerer, P. E. Verboket, K. Eyer, K. Jefimovs, R. Zenobi and P. S. Dittrich, Anal. Chem., 2013, 85, 1285–1289 CrossRef PubMed.
- F. L. Geyer, E. Ueda, U. Liebel, N. Grau and P. A. Levkin, Angew. Chem., Int. Ed., 2011, 50, 8424–8427 CrossRef CAS PubMed.
- W. Feng, L. Li, E. Ueda, J. Li, S. Heißler, A. Welle, O. Trapp and P. A. Levkin, Adv. Mater. Interfaces, 2014, 1, 1400269 CrossRef.
- W.-W. Liu, Y. Zhu, Y.-M. Feng, J. Fang and Q. Fang, Anal. Chem., 2017, 89, 822–829 CrossRef CAS PubMed.
- L. Xu, K.-C. Chang, E. M. Payne, C. Modavi, L. Liu, C. M. Palmer, N. Tao, H. S. Alper, R. T. Kennedy, D. S. Cornett and A. R. Abate, Nat. Commun., 2021, 12, 6803 CrossRef CAS PubMed.
- L. Xu, X. Li, W. Li, K.-C. Chang, H. Yang, N. Tao, P. Zhang, E. M. Payne, C. Modavi, J. Humphries, C.-W. Lu, A. R. Abate, L. Xu, X. Li, K.-C. Chang, P. Zhang, C. Modavi, A. R. Abate, W. Li, J. Humphries, C.-W. Lu, H. Yang, E. M. Payne, A. R. A. Chan and Z. Biohub, Adv. Mater., 2022, 34, 2108194 CrossRef CAS PubMed.
- S. Bachler, M. Ort, S. D. Krämer and P. S. Dittrich, Anal. Chem., 2021, 93, 5137–5144 CrossRef CAS PubMed.
- S. Bachler, D. Haidas, M. Ort, T. A. Duncombe and P. S. Dittrich, Commun. Biol., 2020, 3, 769 CrossRef CAS PubMed.
- J. W. Khor, U. N. Lee, J. Berthier, E. Berthier and A. B. Theberge, Adv. Mater. Interfaces, 2023, 10, 2202234 CrossRef.
- W. Lei, K. Demir, J. Overhage, M. Grunze, T. Schwartz and P. A. Levkin, Adv. Biosyst., 2020, 4, 2000073 CrossRef CAS PubMed.
- Y. Zhang, Y. Zhu, B. Yao and Q. Fang, Lab Chip, 2011, 11, 1545 RSC.
- M. Pabst, S. R. Fagerer, R. Köhling, S. K. Küster, R. Steinhoff, M. Badertscher, F. Wahl, P. S. Dittrich, K. Jefimovs and R. Zenobi, Anal. Chem., 2013, 85, 9771–9776 CrossRef CAS PubMed.
- H. Cui, X. Wang, J. Wesslowski, T. Tronser, J. Rosenbauer, A. Schug, G. Davidson, A. A. Popova and P. A. Levkin, Adv. Mater., 2021, 33, 2006434 CrossRef CAS PubMed.
- D. Park, M. Kang, J. W. Choi, S. M. Paik, J. Ko, S. Lee, Y. Lee, K. Son, J. Ha, M. Choi, W. Park, H. Y. Kim and N. L. Jeon, Lab Chip, 2018, 18, 2013–2022 RSC.
- R. H. Cole, S.-Y. Tang, C. A. Siltanen, P. Shahi, J. Q. Zhang, S. Poust, Z. J. Gartner and A. R. Abate, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 8728–8733 CrossRef CAS PubMed.
- B. Hadwen, G. R. Broder, D. Morganti, A. Jacobs, C. Brown, J. R. Hector, Y. Kubota and H. Morgan, Lab Chip, 2012, 12, 3305 RSC.
- W. Wang, J. Chen and J. Zhou, Appl. Phys. Lett., 2016, 108, 243701 CrossRef.
- A. Yusof, H. Keegan, C. D. Spillane, O. M. Sheils, C. M. Martin, J. J. O'Leary, R. Zengerle and P. Koltay, Lab Chip, 2011, 11, 2447 RSC.
- M. Brehm, S. Heissler, S. Afonin and P. A. Levkin, Small, 2020, 16, 1905971 CrossRef CAS PubMed.
- Z. Dong and Q. Fang, TrAC, Trends Anal. Chem., 2020, 124, 115812 CrossRef CAS.
- P. Zhang, C. Liu, C. Modavi, A. Abate and H. Chen, Trends Biotechnol., 2023 DOI:10.1016/j.tibtech.2023.09.001.
- Y. Zhu, Y.-X. Zhang, W.-W. Liu, Y. Ma, Q. Fang and B. Yao, Sci. Rep., 2015, 5, 9551 CrossRef PubMed.
- A. Li, H. Li, Z. Li, Z. Zhao, K. Li, M. Li and Y. Song, Sci. Adv., 2020, 6, eaay5808 CrossRef CAS PubMed.
- Q. Sun, D. Wang, Y. Li, J. Zhang, S. Ye, J. Cui, L. Chen, Z. Wang, H. J. Butt, D. Vollmer and X. Deng, Nat. Mater., 2019, 18, 936–941 CrossRef CAS PubMed.
- D. Haidas, M. Napiorkowska, S. Schmitt and P. S. Dittrich, Anal. Chem., 2020, 92, 3810–3818 CrossRef CAS PubMed.
- Y. Ma, J. Z. Pan, S. P. Zhao, Q. Lou, Y. Zhu and Q. Fang, Lab Chip, 2016, 16, 4658–4665 RSC.
- P. Garstecki, S. T. S. Kaminski, P. Garstecki and T. S. Kaminski, Chem. Soc. Rev., 2017, 46, 6210–6226 RSC.
- Y. Zhu, L.-N. Zhu, R. Guo, H.-J. Cui, S. Ye and Q. Fang, Sci. Rep., 2014, 4, 5046 CrossRef CAS PubMed.
- G. Chen, B. Ji, Y. Gao, C. Wang, J. Wu, B. Zhou and W. Wen, Sens. Actuators, B, 2019, 286, 181–190 CrossRef CAS.
- Y. Sun, X. Chen, X. Zhou, J. Zhu and Y. Yu, Lab Chip, 2015, 15, 2429–2436 RSC.
- J. Li, N. S. Ha, T. Liu, R. M. van Dam and C.-J. Kim, Nature, 2019, 572, 507–510 CrossRef CAS PubMed.
- Z. Yuan, C. Lu, C. Liu, X. Bai, L. Zhao, S. Feng and Y. Liu, Sci. Adv., 2023, 9, eadg2352 CrossRef PubMed.
- F. Wang, M. Liu, C. Liu, C. Huang, L. Zhang, A. Cui, Z. Hu and X. Du, Natl. Sci. Rev., 2023, 10, nwac164 CrossRef CAS PubMed.
- L. Sun, F. Bian, Y. Wang, Y. Wang, X. Zhang and Y. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 4527–4532 CrossRef CAS PubMed.
- Y. Zhang and T. H. Wang, RSC Adv., 2019, 9, 21741–21747 RSC.
- A. Alcinesio, O. J. Meacock, R. G. Allan, C. Monico, V. Restrepo Schild, I. Cazimoglu, M. T. Cornall, R. Krishna Kumar and H. Bayley, Nat. Commun., 2020, 11, 2105 CrossRef CAS PubMed.
- O. Frey, P. M. Misun, D. A. Fluri, J. G. Hengstler and A. Hierlemann, Nat. Commun., 2014, 5, 4250 CrossRef CAS PubMed.
- M. A. Holden, D. Needham and H. Bayley, J. Am. Chem. Soc., 2007, 129, 8650–8655 CrossRef CAS PubMed.
- M. S. Friddin, G. Bolognesi, Y. Elani, N. J. Brooks, R. V. Law, J. M. Seddon, M. A. A. Neil and O. Ces, Soft Matter, 2016, 12, 7731–7734 RSC.
- T. Wauer, H. Gerlach, S. Mantri, J. Hill, H. Bayley and K. T. Sapra, ACS Nano, 2014, 8, 771–779 CrossRef CAS PubMed.
- R. Strutt, F. Sheffield, N. E. Barlow, A. J. Flemming, J. D. Harling, R. V. Law, N. J. Brooks, L. M. C. Barter and O. Ces, Lab Chip, 2022, 22, 972–985 RSC.
- R. Krishna Kumar, T. A. Meiller-Legrand, A. Alcinesio, D. Gonzalez, D. A. I. Mavridou, O. J. Meacock, W. P. J. Smith, L. Zhou, W. Kim, G. S. Pulcu, H. Bayley and K. R. Foster, Nat. Commun., 2021, 12, 857 CrossRef CAS PubMed.
- J. Govey-Scotland, L. Johnstone, C. Myant and M. S. Friddin, Lab Chip, 2023, 23, 5068–5080 RSC.
- F. De Angelis, F. Gentile, F. Mecarini, G. Das, M. Moretti, P. Candeloro, M. L. Coluccio, G. Cojoc, A. Accardo, C. Liberale, R. P. Zaccaria, G. Perozziello, L. Tirinato, A. Toma, G. Cuda, R. Cingolani and E. Di Fabrizio, Nat. Photonics, 2011, 5, 682–687 CrossRef CAS.
- X. Song, T. Xu, Y. Song, X. He, D. Wang, C. Liu and X. Zhang, Talanta, 2020, 218, 121206 CrossRef CAS PubMed.
- S. Devineau, M. Anyfantakis, L. Marichal, L. Kiger, M. Morel, S. Rudiuk and D. Baigl, J. Am. Chem. Soc., 2016, 138, 11623–11632 CrossRef CAS PubMed.
- J. Y. Jung and H. Y. Kwak, Anal. Chem., 2007, 79, 5087–5092 CrossRef CAS PubMed.
- H. Zargartalebi, S. H. Hejazi and A. Sanati-Nezhad, Nat. Commun., 2022, 13, 3085 CrossRef CAS PubMed.
- Y. Zhu, G. Clair, W. B. Chrisler, Y. Shen, R. Zhao, A. K. Shukla, R. J. Moore, R. S. Misra, G. S. Pryhuber, R. D. Smith, C. Ansong and R. T. Kelly, Angew. Chem., Int. Ed., 2018, 57, 12370–12374 CrossRef CAS PubMed.
- Y. Zhu, P. D. Piehowski, R. Zhao, J. Chen, Y. Shen, R. J. Moore, A. K. Shukla, V. A. Petyuk, M. Campbell-Thompson, C. E. Mathews, R. D. Smith, W.-J. Qian and R. T. Kelly, Nat. Commun., 2018, 9, 882 CrossRef PubMed.
- Y. Su, Y. Zhu and Q. Fang, Lab Chip, 2013, 13, 1876–1882 RSC.
- C. RamalloGuevara, D. Paulssen, A. A. Popova, C. Hopf and P. A. Levkin, Adv. Biol., 2021, 5, 2000279 CrossRef CAS PubMed.
- S. E. Bell, I. Park, S. S. Rubakhin, R. Bashir, Y. Vlasov and J. V. Sweedler, ACS Meas. Sci. Au, 2021, 1, 147–156 CrossRef CAS PubMed.
- S. K. Küster, M. Pabst, K. Jefimovs, R. Zenobi and P. S. Dittrich, Anal. Chem., 2014, 86, 4848–4855 CrossRef PubMed.
- S. K. Küster, M. Pabst, R. Zenobi and P. S. Dittrich, Angew. Chem., Int. Ed., 2015, 54, 1671–1675 CrossRef PubMed.
- M. V. Gomez, S. Baas and A. H. Velders, Nat. Commun., 2023, 14, 3885 CrossRef CAS PubMed.
- I. Swyer, S. von der Ecken, B. Wu, A. Jenne, R. Soong, F. Vincent, D. Schmidig, T. Frei, F. Busse, H. J. Stronks, A. J. Simpson and A. R. Wheeler, Lab Chip, 2019, 19, 641–653 RSC.
- J. L. Garcia-Cordero and Z. H. Fan, Lab Chip, 2017, 17, 2150–2166 RSC.
- J. J. Wiedmann, Y. N. Demirdögen, S. Schmidt, M. A. Kuzina, Y. Wu, F. Wang, B. Nestler, C. Hopf and P. A. Levkin, Small, 2023, 19, 2204512 CrossRef CAS PubMed.
- A. R. Liberski, J. T. Delaney and U. S. Schubert, ACS Comb. Sci., 2011, 13, 190–195 CrossRef CAS PubMed.
- C. A. Siltanen, R. H. Cole, S. Poust, L. Chao, J. Tyerman, B. Kaufmann-Malaga, J. Ubersax, Z. J. Gartner and A. R. Abate, Sci. Rep., 2018, 8, 7913 CrossRef PubMed.
- T. A. Duncombe, A. Ponti, F. P. Seebeck and P. S. Dittrich, Anal. Chem., 2021, 93, 13008–13013 CrossRef CAS PubMed.
- S. Ladeveze, P. J. Zurek, T. S. Kaminski, S. Emond and F. Hollfelder, ACS Catal., 2023, 13, 10232–10243 CrossRef CAS PubMed.
- E. Ueda, F. L. Geyer, V. Nedashkivska and P. A. Levkin, Lab Chip, 2012, 12, 5218 RSC.
- A. Ruszczak, P. Jankowski, S. K. Vasantham, O. Scheler and P. Garstecki, Anal. Chem., 2023, 95, 1574–1581 CAS.
- C. Li, S. McCrone, J. W. Warrick, D. R. Andes, Z. Hite, C. F. Volk, W. E. Rose and D. J. Beebe, Lab Chip, 2023, 23, 2005–2015 RSC.
- Y. Zhou, Y. Pang and Y. Huang, Anal. Chem., 2012, 84, 2576–2584 CrossRef CAS PubMed.
- D. Verardo, B. Adelizzi, D. A. Rodriguez-Pinzon, N. Moghaddam, E. Thomée, T. Loman, X. Godron and A. Horgan, Sci. Adv., 2023, 9, eadi0263 CrossRef CAS PubMed.
- L. Mannocci, Y. Zhang, J. Scheuermann, M. Leimbacher, G. De Bellis, E. Rizzi, C. Dumelin, S. Melkko and D. Neri, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17670–17675 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |