Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Making the InChI FAIR and sustainable while moving to inorganics†‡
Gerd Blanke *a,
Jan Brammerb,
Djordje Baljozovic
*a,
Jan Brammerb,
Djordje Baljozovic b,
Nauman Ullah Khan
b,
Nauman Ullah Khan b,
Frank Lange
b,
Frank Lange b,
Felix Bänschc,
Clare A. Tovee
b,
Felix Bänschc,
Clare A. Tovee d,
Ulrich Schatzschneider
d,
Ulrich Schatzschneider e,
Richard M. Hartshorn
e,
Richard M. Hartshorn f and
Sonja Herres-Pawlis
f and
Sonja Herres-Pawlis *b
*b
aStructurePendium GmbH, Essen, Germany. E-mail: gerd.blanke@structurependium.com
bInstitut für Anorganische Chemie, Landoltweg 1a, 52074 Aachen, Germany. E-mail: sonja.herres-pawlis@ac.rwth-aachen.de
cBeilstein-Institut zur Förderung der Chemischen Wissenschaften, Trakehner Straße 7-9, 60487 Frankfurt am Main, Germany
dCambridge Crystallographic Data Centre, 12 Union Road, Cambridge, CB2 1EZ, UK
eInstitut für Anorganische Chemie, Julius-Maximilians-Universität Würzburg, Am Hubland, 97074 Würzburg, Germany
fSchool of Physical and Chemical Sciences, University of Canterbury, Christchurch, New Zealand
First published on 4th September 2024
Abstract
The InChI (International Chemical Identifier) standard stands as a cornerstone in chemical informatics, facilitating the structure-based identification and exchange chemical information about compounds across various platforms and databases. The InChI as a unique canonical line notation has made chemical structures searchable on the internet at a broad scale. The largest repositories working with InChIs contain more than 1 billion structures. Central to the functionality of the InChI is its codebase, which orchestrates a series of intricate steps to generate unique identifiers for chemical compounds. Up to now, these steps have been sparsely documented and the InChI algorithm had to be seen as a black box. For the new v1.07 release, the code has been analyzed and the major steps documented, more than 3000 bugs and security issues, as well as nearly 60 Google OSS-Fuzz issues have been fixed. New test systems have been implemented that allow users to directly test the code developments. The move to GitHub has not only made the development more transparent but will also enable external contributors to join the further development of the InChI code. Motivation for this modernisation was the urgency to treat molecular inorganic compounds by the InChI in a meaningful way. Until now, no classic string representation fulfills this need of molecular inorganic chemistry. Currently bonds to metal centers are by definition disconnected which makes most inorganic InChIs meaningless at the moment. Herein, we propose new routines to remedy this problem in the representation of molecular inorganic compounds by the InChI.
Introduction
The rapid development of machine learning (ML) has fueled the digitization of chemistry at a fast pace over the last decade. For many digital purposes, the typical graphical representation of chemical substances has to be converted into a simpler machine-readable molecular representation (Fig. 1).While Molfiles are two dimensional Chemical Tables1 that cannot be canonicalized because of the embedded atom coordinates, line notations encode basic molecular structural information into a one-dimensional string of characters that makes chemical structures accessible for machine learning.2 The most common line notations are SMILES and InChI.
The Simplified Molecular Input Line Entry System (SMILES) is very common in computational chemistry.3–5 “SMILES is a true language, albeit with a simple vocabulary (atom and bond symbols) and only a few grammar rules. SMILES representations of structure can in turn be used as “words” in the vocabulary of other languages designed for storage of chemical information (information about chemicals) and chemical intelligence (information about chemistry).”6 The “words” consist of characters representing atoms according to the symbols of the chemical elements, bond types connecting each of the atoms, and numbers indicating ring closures. Each chemical structure may be represented by multiple equivalent SMILES strings; canonical SMILES are supposed to offer a unique representation for each compound. SMILES are human-readable and machine-digestible but suffer from many shortcomings such as the existence of multiple SMILES “dialects”, vendor specific canonicalization algorithms for SMILES, the non-ideal treatment of tautomers and inorganic compounds, and the limited functionality to represent non-tetrahedral stereochemistry.
The IUPAC International Chemical Identifier (InChI) is a structure-based canonical chemical identifier, i.e. each chemical structure can only be represented by one InChI specific for this structure and every InChI identifies just one molecule.7 Hereby, identical structures can be recognised as such, and chemical data of structures can be linked e.g. in repositories, in databases, or on the internet.
InChI encodes chemical structures into a layered line notation. Unlike SMILES, InChI only represents the pure connectivity of a molecule, but not the bond order between the atoms. Instead, all non-hydrogen ligands of each atom are explicitly connected while the hydrogen atoms are represented in a separate hydrogen layer that comprises all implicit and explicit hydrogens of the chemical structure.
The InChI is non-proprietary and open source. Hence, it has been adopted in many chemical information resources and software programs. By design, anyone can compute the InChI code for a chemical compound, either by downloading the freely available InChI software,8 by working with an appropriate structure drawing tool, or by using web resources such as the PubChem Sketcher9 or the free InChI web demo.10 Furthermore, many open-educational resources are available for instructors.11 Especially with regard to the growing importance of the FAIR data principles12 in chemistry and chemical publishing,13 the InChI gains an important role by providing a straight-forward mechanism for connecting chemical knowledge across databases and other scientific resources.
In 2005, the first version of InChI was made publicly available.7 Many further versions followed.14 Moreover, additional developments cover reactions (Reaction-InChIs/RInChI, RInChI 1.1 published in 202415), mixtures (Mixture-InChI/MInChI, prototype16), and nanomaterials (Nanomaterial-InChIs/NInChI17). InChI v1.06 is the current version which is used by databases such as PubChem and EBI UniChem.
In order to prevent parallel developments or dialects (which happens in other molecular representations), the development of the InChI is guarded by the InChI Trust18 and the IUPAC InChI committee.19 This ensures a clean versioning and generates trust in the databases worldwide. Over recent years, many working groups for the further development of the InChI emerged, driven by volunteers from all subdisciplines of chemistry, to advance the InChI development in molecular inorganics, stereochemistry, concerning mixtures, polymers, Markush structures, and many more.20 All new developments are agreed upon in the working groups and then in the IUPAC subcommittee. The subcommittee and the InChI Trust decide on the implementation strategy, versioning, testing phases, and release criteria that must be reached to let the new version become the standard version.
In today’s world, code development is no longer done by a single programmer but has become a team effort using modern tools of connected software development such as GitHub. Thus, in order to enhance the sustainability of the InChI and to manage the multiple requirements as well as the exchange between InChI working groups, the further InChI development has been set on a new basis, enabling direct digital collaboration worldwide. This is accomplished by cleaning up the “old” code, bringing it to GitHub,21 and changing from the IUPAC/InChI-Trust License to the MIT license. The key steps of this process which yield the new version 1.07 are described here, along with an outline of the approach that is envisaged for treating coordination and organometallic compounds in the InChI framework. Besides the utilization of GitHub, also a new open testing environment and a more comprehensive web demo have been set up.
Results achieved with the most recent InChI release version 1.07
In order to bring the InChI code into a modern coding environment, the following steps have been carried out:• The InChI development has been moved to GitHub.21
• The code fragments that existed after the former developer Igor Pletnev passed away were fused to a new code base.
• Several thousand bugs, warnings and hints have been fixed, and the total number of Google® OSS-Fuzz project22 reports has been reduced drastically.
• The architecture of the code has been opened and better documented to allow the participation of additional open source developers.
• The core of the InChI programming (canonicalization and connectivity string creator) are kept as they are. Structures are normalized to fit into the existing canonicalization process. Changes to the InChIs of organic compounds are avoided.
• The control and test of code enhancements now uses standard procedures.
• Extensive code testing uses the PubChem substance database with 300 million structures and the PubChem 3D compounds for regression and invariance tests.
• To gain more transparency of the InChI development, the newly introduced web demo provides InChI calculations via web browsers based on the most recent release version of InChI or the latest developmental release.
• Enhanced documentation covering the chemical representation and more technical details helps the users.
The work with GitHub has opened a new support stream to the InChI Trust that is responsible for the technical development of the InChI. Up to now, only membership fees could be used to maintain and develop the InChI. The open environment makes it possible now to work with contributions in kind by organizations that are legally not allowed to become an official member of the trust but are able to let their developers work on the code. That changes the role of the Trust that now has to oversee the different developments and has to take care of a unique code concept integrating all contributed sources.
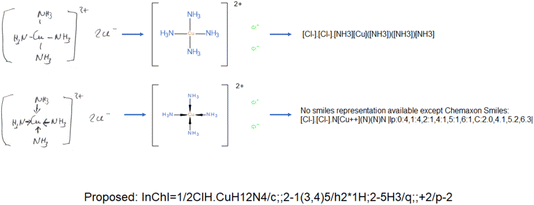
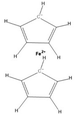
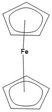
Inorganics and organometallics lack FAIR data repositories. The Findability, Accessibility, Interoperability and Reproducibility of data for inorganics and organometallics is very limited because of missing well-working chemical representations and identifiers. Not really fitting tools has led to multiple workarounds to store these substance classes in databases. Ferrocene as a typical representative in this area is found very frequently in multiple versions in the same database because the structure-based duplicate checks are not able to recognize the different structural depiction formats as representing the same compound (Fig. 1). In 2011, haptic and coordinative bonds as zero-order-bonds were introduced into the Molfile format23 and led to an improved representation of inorganics and organometallics. However these enhancements have only been transferred into SMILES and InChIs on a very limited base. Most of the SMILES dialects do not understand them at all.
Within the InChI community the demands on the identification of inorganics and organometallics have increased in the last few years when it became clear that only a unique identifier guarantees findable, accessible, interoperable and reproducible data, for instance in lab notebooks and databases.24 With the long-term goal to bring ML to molecular inorganic chemistry, the IUPAC funded InChI subgroup is working on a solution which allows the meaningful handling of molecular inorganic compounds including both classical coordination complexes as well as organometallic compounds.
InChI development for molecular inorganic (coordination and organometallic) compounds
While organic chemistry and solid-state chemistry can be easily handled by machine learning methods,25 the investigation of molecular inorganic compounds by ML methods is very limited because of the insufficient string representation of coordination compounds and organometallics as Table 1 illustrates for the example of ferrocene. Neither the SMILES versions nor the “old” InChI can handle the bonds in ferrocene appropriately and the connectivity is lost. Only the TUCAN representation makes an exception here with an identical line notation for the first two depictions.26 As sandbox, the TUCAN demonstrates how inorganic molecules can be handled when the metal bonds stay connected.| Depictions | Representation types | Smiles35 | ChemaxonSmiles35 | InChI10 | Proposed new standard InChI10 |
|---|---|---|---|---|---|
 |
PubChem CID 7611 | [Fe+2].[cH-]1cccc1.[cH-]2cccc2 | [Fe++].[H][C-]1C([H])=C([H])C([H])=C1[H].[H][C-]1C([H])=C([H])C([H])=C1[H]|c:7,17,t:3,13,lp:2:1,12:1| | InChI=1S/2C5H5.Fe/c2*1-2-4-5-3-1;/h2*1-5H;/q2*-1;+2 | InChI=1S/C10H10Fe/c1-2-4-5-3(1)11(1,2,4,5)6-7(11)9(11)10(11)8(6)11/h1-10H |
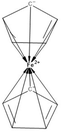 |
Coordinative bonds34 | [Fe++].[CH-]1C=CC=C1.[CH-]1C=CC=C1 | [Fe++]123456789C%10=C1[CH-]2C3=C4%10.C51=C6[CH-]7C8=C91|lp:3:1,8:1,C:4.6,5.8,3.4,2.2,1.0,9.16,8.14,7.12,6.10,10.18| | Unrecognized bond type | |
 |
Haptic bonds34 | [Fe].[CH-]1C=CC=C1.[CH-]1C=CC=C1 | *[Fe]*.[CH-]1C=CC=C1.[CH-]1C=CC=C1|c:3,5,8,10,lp:3:1,8:1,m:0:3.4.5.7.6,2:8.9.10.12.11,C:0.0,2.1| | Unrecognized bond type |
Currently, metal–ligand bonds are normally disconnected by the InChI algorithm because it was developed with a focus on organic compounds. In that approach, interactions with metal atoms by default were considered to be ionic in nature, and consequently all connectivity between organic fragments and metal centers were removed by the algorithm. While this is adequate for consistent treatment of organic molecules and classical ions, it creates serious problems for the treatment of many inorganic species, particularly coordination and organometallic compounds, where there is stereochemical information associated with the connections to the metal atom/ion, or where binding to a central atom introduces stereochemical elements to a ligand, for example by reducing the symmetry present in the absence of the metal atom. Such disconnections led to loss of all stereochemical information associated with the presence of ligands to a central atom. Furthermore, there are problems with the proper representation of haptic bonding in sandwich and half-sandwich compounds. Until now, most of the line notations only handle covalent bonds.
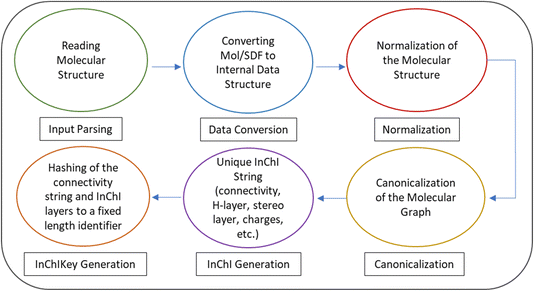
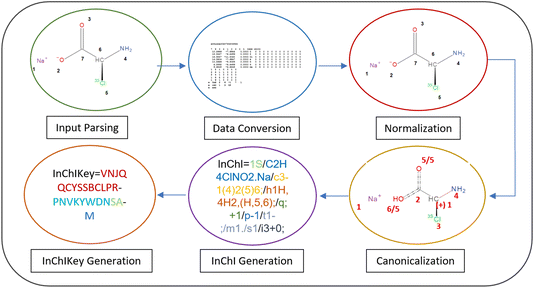
The InChI workflow
Fig. 2 illustrates the overarching code flow within the InChI codebase, employing arrows to delineate the sequential progression of operations.In the normalization process, one of the key transformations/adjustments applied to the molecular structure is tautomer enumeration. Tautomers are constitutional isomers of organic compounds that interconvert by chemical reactions, most often involving the transfer of one or more hydrogen atoms between different heavy atom centers (prototropic tautomerism).27 The InChI normalization algorithm involves enumerating tautomeric forms and selecting the most appropriate representation based on established rules and guidelines. This ensures that the resulting InChI represents the most stable and chemically relevant tautomeric form. The standard InChI supports only limited tautomerisms, however, additional tautomerisms are available in the engineering mode.28 By applying this normalization transformation, the InChI ensures that diverse molecular structures are represented in a reproducible and consistent manner, thereby facilitating accurate interpretation and comparison across different chemical compounds.
(a) Stereochemical representation: InChI standardizes the representation of stereochemical information, including stereo centers and double bond geometries. This involves assigning stereochemistry descriptors such as “R” and “S” to chiral centers and specifying E/Z configurations for double bonds. By adhering to consistent stereochemical conventions, this ensures that the resulting InChI accurately reflects the three-dimensional arrangement of atoms in the molecule.30 Greater stereochemical complexity is one of the challenges that will have to be met for coordination and organometallic compounds.
(b) Protonation and deprotonation: The protonation state of functional groups in a molecule can significantly impact its chemical properties and reactivity. Pre-canonicalization steps include adjusting the protonation and deprotonation states of functional groups to reflect the most probable physiological conditions or experimental settings. This ensures that the resulting InChI provides a relevant representation of the molecular structure under consideration.31 Recognising and representing the ways that this can interact with elements that can exhibit variable oxidation states is particularly important in inorganic systems.
(c) Isotopic composition: Isotopic substitution can occur naturally or artificially in chemical compounds, leading to variations in molecular mass and properties. Isotopic composition accounts for the presence of non-natural isotopes or abundances. This ensures that the resulting InChI accurately reflects the isotope profile of the molecule, enabling precise identification and characterization. The InChI technical document briefly explains how InChI manages isotopes.32
After the above mentioned steps, the subsequent canonicalization steps are undertaken to ensure the generation of a unique canonical representation.
(a) Atom numbering: After the canonicalization has created a unique number scheme InChI re-numbers the atom number according to the order in the chemical formula without taking the hydrogen atoms into account. These atom numbers are used in the connectivity string of InChI.
(b) Bond stereochemistry: Canonicalization standardizes the representation of stereochemistry by applying consistent rules for assigning E/Z configurations to double bonds and specifying cis/trans relationships for cyclic systems. By resolving ambiguities in bond stereochemistry, canonicalization ensures that equivalent structures yield identical canonical forms.31
(c) Canonicalization algorithms: InChI employs sophisticated algorithms, such as canonical labeling and graph isomorphism, to generate a unique canonical representation of the molecular graph. These algorithms ensure that the resulting identifiers are invariant under permutations of atom labels and bond orientations, thereby guaranteeing the uniqueness of the generated InChI identifiers.33
• The stereochemical layer consists of information regarding tetrahedral stereochemistry and double bonds.
• The isotope layer accounts for isotope substitutions.
• The charge layer captures information about formal charges on atoms.
Together, these layers ensure a comprehensive representation of a molecular structure, facilitating precise interpretation and comparison across different chemical compounds.
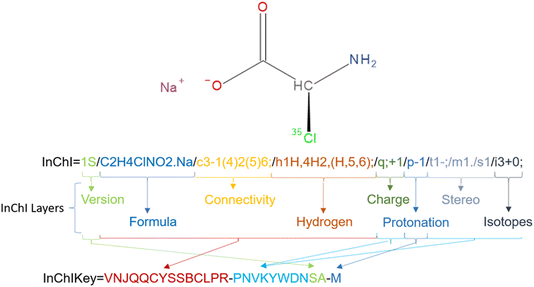
Take sodium (2S)-2-amino-2-(35Cl)chloranyl-acetate as an example (Fig. 3). Behind the “InChI=” prefix the digit 1 denotes the version of the InChI software and the character S denotes that it is a Standard InChI. The next part of the string is the formula of the chemical compound given as “/C2H4ClNO2.Na” where the dot “.” divides separated fragments from each other. The counting in the next layer – the connectivity string – is based on the order of the elements in the formula with the hydrogens being neglected: /c3-1(4)2(5)6. 3 corresponds to the Cl atom, 1 and 2 are the C atoms, 4 is the N atom, and 5 and 6 represent the oxygen atoms. Therefore the main branch runs from the Cl atom (3) to the C atoms 1 and 2 and ends at the O atom 6. To the first C atom (1), the N atom is linked while a branch to the second O atom (5) takes off from the second C atom (2). InChIs know about connectivity but not about the bond order. Therefore, the environment of each non-H atom must be fully described. That includes the full description of the H atom environment of each heavy atom that is defined in the so-called hydrogen layer/h. In the example above the first atom (C) has 1 H atom, atom 4 (N atom) 2 H atoms and the two O atoms 5 and 6 share 1 H atom because you cannot localize the H atom to one specific oxygen in a carboxylic acid group. Note that the acid part of the molecule is described as a neutral fragment in the string so that in the charge layer/c only the Na cation is characterized by its positive charge “/q; +1”. To regain the negative charge in the acid component the protonation layer “/p-1” subtracts a proton from the molecule leaving the necessary negative charge behind. The first C atom is a tetrahedral stereocenter whose parity is determined to be −1, denoted as “/t1-”. “/m1” indicates that the InChI uses the inverse arrangement of the center while “/s1” displays that absolute stereochemistry was requested. The final layer represents the isotope 35Cl corresponding to atom 3 that is marked by the atomic mass 35 with “/i3+0” with 35 as lowest isotopic mass of chlorine in InChI. For more syntax details see the InChI Technical Manual.33
On the other hand, the InChIKey as a condensed/hashed form of the full InChI string is the most commonly used InChI. It is limited to 27 alphabetic characters that are particularly useful for rapid searching and indexing in chemical databases.14,33 The chemical formula of the molecule, the connectivity layer, hydrogen positions, and the protonation state of the molecule are encoded within the first 14 characters by building one string out of those 4 layers that is hashed by using the cryptographic SHA-2 256-bit hash function using base-26 encoding. The returned uppercase string is cut off behind the 14th character (Fig. 3). These are followed by a hyphen and another string which consists of 10 characters: the first eight which encode the features that supplement the core data (charges, stereochemistry, isotopic layer), and the remaining two which indicate whether the InChI string is a Standard InChI “S”, Non-standard “N” or Beta “B”, as well as the InChI software version number with A for version 1. The (de)protonation state is indicated by the final character of the InChIKey.34 In the example of Fig. 3 it is the “M” representing the deprotonation by 1 proton as described above. (N represents no-deprotonation, O means that one proton is added). 33 The InChIKey serves as a global identifier, facilitating seamless integration and interoperability across diverse chemical databases and applications.31 The full process is visualised for sodium (2S)-2-amino-2-(35Cl)chloranyl-acetate in Fig. 4.
Requirements for the InChI FAIRification of inorganics and organometallics
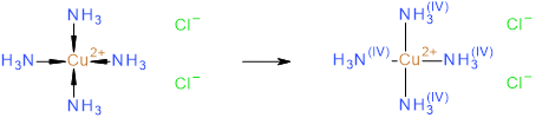
The implementation of the additional bond types to represent inorganic and organometallic compounds into the current InChI workflow (Fig. 2) requires that the InChIs of organic compounds must not be changed.The canonicalization and unique string generation processes (step 4 and 5) of the InChI algorithm are very sensitive to any code changes and should not be altered. The canonicalization process understands standard covalent bond systems only so that the molfiles of structures with haptic and “coordinative” bonds (see ref. 35 for details of the definition of haptic and coordinative bonds in the context of Molfiles) must be normalized by exchanging these specific bonds by single bonds while the valences of the linked atoms must be adapted accordingly to keep the hydrogen count as defined by the structure depiction. Fig. 5 demonstrates this InChI internal structure normalization of coordinative bonds.
Note that only the hydrogens of the non-metal elements are internally handled by atom valence counts. Any hydrogen atom that is directly bound to a metal atom must be drawn explicitly to be taken into account.
On the other hand, salts are represented by ionic bonds. The normalization process has to determine which bonds to keep and which ones to disconnect based on fixed rule sets inspired by the electronegativity differences between the different elements.
The proposed set of “disconnection rules” is summarized in Fig. 6. In general, these rules are intended to result in disconnection of simple salts, which are present in the solid state either as ionic compounds or coordination polymers with a structure not reflected by the sum formula, but try to keep all other metal–ligand connections in place.
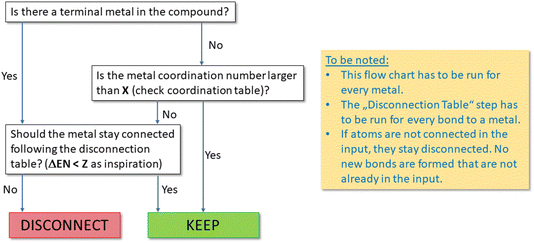 | ||
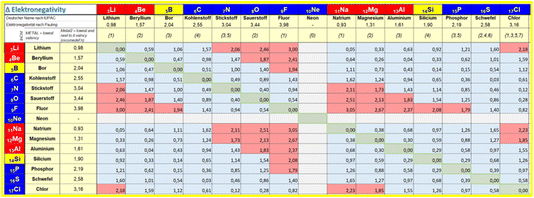
| Fig. 6 Flow chart for the preprocessing step when metals are present in a regarded compound. X = standard valence of the metals collected in a separate lookup table; ΔEN = electronegativity difference collected in a separate lookup table (see Fig. 7). | ||
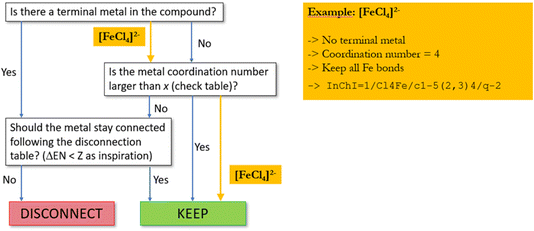
In an iterative process, first all terminal metal atoms (i.e. metal atoms that are connected to only one other atom) are checked and disconnected according to the lookup table of Fig. 7 based on electronegativity differences. This will ensure that, for example, all metal–metal bonds will be preserved. In a second step, metallic atoms bound to more than one ligand are examined. If the coordination number of such an atom is larger than a threshold value individually defined for each element, no bonds will be disconnected, while for low coordination numbers, a procedure similar to the one described above for terminal metals will decide on whether to keep or disconnect a bond based on differences in electronegativity. If only one metal–ligand bond is found to be kept, all others will also be retained and no disconnection carried out.
For example, assuming a threshold value of two for iron, all bonds in [FeCl4]2− will be retained (see Fig. 8) while for FeCl2, when represented as Cl–Fe–Cl, the two metal–ligand bonds will be disconnected.
Further fine-tuning is currently still under way to ensure that distinct molecular compounds with metals featuring unusually low coordination numbers, as for example stabilized by bulky ligands, are not disconnected. Other difficult cases are organolithium and organomagnesium compounds like Grignard-type ones. Here, it is proposed, by careful choice of the disconnection rules, to retain the metal–carbon bonds but disconnect the metal–halide ones, thus disconnecting RMgX into the fragments RMg and X and keeping RLi intact. Since under special conditions, for example in the gas phase, sodium chloride can also be present as molecular compounds Na–Cl and clusters [Na2Cl2], an optional parameter will allow users to deliberately generate an InChI for a fully connected structure, in which the disconnection algorithm is bypassed.
As there is no algorithmic way to “re-connect” individual atoms or larger fragments of a molecule, the InChI will not attempt to add any bonds that are not present in the input or, to put it in a different way, “what was already disconnected in the input will stay disconnected”.
We provide some additional examples of how the normalization might be applied to different compounds in the ESI (Fig. S7 and S8‡), and invite all stakeholders to consider interesting compounds and how they would be treated by this algorithm. Therefore, the InChI Web Demo interface has been created for user testing purposes (Fig. 9). This web application instantly calculates the InChI, InChIKey and AuxInfo from a drawn chemical structure or from a Molfile. Vice versa, structures can be created out of InChI or AuxInfo strings. The equivalent functionality is also available for the Reaction InChI (RInChI). We emphasize that all computations are performed within the user’s web browser making it a suitable tool to process undisclosed chemical structures. More details about the InChI Web Demo are provided in the ESI.‡
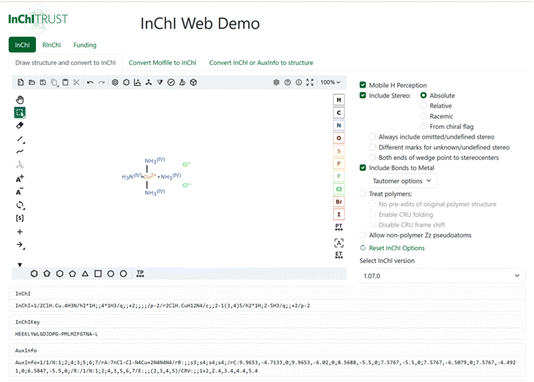 | ||
| Fig. 9 Screenshot of the InChI web demo demonstrating the drawing of a chemical structure, the immediate generation of InChI, InChIKey and AuxInfo and the interactive selection of the InChI parameters and version.10 | ||
Make InChI FAIRer by improving the documentation
The InChI Technical Manual will be divided into two distinct documents: one focusing on technical details and the other on chemical principles. This restructuring aims to enhance both sections to accommodate planned updates. The Chemical Manual will provide chemists with a deeper understanding of InChI strings and their variations, addressing topics such as stereochemistry, tautomerism, polymers, and organometallic compounds. Meanwhile, the Technical Manual will offer software developers detailed information on algorithms, data structures, and coding considerations for efficient InChI integration.The Chemical Manual will be a valuable resource for chemistry educators and students, helping them understand the logic behind the InChI algorithm and chemical structure representation. The Technical Manual, on the other hand, will support developers in incorporating InChI into new tools and workflows, thereby promoting wider adoption of the standard. High-level code documentation included in the technical manual will demystify the InChI algorithm, making it easier for developers to grasp the logic behind the code and streamline the integration process.
By separating the InChI Technical Manual into dedicated chemical and technical documents, users will benefit from improved accessibility and clarity. This approach will facilitate ongoing development and encourage contributions from other developers, ultimately increasing the utility and adoption of the InChI standard.
In conclusion, the split into two specialized documents enhances the InChI Technical Manual’s value for both current and future users. This change supports the broader adoption of InChI, making it a more powerful tool for chemical information management and ensuring its continued relevance and effectiveness.
Conclusion
The InChI plays a crucial role in making data FAIR (Findable, Accessible, Interoperable, and Reusable) in the realm of chemistry and related fields.• Findable: InChI provides a unique and persistent identifier for chemical compounds. When included in datasets or publications, it facilitates the easy location of chemical information, as researchers can search for compounds using their InChI.
• Accessible: By providing a standardized identification of chemical structures, InChI ensures that chemical data is accessible to both humans and machines. This facilitates sharing and dissemination of chemical information across different platforms and databases.
• Interoperable: InChI promotes interoperability by enabling seamless integration of chemical data from diverse sources. Because InChI is a standardized format, software tools and databases can easily exchange and process chemical information without compatibility issues.
• Reusable: InChI enhances the reusability of chemical data by enabling precise identification and comparison of chemical compounds. Researchers can confidently use data containing InChI identifiers, knowing that they can accurately reference and replicate chemical structures.
Overall, the adoption of InChI contributes significantly to the FAIR principles by ensuring that chemical data is easily discoverable, accessible, interoperable, and reusable across different scientific disciplines and research endeavors.
The new InChI version 1.07 paves the way to a more sustainable and transparent code development. It has moved to the GitHub environment for more facile community contributions. Moreover, it features an efficient testing environment and a new web demo. Several thousand code errors were fixed and new documentation added. The current InChI version 1.07 has been approved by IUPAC’s Committee on Publications and Cheminformatics Data Standards (CPCDS) and enables efficient code development for the next steps, since many chemical subdisciplines have special requirements for the InChI. Hereby, the InChI fulfills its role as the International Chemical Identifier for all purposes, ranging from “simple” molecular representation over web-based compound searches and database applications to machine learning.
The currently implemented extensions of the next InChI version for inorganics and organometallics will enhance the data handling of these substance classes and make the data management FAIR. Unlike in the current moment, data for inorganic and organometallic compounds will become Findable, Accessible, Interoperable and Reusable by providing an enhanced InChI that will offer a unique identification pattern for the inorganics. Furthermore, as an outcome of these tests, we value comments on the enhancements for inorganics and organometallics to let us improve our work.
In the next upcoming InChI releases, we will work on the stereochemistry of inorganics and organometallics beside the introduction of general stereochemistry enhancements like the implementation of the “MDL enhanced stereochemistry”35 and the stereochemistry of atropisomers.
In the area of InChI applications we are working on the implementation of Mixture InChIs (MInChIs) and Nano-InChIs (NInChIs). MInChIs address the needs of unique identification of mixtures and formulations (e.g. alloys) while NInChIs will identify nano-materials.
Data availability
All documentation, binaries/API, instructions for compiling InChI software from the source code, a comprehensive list of known issues as well as many other important technical details can be found on the landing page of the InChI GitHub repository:21 https://github.com/IUPAC-InChI/InChI.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the VolkswagenStiftung for generous funding in the Momentum framework. The authors acknowledge the German Research Foundation (DFG) for funding under the project number 441958208 (NFDI4Chem) as well as the funding and support within the framework of the DALIA project with funding code 16DWWQP07B, funded by the Federal Ministry of Education and Research (BMBF) and the funding measure from the EU’s Capacity Building and Resilience Facility. Moreover, we thank the InChI Trust.References
- CTFile Formats, BIOVIA Chemistry 2024, ©2023 Dassault Systèmes.
- Chemoinformatics: Basic Concepts and Methods, ed. T. Engel and J. Gasteiger, Wiley-VCH, Weinheim, 1st edn, 2018 Search PubMed.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36, DOI:10.1021/ci00057a005.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1990, 30, 237–243, DOI:10.1021/ci00067a005.
- D. Weininger, A. Weininger and J. L. Weininger, J. Chem. Inf. Comput. Sci., 1989, 29, 97–101, DOI:10.1021/ci00062a008.
- Daylight Chemical Information Systems, Inc., Daylight Theory: SMILES, accessed on 17 July 2024, https://daylight.com/dayhtml/doc/theory/theory.smiles.html Search PubMed.
- J. M. Goodman, I. Pletnev, P. Thiessen, E. Bolton and S. R. Heller, J. Cheminf., 2021, 13, 40, DOI:10.1186/s13321-021-00517-z.
- GitHub, IUPAC-InChI/InChI Releases, accessed on 17 July 2024, https://github.com/IUPAC-InChI/InChI/releases Search PubMed.
- W. D. Ihlenfeldt, E. E. Bolton and S. H. Bryant, J. Cheminf., 2009, 1, 20, DOI:10.1186/1758-2946-1-20.
- InChI Trust, InChI Web Demo, accessed on 17 July 2024, https://iupac-inchi.github.io/InChI-Web-Demo/ Search PubMed.
- A. P. Cornell, S. Kim, J. Cuadros, E. C. Bucholtz, H. E. Pence, R. Potenzone and R. E. Belford, Chemistry Teacher International, 2024, 6, 77–91, DOI:10.1515/cti-2023-0009.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018, DOI:10.1038/sdata.2016.18.
- The WorldFAIR Project, accessed on 17 July 2024, https://worldfair-project.eu/ Search PubMed.
- (a) W. A. Warr, J. Comput.-Aided Mol. Des., 2015, 29, 681–694, DOI:10.1007/s10822-015-9854-3; (b) A. D. McNaught and S. R. Heller, in Principles of Chemical Nomenclature – A Guide to IUPAC Recommendations, ed. G. J. Leigh, RSC Publishing, Cambridge, 2011, pp. 190–194 Search PubMed; (c) S. Heller, A. McNaught, S. Stein, D. Tchekhovskoi and I. Pletnev, J. Cheminf., 2013, 5, 7, DOI:10.1186/1758-2946-5-7; (d) R. Boucher, S. Heller and A. McNaught, Chem. Int., 2017, 39, 47, DOI:10.1515/ci-2017-0316.
- GitHub, RInChI, accessed on 17 July 2024, https://github.com/IUPAC-InChI/RInChI Search PubMed.
- GitHub, MInChI: Mixtures InChI, accessed on 17 July 2024, https://github.com/IUPAC/MInChI Search PubMed.
- K. Blekos, K. Chairetakis, I. Lynch and E. Marcoulaki, J. Cheminf., 2023, 15, 44, DOI:10.1186/s13321-022-00669-6.
- InChI Trust – InChI: structure-based chemical identifier, accessed on 17 July 2024, https://www.inchi-trust.org/ Search PubMed.
- International Union of Pure and Applied Chemistry, The IUPAC International Chemical Identifier (InChI), accessed on 17 July 2024, https://iupac.org/who-we-are/divisions/division-details/inchi/ Search PubMed.
- InChI Trust, InChI Working Groups, accessed on 17 July 2024, https://www.inchi-trust.org/inchi-working-groups/ Search PubMed.
- GitHub, Official home of the InChI, accessed on 17 July 2024, https://github.com/IUPAC-InChI/InChI Search PubMed.
- Google, OSS-Fuzz, accessed on 17 July 2024, https://google.github.io/oss-fuzz/ Search PubMed.
- Accelrys Software Inc., CTFile Formats, 2011 Search PubMed.
- S. Herres-Pawlis, F. Bach, I. J. Bruno, S. J. Chalk, N. Jung, J. C. Liermann, L. R. McEwen, S. Neumann, C. Steinbeck, M. Razum and O. Koepler, Angew. Chem., Int. Ed., 2022, 61, e202203038, DOI:10.1002/anie.202203038.
- Machine Learning in Chemistry: the Impact of Artificial Intelligence, ed. H. M. Cartwright, The Royal Society of Chemistry, 2020 Search PubMed.
- J. C. Brammer, G. Blanke, C. Kellner, A. Hoffmann, S. Herres-Pawlis and U. Schatzschneider, J. Cheminf., 2022, 14, 66, DOI:10.1186/s13321-022-00640-5.
- R. A. Sayle, J. Comput.-Aided Mol. Des., 2010, 24, 485–496, DOI:10.1007/s10822-010-9329-5.
- M. C. Nicklaus, Tautomers in InChI, NIH InChI Workshop, March 22–24 2021, https://www.inchi-trust.org/wp/wp-content/uploads/2022/11/Day_1_Nicklaus_Tautomerism_2021-03-21A.pdf Search PubMed.
- InChI Trust, InChI Technical FAQ – Stereochemistry, accessed on 17 July 2024, https://www.inchi-trust.org/technical-faq-2/#4.8 Search PubMed.
- InChI Trust, InChI Technical FAQ – Stereochemistry, accessed on 17 July 2024, https://www.inchi-trust.org/technical-faq-2/#8 Search PubMed.
- S. R. Heller, A. McNaught, I. Pletnev, S. Stein and D. Tchekhovskoi, J. Cheminf., 2015, 7, 23, DOI:10.1186/s13321-015-0068-4.
- InChI Trust, InChI Technical FAQ – Isotopes, accessed on 17 July 2024, https://www.inchi-trust.org/technical-faq-2/#9 Search PubMed.
- S. E. Stein, S. R. Heller, D. V. Tchekhovskoi and I. V. Pletnev, IUPAC International Chemical Identifier (InChI), InChI Version 1, Software Version 1.06, Technical Manual, InChI Trust, Cambridge UK, 15 December 2020.
- R. Boucher, S. Heller, R. Kidd, A. McNaught and I. Pletnev, What on Earth is InChI?, accessed on 17 July 2024, https://iupac.org/100/stories/what-on-earth-is-inchi/ Search PubMed.
- Dassault Systèmes, Biovia Chemistry 2024 – Chemical Representation, 2023 Search PubMed.
Footnotes |
| † Dedicated to Prof. Dr Igor Pletnev. |
| ‡ Electronic supplementary information (ESI) available: Additional information on programming details, on the interactive user interfaces (WInChI and web demo) and the testing. See DOI: https://doi.org/10.1039/d4fd00145a |
| This journal is © The Royal Society of Chemistry 2024 |