Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Visual monitoring of biocatalytic processes using small molecular fluorescent probes: strategies-mechanisms-applications
Guang
Chen
 a,
Jie
Xu
a,
Siyue
Ma
a,
Xinrui
Ji
*c,
Jared B.
Carney
d,
Chao
Wang
a,
Xiaoyong
Gao
e,
Pu
Chen
c,
Baolei
Fan
*b,
Ji
Chen
e,
Yanfeng
Yue
a,
Jie
Xu
a,
Siyue
Ma
a,
Xinrui
Ji
*c,
Jared B.
Carney
d,
Chao
Wang
a,
Xiaoyong
Gao
e,
Pu
Chen
c,
Baolei
Fan
*b,
Ji
Chen
e,
Yanfeng
Yue
 *d and
Tony D.
James
*d and
Tony D.
James
 *fg
*fg
aThe Youth Innovation Team of Shaanxi Universities, Shaanxi Key Laboratory of Chemical Additives for Industry, College of Chemistry and Chemical Engineering, Shaanxi University of Science & Technology, Xi’an, 710021, China
bHubei University of Science and Technology, No. 88, Xianning Avenue, Xianan District, Xianning 437000, China. E-mail: fanb1980@163.com
cDepartment of Chemical Engineering and Waterloo Institute for Nanotechnology, University of Waterloo, 200 University Avenue West, Waterloo, Ontario N2L 3G1, Canada. E-mail: X62ji@uwaterloo.ca
dDepartment of Chemistry, Delaware State University, Dover, Delaware 19901, USA. E-mail: yyue@desu.edu
eJiangsu Simba Biological Medicine Co., Ltd. Gaogang Distrct Qidizhihui Park, Taizhou City, China
fDepartment of Chemistry, University of Bath, Bath BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
gSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
First published on 14th February 2024
Abstract
Real-time monitoring of biocatalytic-based processes is significantly improved and simplified when they can be visualized. Visual monitoring can be achieved by integrating a fluorescent unit with the biocatalyst. Herein, we outline the design strategies of fluorescent probes for monitoring biocatalysis: (1) probes for monitoring biocatalytic transfer: γ-glutamine is linked to the fluorophore as both a recognition group and for intramolecular charge transfer (ICT) inhibition; the probe is initially in an off state and is activated via the transfer of the γ-glutamine group and the release of the free amino group, which results in restoration of the “Donor–π–Acceptor” (D–π–A) system and fluorescence recovery. (2) Probes for monitoring biocatalytic oxidation: a propylamine is connected to the fluorophore as a recognition group, which cages the hydroxyl group, leading to the inhibition of ICT; propylamine is oxidized and subsequently β-elimination occurs, resulting in exposure of the hydroxyl group and fluorescence recovery. (3) Probes for monitoring biocatalytic reduction: a nitro group attached to a fluorophore as a fluorescence quenching group, this is converted to an amino group by catalytic reduction, resulting in fluorescence recovery. (4) Probes for monitoring biocatalytic hydrolysis: β-D-galactopyranoside or phosphate acts as a recognition group attached to hydroxyl groups of the fluorophore; the subsequent biocatalytic hydrolysis reaction releases the hydroxyl group resulting in fluorescence recovery. Following these 4 mechanisms, fluorophores including cyanine, coumarin, rhodamine, and Nile-red, have been used to develop systems for monitoring biocatalytic reactions. We anticipate that these strategies will result in systems able to rapidly diagnose and facilitate the treatment of serious diseases.
Introduction
Biocatalysis accelerates the transformation of substances in the body. Enzymes involved in biocatalysis are divided into oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases. While, many molecular fluorescent probes for biocatalytic processes have been developed, with this highlight we will concentrate on four important types of probes associated with cancer and aging: (1) biocatalytic transfer reactions: the γ-glutamine group is catalytically transferred from glutathione (GSH) to an amino acceptor. This reaction regulates GSH levels and cellular oxidative stress. Transferases are also important enzymes involved in tumour expression.1,2 (2) Biocatalytic oxidation reactions: monoamines and exogenous drugs are catalytically oxidized and then inactivated. This process is involved in drug metabolism and maintains the homeostasis of neurotransmitters, and has also been found to be associated with age-related neurological diseases.3 (3) Biocatalytic reduction reactions: under anoxic conditions, nitro compounds are reduced. This process is often used to assess the hypoxic state of cells and diagnose diseases such as cancer.4–6 (4) Biocatalytic hydrolysis reactions: β-galactoside produces galactose through biocatalytic hydrolysis. This reaction is used for the diagnosis and detection of tumours as well as the detection of senescent cells.7–9Given the significant role that biocatalysis plays in biological systems, visual detection is a particularly useful approach since it facilitates the facile evaluation of biomechanisms and the kinetics of biological process. Moreover, the development of visible molecular tools is essential for rapid disease diagnosis and treatment. Although bioanalytical techniques including colorimetric and electrochemical methods, etc., are widely used for bioanalysis, these techniques lack the advantages of real-time visualization in situ.10–12 As such, it is important to find a visible method for in situ biocatalysis monitoring. In recent years, fluorescence imaging technologies have been shown to exhibit sensitive and fast response, enabling visible real-time bioanalysis. Kovačević et al. and Li et al. have used fluorescent protein labelling and a metal organic framework (MOF) nanosheet sensor to detect biocatalytic glucose oxidation.13,14
While Huang et al. have developed the first monoamine oxidases (MAO)-A-specific two-photon fluorogenic probe.15 Due to their unique properties, fluorescent probes for the analysis of biocatalysts have been rapidly developed. As such it is now appropriate to summarize the strategies and working mechanisms for these probes and to highlight representative fluorescent probes for the visual detection of biocatalysis (Fig. 1). In addition we have separated them into 4 categories according to the types of biocatalytic reaction involved. Design strategies, working mechanisms, and biological applications are: (1) probes for biocatalytic transfer: amino, γ-glutamine or glucoside are used as the recognition group that is linked to the fluorophore. The fluorophore can be cyanine, naphthalimide, anthracene derivatives and indole-quinolines. For example, with an amine, when the probe is activated by the biocatalyst, a carbonyl group is formed, creating a “Donor–π–Acceptor” (D–π–A) system, and a fluorescence output. Similarly, after the biocatalytic transfer of the γ-glutamine group, the amino group is released. Thus, the “D–π–A” effect of the probe is increased, thereby resulting in enhanced fluorescence. (2) Probes for biocatalytic oxidation: glucose, tyrosine or propylamine can be used as biocatalytic recognition sites, with fluorophores including cyanine, rhodamine and nano-quantum dots. The fluorescence mechanism includes the H2O2 assisted strategy (H2O2 generated during the biocatalytic process can enhance/suppress the luminescence of nano-quantum dots), and the switch of intramolecular charge transfer (ICT) (before and after the process, hydroxyl groups are caged and released, leading to enhancement of ICT and fluorescence recovery). (3) Probes for biocatalytic reduction: involving nitro/p-nitrobenzyl group and azo groups integrated with cyanine, BODIPY, Nile-red or naphthalimide cores. For example, when a nitro group is reduced to an amino group, fluorescence is restored due to the removal of the quenching effect by the nitro group; similarly, when a p-nitrobenzyl group is reduced, a 1,6-rearrangement elimination occurs, which results in the recovery of fluorescence. Similarly, when the azo group is specifically reduced to an amino group the ICT process from the amino group (D) to the fluorophore (A) is enabled, resulting in enhanced fluorescence emission. (4) Probes for biocatalytic hydrolysis: where galactoside or phosphate moieties are added to fluorophores including BODIPY, naphthalimide and natural quercetin. Here, when biocatalytic hydrolysis occurs, the galactopyranoside or the phosphate group is removed, releasing the hydroxyl group, and enhancing electron-donation to the fluorophore, resulting in fluorescence recovery. In summary, we have compiled probe construction strategies based on various fluorescent groups such as rhodamine, naphthalimide, Nile red, BODIPY and cyanine for the visualization of 4 classes of biocatalysts. Among them, rhodamine, naphthalimide, Nile red and BODIPY are widely used since these fluorophores are easily modified. However, the shorter emission wavelength limits the biological visualization application of these fluorophores. Therefore, as a near-infrared (NIR) dye, cyanine can break this limitation by enabling deeper tissue penetration. These probes play important roles in monitoring biological processes. As such, we anticipate that such probes will contribute to a deeper understanding of biocatalysis in biomedicine.
Biocatalytic transfer
Transferases are one of the six main enzymes that play an important role in living organisms. Here we used γ-glutamyl transpeptidase (GGT) as an example. GSH as an endogenous antioxidant and can regulate the oxidative stress state of cells. Cysteine is an essential amino acid in protein synthesis in vivo and plays an important role in the proliferation and metastasis of cancer cells. GGT has been found to be significantly enhanced in ovarian cancer, cervical cancer, liver cancer and other malignant tumours. Therefore, GGT is a suitable biomarker for tumour diagnosis. Visual monitoring of this process enables exploration of the catalytic mechanism and facilitates the diagnosis and prevention of clinical diseases.Biocatalytic transamination
Chiral amines are important molecules widely used in the chemical industry.16 Synthetic methods for chiral amines using ω-transaminase (ω-TA) have become particularly important due to the green synthetic credentials of the process. Therefore, it is necessary to develop an efficient biocatalytic screening platform for large-scale industrial production of chiral amines.17–19 ω-TA relies on pyridoxal phosphate (PLP) to achieve transamination. The probe is composed of an amino group, electron-donating groups and a π-conjugated group which is the linker. When the probe is catalyzed by ω-TA, the amino group is transferred to the PLP, and the probe is converted into a carbonyl group. Therefore, a D–π–A structure is formed between the electron-donating group (D) and carbonyl (A) which results in enhanced fluorescence (Fig. 2). | ||
| Fig. 2 Schematic diagram of “D–π–A” structure fluorophore used for detecting biocatalytic transamination. | ||
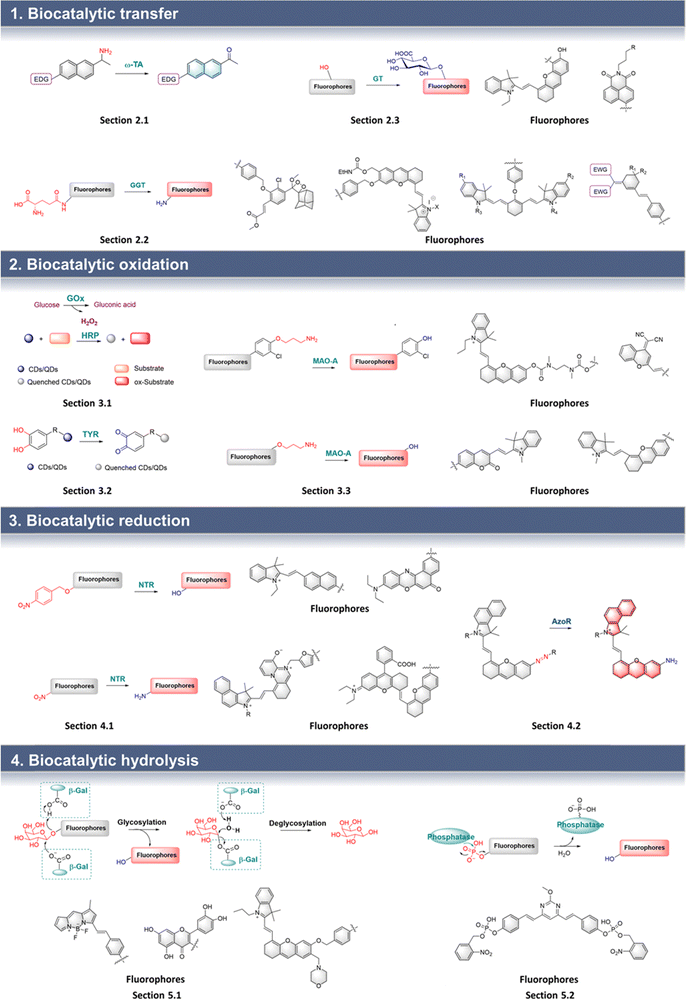
Based on this strategy, Wang et al. developed probe 1 for the efficient screening of transamination biocatalysts (Fig. 3).20 Probe 1 uses a naphthalene as the fluorophore, and the propan-2-amino group as the recognition group. Once probe 1 is recognized by ω-TA, the amino group is transferred to PLP, followed by the formation of a carbonyl group. Thus, the “D–π–A” interaction of a carbonyl group and an amino group occurs, resulting in the formation of fluorescent AN. Therefore, the efficient screening of ω-TA can be achieved by the monitoring of AN fluorescence emission. Probe 1 exhibits the advantages of low background interference, high sensitivity, and wide dynamic range. Therefore, probe 1 was used for the efficient screening of ω-TA variants, where variant M5 was shown to exhibit 3.2 times enhanced biocatalytic activity. In addition, the remarkable activity of M5 makes it a potential biocatalyst for the synthesis of (R)-amines.
 | ||
| Fig. 3 (A) The structure and response mechanism of probe 1 and probe 2. (B) Kinetic curves of wild-type and variants characterized by probe 1 (0–0.8 mM) (reprinted with permission from ref. 20 Copyright © 2020, Springer Nature). | ||
A similar structure was used by Fessner et al. to develop probe 2 for high throughput detection (Fig. 3).21 When probe 2 is biocatalyzed, a carbonyl group is generated, resulting in the formation of a new “D–π–A” structure, which in turn causes MN to fluoresce. Probe 2 was used to evaluate the biocatalytic process.
Biocatalytic glutamine transfer
GGT can transfer the γ-glutamine group from GSH to water, amino acids and peptides.22 It has been shown to be overexpressed in cancer cells. As such, the visual detection of biocatalytic γ-glutamine transfer is useful for the diagnosis and treatment of cancer.23–26 With these probes, the γ-glutamine group acts as the recognition group, and the fluorophore is attached either directly or using a linker.The γ-glutamine can usually be directly attached to fluorophores, using N atoms serving as both fluorescent and recognition groups, participating in both the biocatalytic recognition and changes in fluorescence signals. The probe is non-fluorescent due to the caging by the γ-glutamine group. Subsequently, the glutamine group is recognized and cleaved by GGT, and the amino group is exposed, resulting in a significant fluorescence response.
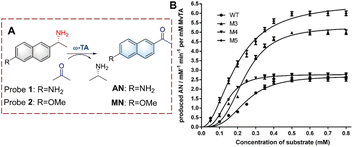
Using this strategy, Lv et al. developed probe 3 for monitoring biocatalytic glutamine transfer in vivo (Fig. 4).27 The cyanine fluorophore of probe 3 is a NIR dye and exhibits the advantages of reducing background interference and enables the deep penetration of tissues. The γ-glutamine group was added to cage the amine of cyanine. Therefore, the fluorescence of probe 3 is quenched. When probe 3 is recognized by GGT, the γ-glutamine group is transferred, thereby releasing the amino group, resulting in the recovery of the fluorescence. Since probe 3 exhibited a rapid and selective response it was suitable for imaging pulmonary fibrosis cells in mice and confirmed that the expression levels of GGT were directly related to pulmonary fibrosis. Therefore, providing visual evidence for research on the pathogenesis of idiopathic pulmonary fibrosis.
 | ||
| Fig. 4 (A) The structure and response mechanism of probe 3. (B) (a) Images of probe 3 (100 μM, 30 min) in pulmonary fibrosis (bleomycin) and normal mice (control). (b) Images of isolated organs in (a) (reprinted with permission from ref. 27 Copyright © 2020, Elsevier). | ||
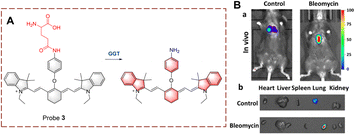
Kong et al. developed probe 4 for the visual tracking of endogenous GGT in vivo (Fig. 5).28 The structure of probe 4 is 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile, linked to a benzene through a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond. A γ-glutamine group is attached as the recognition group. Probe 4 is non-fluorescent because both the cyano and γ-glutamine groups are electron-withdrawing groups. However, when the γ-glutamine group is transferred, and the amino group is released as a donor (D). Thus, probe 4 exhibits strong fluorescence due to the release of the “D–π–A” interaction, in which the cyano group acts as electron acceptor (A). Probe 4 has the advantages of large Stokes shift (213 nm), high sensitivity (LOD = 0.024 U L−1), high specificity, high imaging resolution and low biological toxicity. Due to these advantages, probe 4 was used for the real-time imaging of GGT in tumour-bearing mice. Providing an effective fluorescence tool for pathological research and diagnosis of GGT-related diseases in vivo.
C bond. A γ-glutamine group is attached as the recognition group. Probe 4 is non-fluorescent because both the cyano and γ-glutamine groups are electron-withdrawing groups. However, when the γ-glutamine group is transferred, and the amino group is released as a donor (D). Thus, probe 4 exhibits strong fluorescence due to the release of the “D–π–A” interaction, in which the cyano group acts as electron acceptor (A). Probe 4 has the advantages of large Stokes shift (213 nm), high sensitivity (LOD = 0.024 U L−1), high specificity, high imaging resolution and low biological toxicity. Due to these advantages, probe 4 was used for the real-time imaging of GGT in tumour-bearing mice. Providing an effective fluorescence tool for pathological research and diagnosis of GGT-related diseases in vivo.
 | ||
| Fig. 5 (A) The structure and response mechanism of probe 4 and probe 5. (B) Image of probe 4 (50 μM) or DON (GGT inhibitor, 2 mM) before probe 4 (50 μM) in HepG-2 tumour bearing mice (reprinted with permission from ref. 28 Copyright © 2019, Elsevier). | ||
Similarly, Peng et al. developed probe 5 (Fig. 5).29 Probe 5 consists of a conjugated 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF).30 And the γ-glutamine group is directly attached to the core via an amine. Since both the cyano and γ-glutamine are electron-withdrawing groups, the fluorescence of probe 5 is in an off state. When probe 5 is recognized and undergoes biocatalysis by GGT, an amino group is generated. As such probe 5 emits fluorescence due to the released “D–π–A” interaction. In addition, probe 5 exhibits low detection limit (0.014 mU mL−1) and fast response (Te = 14 min). Probe 5 was used to distinguish normal cells from cancer cells, therefore, providing visual guidance for clinical tumour resection.
In addition, using a linker to connect the γ-glutamine group and fluorophores reduces the steric hindrance of the probe and facilitates the interaction between the probe and the enzyme, which is hindered when the receptor is directly connected. Due to a covalent connection between the linker and the phenolic group of the fluorophore the hydroxyl groups are caged and the fluorescence is quenched. However, when the recognition group is triggered, the linker also detaches, and the probe exhibits bright fluorescence, enabling the visualization of the biocatalytic process.
Inspired by the above strategy, Xie et al. developed probe 6 for the monitoring of the glutamine transfer reaction (Fig. 6).31 Probe 6 is non-fluorescent due to the HD being caged (locked) by p-aminobenzyl alcohol (PABA). PABA is a self-immolative linker that reduces the steric hindrance between probe 6 and GGT, while also enabling the release of activated fluorophores. When probe 6 undergoes biocatalytic transfer, the γ-glutamine is cleaved, resulting in exposure of the amino group. Subsequently, PABA undergoes 1,6-elimination generating intermediate (6-I). This intermediate is unstable and converts into (6-II) by the release of the ethyl carbamate. Subsequently, intermediate (6-II) is attacked by a nucleophilic residue of GGT, and covalently immobilized and the fluorescence signal is turned on. Compared with other small molecule probes, probe 6 reduces the impact of diffusion phenomenon, thus significantly enhancing the sensitivity. As such, probe 6 could be used for the real-time imaging of HepG-2 cells and tumour-bearing mice.
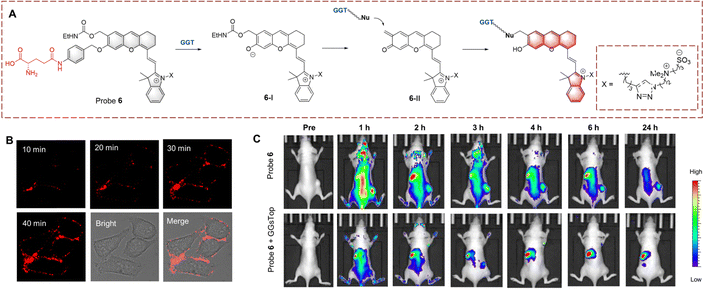 | ||
| Fig. 6 (A) The structure and response mechanism of probe 6. (B) Fluorescence image of probe 6 (1 μM) in HepG-2 cells. (C) Images of probe 6 or probe 6 with GGsTop (GGT inhibitor, 5 mM) in U87MG tumour bearing mice (reprinted with permission from ref. 31 Copyright © 2020, American Chemical Society). | ||
Ye et al. have also used PABA as a linker to synthesize probe 7 based on Schaap's phenoxy-dioxetane (Fig. 7).32 Similarly, the large fluorophore Schaap's phenoxy dioxetane is kept an appropriate distance from the active site of GGT by the PABA linker. When the γ-glutamine group is transferred, PABA within (7-I) undergoes self-elimination generating intermediates (7-II). 7-II then undergoes chemiexcitation to generate (7-III) and chemiluminescence. Probe 7 exhibits the advantages of low detection limit (16 mU L−1) and high fluorescence turn-on (876-fold). Due to the obvious luminescence change of probe 7, it has been used for the detection of cancer in mice.
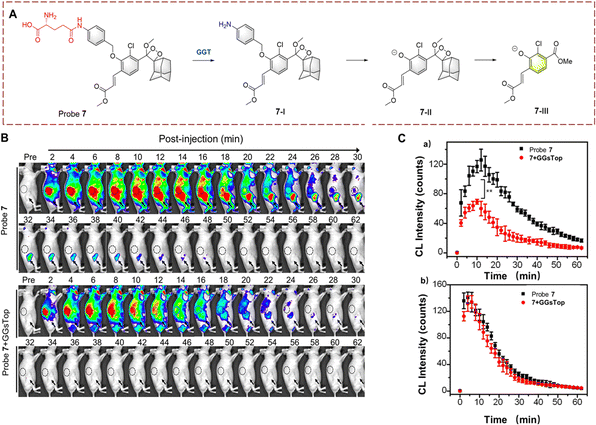 | ||
| Fig. 7 (A) The structure and response mechanism of probe 7. (B) Fluorescence images of probe 7 (100 μM) or probe 7 (100 μM) with GGsTop (10 mM) in U87MG tumour bearing mice. (C) CL intensity curve of tumour (a) and kidney (b) in figure (B) (reprinted with permission from ref. 32 Copyright © 2019, American Chemical Society). | ||
Guo et al. synthesized probe 8 with dipeptide-linker, fluorophore and γ-glutamine (Fig. 8).33 Probe 8 is based on the indole-quinoline (QI) with a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond to enhance the ICT effect. L-Proline-glycine (Pro-Gly) acts a linker, which improves stability of probe 8. Probe 8 was initially non-fluorescent due to caging by the Pro-Gly. Once the γ-glutamine of probe 8 is transferred, the self-cyclization of Pro-Gly is rapid and releases HQI resulting in fluorescence recovery. Probe 8 was shown to target the nucleolus and inhibit RNA Polymerase I transcription. As such probe 8 is a bifunctional molecule suitable for detection and treatment. Probe 8 also provides a new strategy for development of a nucleolar targeted fluorescent probe.
C bond to enhance the ICT effect. L-Proline-glycine (Pro-Gly) acts a linker, which improves stability of probe 8. Probe 8 was initially non-fluorescent due to caging by the Pro-Gly. Once the γ-glutamine of probe 8 is transferred, the self-cyclization of Pro-Gly is rapid and releases HQI resulting in fluorescence recovery. Probe 8 was shown to target the nucleolus and inhibit RNA Polymerase I transcription. As such probe 8 is a bifunctional molecule suitable for detection and treatment. Probe 8 also provides a new strategy for development of a nucleolar targeted fluorescent probe.
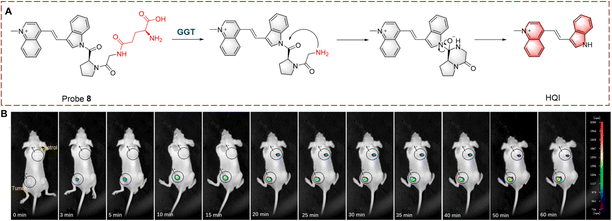 | ||
| Fig. 8 (A) The structure and response mechanism of probe 8. (B) Image of probe 8 in tumour-bearing mice (reprinted with permission from ref. 33 Copyright © 2022, Elsevier). | ||
Given that probes for in vivo monitoring suffer from inaccurate imaging due to irregular distribution, ratiometric probes have been developed to overcome differences in concentration suitable for monitoring the transamination reaction. Changes in the electron distribution of the probe, results in a red wavelength shift, which enables the ratiometric visualization of biocatalytic glutamine transfer.
Ahn et al. developed probe 9, for monitoring GGT on cell membranes (Fig. 9).34 The naphthalene ring acts as the fluorophore and the γ-glutamine acts as the recognition site. Probe 9 immobilizes in the cell membrane using a long-chain alkane and electrostatic interactions. When probe 9 is recognized, the γ-glutamine group is transferred, leading to the generation of an amino group. The product has stronger electron donating ability, resulting in fluorescence changes from green to red. Probe 9 was used the imaging of different cell lines and tissues using the wavelength change. In addition, the levels of GGT in cancer cells was found to be significantly higher than that for normal cells.
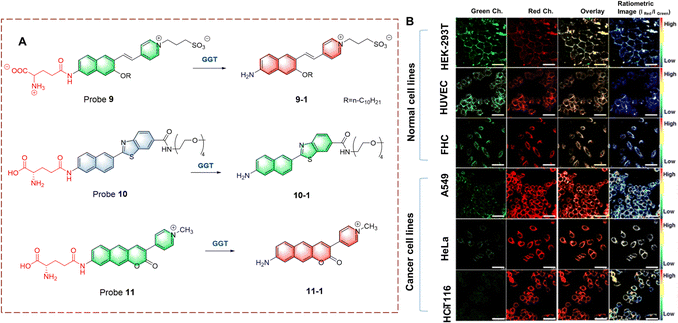 | ||
| Fig. 9 (A) The structure and response mechanism of probe 9, probe 10 and probe 11. (B) Fluorescence images of normal cell lines and cancer cell lines with probe 9 (5.0 μM) (reprinted with permission from ref. 34 Copyright © 2020, American Chemical Society). | ||
Kim et al. developed a ratiometric probe 10 for live cell imaging (Fig. 9).35 Probe 10 was based on a benzothiazole attached to a naphthalene, to enhance the conjugation and fluorescence quantum yield. With γ-glutamine as a recognition group, probe 10 emits blue fluorescence. However, when probe 10 undergoes γ-glutamine transfer, the electron-donating amino group is released. This biocatalytic process changes the electron distribution of probe 10, resulting in a red-shift of the wavelength (blue to green). In addition, oligoethylene glycol enhances the solubility of probe 10 in a physiological environment. Probe 10 exhibits excellent GGT selectivity and high fluorescence efficiency. Therefore, probe 10 could be used to quantitatively image GGT in human colon and cancer tissue.
Ahn et al. have developed a ratiometric probe 11 which is not affected by environmental conditions (Fig. 9).36 With benzo coumarin as the fluorophore, probe 11 emits green fluorescence. However, following transfer of the γ-glutamine group the wavelength is red-shifted (to red). Importantly, probe 11 is not affected by pH, viscosity and polarity changes, which solves problems associated with signal fluctuations. Therefore, probe 11 could be used for imaging HeLa cells and cancerous tissue. In addition, probe 11 exhibits potential as a quantitative analytic tool for complex biological systems.
Biocatalytic glucoside transfer
UDP-glucuronosyltransferases (UGTs) can catalyze the transfer of glucuronic acid to receptors under the conditions of UDP-glucuronic acid (UDPGA). This process plays an important role in the metabolic clearance of clinical drugs in vivo.37–40 In addition, using glucosyltransferases (GTs) to achieve glycosylation has become an emerging method for industrial glycoside production.41Ma et al. developed probe 12 for the real-time monitoring of glucoside transfer (Fig. 10).42 The hemi cyanine acts as the fluorophore. To simultaneously preserve the fluorescence site of the hemi cyanine (6-OH) and the recognition site of UGT1A1 (5-OH), a “molecular splicing strategy” was used to synthesize probe 12. Due to quenching by the O-diphenol, probe 12 was non-fluorescent. However, under biocatalysis by UGT1A1, the fluorescence is restored. Probe 12 is a highly selective near-infrared fluorescent probe. Therefore, probe 12 could be used for real-time UGT imaging of living cells and mice.
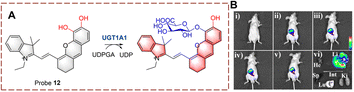 | ||
| Fig. 10 (A) The structure and response mechanism of probe 12. (B) Image of probe 12 (10 μM, 0–30 min) in mice and isolated organs (reprinted with permission from ref. 42 Copyright © 2021, John Wiley and Sons). | ||
Ma et al. developed probe 13, as a two-photon ratiometric fluorescent probe to detect GTs (Fig. 11).43 The naphthalimide is used as the fluorophore, and the hydroxyl was used as the glycosylation site. Naphthalimide is a fluorescent group with high quantum yield, large Stokes shift, and good photostability, which is widely used in the field of fluorescence. In the presence of uridine diphosphate glucose (UDPG), a glucose group was transferred from UDPG to the phenolic hydroxyl group. After glycosylation the phenolic hydroxyl group is replaced by a glucose group, which causes a significant blue shift in the fluorescence of the glycosylation product (13-1). In addition, the fluorescence quantum yield of 13-1 is much higher than that of probe 13, leading to an increase in fluorescence intensity. Using this platform, two fungi with glycosylation ability were successfully screened.
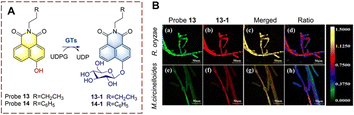 | ||
| Fig. 11 (A) The structure and response mechanism of probe 13 and probe 14. (B) Confocal microscopy images of probe 13 (50 μM, 8 h) and 13-1 in R. oryzae and M. circinelloides (reprinted with permission from ref. 43 Copyright © 2018, American Chemical Society). | ||
Using a similar strategy, James et al. developed probe 14 for screening GTs inhibitors for the prevention of dental caries (Fig. 11).44 After biocatalytic glucosyl transfer the product (14-1) exhibits blue fluorescence emission, distinct from the yellow fluorescence of probe 14. Probe 14 could be used for real-time detection and imaging of GTs of cariogenic bacteria. In addition, inhibitors were screened from green tea for oral treatments.
Biocatalytic oxidation
Biocatalytic oxidation plays an important role in biological systems. The oxidation of glucose is the foundation of energy metabolism in organisms and an important pathway for obtaining energy. Carbon dioxide and water produced by its biocatalytic oxidation play a crucial role in maintaining the homeostasis of organisms. In addition, tyrosinase (TYR) directly affects the synthesis of melanin and participates in various physiological functions in the body, especially those involving antioxidants. The oxidation of monoamine substances in living organisms relies on MAOs. Research has found that mental illnesses such as depression, Parkinson's syndrome, and Alzheimer's disease are closely related to the activity of MAOs. Therefore, visual monitoring of the biocatalytic oxidation process is of great significance for the study of organisms and disease treatment.Biocatalytic glucose oxidation
Many methods for detecting glucose biocatalytic oxidation use the H2O2 monitoring strategy. Detecting the H2O2 released during the oxidation process of glucose using glucose oxidase (GOD) can be used to monitor the biocatalytic process.45 In addition sensors using carbon dots (CDs) and quantum dots (QDs) have been widely developed due to their advantages of simple operation and easy signal acquisition.46–48 Liu et al. have developed a sensor DEMBs, consisting of two parts: rhodamine-modified silica microbeads (MBs) and CdTe (QDs) (Fig. 12).49 MBs is the core and the QDs are the signal unit. When glucose is oxidized by oxidases, H2O2 is produced to quench the red fluorescence of the CdTe, while the green fluorescence of the rhodamine remains unchanged. When the biocatalytic reaction occurs, the DEMBs exhibit a specific red to green change in the fluorescence ratio. In addition, a colour change can be observed using naked eyes. As such, DEMBs have been used for the detection of glucose in serum. Significantly, these DEMBs can be used as general fluorescent sensors to detect any biocatalytic process that can produce H2O2.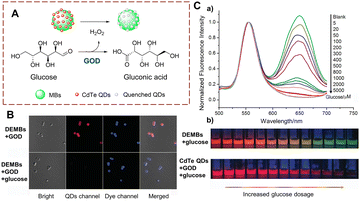 | ||
| Fig. 12 (A) The structure and response mechanism of DEMBs. (B) Confocal images of DEMBs (6 mg mL−1). (C) (a) Normalized fluorescence emission spectra of DEMBs (with GOD) treated with different concentrations of glucose; (b) confocal images of DEMBs and CdTe QDs treated with different concentrations of glucose (reprinted with permission from ref. 49 Copyright © 2016, John Wiley and Sons). | ||
Zhang et al. have developed a colorimetric fluorescent probe hemin@CDs (Fig. 13).50 The CDs act as the probes for biocatalytic glucose oxidation by-product hydrogen peroxide. Meanwhile, the CDs support reduces the aggregation of hemin in the aqueous phase. In the presence of 4-aminoantipyrine (4-AAP), phenol and H2O2, the hemin@CDs catalyses a coupling reaction to produce a pink compound. Hemin@CDs exhibits the advantages of low-cost, rapid detection, and low detection limit. Hemin@CDs was successfully used to detect glucose and xanthine.
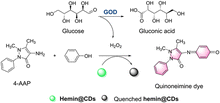 | ||
| Fig. 13 The response mechanism of hemin@CDs (reprinted with permission from ref. 50 Copyright ©, 2020, Springer Nature). | ||
Biocatalytic tyrosine oxidation
Gold nanoclusters (AuNCs) have the advantages of resistance to photobleaching, good biocompatibility and easy handling. Therefore, they represent an excellent material for developing fluorescent sensors. Lu et al. developed a detection strategy using AuNCs to monitor TYR (Fig. 14).51 After biocatalytic oxidation of tyrosine, a quinone is produced, and a cross-linking reaction forms melanoid oligomers. The oligomers continue to form a polymer spontaneously on the surface of AuNCs. The melanin-like polymer can quench the fluorescence of the AuNCs. However, this quenching can be significantly inhibited by H2O2. This method has the advantages of high selectivity and simplicity. As such the AuNCs can be used to detect cholesterol and acetylcholine sensitively.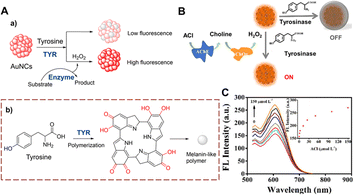 | ||
| Fig. 14 (A) The response mechanism of AuNCs (a) and the generation of melanin-like polymer (b). (B) The illustration of AuNCs for ACl. (C) The fluorescence intensity spectra of AuNCs with different concentrations of ACl (0–150 μmol L−1) (reprinted with permission from ref. 51 Copyright © 2019, Elsevier). | ||
QDs are also used for the detection of TYR.52–55 Using antibody linked TYR, Ma et al. developed dopamine functionalized DAs-QDs that indirectly measures alpha-fetoprotein (AFP) (Fig. 15).56 The sensor DAs-QDs is composed of CdSe/ZnS QDs and N-(3,4-dihydroxyphenethyl)-2-mercaptopentanamide (DAs). With AFP, the ortho-dihydroxyl group of the DAs is oxidized to quinones on the surface of the QDs. The quinone acts as an electron acceptor which can quench the QDs fluorescence. This fluorescence detection strategy exhibits low detection limit (10 pm), while being fast and simple. It provides an efficient method for AFP detection, which has the potential for clinical applications.
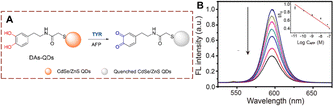 | ||
| Fig. 15 (A) The structure and response mechanism of DAs-QDs. (B) The fluorescence intensity spectra of DAs-QDs to different concentrations of AFP (0–100 nM) (reprinted with permission from ref. 56 Copyright © 2016, American Chemical Society). | ||
Biocatalytic monoamine oxidation
In the human body, biocatalytic oxidation of dopamine and phenethylamine is the primary mode of amine metabolism. MAO-A and MAO-B play key roles in this biocatalytic process.57 Unfortunately, aberrant expression of MAOs damages nerve cells, which induces Parkinson's disease, Alzheimer's disease, depression and cancer.58–61 Therefore, the development of visual methods to detect MAOs is of significance for the clinical diagnosis and treatment of neurological diseases.Clorgyline as an inhibitor of MAO-A, has been widely used in the clinic. Thus, probes based on clorgyline have become useful molecular tools exhibiting the dual function of visualization and treatment.
Ma et al. have developed probe 15 by introducing a clorgyline segment into a hemi cyanine (Fig. 16).62 To increase the flexibility of probe 15, N,N′-dimethyl-1,2-ethanediamine carbamate has been introduced between the clorgyline segment and the hemi cyanine. When probe 15 is biocatalyzed, the amino is oxidatively removed. Subsequently, the N,N′-dimethyl-1,2-ethylenediamine carbamate acts as a self-immolating linker and dissociates from probe 15, exposing the hydroxyl. ICT is recovered and the fluorophore is released. Probe 15 is a near-infrared fluorescence probe with high selectivity and sensitivity (LOD = 4.5 ng mL−1). As such, probe 15 can be used for MAO-A imaging in cells, zebrafish and mice.
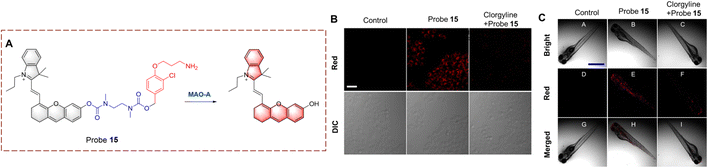 | ||
| Fig. 16 (A) The structure and response mechanism of probe 15. (B) Fluorescence images and the differential interference contrast (DIC) images of probe 15 (5 μM, 1 h) in HeLa cells. (C) Fluorescence images of probe 15 (5 μM, 1 h) in zebrafish (reprinted with permission from ref. 62 Copyright © 2021, American Chemical Society). | ||
Using a similar strategy, Qin et al. have developed probe 16 (Fig. 17).63 The dicyanomethylene (DCM) acts as the fluorophore which is linked to clorgyline through the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond. Since the propylamine is attached to the hydroxyl, the ICT process is inhibited, resulting in the quenched fluorescence of probe 16. When probe 16 is catalytically oxidized, the propylamine is removed and the phenolic hydroxyl group is recovered. Which results in a “D–π–A” structure that emits strong fluorescence. Probe 16 has the advantages of high selectivity, low detection limits (2.6 ng mL−1), and rapid response (60 min).
C bond. Since the propylamine is attached to the hydroxyl, the ICT process is inhibited, resulting in the quenched fluorescence of probe 16. When probe 16 is catalytically oxidized, the propylamine is removed and the phenolic hydroxyl group is recovered. Which results in a “D–π–A” structure that emits strong fluorescence. Probe 16 has the advantages of high selectivity, low detection limits (2.6 ng mL−1), and rapid response (60 min).
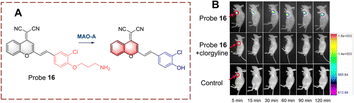 | ||
| Fig. 17 (A) The structure and response mechanism of probe 16. (B) Fluorescence images of probe 16 in tumour-bearing mice (reprinted with permission from ref. 63 Copyright © 2019, Royal Society of Chemistry). | ||
Qin et al. developed a series of probes and using a screening approach determined that probe 17 was suitable for biocatalytic monoamine oxidation detection in vivo (Fig. 18).64 Probe 17 is based on a dihydroxanthene (DH) which is linked to a 1,2,3,3-tetramethyl-3H-indol-1-ium. Probe 17 exhibits weak fluorescence, since the ICT process is inhibited by a propylamine which is attached to the hydroxyl. When the propylamine is catalytically oxidized, a carbonyl group is generated, followed by a β-elimination which exposes the hydroxyl on the anthracene and the fluorescence recovers due to ICT from the hydroxyl. Thus, probe 17 is suitable for monitoring biocatalytic oxidation by MAO-A. In addition, probe 17 was used for real-time detection of monoamine oxidation in mouse liver fibrosis tissue.
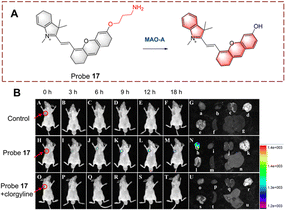 | ||
| Fig. 18 (A) The structure and response mechanism of probe 17. (B) Fluorescence images of probe 17 (50 μM) in SH-SY5Y tumour-bearing mice and isolated organs (reprinted with permission from ref. 64 Copyright © 2022, American Chemical Society). | ||
Ma et al. developed probe 18 for visualizing biocatalytic monoamine oxidation in living cells (Fig. 19).65 The hemi cyanine hybrid coumarin acts as the fluorophore and propylamine acts as the recognition group. Probe 18 is non-fluorescent because propylamine is linked to the hydroxyl group of coumarin inhibiting ICT. When probe 18 undergoes biocatalysis, the amino group is oxidized to a carbonyl group, and subsequently, elimination occurs resulting in release of the coumarin hydroxyl. Thus, due to the recovery of the ICT process, probe 18 exhibits enhanced fluorescence in the mitochondria. Probe 18 was used as an effective strategy for the development of MAO-A inhibitors.
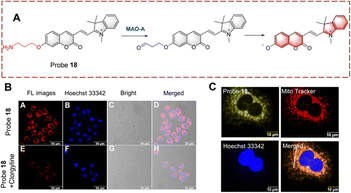 | ||
| Fig. 19 (A) The structure and response mechanism of probe 18. (B) Image of probe 18 in HeLa cells. (C) Mitochondrial localization imaging of probe 18 in HeLa cells (reprinted with permission from ref. 65 Copyright © 2022, Elsevier). | ||
Tang et al. developed probe 19 for the rapid detection of biocatalytic amine oxidation (Fig. 20).66 When probe 19 is oxidized, the amino is converted to an aldehyde group. Then, the aldehyde group reacts with the amino on the adjacent benzene to form a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond. This reaction enhances the degree of conjugation in the system, resulting in strong fluorescence. Probe 19 has the remarkable advantages of simple preparation, fast detection (<10 min) and low detection limit (0.02 ng μL−1). In addition, probe 19 was used for in situ imaging of deep tissue in mice with liver fibrosis, making it suitable for early diagnostic applications.
N bond. This reaction enhances the degree of conjugation in the system, resulting in strong fluorescence. Probe 19 has the remarkable advantages of simple preparation, fast detection (<10 min) and low detection limit (0.02 ng μL−1). In addition, probe 19 was used for in situ imaging of deep tissue in mice with liver fibrosis, making it suitable for early diagnostic applications.
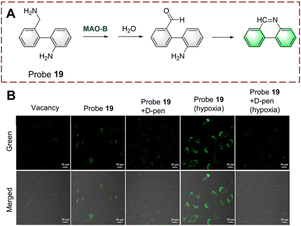 | ||
| Fig. 20 (A) The structure and response mechanism of probe 19. (B) Fluorescence images of probe 19 or probe 19 with D-pen (inhibitor) in LX-2 cells (reprinted with permission from ref. 66 Copyright © 2021, American Chemical Society). | ||
Biocatalytic reduction
Since cancer cells need to consume a large amount of oxygen for proliferation, hypoxia has become one of the main characteristics of the tumour microenvironment (TME).67–70 Hypoxic conditions cause reductases such as nitroreductase (NTR) and azoreductase (AzoR) to be overexpressed in cancer cells.71–76 Thus, the visual monitoring of biocatalytic nitro and azo-reductions are important for cancer diagnosis and treatment.Biocatalytic nitro reduction
Probes for monitoring biocatalytic nitro reduction have been developed with nitro as the recognition group and cyanine, naphthalimide, rhodamine and Nile-red as the fluorophores. Under the biocatalysis of NTR, the nitro group is recognized and reduced to an amino group. Due to this transformation from electron acceptor to donor, the fluorescence of the probe is turned on, enabling the visual detection of the biocatalytic nitro reduction.Ge et al. have developed probe 20 with dual targeting of the mitochondria and lysosomes (Fig. 21).77 With probe 20, a benzo[e]indol aromatic azonia skeleton cyanine acted as the fluorophore, and nitrofuran acts as the recognition group. Significantly, the benzo[e]indol cation not only has the ability to target the mitochondria, but also can enhance the fluorescence intensity. The fluorescence of probe 20 is quenched due to the presence of the nitro group. Probe 20 exhibits good selectivity, fast response (180 s) and high sensitivity (LOD = 3.2 ng mL−1). Therefore, probe 20 could be used to image HeLa cells. Additionally, the probe exhibited dual-targeting and could be used to detect NTR in the mitochondria and lysosomes.
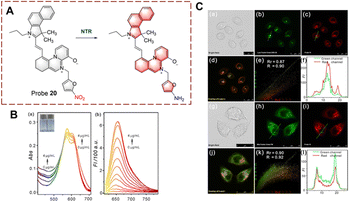 | ||
| Fig. 21 (A) The structure and response mechanism of probe 20. (B) The absorbance spectra (a) and fluorescent spectra (b) of probe 20. (C) Confocal fluorescence images of probe 20, LysoTracker Green DND-26 or Mito Tracker Green FM in HeLa cells (reprinted with permission from ref. 77 Copyright © 2020, Elsevier). | ||
Ma et al. developed probe 21 for imaging the biocatalytic nitro reduction in the second near-infrared window (NIR-II) (Fig. 22).78 Compared to NIR-I based probes, NIR-II based probes exhibit higher signal-to-noise ratio and deeper tissue penetration. Probe 21 is based on rhodamine hybrid polymethine. Initially, probe 21 exhibits no fluorescence due to the quenching effect of the nitro group. When the nitro group is catalytically reduced to an amino group, the fluorescence of probe 21 is recovered. Compared with other NIR-II probes, probe 21 displays a very low detection limit (3.2 ng mL−1). Therefore, probe 21 could be used for tumour imaging in vivo due to its excellent deep tissue penetration.
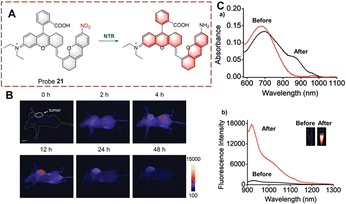 | ||
| Fig. 22 (A) The structure and response mechanism of probe 21. (B) Images of probe 21 in A549 tumour-bearing mice. (C) The absorbance spectra (a) and fluorescent spectra (b) of probe 21 before and after reduction (reprinted with permission from ref. 78 Copyright © 2021, Royal Society of Chemistry). | ||
NTR has become a common marker for hypoxia in living cells.79,80 In addition, a decrease in intracellular oxygen can lead to changes in adenosine triphosphate (ATP).81 Therefore, ATP is expected to become a marker that, together with NTR, illustrates the hypoxic state of cells. Based on this strategy, Ma et al. developed probe 22 for the simultaneous detection of NTR and ATP as markers for cellular hypoxia (Fig. 23).82 Probe 22 was designed based on the rhodamine/1,8-naphthalimide hybrid structure. Diethylenetriamine and the nitro are the recognition elements for ATP and NTR, respectively. Due to the quenching effect of the nitro group and the formation of the spiroxamine, probe 22 exhibits weak fluorescence. However, when probe 22 is reduced, the nitro group is converted to an amine group. Moreover, in the presence of ATP, the spirolactam ring can be opened and probe 22 exhibits strong fluorescence. As a dual-function probe for the detection of NTR and ATP, probe 22 was used for anoxic cell imaging. Due to the synergistic effect of NTR and ATP, probe 22 is a more accurate tool for assessing hypoxia.
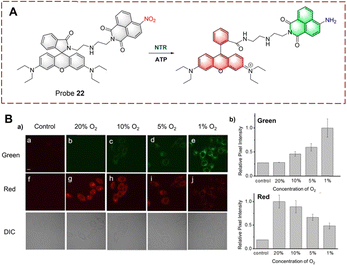 | ||
| Fig. 23 (A) The structure and response mechanism of probe 22. (B) Confocal fluorescence images of probe 22 in HeLa cells (a); relative pixel intensity of green channel and red channel (b) (reprinted with permission from ref. 82 Copyright © 2018, Royal Society of Chemistry). | ||
Hong et al. developed probe 23 for imaging in hypoxic mice (Fig. 24).83 The system consists of a BF2-chelated azadipyrromethane (BODIPY) as the fluorophore, and is linked to a p-nitrobenzyl group which can responded to NTR. The fluorescence is quenched due to the electron withdrawing effect of the p-nitrobenzyl group. When probe 23 undergoes reduction, the nitro group is converted to an electron-donating amino group. Then a 1,6-rearrangement elimination reaction occurs which results in the cleavage of the ether bond. The p-nitrobenzyl group is cleaved and the fluorophore is released, resulting in recovery of the fluorescence. Probe 23 exhibits the advantages of high sensitivity (LOD = 1.5 ng mL−1) and rapid response (<5 min). Therefore, probe 23 could be used for tumour imaging in vitro and in vivo. Furthermore, probe 23 has the potential to be used in preoperative diagnosis of tumours.
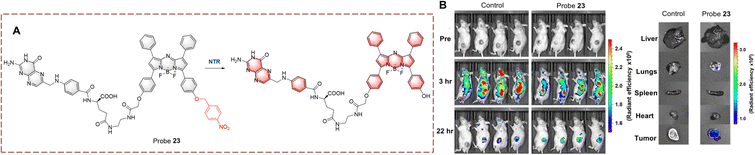 | ||
| Fig. 24 (A) The structure and response mechanism of probe 23. (B) Images of probe 23 in CT26 tumour-bearing mice and isolated organs (reprinted with permission from ref. 83 Copyright © 2021, American Chemical Society). | ||
Peng et al. developed a two-photon (TP) probe 24 for bioimaging the biocatalytic nitro reduction in mice (Fig. 25).84 Probe 24 is a Nile-red derivative, and such fluorophores have previously been used as the TP fluorophore for other targets. TP fluorophores have the advantages of deep tissue penetration, lower background interference, and lower phototoxicity. With probe 24, the p-nitrobenzyl group and the fluorophore are connected by an ether bond. When probe 24 undergoes a biocatalytic nitro reduction reaction, the nitro group is reduced to an amino group, and a 1,6-rearrangement elimination reaction occurs, which results in release of the Nile-red fluorophore. Probe 24 exhibits a 45-fold fluorescence enhancement at 655 nm. Additionally, probe 24 was used to image mouse liver sections and for in vivo imaging due to its deep tissue penetrating ability.
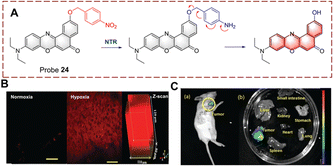 | ||
| Fig. 25 (A) The structure and response mechanism of probe 24. (B) TP imaging and confocal Z-scan images of probe 24 in mouse liver slices. (C) Fluorescence imaging of probe 24 in 4T1 tumour-bearing mice (a) and isolated organs (b) (reprinted with permission from ref. 84 Copyright © 2019, Royal Society of Chemistry). | ||
Based on the same mechanism, the Chen group developed probe 25 containing a hemi cyanine (Fig. 26).85 Probe 25 exhibits weak fluorescence due to the quenching effect of the p-nitrophenzyl group. When probe 25 participates in the biocatalytic reduction reaction, the nitro group is reduced to an amino group, this is followed by a 1,6-rearrangement elimination reaction. Probe 25 is characterized by simple synthesis, high yield, wide detection range (0–20 μg mL−1) and low detection limit (26 ng mL−1). Based on these advantages, probe 25 was used to detect NTR in cells and mice under hypoxic conditions. Significantly, probe 25 was used for monitoring NTR under hyperbaric oxygen (HBO) treatment. As such the authors believe that probe 25 will become a pre-evaluation tool for surgery.
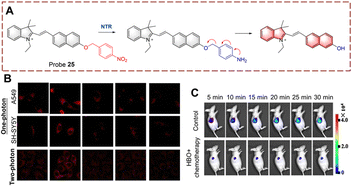 | ||
| Fig. 26 (A) The structure and response mechanism of probe 25. (B) Confocal microscopic images of probe 25 in SH-SY5Y cells and A549 cells during tumour treatment. (C) Image of probe 25 in A549 tumour-bearing mice (reprinted with permission from ref. 85 Copyright © 2020, Elsevier). | ||
Biocatalytic azo reduction
Using the azo group as the recognition group, probes for detecting biocatalytic azo reduction have been developed.86,87 The azo group can quench the fluorescence of a fluorophore, however, when the probe is reduced by a biocatalyst AzoR, the azo group is transformed into an electron donating amino group. As such this biocatalytic reaction results in a significant increase in the fluorescence of the probe.Li developed probe 26 for monitoring biocatalytic azo reduction in vivo (Fig. 27).88 A hemi cyanine acts as the fluorophore, and the azo group acts as both the recognition site and the quenching group. When probe 26 is reduced, the azo group is cleaved and an amino group is generated. The ICT process from amino to hemi-cyanine is then enabled resulting in fluorescence enhancement. Specifically, probe 26 is a NIR probe exhibiting high fluorescence enhancement (17-fold), high sensitivity (LOD = 0.017 μg mL−1) and low biotoxicity. Therefore, probe 26 was used to monitor AzoR in mice with acute and chronic ulcerative colitis (UC). In addition, probe 26 was the first probe able to visualize changes of AzoR in vivo.
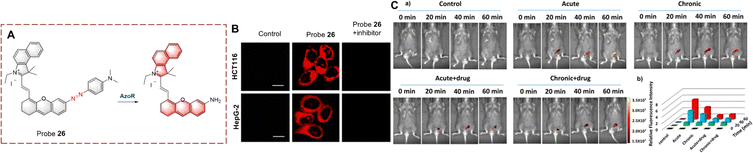 | ||
| Fig. 27 (A) The structure and response mechanism of probe 26. (B) Fluorescence imaging of probe 26 in HCT116 cells and HepG-2 cells. (C) (a) Fluorescence imaging of probe 26 in mice; (b) relative fluorescent intensity of (a) (reprinted with permission from ref. 88 Copyright © 2019, American Chemical Society). | ||
Biocatalytic hydrolysis
Hydrolases are biocatalytic enzymes found in living organisms that can accelerate the hydrolysis of large molecules into small molecules. They play an important role in the metabolism and decomposition processes of organisms. For example, small molecules are more easily absorbed and transformed by organisms, and metabolic waste can also be hydrolyzed and more easily excreted to maintain cellular balance.Biocatalytic galactoside hydrolysis
β-Galactosidase is a hydrolase that catalyzes the hydrolysis of β-D-galactosides thus releasing terminal β-D-galactose units. This biocatalytic process is involved in cellular senescence and ovarian cancer.89,90 Therefore, visual monitoring of biocatalytic galactoside hydrolysis is important to uncover its role in disease development. Probes are usually constructed containing a β-D-galactopyranoside and an appropriate fluorophore (i.e. naphthalimide, cyanine, and coumarin) for visual detection. The β-D-galactopyranoside is the recognition group linked to the fluorophore as a β-D-galactoside. Then under the biocatalytic action of β-galactosidase (β-Gal), the glycosidic group is cleaved, and the fluorophore is released with enhanced fluorescence.Lin developed a ratiometric probe 27 for the detection of biocatalytic glycoside hydrolysis in cancer cells (Fig. 28).91 Probe 27 uses a fluorescence resonance energy transfer (FRET) mechanism, with 7-diethylaminocoumarin and 4-hydroxy-1,8-naphthalimide as the donor (D) and acceptor (A). β-D-Galactopyranoside acts as a recognition group attached to the hydroxyl group of the naphthalimide, which results in reduced ICT and FRET. In this case, the blue fluorescence of probe 27 is from the coumarin. When probe 27 is hydrolyzed, the glycosidic bond is broken, and the hydroxyl group is released. The ICT process from the hydroxyl to naphthalimide is enabled, and the fluorescence is enhanced. While the absorption of the naphthalimide is red-shifted due to ICT, and FRET occurs with the coumarin, and probe 27 emits yellow fluorescence. Probe 27 is characterized by fast response (<20 s), high sensitivity (LOD = 0.081 U mL−1), high biocatalytic efficiency and good biocompatibility. As such, probe 27 was used to image live ovarian cancer cells (OVCAR-3 cells).
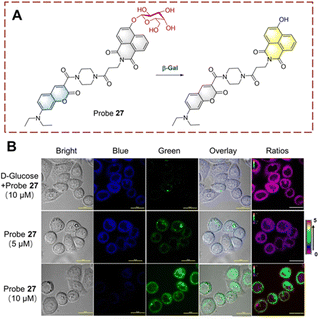 | ||
| Fig. 28 (A) The structure and response mechanism of probe 27. (B) Fluorescent imaging of probe 27 in OVCAR-3 cells (reprinted with ref. 91 Copyright © 2019, American Chemical Society). | ||
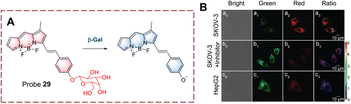
Liu et al. have developed probe 28 which exhibits lysosomal targeting (Fig. 29).92 Probe 28 is based on a hemi cyanine fluorophore, with β-D-galactopyranoside recognition group. The lysosome-targeting ability of probe 28 is due to the morpholine group. Initially, probe 28 is non-fluorescent because the hydroxyl group is caged by the β-D-galactopyranoside. When probe 28 targets the lysosome to participate in biocatalytic processes, the β-D-galactopyranoside is then cleaved. Resulting in the production of a phenolate intermediate which undergoes a 1,6-elimination reaction. As such, probe 28 emits strong fluorescence due to the recovery of the ICT process. Since probe 28 exhibits rapid response (<1 min). it could be used for the visual detection of endogenous β-Gal in ovarian cancer cells.
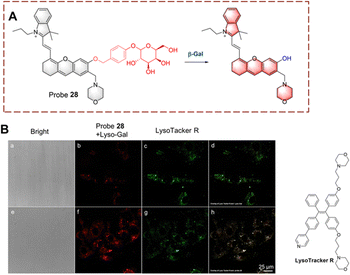 | ||
| Fig. 29 (A) The structure and response mechanism of probe 28. (B) Fluorescence images of probe 28 or LysoTracker R in SKOV-3 cells (reprinted with ref. 92 Copyright © 2020, American Chemical Society). | ||
Zhu et al. developed a ratiometric probe 29 for monitoring biocatalytic galactoside hydrolysis (Fig. 30).93 Probe 29 is based on BODIPY and the hydroxyl group is conjugated through a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond. This conjugated system has been shown to have a remarkable ICT effect. In probe 29, β-D-galactopyranoside is used as the recognition group masking the hydroxyl group. When probe 29 is catalytically hydrolyzed, the β-D-galactopyranoside is cleaved and converted to the phenolate intermediate. As a result, the hydroxyl group is exposed, and the ICT process is turned on with a significant red shift (575 nm to 730 nm). During the hydrolysis of probe 29, a shift in the absorption peak from 560 nm to 620 nm occurs resulting in a colour change from pink to dark blue. Making it possible to observe the activity of the β-Gal using naked eyes.
C bond. This conjugated system has been shown to have a remarkable ICT effect. In probe 29, β-D-galactopyranoside is used as the recognition group masking the hydroxyl group. When probe 29 is catalytically hydrolyzed, the β-D-galactopyranoside is cleaved and converted to the phenolate intermediate. As a result, the hydroxyl group is exposed, and the ICT process is turned on with a significant red shift (575 nm to 730 nm). During the hydrolysis of probe 29, a shift in the absorption peak from 560 nm to 620 nm occurs resulting in a colour change from pink to dark blue. Making it possible to observe the activity of the β-Gal using naked eyes.
 | ||
| Fig. 30 (A) The structure and response mechanism of probe 29. (B) Fluorescence images of probe 29 in SKOV-3 and HepG-2 cells (reprinted with permission from ref. 93 Copyright © 2019, Royal Society of Chemistry). | ||
Guo et al. has extracted hyperoside from Hedyotis diffusa as a probe 30 (Fig. 31).94 The β-galactoside is hydrolyzed and cleaved after being recognized by β-Gal. Subsequently, the hydrolyzed product quercetin was released and aggregates in situ. Quercetin was shown to be an aggregation-induced emission luminophore (AIEgen) with excited-state intramolecular proton transfer (ESIPT) properties.95,96 The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and hydroxyl in quercetin act as proton acceptor and donor respectively, so intramolecular hydrogen bonds are easily formed. However, probe 30 does not exhibit aggregative luminescence since the hydroxyl group is caged by the β-galactoside. Probe 30 is a natural substance that is easy to obtain and exhibits the advantages of high selectivity, high sensitivity (0.013 U mL−1), long-term imaging (8 h) and low toxicity. As such, probe 30 has been used to perform transient and long-term imaging in human ovarian cancer cells.
O and hydroxyl in quercetin act as proton acceptor and donor respectively, so intramolecular hydrogen bonds are easily formed. However, probe 30 does not exhibit aggregative luminescence since the hydroxyl group is caged by the β-galactoside. Probe 30 is a natural substance that is easy to obtain and exhibits the advantages of high selectivity, high sensitivity (0.013 U mL−1), long-term imaging (8 h) and low toxicity. As such, probe 30 has been used to perform transient and long-term imaging in human ovarian cancer cells.
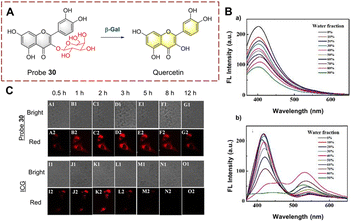 | ||
| Fig. 31 (A) The structure and response mechanism of probe 30. (B) The fluorescence intensity spectra of probe 30 (a) and quercetin (b) (THF/H2O). (C) Images of probe 30 and ICG in SKOV-3 cells (reprinted with permission from ref. 94 Copyright © 2020, Royal Society of Chemistry). | ||
Biocatalytic phosphate hydrolysis
In humans, the biocatalytic hydrolysis of phosphate groups in proteins is of great significance for disease diagnosis. This biocatalytic process relies on phosphatases.97–99 Therefore, the visual detection of phosphate hydrolysis is significant for a full understanding of the mechanism of biocatalysts and improved treatment strategies.Yao et al. have developed YP4 for the detection and imaging of phosphatase activity (Fig. 32).100YP4 is constructed using a two-photon fluorophore (Y1). The phosphate group acts as recognition group. While the phenol hydroxyl is caged by electron-withdrawing phosphate, resulting in the reduction of the conjugated π-electron system, thus YP4 is non fluorescent. YP4 consist of two units a fluorescent group connected to a cell penetrating peptide CPP with masking peptide attached. Initially YP4 cannot enter cells but on cleavage of the masking peptide by matrix metalloproteases (MMPs), YP4 can be absorbed into cells. After YP4 enters the cell, and subsequent UV irradiation can remove the 2-nitrobenzyloxy, thus exposing the phosphate group. The phosphatase then removes the phosphate group and the fluorescence is restored. YP4 exhibits good photophysical properties, deep tissue penetration (110 μm) and low background interference. Therefore, YP4 has been used to image phosphatase in cancer cells and Drosophila cerebrum. Importantly, YP4 exhibits two-photon photophysical properties for the detection of endogenous phosphatase activity.
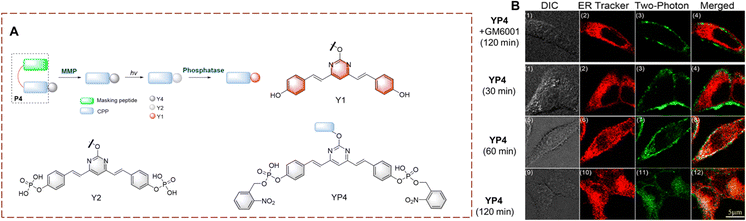 | ||
| Fig. 32 (A) The structure and response mechanism of YP4, Y2 and Y1. (B) Two-photon fluorescence microscopy (TPFM) of YP4 or YP4 with GM6001 (MMP inhibitor) in HeLa cells (reprinted with permission from ref. 100 Copyright © 2012, American Chemical Society). | ||
Conclusions
Fluorescence techniques enable the visual monitoring of biocatalysis, which enables the understanding of catalytic mechanisms in biological systems. With this highlight we have outlined strategies for monitoring biocatalysts including (1) biocatalytic γ-glutamine transfer: linking a γ-glutamine to fluorophores including cyanine, coumarin and naphthalimide, quenches the fluorescence; then once the glutamine group is transferred, the amino group formed recovers the fluorescence. (2) Biocatalytic monoamine oxidation: attaching propylamine to fluorophores including cyanine and rhodamine, masks the hydroxyl and blocks ICT, after oxidation, a carbonyl group is produced with concomitant β-elimination of PABA facilitating the recovery of ICT and enhanced fluorescence. (3) Biocatalytic nitro reduction: linking a nitro group or p-nitrobenzyl group to fluorophores including cyanine, BODIPY, Nile-red and naphthalimide, the fluorescence is quenched. When the nitro group is reduced, the quenching effect is removed and fluorescence is recovered. (4) Biocatalytic β-galactoside hydrolysis: attaching a β-D-galactopyranoside to a fluorophore including BODIPY, coumarin-naphthalimide and natural quercetin, generates a masked hydroxyl group generating a ratiometric fluorescence system. Using these strategies, monitoring biological and pathological processes in cells or in vivo becomes possible. We anticipate that in the future, biocatalytic probes will become ubiquitous in disease monitoring and treatment and help contribute to the rapid development and advancement of precision medicine.Author contributions
Guang Chen, Jie Xu, Siyue Ma and Chao Wang wrote and edited the original draft. Xiaoyong Gao and Ji Chen contributed to the scientific illustrations in the manuscript. Xinrui Ji, Jared B. Carney and Pu Chen created an outline for the review paper. Tony D. James, Yanfeng Yue and Baolei Fan conceived the topic and revised the manuscript. All authors contributed to the final checking of the manuscript.Conflicts of interest
TDJ acts as an academic consultant for Jiangsu Simba Biological Medicine Co., Ltd.Acknowledgements
This work supported by the National Natural Science Foundation of China (22174090); the Natural Science Basic Research Program of Shaanxi (2022JM-089); Long-term Project of high-level talents innovation in Shaanxi Province (Guang Chen); XJ and PC wishes to thank the High-end project of National Foreign Expert Program (G2021041002L); SM thanks the fellowship of China Postdoctoral Science Foundation (2022M711994); Key R&D Program of Shaanxi Province (2022GY-203); Science and Technology Plan Project of Weiyang District (202114); Science and Technology Plan Project of Xi’an (21NYYF0057). TDJ wishes to thank the University of Bath and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University (2020ZD01) for support. BF wishes to thank the Key R&D Plan of Hubei Province for local special support in the field of general health (2022BCE066).References
- H. Yan, P. Xu, H. Ma, Y. Li, R. Zhang, H. Cong, B. Yu and Y. Shen, Biomaterials, 2023, 301, 122213 CrossRef CAS PubMed.
- S. Yardim Akaydin, E. Miser Salihoglu, D. Gelen Gungor, H. Karanlik and S. Demokan, Eur. J. Breast Health, 2020, 16, 72–76 CrossRef PubMed.
- Y. Santin, J. Resta, A. Parini and J. Mialet-Perez, Ageing Res. Rev., 2021, 66, 101256 CrossRef CAS PubMed.
- S. Wang, X.-F. Zhang, H.-S. Wang, J. Liu, S.-L. Shen and X.-Q. Cao, Talanta, 2023, 252, 123834 CrossRef CAS PubMed.
- Y. Wang, X. Han, X. Zhang, L. Zhang and L. Chen, Analyst, 2020, 145, 1389–1395 RSC.
- Z. Tang, Z. Yan, L. Gong, L. Zhang, X. Yin, J. Sun, K. Wu, W. Yang, G. Fan, Y. Li and H. Jiang, Anal. Chem., 2022, 94, 14778–14784 CrossRef CAS PubMed.
- Y. Kim, H. Li, J. Choi, J. Boo, H. Jo, J. Y. Hyun and I. Shin, Chem. Soc. Rev., 2023, 52, 7036–7070 RSC.
- P. K. Hashim, H. M. Dokainish and N. Tamaoki, Org. Biomol. Chem., 2023, 21, 6120–6123 RSC.
- B. Feng, F. Chu, X. Huang, Y. Fang, M. Liu, M. Liu, F. Chen and W. Zeng, Sens. Actuators, B, 2023, 396, 134541 CrossRef CAS.
- G. Cabrera, T. Linares, M. E. de la Calle, D. Cantero, A. Valle and J. Bolivar, Int. J. Mol. Sci., 2020, 21, 8523 CrossRef CAS PubMed.
- K. Norvaisa, M. Kielmann and M. O. Senge, ChemBioChem, 2020, 21, 1793–1807 CrossRef CAS PubMed.
- S. R. B. R. A. A. Husain, S. Chatterjee, I. Khan and Z. Lin, J. Mater. Chem. B, 2020, 8, 3192–3212 RSC.
- G. Kovacevic, R. Ostafe, A. M. Balaz, R. Fischer and R. Prodanovic, J. Biosci. Bioeng., 2019, 127, 30–37 CrossRef CAS PubMed.
- D. Ning, Q. Liu, Q. Wang, X.-M. Du, W.-J. Ruan and Y. Li, Sens. Actuators, B, 2019, 282, 443–448 CrossRef CAS.
- H. Fang, H. Zhang, L. Li, Y. Ni, R. Shi, Z. Li, X. Yang, B. Ma, C. Zhang, Q. Wu, C. Yu, N. Yang, S. Q. Yao and W. Huang, Angew. Chem., Int. Ed., 2020, 59, 7536–7541 CrossRef CAS PubMed.
- S. Wu, R. Snajdrova, J. C. Moore, K. Baldenius and U. T. Bornscheuer, Angew. Chem., Int. Ed., 2020, 60, 88–119 CrossRef PubMed.
- D. A. Holland-Moritz, M. K. Wismer, B. F. Mann, I. Farasat, P. Devine, E. D. Guetschow, I. Mangion, C. J. Welch, J. C. Moore, S. Sun and R. T. Kennedy, Angew. Chem., Int. Ed., 2020, 59, 4470–4477 CrossRef CAS PubMed.
- Q. Meng, C. Ramirez-Palacios, N. Capra, M. E. Hooghwinkel, S. Thallmair, H. J. Rozeboom, A. W. H. Thunnissen, H. J. Wijma, S. J. Marrink and D. B. Janssen, ACS Catal., 2021, 11, 10733–10747 CrossRef CAS PubMed.
- C. Wang, K. Tang, Y. Dai, H. Jia, Y. Li, Z. Gao and B. Wu, ACS Omega, 2021, 6, 17058–17070 CrossRef CAS PubMed.
- F. Cheng, X. L. Chen, C. Xiang, Z. Q. Liu, Y. J. Wang and Y. G. Zheng, Appl. Microbiol. Biotechnol., 2020, 104, 2999–3009 CrossRef CAS PubMed.
- T. Scheidt, H. Land, M. Anderson, Y. Chen, P. Berglund, D. Yi and W.-D. Fessner, Adv. Synth. Catal., 2015, 357, 1721–1731 CrossRef CAS.
- K. Takemura, P. G. Board and F. Koga, Antioxidants, 2021, 10, 549 CrossRef CAS PubMed.
- F. Liu, D. Zhu, Y. Li, M. Kong, Y. Li, J. Luo and L. Kong, Sens. Actuators, B, 2022, 363, 131838 CrossRef CAS.
- Q. Liu, J. Yuan, R. Jiang, L. He, X. Yang, L. Yuan and D. Cheng, Anal. Chem., 2023, 95, 2062–2070 CrossRef CAS PubMed.
- R. Obara, M. Kamiya, Y. Tanaka, A. Abe, R. Kojima, T. Kawaguchi, M. Sugawara, A. Takahashi, T. Noda and Y. Urano, Angew. Chem., Int. Ed., 2020, 60, 2125–2129 CrossRef PubMed.
- S. M. Usama, F. Inagaki, H. Kobayashi and M. J. Schnermann, J. Am. Chem. Soc., 2021, 143, 5674–5679 CrossRef CAS PubMed.
- N. He, Y. Wang, Y. Huang, X. Wang, L. Chen and C. Lv, Sens. Actuators, B, 2020, 322, 128565 CrossRef CAS.
- F. Liu, Z. Wang, T. Zhu, W. Wang, B. Nie, J. Li, Y. Zhang, J. Luo and L. Kong, Talanta, 2019, 191, 126–132 CrossRef CAS PubMed.
- H. Li, Q. Yao, F. Xu, N. Xu, R. Duan, S. Long, J. Fan, J. Du, J. Wang and X. Peng, Biomaterials, 2018, 179, 1–14 CrossRef CAS PubMed.
- P. Siarkiewicz, R. Michalski, A. Sikora, R. Smulik-Izydorczyk, M. Szala, A. Grzelakowska, J. Modrzejewska, A. Bailey, J. E. Nycz, B. Kalyanaraman, J. G. Malecki, J. Zielonka and R. Podsiadły, Dyes Pigm., 2021, 192, 109405 CrossRef CAS.
- Y. Li, C. Xue, Z. Fang, W. Xu and H. Xie, Anal. Chem., 2020, 92, 15017–15024 CrossRef CAS PubMed.
- R. An, S. Wei, Z. Huang, F. Liu and D. Ye, Anal. Chem., 2019, 91, 13639–13646 CrossRef CAS PubMed.
- H. Xue, J. Lu, H. Yan, J. Huang, H. B. Luo, M. S. Wong, Y. Gao, X. Zhang and L. Guo, Talanta, 2022, 237, 122898 CrossRef CAS PubMed.
- Y. J. Reo, M. Dai, Y. J. Yang and K. H. Ahn, Anal. Chem., 2020, 92, 12678–12685 CrossRef CAS PubMed.
- Y. J. Kim, S. J. Park, C. S. Lim, D. J. Lee, C. K. Noh, K. Lee, S. J. Shin and H. M. Kim, Anal. Chem., 2019, 91, 9246–9250 CrossRef CAS PubMed.
- Y. J. Reo, Y. W. Jun, S. Sarkar, M. Dai and K. H. Ahn, Anal. Chem., 2019, 91, 14101–14108 CrossRef CAS PubMed.
- Y.-B. Luo, Y.-Y. Hou, Z. Wang, X.-M. Hu, W. Li, Y. Li, Y. Liu, T.-J. Li and C.-Z. Ai, Comput. Biol. Med., 2022, 149, 105959 CrossRef CAS PubMed.
- X.-F. Zhai, Y. Yi, R. Yu, Y. Kuang, S. Shaker, H.-F. Su, G. Ye, C.-R. Liu, X. Qiao, L. Liang and M. Ye, Sens. Actuators, B, 2022, 364, 131826 CrossRef CAS.
- Z. Tian, J. Wang, Y. Gao, X. Huo, Z. Yu, Y. Wang, C. Wang, L. Feng, J. Cui and X. Tian, Sens. Actuators, B, 2022, 369, 132342 CrossRef CAS.
- X.-F. Zhai, J.-J. Fan, Y. Yi, M. Zhang, X. Yuan, X. Qiao, L. Liang and M. Ye, Chem. Eng. J., 2023, 463, 142382 CrossRef CAS.
- A. Andreu, M. Ćorović, C. Garcia-Sanz, A. S. Santos, A. Milivojević, C. Ortega-Nieto, C. Mateo, D. Bezbradica and J. M. Palomo, Catalysts, 2023, 13, 1359 CrossRef CAS.
- X. Tian, T. Liu, Y. Ma, J. Gao, L. Feng, J. Cui, T. D. James and X. Ma, Angew. Chem., Int. Ed., 2021, 60, 24566–24572 CrossRef CAS PubMed.
- L. Feng, P. Li, J. Hou, Y. L. Cui, X. G. Tian, Z. L. Yu, J. N. Cui, C. Wang, X. K. Huo, J. Ning and X. C. Ma, Anal. Chem., 2018, 90, 13341–13347 CrossRef CAS PubMed.
- L. Feng, Q. Yan, B. Zhang, X. Tian, C. Wang, Z. Yu, J. Cui, D. Guo, X. Ma and T. D. James, Chem. Commun., 2019, 55, 3548–3551 RSC.
- H. Zhao, X. Yuan, X. Yang, F. Bai, C. Mao and L. Zhao, Inorg. Chem., 2021, 60, 15485–15496 CrossRef CAS PubMed.
- X. Li, S. Zhao, B. Li, K. Yang, M. Lan and L. Zeng, Coord. Chem. Rev., 2021, 431, 213686 CrossRef CAS.
- C. Niu, Z. Yao and S. Jiang, Sci. Total Environ., 2023, 882, 163565 CrossRef CAS PubMed.
- Q. Yang, C. Li, J. Li, M. Arabi, X. Wang, H. Peng, H. Xiong, J. Choo and L. Chen, J. Mater. Chem. C, 2020, 8, 5554–5561 RSC.
- X. Lu, P. Wang, Y. Wang, C. Liu and Z. Li, Adv. Mater. Technol., 2016, 1, 1600024 Search PubMed.
- L. Su, Y. Cai, L. Wang, W. Dong, G. Mao, Y. Li, M. Zhao, Y. Ma and H. Zhang, Mikrochim. Acta, 2020, 187, 132 CrossRef CAS PubMed.
- X. Yan, H. Li, R. Jin, X. Zhao, F. Liu and G. Lu, Sens. Actuators, B, 2019, 279, 281–288 CrossRef CAS.
- X. Cheng, Y. Chai, J. Xu, L. Wang, F. Wei, G. Xu, Y. Sun, Q. Hu and Y. Cen, Sens. Actuators, B, 2020, 309, 127765 CrossRef CAS.
- Y. Liu, Y. Zhang, X. Zhang, W. Zhang, X. Wang, Y. Sun, P. Ma, Y. Huang and D. Song, Sens. Actuators, B, 2021, 344, 130234 CrossRef CAS.
- S. Li, R. Zhao, M. Ma, G. Fu, S. Mu, T. Han, X. Liu and H. Zhang, Sens. Actuators, B, 2022, 370, 132438 CrossRef CAS.
- M. Wang, J.-L. Xie, J. Li, Y.-Y. Fan, X. Deng, H.-L. Duan and Z.-Q. Zhang, ACS Sens., 2020, 5, 1634–1640 CrossRef CAS PubMed.
- W. H. Zhang, W. Ma and Y. T. Long, Anal. Chem., 2016, 88, 5131–5136 CrossRef CAS PubMed.
- H. U. Cho, S. Kim, J. Sim, S. Yang, H. An, M. H. Nam, D. P. Jang and C. J. Lee, Exp. Mol. Med., 2021, 53, 1148–1158 CrossRef CAS PubMed.
- W. Y. Kim, M. Won, A. Salimi, A. Sharma, J. H. Lim, S.-H. Kwon, J.-Y. Jeon, J. Y. Lee and J. S. Kim, Chem. Commun., 2019, 55, 13267–13270 RSC.
- Y. Shu, Y. Liu, Y. Gao and J. Li, Sens. Actuators, B, 2023, 390, 133912 CrossRef CAS.
- H. Fang, H. Zhang, L. Li, Y. Ni, R. Shi, Z. Li, X. Yang, B. Ma, C. Zhang, Q. Wu, C. Yu, N. Yang, S. Q. Yao and W. Huang, Angew. Chem., 2020, 132, 7606–7611 CrossRef.
- Y. Mei, Z. Liu, M. Liu, J. Gong, X. He, Q.-W. Zhang and Y. Tian, Chem. Commun., 2022, 58, 6657–6660 RSC.
- J. Shang, W. Shi, X. Li and H. Ma, Anal. Chem., 2021, 93, 4285–4290 CrossRef CAS PubMed.
- Z. Yang, W. Li, H. Chen, Q. Mo, J. Li, S. Zhao, C. Hou, J. Qin and G. Su, Chem. Commun., 2019, 55, 2477–2480 RSC.
- Z. M. Yang, Q. Y. Mo, J. M. He, D. L. Mo, J. Li, H. Chen, S. L. Zhao and J. K. Qin, ACS Sens., 2020, 5, 943–951 CrossRef CAS PubMed.
- J. Wu, C. Han, X. Cao, Z. Lv, C. Wang, X. Huo, L. Feng, B. Zhang, X. Tian and X. Ma, Anal. Chim. Acta, 2022, 1199, 339573 CrossRef CAS PubMed.
- N. Fan, C. Wu, Y. Zhou, X. Wang, P. Li, Z. Liu and B. Tang, Anal. Chem., 2021, 93, 7110–7117 CrossRef CAS PubMed.
- L. Gai, Y. Liu, Z. Zhou, H. Lu and Z. Guo, Coord. Chem. Rev., 2023, 481, 215041 CrossRef CAS.
- K. Mortezaee and J. Majidpoor, Life Sci., 2021, 286, 120057 CrossRef CAS PubMed.
- R. C. Augustin, G. M. Delgoffe and Y. G. Najjar, Cancers, 2020, 12, 3802 CrossRef CAS PubMed.
- A. Vito, N. El-Sayes and K. Mossman, Cells, 2020, 9, 992 CrossRef CAS PubMed.
- S. Wang, W. Tan, W. Lang, H. Qian, S. Guo, L. Zhu and J. Ge, Anal. Chem., 2022, 94, 7272–7277 CrossRef CAS PubMed.
- J. Zhou, S. Fang, D. Zhang, Y. Qu, L. Wang, S. Pan, L. Li, J. Li, W. Du and Q. Wu, Sens. Actuators, B, 2023, 390, 134015 CrossRef CAS.
- K.-C. Yan, J. E. Gardiner, A. C. Sedgwick, N. Thet, R. A. Heylen, T. D. James, A. T. A. Jenkins and X.-P. He, Chem. Commun., 2023, 59, 8278–8281 RSC.
- L.-L. Wu, Q. Wang, Y. Wang, N. Zhang, Q. Zhang and H.-Y. Hu, Chem. Sci., 2020, 11, 3141–3145 RSC.
- L. Fan, Q. Zan, B. Lin, X. Wang, X. Gong, Z. Zhao, S. Shuang, C. Dong and M. S. Wong, Analyst, 2020, 145, 5657–5663 RSC.
- L. Guo, P. Wang, H. Chen, X. Fan, H. L. Zhu and Z. Li, Mater. Today Chem., 2022, 25, 100928 CrossRef CAS.
- X.-L. Sha, X.-Z. Yang, X.-R. Wei, R. Sun, Y.-J. Xu and J.-F. Ge, Sens. Actuators, B, 2020, 307, 127653 CrossRef.
- X. Zhang, X. Li, W. Shi and H. Ma, Chem. Commun., 2021, 57, 8174–8177 RSC.
- J. Yan, K. Wang, L. Gui, X. Liu, Y. Ji, J. Lin, M. Luo, H. Xu, J. Lv, F. Tan, L. Lin and Z. Yuan, Anal. Chem., 2023, 95, 14402–14412 CrossRef CAS PubMed.
- B. Hu, N. Song, Y. Cao, M. Li, X. Liu, Z. Zhou, L. Shi and Z. Yu, J. Am. Chem. Soc., 2021, 143, 13854–13864 CrossRef CAS PubMed.
- S. Chen, D. Yu, W. Zhong, J. Liu, J. Liu, B. Liu, J. Zheng and R. Yang, Chem. Commun., 2021, 57, 7786–7789 RSC.
- Y. Fang, W. Shi, Y. Hu, X. Li and H. Ma, Chem. Commun., 2018, 54, 5454–5457 RSC.
- S. Karan, M. Y. Cho, H. Lee, H. Lee, H. S. Park, M. Sundararajan, J. L. Sessler and K. S. Hong, J. Med. Chem., 2021, 64, 2971–2981 CrossRef CAS PubMed.
- K. H. Gebremedhin, Y. Li, Q. Yao, M. Xiao, F. Gao, J. Fan, J. Du, S. Long and X. Peng, J. Mater. Chem. B, 2019, 7, 408–414 RSC.
- Y. Wang, L. Zhang, Y. Huang, X. Wang, L. Zhang and L. Chen, Sens. Actuators, B, 2020, 310, 127755 CrossRef CAS.
- Y. Tian, Y. Li, W.-X. Wang, W.-L. Jiang, J. Fei and C.-Y. Li, Anal. Chem., 2020, 92, 4244–4250 CrossRef CAS PubMed.
- X.-b Zhao, W. Ha, K. Gao and Y.-p Shi, Anal. Chem., 2020, 92, 9039–9047 CrossRef CAS PubMed.
- Y. Tian, Y. Li, W. L. Jiang, D. Y. Zhou, J. Fei and C. Y. Li, Anal. Chem., 2019, 91, 10901–10907 CrossRef CAS PubMed.
- Y. Gao, Y. Hu, Q. Liu, X. Li, X. Li, C. Y. Kim, T. D. James, J. Li, X. Chen and Y. Guo, Angew. Chem., Int. Ed., 2021, 60, 10756–10765 CrossRef CAS PubMed.
- Y. Yao, Y. Zhang, C. Yan, W.-H. Zhu and Z. Guo, Chem. Sci., 2021, 12, 9885–9894 RSC.
- X. Kong, M. Li, B. Dong, Y. Yin, W. Song and W. Lin, Anal. Chem., 2019, 91, 15591–15598 CrossRef CAS PubMed.
- X. Li, Y. Pan, H. Chen, Y. Duan, S. Zhou, W. Wu, S. Wang and B. Liu, Anal. Chem., 2020, 92, 5772–5779 CrossRef CAS PubMed.
- L. Shi, C. Yan, Y. Ma, T. Wang, Z. Guo and W. H. Zhu, Chem. Commun., 2019, 55, 12308–12311 RSC.
- R. Long, C. Tang, Z. Yang, Q. Fu, J. Xu, C. Tong, S. Shi, Y. Guo and D. Wang, J. Mater. Chem. C, 2020, 8, 11860–11865 RSC.
- D. Xiao, M. Jiang, X. Luo, S. Liu, J. Li, Z. Chen and S. Li, ACS Sustainable Chem. Eng., 2021, 9, 4139–4145 CrossRef CAS.
- X.-M. Cai, Y. Lin, Y. Li, X. Chen, Z. Wang, X. Zhao, S. Huang, Z. Zhao and B. Z. Tang, Nat. Commun., 2021, 12, 1773 CrossRef CAS PubMed.
- X. Zhang, C. Ren, F. Hu, Y. Gao, Z. Wang, H. Li, J. Liu, B. Liu and C. Yang, Anal. Chem., 2020, 92, 5185–5190 CrossRef CAS PubMed.
- S. M. Stanford and N. Bottini, Nat. Rev. Drug Discovery, 2023, 22, 273–294 CrossRef CAS PubMed.
- X. Gao, G. Ma, C. Jiang, L. Zeng, S. Jiang, P. Huang and J. Lin, Anal. Chem., 2019, 91, 7112–7117 CrossRef CAS PubMed.
- L. Li, J. Ge, H. Wu, Q. H. Xu and S. Q. Yao, J. Am. Chem. Soc., 2012, 134, 12157–12167 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |