An esterase-cleavable persulfide donor with no electrophilic byproducts and a fluorescence reporter†
Bharat S.
Choudhary‡
 a,
T. Anand
Kumar‡
a,
T. Anand
Kumar‡
 a,
Akshi
Vashishtha
a,
Akshi
Vashishtha
 b,
Sushma
Tejasri
b,
Sushma
Tejasri
 a,
Amal S.
Kumar
a,
Amal S.
Kumar
 a,
Rachit
Agarwal
a,
Rachit
Agarwal
 b and
Harinath
Chakrapani
b and
Harinath
Chakrapani
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Pune 411 008, Maharashtra, India. E-mail: harinath@iiserpune.ac.in
bDepartment of Bioengineering, Indian Institute of Science, Bengaluru 560 012, Karnataka, India
First published on 10th January 2024
Abstract
Hydrogen sulfide (H2S) and associated sulfur species known as persulfide or sulfane sulfur are considered among the first responders to oxidative stress. However, tools that reliably generate these species without any potentially toxic byproducts are limited, and even fewer report the generation of a persulfide. Here, using a latent fluorophore embedded with N-acetylcysteine persulfide, we report a new tool that is cleaved by esterase to produce a persulfide as well as a fluorescence reporter without any electrophilic byproducts. The rate of formation of the fluorescence reporter is nearly identical to the rate of formation of the persulfide suggesting that the use of this probe eliminates the need for secondary assays that report persulfide formation. Symptomatic with persulfide generation, the newly developed donor was able to protect chondrocyte cells from oxidative stress.
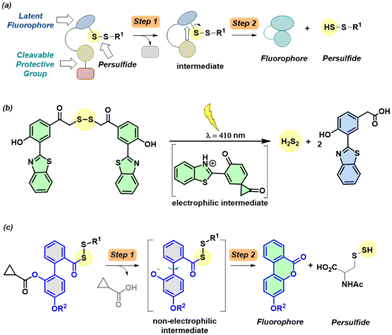
The gasotransmitter hydrogen sulfide (H2S) has emerged as an important mediator of numerous cellular processes and a possible therapeutic agent in a number of diseases including progressive degenerative diseases associated with inflammation such as Parkinson's, Alzheimer's and osteoarthritis.1–4 Recently, an oxidative post-translation modification induced by H2S, known as protein persulfidation4–6 where a cysteine (RSH) is modified to a persulfide (RS-SH), has been shown to have protective effects in cells under oxidative stress.7 For example, KEAP1 an oxidative stress sensor protein forms a complex with Nrf2 under normal conditions, where Nrf2 is ubiquitinated by the Culin3-KEAP1 E3 ubiquitin ligase complex leading to proteasomal degradation. Under oxidative stress conditions, persulfidation of KEAP1 leads to the dissociation of the KEAP1-Nrf2 complex. The liberated Nrf2 then translocates to the nucleus and promotes the activation of antioxidant responsive elements (AREs).8–10 While many tools have been developed to reliably generate persulfides, including those activated by enzymes,11–15 light,16,17 reactive oxygen species (ROS),18–21 pH,22,23 fluoride22 and peroxynitrite.24 One possible limitation with some of these approaches is the generation of electrophilic byproducts such as a quinone methide13,14,18,21,24 or an aldehyde;11,16,17,20 such byproducts may contribute to electrophilic stress.25,26 Also, there is a need for secondary assays to report persulfide produced by these donors. To overcome the need for secondary assays to report persulfide generation, a latent fluorophore strategy can be employed. Upon activation by particular stimuli, the protective group gets cleaved (Step 1) leading to the formation of an intermediate, which subsequently dissociates (Step 2) to release persulfide along with a fluorescence reporter (Fig. 1a).
Recently, a donor that generates persulfide (H2S2) as well as a fluorescence reporter upon activation by light were reported (Fig. 1b).17 However, the applicability of this tool may be limited by the generation of electrophilic byproducts as well as the limited use of light-based tools in the cellular experiments. Here, we developed a tool that generates persulfide along with a fluorophore that reports the generation of persulfide.
To achieve this goal, we considered lactonization as the key step in persulfide generation (Fig. 1c). Hence, a biphenyl-based system was chosen here, cleavage of the ester by esterase (Es) should lead to lactonization, which generates a lactone that belongs to a class of known fluorophores.27,28 Lactonization in such systems is reported to be quite rapid;29 hence, the persulfide generation will likely occur during lactonization leading to nearly concomitant release of the persulfide and the production of a fluorescence signal.
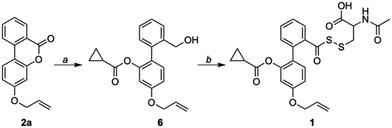
Compound 2a (Scheme 1) was synthesized following a reported protocol.30 Reduction of lactone by LAH, silyl protection of the resulting alcohol, and esterification with cyclopropyl carboxylic acid (CPCA) followed by silyl group deprotection afforded compound 6.
Oxidation of a primary alcohol in two steps gave carboxylic acid 8, which was then converted to its corresponding thiocarboxylic acid using Lawesson's reagent and finally coupled with NAC-pyridyl disulfide (NAC-SS-Py) 9 to provide the persulfide donor 1 (Scheme S1, ESI†). This compound with an allyl group was used for initial experiments, since the lactone (2b) without the allyl group had diminished fluorescence signal when compared with 2a (see ESI,† Fig. S1). We hence proceeded to use 1 which has an allyl group for our analysis.
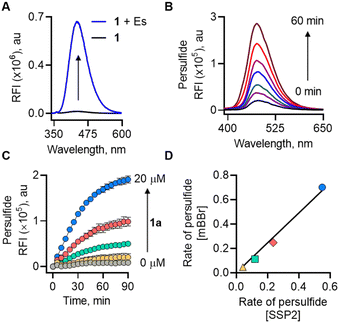
We first examined the formation of lactone (2a) from 1 (20 μM) by monitoring the change in fluorescence in the presence of Es (1 U mL−1). The compound was itself non-fluorescent, but addition of Es (1 U mL−1) led to a significant increase in fluorescence intensity (48-fold) attributable to the formation of 2a (λex = 320 nm; λem = 432 nm;) after incubation for 1 h in pH 7.4 buffer (Fig. 2a). The formation of 2a was independently confirmed by mass spectrometry analysis (m/z 253.0864; see ESI,† Fig. S2). Furthermore, dose-dependent studies demonstrated that the fluorescence gradually increased over time and plateaued after 1 h (see ESI,† Fig. S3). Curve fitting for lactone formation to a first-order exponential equation yielded rate constants ranging from 0.046 to 0.073 min−1 (see ESI,† Table S1). These results supported that 1 underwent lactonization to produce 2a following esterase-mediated hydrolysis. We tested the ability of 1 to be cleaved under cell culture conditions and found that 1 was cleaved in cell lysates to produce a fluorescence signal (see ESI,† Fig. S4). Compound 1 was well tolerated by MEF cells up to 100 μM (see ESI,† Fig. S5). To ascertain if 1 permeates cells to generate 2a, MEF cells were treated with 1 followed by confocal imaging after 4 h, since this class of fluorophores is known to be compatible with two-photon imaging31 (λex = 700 nm; see ESI,† Fig. S6). Together, these results support the suitability of 1 in cellular assays.
Next, a series of experiments were conducted to directly trap the persulfide release from 1 using electrophilic trapping agents and persulfide-specific fluorescent probes. First, a monobromobimane (mBBr)-based fluorescence enhancement assay was used.17 Here, when 1 was co-incubated with Es and mBBr, we found a distinct time-dependent increase in fluorescence (λex = 380 nm; λem = 455 nm) (Fig. 2b). This fluorescence was attributed to the formation of NAC-SS-bimane adduct (M + Na+; m/z 408.0663) (Scheme S2, ESI†), which was further confirmed by MS analysis (see ESI,† Fig. S7). A dose-dependent study of 1 with mBBr showed a good linear relationship (see ESI,† Fig. S8).
Independently, we also measured the rate of persulfide generation from 1 using a well-established fluorogenic probe, sulfane sulfur probe (SSP2).32 Again, we observed a significant increase in fluorescence intensity (λex = 482 nm; λem = 518 nm), corresponding to the generation of sulfane sulfur (Fig. 2c). Under these experimental conditions, the rate of persulfide formation in both mBBr and SSP2 assays showed an excellent correlation (R2 = 0.99) (Fig. 2d).
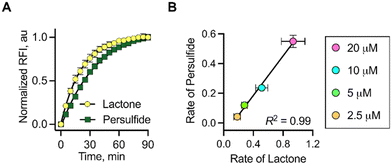
The time course for the formation of 2a as well as NAC persulfide at various concentrations of 1 was independently monitored by the change in fluorescence signal. When followed over 90 min, the time courses of these events were similar. The rate for lactone formation was found to be marginally higher in magnitude (2–3 fold) than the rate of persulfide generation (see ESI,† Table S1). This is expected since the lactonization occurs at a faster rate compared to the rate of formation of NAC persulfide covalent adducts that occurs through a two-step process rather than a single step. To establish a correlation between the lactone (2a) and persulfide formation (SSP2) from 1, the maximum fluorescence change at 90 min was considered as the plateau was reached. These values were normalized and then plotted. An excellent correlation between the two parameters was observed, suggesting that the release of fluorophore and NAC persulfide occurs nearly concurrently following esterase-mediated cleavage and subsequent lactonization (Fig. 3a). Similarly, the rates of persulfide generation as determined by SSP2 and rates of lactone 2a formation showed a good linear relationship at concentrations ranging from 2.5 to 20 μM (Fig. 3b). Taken together, these data show that the fluorescence signal produced can be used as a proxy for persulfide production.
Lastly, two additional assays were carried out to demonstrate persulfide and hydrogen sulfide formation. A standard LC/MS assay was conducted using an established HPE-IAM electrophile as a persulfide trapping agent (Scheme S3, ESI†).33 Upon co-incubation of 1 in the presence of Es (1 U mL−1) and HPE-IAM, a new peak attributable to a persulfide adduct, NAC-SS-HPE-AM (m/z 373.0891), was observed, indicating the release of NAC persulfide (see ESI,† Fig. S9 and S10). Under these conditions, we also observe Bis-S-HPE-AM (m/z 389.1537), presumably due to the reaction of HPE-IAM with H2S formed by the decomposition of NAC persulfide through disproportionation (see ESI,† Fig. S9 and S11). Lastly, we measured H2S formation from 1 using the standard methylene blue (MB) colorimetric assay.34 This assay revealed the formation of H2S from 1 in the presence of Es and excess amounts of DTT (see ESI,† Fig. S12). To independently verify H2S release, we employed a lead acetate assay. Addition of an aliquot of the reaction mixture containing 1 and Es to lead acetate paper resulted in a dark coloration, indicating the formation of lead sulfide (see ESI,† Fig. S13).
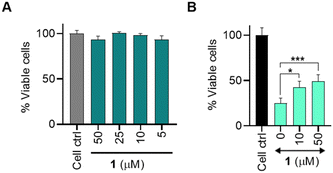
Osteoarthritis (OA) is the most common joint disease and is characterized by a gradual loss of articular cartilage and joint hypertrophy.35 Chronic inflammation and oxidative stress contribute to the onset and progression of OA.36,37 We considered the antioxidant property of persulfide as a possible therapeutic strategy to alleviate oxidative stress and inflammation in chondrocytes. Persulfides are known to exhibit potent antioxidant properties, superior to thiols and H2S against oxidative stress.7 We first measured the cytotoxicity of 1 in the human chondrocyte C28/I2 cells and found that the compound was well tolerated up to 50 μM and the LD50 was found to be ∼500 μM (Fig. 4a and see ESI,† Fig. S14). This result is consistent with no electrophilic byproducts being formed during the persulfide generation step. Next, we determined the cytoprotective effects of 1 in C28/I2 cells against oxidative stress induced by a cell-permeable ROS generator MGR-1 (Scheme S4, ESI†).38 Cells were preincubated with 1 for 3 h and then were exposed to MGR-1 (15 μM), after which cell viability was assessed. As expected, exposure of cells to MGR-1 resulted in a drastic reduction in cell viability. Pre-treatment of cells with 1 demonstrated significant protective effects from MGR-1 induced oxidative stress in a concentration-dependent manner (Fig. 4b). A similar result was recorded when 1 was co-treated with MGR-1 or added after exposure to MGR-1 (see ESI,† Fig. S15). To understand if 1 acted through diminishing ROS levels, a standard H2-DCFDA assay was performed in MEF cells (see ESI,† Fig. S16). These results indicate that 1 was capable of protecting as well as rescuing cells from oxidative stress.
The central characteristic of osteoarthritis (OA) is the degradation of the extracellular matrix (ECM). When chondrocytes are densely seeded (micromass) and exposed to specific growth factors, they release extracellular matrix (ECM) components, such as sulphated glycosaminoglycans (sGAG).39 Next, we employed micromass cultures derived from the C28/I2 cell line,40,41 which can be considered as an in vitro mimic of the cartilage39,41 to assess the ability of 1 in sustaining sGAG production in the presence of oxidative stress induced by MGR-1. sGAG production was measured using Alcian blue staining as described.39–41 When exposed to MGR-1 alone, there was a significant decrease in absorbance compared to the untreated group. However, micromasses co-treated with MGR-1 and 1 displayed an increased absorbance, similar to the untreated samples, indicating that the cells were capable of sustaining sGAG production even under oxidative stress when co-treated with 1 (see ESI,† Fig. S17). These results support the ability of persulfides generated by 1 to counter oxidative stress in a model relevant to OA. Further in vitro and in vivo experiments towards exploiting the therapeutic potential of persulfide generating systems for the treatment of OA are indicated.
In summary, we report a new probe that is cleaved by esterase to produce persulfide as well as a fluorescence signal. No electrophilic byproducts were formed during this reaction (see ESI,† Fig. S18), and the rate of persulfide generation closely correlated with the rate of generation of a fluorescence signal. Hence, the use of this new tool eliminates the need for independent assays to detect persulfides.
Financial support was provided by the Science and Engineering Research Board (CRG/2019/002900), Department of Biotechnology (HC, BH/HRD/NBM-NWB/39/2020-21) and IISER Pune, and the DST Fund for Improvement of S&T Infrastructure (SR/FST/LSII-043/2016) to the IISER Pune Biology Department for setting up the Biological Mass Spectrometry Facility. The manuscript was written with inputs from all authors. BSC, TAK, AV, ST and ASK carried out all experiments under the supervision of RA and HC.
Conflicts of interest
There are no conflicts to declare.Notes and references
- T. V. Mishanina, M. Libiad and R. Banerjee, Nat. Chem. Biol., 2015, 11, 457–464 CrossRef CAS PubMed.
- M. R. Filipovic, J. Zivanovic, B. Alvarez and R. Banerjee, Chem. Rev., 2018, 118, 1253–1337 CrossRef CAS PubMed.
- S. Nasi, D. Ehirchiou, A. Chatzianastasiou, N. Nagahara, A. Papapetropoulos, J. Bertrand, G. Cirino, A. So and N. Busso, Arthritis Res. Ther., 2020, 22, 49 CrossRef CAS PubMed.
- A. K. Mustafa, M. M. Gadalla, N. Sen, S. Kim, W. Mu, S. K. Gazi, R. K. Barrow, G. Yang, R. Wang and S. H. Snyder, Sci. Signaling, 2009, 2, ra72 Search PubMed.
- C. Yang, N. O. Devarie-Baez, A. Hamsath, X. Fu and M. Xian, Antioxid. Redox Signaling, 2020, 33, 1092–1114 CrossRef CAS PubMed.
- H. Kimura, Br. J. Pharmacol., 2020, 177, 720–733 CrossRef CAS PubMed.
- T. Ida, T. Sawa, H. Ihara, Y. Tsuchiya, Y. Watanabe, Y. Kumagai, M. Suematsu, H. Motohashi, S. Fujii, T. Matsunaga, M. Yamamoto, K. Ono, N. O. Devarie-Baez, M. Xian, J. M. Fukuto and T. Akaike, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7606–7611 CrossRef CAS PubMed.
- J. W. Kaspar, S. K. Niture and A. K. Jaiswal, Free Radical Biol. Med., 2009, 47, 1304–1309 CrossRef CAS PubMed.
- S. Koike, Y. Ogasawara, N. Shibuya, H. Kimura and K. Ishii, FEBS Lett., 2013, 587, 3548–3555 CrossRef CAS PubMed.
- G. Yang, K. Zhao, Y. Ju, S. Mani, Q. Cao, S. Puukila, N. Khaper, L. Wu and R. Wang, Antioxid. Redox Signaling, 2013, 18, 1906–1919 CrossRef CAS PubMed.
- Y. Zheng, B. Yu, Z. Li, Z. Yuan, C. L. Organ, R. K. Trivedi, S. Wang, D. J. Lefer and B. Wang, Angew. Chem., Int. Ed., 2017, 56, 11749–11753 CrossRef CAS PubMed.
- Z. Yuan, Y. Zheng, B. Yu, S. Wang, X. Yang and B. Wang, Org. Lett., 2018, 20, 6364–6367 CrossRef CAS PubMed.
- K. M. Dillon, R. J. Carrazzone, Y. Wang, C. R. Powell and J. B. Matson, ACS Macro Lett., 2020, 9, 606–612 CrossRef CAS PubMed.
- K. M. Dillon, H. A. Morrison, C. R. Powell, R. J. Carrazzone, V. M. Ringel-Scaia, E. W. Winckler, R. M. Council-Troche, I. C. Allen and J. B. Matson, Angew. Chem., Int. Ed., 2021, 60, 6061–6067 CrossRef CAS PubMed.
- P. Bora, M. B. Sathian and H. Chakrapani, Chem. Commun., 2022, 58, 2987–2990 RSC.
- A. Chaudhuri, Y. Venkatesh, J. Das, M. Gangopadhyay, T. K. Maiti and N. D. P. Singh, J. Org. Chem., 2019, 84, 11441–11449 CrossRef CAS PubMed.
- A. Chaudhuri, Y. Venkatesh, B. C. Jena, K. K. Behara, M. Mandal and N. D. P. Singh, Org. Biomol. Chem., 2019, 17, 8800–8805 RSC.
- C. R. Powell, K. M. Dillon, Y. Wang, R. J. Carrazzone and J. B. Matson, Angew. Chem., Int. Ed., 2018, 57, 6324–6328 CrossRef CAS PubMed.
- R. A. Hankins, S. I. Suarez, M. A. Kalk, N. M. Green, M. N. Harty and J. C. Lukesh, Angew. Chem., Int. Ed., 2020, 59, 22238–22245 CrossRef CAS PubMed.
- P. Bora, P. Chauhan, S. Manna and H. Chakrapani, Org. Lett., 2018, 20, 7916–7920 CrossRef CAS PubMed.
- Y. Wang, K. M. Dillon, Z. Li, E. W. Winckler and J. B. Matson, Angew. Chem., Int. Ed., 2020, 59, 16698–16704 CrossRef CAS PubMed.
- J. Kang, S. Xu, M. N. Radford, W. Zhang, S. S. Kelly, J. J. Day and M. Xian, Angew. Chem., Int. Ed., 2018, 57, 5893–5897 CrossRef CAS PubMed.
- V. S. Khodade, B. M. Pharoah, N. Paolocci and J. P. Toscano, J. Am. Chem. Soc., 2020, 142, 4309–4316 CrossRef CAS PubMed.
- Y. Xu, B. Xu, J. Wang, H. Jin, S. Xu, G. Wang and L. Zhen, Chem. – Eur. J., 2022, 28, e202200540 CrossRef CAS PubMed.
- D. C. Thompson, J. A. Thompson, M. Sugumaran and P. Moldéus, Chem. – Biol. Interact., 1993, 86, 129–162 CrossRef CAS PubMed.
- K. Ali, P. Mishra, A. Kumar, D. N. Reddy, S. Chowdhury and G. Panda, Chem. Commun., 2022, 58, 6160–6175 RSC.
- A. Fallah, B. Noshadi, M. Gazi and H. O. Gülcan, J. Fluoresc., 2020, 30, 113–120 CrossRef CAS PubMed.
- X. Peng, J. Gao, Y. Yuan, H. Liu, W. Lei, S. Li, J. Zhang and S. Wang, Bioconjugate Chem., 2019, 30, 2828–2843 CrossRef CAS PubMed.
- M. Caswell and G. L. Schmir, J. Am. Chem. Soc., 1980, 102, 4815–4821 CrossRef CAS.
- A. T. Franks and K. J. Franz, Chem. Commun., 2014, 50, 11317–11320 RSC.
- P. D. McFadden, K. Frederick, L. A. Argüello, Y. Zhang, P. Vandiver, N. Odegaard and D. A. Loy, ACS Appl. Mater. Interfaces, 2017, 9, 10061–10068 CrossRef CAS PubMed.
- W. Chen, C. Liu, B. Peng, Y. Zhao, A. Pacheco and M. Xian, Chem. Sci., 2013, 4, 2892–2896 RSC.
- H. A. Hamid, A. Tanaka, T. Ida, A. Nishimura, T. Matsunaga, S. Fujii, M. Morita, T. Sawa, J. M. Fukuto, P. Nagy, R. Tsutsumi, H. Motohashi, H. Ihara and T. Akaike, Redox Biol., 2019, 21, 101096 CrossRef PubMed.
- A. K. Sharma, M. Nair, P. Chauhan, K. Gupta, D. K. Saini and H. Chakrapani, Org. Lett., 2017, 19, 4822–4825 CrossRef CAS PubMed.
- J. Martel-Pelletier, Osteoarthr. Cartil., 1998, 6, 374–376 CrossRef CAS PubMed.
- M. Y. Ansari, N. Ahmad and T. M. Haqqi, Biomed. Pharmacother., 2020, 129, 110452 CrossRef CAS PubMed.
- L. Liu, P. Luo, M. Yang, J. Wang, W. Hou and P. Xu, Front. Mol. Biosci., 2022, 9, 1001212 CrossRef CAS PubMed.
- D. S. Kelkar, G. Ravikumar, N. Mehendale, S. Singh, A. Joshi, A. K. Sharma, A. Mhetre, A. Rajendran, H. Chakrapani and S. S. Kamat, Nat. Chem. Biol., 2019, 15, 169–178 CrossRef CAS PubMed.
- K. V. Greco, A. J. Iqbal, L. Rattazzi, G. Nalesso, N. Moradi-Bidhendi, A. R. Moore, M. B. Goldring, F. Dell’Accio and M. Perretti, Biochem. Pharmacol., 2011, 82, 1919–1929 CrossRef CAS PubMed.
- K. M. Dhanabalan, A. A. Dravid, S. Agarwal, R. K. Sharath, A. K. Padmanabhan and R. Agarwal, Bioeng. Transl. Med., 2023, 8, e10298 CrossRef CAS PubMed.
- C. De Bari, F. Dell’Accio and F. P. Luyten, Arthritis Rheum., 2001, 44, 85–95 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc04948e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |