Unravelling microRNA regulation and miRNA–mRNA regulatory networks in osteogenesis driven by 3D nanotopographical cues†
Gowri Manohari
Balachander‡
 *a,
Sagar
Nilawar‡
b,
Sai Rama Krishna
Meka§
b,
Lopamudra Das
Ghosh¶
*a,
Sagar
Nilawar‡
b,
Sai Rama Krishna
Meka§
b,
Lopamudra Das
Ghosh¶
 b and
Kaushik
Chatterjee
b and
Kaushik
Chatterjee
 *b
*b
aSchool of Biomedical Engineering, Indian Institute of Technology (BHU) Varanasi, Varanasi-221005, India. E-mail: gowribalachander.bme@iitbhu.ac.in; Tel: +91-542-7165134
bDepartment of Materials Engineering, Indian Institute of Science, Bangalore-560012, India. E-mail: kchatterjee@iisc.ac.in; Tel: +91-80-22933408
First published on 21st December 2023
Abstract
Three-dimensional (3D) culturing of cells is being adopted for developing tissues for various applications such as mechanistic studies, drug testing, tissue regeneration, and animal-free meat. These approaches often involve cost-effective differentiation of stem or progenitor cells. One approach is to exploit architectural cues on a 3D substrate to drive cellular differentiation, which has been shown to be effective in various studies. Although extensive gene expression data from such studies have shown that gene expression patterns might differ, the gene regulatory networks controlling the expression of genes are rarely studied. In this study, we profiled genes and microRNAs (miRNAs) via next-generation sequencing (NGS) in human mesenchymal stem cells (hMSCs) driven toward osteogenesis via architectural cues in 3D matrices (3D conditions) and compared with cells in two-dimensional (2D) culture driven toward osteogenesis via soluble osteoinductive factors (OF conditions). The total number of differentially expressed genes was smaller in 3D compared to OF conditions. A distinct set of genes was observed under these conditions that have been shown to control osteogenic differentiation via different pathways. Small RNA sequencing revealed a core set of miRNAs to be differentially expressed under these conditions, similar to those that have been previously implicated in osteogenesis. We also observed a distinct regulation of miRNAs in these samples that can modulate gene expression, suggesting supplementary gene regulatory networks operative under different stimuli. This study provides insights into studying gene regulatory networks for identifying critical nodes to target for enhanced cellular differentiation and reveal the differences in physical and biochemical cues to drive cell fates.
1. Introduction
Culturing cells in three dimensions (3D) is becoming the convention for studying cellular behavior for mechanistic understanding and developing tissues for testing.1–4 Emerging technologies such as animal-free meat and 3D bioprinting are critically dependent on 3D culture. This has been facilitated by the success of 3D models in tissue mimicry5,6 and regenerative and precision medicine.7,8 3D cultures could be developed scaffold-free such as organoids, or they could be developed on natural or synthetic matrices. Whereas the former derives from self-assembly and allows the natural patterning of cells,9 the latter provides a variety of engineering controls such as confinement, stresses, and stiffness. These biophysical stimuli can be used and controlled to achieve targeted and accelerated tissue development.10Cells react to a 3D substrate by changing their shape,11 force fields12 and cell–cell/matrix adhesions,13 which in turn alter several signaling processes inside the cells in an outside-in manner.14 These actuate a series of bio-chemical signaling processes inside the cells that translate to gene and protein expression changes, which influence cellular behavior.15,16 Gene expression changes upon growth in 3D have been extensively studied.17,18 However, the changes in the microRNAs (miRNAs) that control mRNA translation in response to growth in 3D have been less explored.19 miRNAs are short non-coding RNAs, typically of length 18–22 nucleotides that bind to complementary sequences in the mRNA and prevent the access of the ribosome to the mRNA, thereby negatively modulating the translation of the mRNA. The level of complementarity dictates the level of translational repression; complete complementary sequences lead to mRNA degradation.20 miRNAs were first discovered to be involved in the temporal control of gene expression in larvae development and since then, several other functions have been reported.21 miRNAs are critically implicated in the differentiation of stem cells and organogenesis and in protecting the developmental robustness of the system.22 It is probable that such post-transcriptional controls are also perturbed when cells are cultured in 3D and such changes can achieve significant effects on protein expression and phenotypic outcomes even without major changes in gene expression.
We aim to understand how miRNAs are differentially regulated when stem cells undergo differentiation solely driven by biophysical cues from a 3D substrate compared to differentiation driven by supplementation of growth factors. We hypothesize that an alternate set of miRNAs operates under these two different but functionally equivalent stimuli. To address this, we adopted the experimental strategy from our previous work.23 We cultured human mesenchymal stem cells (hMSCs) on 2D poly(ε-caprolactone) (PCL) films and on 3D nanofibrous PCL matrices fabricated by electrospinning. While the cells cultured on 2D films were cultured in media with osteogenic factors to induce differentiation, the cells on 3D PCL matrices were cultured in growth medium, without supplementation of osteogenic factors. The cells were cultured under these two conditions for 14 days, for which similar levels of mineralization were observed earlier. This indicated that culturing cells on 3D nanofibrous matrices could achieve osteogenic differentiation to the same extent as achieved by osteogenic supplements. We sequenced the mRNAs and miRNAs from these conditions and analyzed their regulation. We integrated the miRNAs with the target gene expression to identify differential sets of miRNAs operative under these conditions and built networks to understand how these differential miRNAs might influence the expression of target genes. These studies have important implications in elucidating molecular mechanisms underlying biophysical regulation of cell fate toward engineering efficient culture platforms.
2. Materials and methods
Bone marrow-derived hMSCs were cultured on 3D nanofibrous scaffolds and 2D spun-coated PCL films, as described previously.23 Fabrication of scaffolds and cell culture methods are described briefly here.2.1 Scaffold fabrication
Nanofibrous 3D PCL scaffolds were fabricated using the electrospinning technique (Espin Nano, India). 12% of the PCL solution was prepared by dissolving PCL pellets (Mn ≈ 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Sigma Aldrich) in trifluoroethanol (TFE, Spectrochem Pvt Ltd) and stirred for 12 h. For spinning, a 2.5 mL syringe with a 22-gauge needle, an operating voltage of 12 kV, a flow rate of 0.5 mL h−1, and a distance of 12 cm between the syringe needle and collector was used. An aluminium sheet was used to collect spun nanofibers. Spin coater (spinNXG-P1) was used to make flat PCL films as 2D substrates. Uniform thin films were prepared using the same PCL solution at 5000 rpm for 40 s.
000, Sigma Aldrich) in trifluoroethanol (TFE, Spectrochem Pvt Ltd) and stirred for 12 h. For spinning, a 2.5 mL syringe with a 22-gauge needle, an operating voltage of 12 kV, a flow rate of 0.5 mL h−1, and a distance of 12 cm between the syringe needle and collector was used. An aluminium sheet was used to collect spun nanofibers. Spin coater (spinNXG-P1) was used to make flat PCL films as 2D substrates. Uniform thin films were prepared using the same PCL solution at 5000 rpm for 40 s.
2.2 Cell culture
hMSCs (Lonza, USA) isolated from a 22-year-old male donor were used in the study. hMSCs were cultured in mesenchymal stem cell (MSC) basal growth medium (GM, Lonza) supplemented with SingleQuot supplements (MSCGM, Lonza, USA) containing serum, L-glutamine, and gentamicin + amphotericin B at 37 °C in a humidified CO2 incubator. The hMSCs were replenished with the growth medium every 48 h until 70–80% confluency was reached. The cells were subcultured using 0.25% trypsin containing 0.5 mM EDTA (Gibco). 2D films and 3D nanofibrous mats were cut into a circular disc of 10 mm diameter to be used in 48 well plates. Before cell seeding, samples were UV sterilized for 1 h. 1 × 104 MSCs were seeded on samples along with 400 μL of growth medium. Cells seeded on 2D spun-coated films were cultured in growth medium (GM) or osteogenic medium (OM). OM was prepared by adding soluble osteoinductive factors (10 nM dexamethasone, 20 mM β-glycerophosphate and 50 μM L-ascorbic acid 2-phosphate) to GM. Cells on 3D nanofibrous matrices were cultured in GM. The medium was refreshed every 3 days. Hereupon, the three conditions are referred with respect to the samples and medium used as 2D GM, 2D OM, and 3D GM.2.3 RNA extraction and NGS ontological analysis
Total RNA was extracted from hMSCs cultured under 2D GM, 2D OM, and 3D GM conditions for 14 days; during this time, the differentiation in the 3D GM was similar to the differentiation under the 2D OM conditions, using the Absolute RNA Miniprep kit (Agilent Technologies, USA). RNA pooled from 6 different samples was used for NGS. The transcriptome sequencing was performed by Genotypic Technology Pvt Ltd (Bangalore, India) through a commercial contract on the Illumina NextSeq500 platform. The quality of the raw data generated was checked using FastQC.24 An in-house Perl script was used to preprocess the reads which included the removal of adapter sequences and low-quality bases (<q30). Tophat-2.0.13,25 which is a splice aligner, was used to align the high-quality data to the reference genome with the default parameters. Cufflinks26 was used to estimate and calculate transcript abundance, which was obtained as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values. Cuffmerge26 was used to further merge together several Cufflinks assemblies by automatically filtering a number of transfrags that are probably artifacts. Cuffdiff was used to find significant changes in transcript expression across the control and treatment samples.26 A merged GTF file produced by Cuffmerge was used as input in Cuffdiff.Two comparisons were carried out between the samples, i.e., 2D OM vs. 2D GM, which is referred to as the effect of osteogenic factors (OF), and 2D GM vs. 3D GM as the effect of geometry/dimension (3D). 2D GM was taken as the control in both comparisons. The condition of log2(fold change) > |1| was applied to categories of differentially regulated genes (DEGs) in sample comparisons. Pathways for each gene were mapped using the PANTHER database.27
2.4 miRNA sequencing, target prediction and integration of miRNA–mRNA
The miRNAs were extracted from total RNA according to the Illumina TruSeq Small library protocol at Genotypic Technology Pvt Ltd. 1 μg of total RNA was used as the starting material. Firstly, the ligation of ‘3’ and ‘5’ adapters was carried out. The ligated products were reverse transcribed and amplified by PCR for 15 cycles and cleaned using polyacrylamide gels. The prepared library was analyzed for quality using a bioanalyzer chip and quantified using a Qubit fluorometer. The sequencing of the library was carried out using the Illumina NextSeq500 platform. Quality of the raw data generated was checked using FastQC.24 srna-workbenchV3.0_ALPHA28 was used to trim the 3′ TruSeq adapter sequences from the raw reads obtained after sequencing. The trimmed reads were filtered based on the length criteria (minimum length 16 bp and maximum 36 bp). Sequences ≥16 bp and ≤36 bp length were considered for further analysis. All reads that qualified QC check were aligned against the Homo sapiens reference genome using bowtie-0.12.9.29 The aligned reads to reference were checked for non-coding RNA (ncRNA) such as ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA) and small nucleolar (snoRNA) contamination. Only reads that did not align to ncRNAs sequences were used for known miRNA prediction.Reads were made unique and hence a read count profile was generated. Further, homology search of these miRNAs was performed against mature human miRNA sequences retrieved from the miRbase-21 database using NCBI-BLAST-2.2.30+.30 Sequences showing a hit against mature miRNA sequences from the miRbase were reported as known. Sequences that did not show any hits against known miRNAs were separated and considered for novel miRNA prediction. These reads were aligned to the reference using bowtie. Novel miRNAs were predicted from the aligned data using Mireap_0.25.31 DGE analysis was carried out using the DESeq tool.32 The differentially expressed miRNAs (DEmiRNAs) were identified between OF and 3D comparisons, keeping 2D GM as the control in both cases. The reads were normalized using the DESeq process, and fold changes were calculated accordingly. Log2(fold change) > |1| was applied to identify DEmiRNAs. Target prediction for the known miRNAs with copy numbers equal to or more than 5 was performed using the miRanda 3.3a tool.33 A minimum free energy ≤−30 and bonding identity >80% were used as cutoffs to filter targets for the miRNAs. Further integration analysis was performed to merge the transcriptome and small-RNA results; an Inhouse Perl script was used to map the regulation of transcripts from transcriptome analysis against the mapped targets. miRNA–mRNA interaction networks were built using the Cytoscape platform.34
3. Results
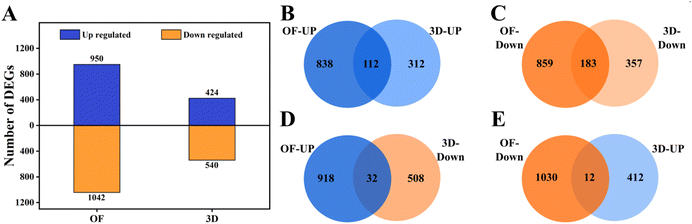
3.1 Differential expression of genes in hMSCs upon osteogenic induction with growth factors versus 3D nanofibrous scaffolds
We sequenced and profiled the genes expressed in hMSCs undergoing osteogenesis via induction using growth factors and under 3D topographical cues on nanofibrous scaffolds, as described above. A total of 950 genes were upregulated and 1042 genes were downregulated in OF (ESI Sheet 1†); 424 genes were upregulated and 540 genes were downregulated in 3D (ESI Sheet 2†) (Fig. 1A). A total of 112 genes were commonly upregulated (Fig. 1B), and a total of 183 genes were commonly downregulated under these two conditions (Fig. 1C). We also observed contrasting regulation of certain genes under these two comparisons; 32 genes that were upregulated in OF were downregulated in 3D (Fig. 1D), and 12 genes that were downregulated were upregulated in 3D (Fig. 1E). Overall, fewer genes were differentially regulated upon growth in 3D compared to exposure to biochemical cues, although both these cues lead to osteogenic differentiation. This suggests that cellular differentiation does not entirely depend on the quantum of gene expression; perhaps other mechanisms are activated and cooperate with transcription to achieve a phenotypic output. Several genes differentially regulated in the OF and 3D conditions have been shown to play important roles in osteogenesis as described in Table 1, which shows that the gene expression profiles from our study corroborate with the literature. Interestingly IGF1, which has shown to be critical for responding to mechanical loading in osteoblasts, is only upregulated in the 3D condition. It has been shown that cells can pull or push the electrospun fibers, thereby stimulating mechanical signals, the modulus of which depends on the direction of the cell-induced load.35 It is plausible that the 3D nanofibrous matrices provide such mechanical signals in addition to the nanotopography that induce osteogenic differentiation of the hMSCs cultured on them.| mRNA | Regulation in OF and 3D | Role in osteogenesis | References |
|---|---|---|---|
| DCN | Upregulated only in OF | Promotes osteogenesis through the ERK1/2 signalling pathway | Adachi et al., 2022![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36 36 |
| CRYAB | Upregulated only in OF | Promotes osteogenesis through the canonical Wnt/β-catenin signalling pathway | Zhu et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
| COL1A1 | Upregulated only in OF | Promotes osteogenesis via the ERK/AKT pathway | Tsai et al., 2010![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 38 |
| COMP | Upregulated only in OF | Promotes osteogenesis by targeting the BMP-2 protein | Ishida et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 39 |
| BMP-6 | Upregulated only in OF | Promotes bone formation through the intramembranous ossification pathways; induces alkaline phosphatase activity and osteocalcin expression | Mizrahi et al., 2013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40,41 40,41 |
| IGF-1 | Upregulated only in 3D | Promotes osteogenesis by conducting mechanical signals via IGF-R1 to activate PI3K/AKT/mTOR signaling | Tian et al., 2018![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 42 |
| MGP | Upregulated only in 3D | Inhibits osteogenesis through the Wnt/β-catenin signalling pathway | Zhang et al., 2019![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
| IL-6 | Downregulated only in OF | Delays osteoblast differentiation by activating STAT3 and thereby inducing the IGFBP5 expression | Peruzzi et al., 2012![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 44 |
| DKK1 | Downregulated only in OF | Inhibits osteogenic differentiation by targeting the canonical Wnt pathway | Chen et al., 2007![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 45 |
3.2 Pathway analyses of differentially regulated genes in osteogenic media and nanofibrous scaffolds
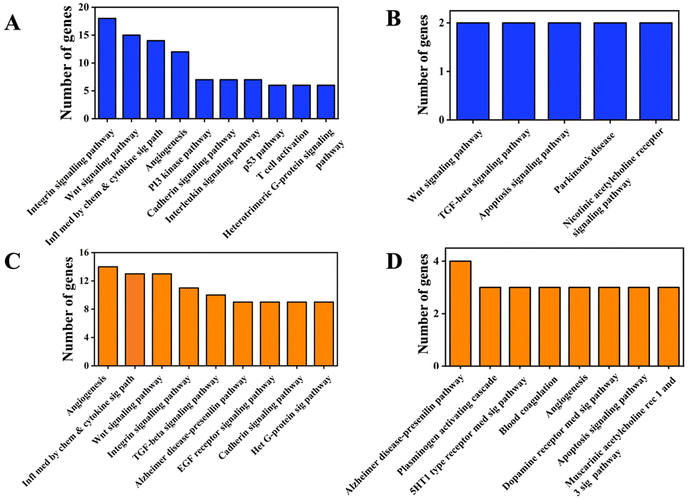
We performed pathway analyses using the Panther database for the differentially expressed genes in OF and 3D comparisons. As expected, we observed genes from the Wnt,46 integrin,47 angiogenesis48 and cadherin49 signaling pathways to be the top hits in the pathway classification for the differentially regulated genes in the OF comparison (Fig. 2A and C). Modulation of these pathways has been documented for osteogenesis differentiation of stem cells,50–52 corroborating our data for growth factor-mediated osteogenic differentiation with the reported literature. However, for the differentially regulated genes in 3D, only a few genes could be classified into some of the aforementioned pathways (Fig. 2B and D). This could be due to the lower number of genes in this group and the stringent criteria used for pathway classification. Also, while growth factor supplementation directly impinges on transcription factor activation and gene expression, growth in 3D could work through other mechanisms that affect osteogenic differentiation.3.3 Differential expression of miRNAs in hMSCs upon osteogenic induction with growth factors versus 3D nanofibrous scaffolds
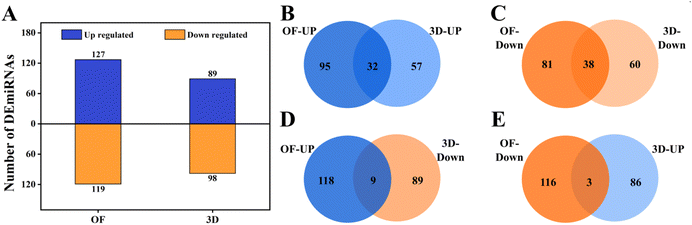
We profiled the miRNAs that were expressed in these conditions as explained in the methods. A total of 127 and 119 known miRNAs were upregulated and downregulated in OF (ESI Sheet 3†); 89 and 98 known miRNAs were upregulated and downregulated in 3D (Fig. 3A) (ESI Sheet 4†). Of these, 32 and 38 miRNAs were commonly upregulated (Fig. 3B) and downregulated (Fig. 3C) in OF and 3D. A set of 9 miRNAs were upregulated in OF and downregulated in 3D (Fig. 3D), while 3 miRNAs (Fig. 3E) had the opposite regulation.Several of the miRNAs that are differentially regulated in the 3D and OF conditions were previously shown in the literature to affect osteogenesis, and we have compiled this in Table 2. This showed that the miRNA profiles from our studies corroborated with the literature on miRNA control in osteogenesis. Interestingly, several of these miRNAs were differentially regulated only in either OF or 3D. A set of 84 novel miRNAs were expressed in the cells cultured in 2D PCL mats in a growth medium, 142 novel miRNAs in the cells in 2D PCL mats in an osteogenic medium, and 81 miRNAs in the cells on 3D nanofibrous scaffolds. The lists of these novel miRNAs (ESI Sheet 5†) and their differential expressions in OF (ESI Sheet 6†) and 3D (ESI Sheet 7†) are given in the ESI.† We have not discussed the novel miRNA expression and regulation in detail as they are not our focus for the current manuscript.
| S. no. | miRNA | Regulation in OF and 3D | Role in osteogenesis | References |
|---|---|---|---|---|
| 1 | miR-1303 | Commonly upregulated | Promotes osteogenesis by targeting the COCH expression | Lin et al., 2023![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 53 53 |
| 2 | miR-106a-5p | Commonly upregulated | Promotes osteogenesis by targeting Fam134a | Wu et al., 2023![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 54 54 |
| 3 | miR-92b-5p | Commonly upregulated | Promotes osteogenesis by targeting ICAM | Li et al., 2019![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 55 |
| 4 | miR-483-3p | Upregulated only in OF | Promotes osteogenesis by targeting DKK2, an antagonist of the Wnt pathway | Zhou et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56 56 |
| 5 | miR-365a-5p | Upregulated only in 3D | Promotes osteogenesis via activation of the Hippo signalling pathway | Kuang et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57 57 |
| 6 | miR-140-3p | Upregulated only in 3D | Promotes osteogenesis by targeting KMT5B | Zhang et al., 2021![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58 58 |
| 7 | miR-141-3p | Commonly downregulated | Inhibits osteogenesis by targeting E2F3 | Xue et al., 2022![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59 59 |
| 8 | miR-146a-5p | Downregulated only in OF | Inhibits osteogenesis by targeting SIRT1 | Zheng et al., 2021![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 60 |
| 9 | miR-216a-3p | Downregulated only in OF | Inhibits osteogenic differentiation by targeting the Wnt3a expression | Liang, Song and Zhang 2022![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 61 |
| 10 | miR-188-3p | Downregulated only in 3D | Inhibits osteogenesis by targeting Beclin-1 mediated autophagy and RUNX1 | Ji et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 62 62 |
| 11 | miR-31-5p | Downregulated only in OF | Inhibits osteogenesis by targeting the SATB2 pathway | Xu et al., 2017![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 63 63 |
| 12 | miR-505-3p | Downregulated only in OF | Inhibits osteogenesis by targeting RUNX2 | Li et al., 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64 64 |
3.4 Integration of miRNA and their targets shows differential miRNAs targeting osteogenic genes under OF and 3D conditions
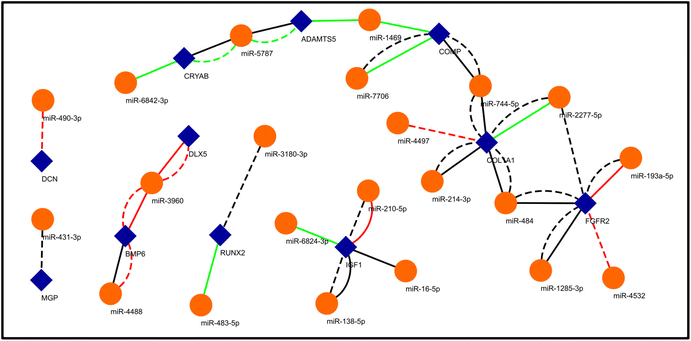
We aimed to understand the miRNA-mediated control of gene regulation in osteogenesis driven by growth factors versus 3D topographical cues. For this, we predicted the targets of the differentially expressed miRNAs in the two conditions and matched them with the genes that are known to be important for osteogenesis (Fig. 4) (ESI Sheet 8†). As the set of genes important for osteogenesis are well characterized, this would reveal how the expressions of such important genes are controlled by different miRNAs under these two conditions to drive osteogenesis. For example, RUNX2 is a transcription factor which is required for commitment of mesenchymal stem cells into osteoblast lineage.65 As depicted in Fig. 4, miR-483-5p and miR-3180-3p are predicted to target RUNX2 and are upregulated in OF and neutrally regulated in 3D conditions, respectively. FGFR2 has been shown to drive the osteogenic differentiation of mesenchymal stem cells.66,67 miR-2277-5p, miR-193a-5p, miR-4532, miR-1285-3p and miR-484 are predicted to target FGFR2. miR-1285-3p and miR-484 are neutrally regulated under OF and 3D conditions, while miR-2277-5p and miR-4532 have neutral and downregulation under 3D conditions, respectively, but are undetected under the OF conditions. The expression of type I collagen is a marker for osteogenic differentiation and it promotes the activation of ERK and AKT pathways that further enhances the proliferation and osteogenesis of MSCs.38 miR-4497, miR-744-5p, miR-2277-5p, miR-484 and miR-214-3p are predicted to target COL1A1. Of these, miR-4497 is downregulated in 3D and undetected under OF conditions. miR-2277-5p is upregulated under OF conditions, but neutrally regulated in 3D, and the rest of the miRNAs are neutrally regulated in the two conditions. Fig. 4 depicts further examples of how osteogenic genes can be differentially targeted by miRNAs under substrate specific conditions.3.5 Unbiased miRNA–mRNA networks predict centres of miRNA control of gene translation
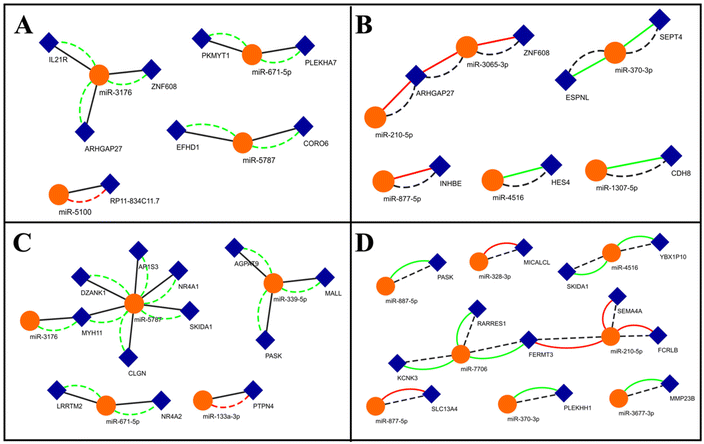
The previous approach shows the miRNA-mediated control of the translation of specific genes implicated in osteogenesis. In order to comprehend the totality of differential regulation of miRNAs in these two conditions and their control on mRNA translation, we followed an unbiased approach. We first screened for commonly up and downregulated genes (log2-fold change) under these conditions. We then matched these genes against the predicted targets of the miRNAs that are up or downregulated in one condition (log2-fold change) and neutrally regulated in the other and developed networks (Fig. 5). While lower cut-offs might provide larger networks, we used high stringency to filter out low-probability interactions. Such networks will provide insights into how miRNAs are differentially expressed (up or downregulated) in a specific condition and how it might affect the expression of its target. For example, miR-5100 is downregulated only in the 3D condition and is predicted to target the long non-coding RNA (lncRNA) RP11-834C11.7 (Fig. 5A). RP11-834C11.7 is commonly upregulated in both these conditions, implying a positive role in osteogenesis. This network suggests that the miR-5100 targeting of RP11 might be exclusively operative only under the 3D conditions in comparison to the OF conditions. miR-877-5p is only downregulated under OF conditions and is predicted to target INHBE, which is commonly upregulated under both these conditions, suggesting that this miRNA–mRNA interaction is exclusive to growth factor-driven osteogenesis (Fig. 5B). miR-5787 is exclusively upregulated under 3D conditions (Fig. 5C). Several of its predicted targets were downregulated under both these conditions (Fig. 5C), implying that they are negatively associated with osteogenesis. While the role of miR-5787 in cancer has been investigated,68 our network shows the induction of this miRNA upon biophysical stimuli and its role in osteogenesis. miR-7706 is upregulated only in OF, while several of its targets are downregulated in both the conditions (Fig. 5D). miR-7706 has previously been shown to be downregulated in rheumatoid arthritis,69 suggesting that it might be more responsive to biochemical cues from the inflammatory response than to substrate-driven mechanical cues. From these networks, we identify several important miRNA nodes that can be central to differential control of gene expression under these conditions.4. Discussion
Gene regulatory networks help us understand the interactions between genes and their regulatory elements, such as promoters, transcription factors, silencers, epigenetic modulators, etc., and the effect of such interactions in determining cell behaviour and responses to stimuli.70 Several experimental and mathematical approaches have been adopted to study these gene regulatory networks.71 Decoding gene regulatory networks is critical to the understanding of cell behavior and responses and for repairing of deregulated controls.72miRNAs have been added to the complex of interactions in the gene regulatory networks. They control the expression of genes through various mechanisms such as direct repression of translation via complementary binding, feedback and feedforward loops and competition with shared miRNAs and shared regulation.73,74 miRNA control in the biochemically guided differentiation of MSCs into osteogenic lineage is known;75–77 however, the miRNA regulation in the matrix-guided differentiation of MSCs into osteogenic lineage is unknown, and the comparison of miRNA regulation under biochemical versus matrix cues have not been attempted.
In this study, we have aimed to understand the miRNA–mRNA gene networks that operate when hMSCs undergo osteogenesis under the influence of growth factors in 2D substrates in comparison to osteogenesis induced by architectural cues in 3D matrices. Our previous study showed that these two different stimuli yield similar cell fates.23 Quantification of osteogenic markers revealed similar mineral deposition in 2D OF and in 3D GM at 14 days in this experimental design. However, at 21 days, the mineral deposition in 2D OF was higher than in 3D GM. Cell crowding due to an increased surface area in 3D could lead to lower layers of cells exposed to nanotopography, while the cells growing on top of other cells are devoid of nanotopographical cues that are driving osteogenesis. Therefore, parameters such as the choice of factors, cell seeding density, etc., can influence the extent of differentiation. However, different studies have independently confirmed that nanotopography alone can induce osteogenesis in hMSCs.78–80 As the focus of the current study is to understand the differential gene expression under OF and 3D conditions and the modulation of miRNAs that control the translation of the expressed genes, we analysed samples at day 14 when the differentiation in these conditions was comparable.
Herein, we performed sequencing of the small RNA and the transcripts to understand their regulation under these conditions. Interestingly, the numbers of differentially expressed transcripts were much lower in 3D compared to those induced by growth factors (Fig. 1). Major signaling factors were differentially regulated in these conditions, suggesting an alternate set of genes transcribed under these conditions. We then looked at the miRNA profiles and identified several miRNAs that were previously implicated in osteogenesis to be differentially regulated in these conditions in a manner to promote osteogenesis (Table 2). We integrated the miRNAs and their predicted targets, from our transcript data, and made networks to understand the miRNA–mRNA interactions under these two conditions. We first sampled genes that are well-studied and known to be critical for osteogenesis and mapped their miRNAs under these two conditions (Fig. 4). We observed that distinct sets of miRNAs that were expressed at different levels were observed in these two conditions. Then, following an unbiased approach, we sampled the entire sets of miRNAs that had distinct regulation in each of these conditions that could target the same genes regulated to the same extent in these conditions (Fig. 5). Our analyses provided two key insights. Firstly, a core set of miRNAs might be operative for osteogenesis that does not depend on the stimulus. Secondly, a stimulus-specific set of miRNAs can be induced, which could either reinforce the core pathways or might provide feedback to them. These stimuli-specific miRNAs might also assist to modulate the translation of osteogenic genes without having to depend solely on transcriptional control, which is a slow process. This might also explain the rapid phenotypic changes observed in 3D models, faster than ones achieved via biochemical signaling, such as the rapid development of lung epithelium under 3D perfusion conditions.81
Identifying miRNAs that majorly control translation in 3D can open newer avenues for research in improving cellular differentiation and identifying potential alternatives for expensive growth factors.82 For example, Mariner et al. transfected hMSCs with miRNAs that were shown to improve alkaline phosphatase activity in a screening assay.83 The cells showed a similar improvement of osteogenesis in 2D and 3D substrates upon transfection with the miRNAs. Although the study exhibited the applications of miRNAs in improving osteogenesis, it did not consider alternate miRNAs operating in 3D, which could be highly effective in osteogenic induction.
This study is a primer to understanding gene regulatory networks when cells are subjected to nanotopographical stimuli. While detailed experimentation is required to prove the specific miRNA–mRNA network and to identify the critical nodes, it nevertheless shows possibilities of different subsets of miRNAs operating to produce the same stimuli and provides avenues for designing and targeting these networks for enhanced differentiation and therapies.
5. Conclusion
We profiled miRNA and transcripts in hMSCs undergoing osteogenic differentiation under the influence of growth factors and via matrix-driven architectural cues, which have previously shown to induce similar levels of osteogenic differentiation. The data from the differential gene and miRNA profiling and integration showed that certain sets of genes and miRNAs were regulated to the same extent under these two conditions, depicting that a core of miRNA–mRNA networks is operative during osteogenesis. We also found a distinct set of genes and miRNAs under either of the conditions, suggesting that other stimuli-specific miRNA–mRNA networks might be operative under these conditions which might be supplementary or provide feedback to the core networks. This study provides insights into a distinct set of miRNAs that can control gene expression in 3D and open avenues for designing targets to improve differentiation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Support from the Department of Science and Technology (DST), Government of India (DST/NM/NB/2018/119(G)) is gratefully acknowledged.References
- L. G. Griffith and M. A. Swartz, Capturing complex 3D tissue physiology in vitro, Nat. Rev. Mol. Cell Biol., 2006, 7(3), 211–224 CrossRef CAS PubMed.
- K.-H. Nam, A. S. T. Smith, S. Lone, S. Kwon and D.-H. Kim, Biomimetic 3D tissue models for advanced high-throughput drug screening, J. Lab. Autom., 2015, 20(3), 201–215 CrossRef CAS PubMed.
- M. Sacchi, R. Bansal and J. Rouwkema, Bioengineered 3D models to recapitulate tissue fibrosis, Trends Biotechnol., 2020, 38(6), 623–636 CrossRef CAS PubMed.
- M. J. Bissell, Goodbye flat biology – time for the 3rd and the 4th dimensions, J. Cell Sci., 2017, 130(1), 3–5 CrossRef CAS PubMed.
- K. M. Yamada and E. Cukierman, Modeling tissue morphogenesis and cancer in 3D, Cell, 2007, 130(4), 601–610 CrossRef CAS PubMed.
- J. H. Kang, J. M. Gimble and D. L. Kaplan, In vitro 3D model for human vascularized adipose tissue, Tissue Eng., Part A, 2009, 15(8), 2227–2236 CrossRef CAS PubMed.
- M. Schindler, A. Nur-E-Kamal, I. Ahmed, J. Kamal, H.-Y. Liu, N. Amor, A. S. Ponery, D. P. Crockett, T. H. Grafe and H. Y. Chung, Living in three dimensions: 3D nanostructured environments for cell culture and regenerative medicine, Cell Biochem. Biophys., 2006, 45, 215–227 CrossRef CAS PubMed.
- I. O. Smith, X. H. Liu, L. A. Smith and P. X. Ma, Nanostructured polymer scaffolds for tissue engineering and regenerative medicine, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1(2), 226–236 CAS.
- C. Xinaris, V. Brizi and G. Remuzzi, Organoid models and applications in biomedical research, Nephron, 2015, 130(3), 191–199 CrossRef PubMed.
- T. G. Kim, H. Shin and D. W. Lim, Biomimetic scaffolds for tissue engineering, Adv. Funct. Mater., 2012, 22(12), 2446–2468 CrossRef CAS.
- G. Kumar, C. K. Tison, K. Chatterjee, P. S. Pine, J. H. McDaniel, M. L. Salit, M. F. Young and C. G. Simon Jr., The determination of stem cell fate by 3D scaffold structures through the control of cell shape, Biomaterials, 2011, 32(35), 9188–9196 CrossRef CAS.
- T. M. Koch, S. Munster, N. Bonakdar, J. P. Butler and B. Fabry, 3D Traction forces in cancer cell invasion, PLoS One, 2012, 7(3), e33476 CrossRef CAS PubMed.
- M. Larsen, V. V. Artym, J. A. Green and K. M. Yamada, The matrix reorganized: extracellular matrix remodeling and integrin signaling, Curr. Opin. Cell Biol., 2006, 18(5), 463–471 CrossRef CAS PubMed.
- D. O. Velez, B. Tsui, T. Goshia, C. L. Chute, A. Han, H. Carter and S. I. Fraley, 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry, Nat. Commun., 2017, 8(1), 1651 CrossRef CAS PubMed.
- B. Weigelt, A. T. Lo, C. C. Park, J. W. Gray and M. J. Bissell, HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment, Breast Cancer Res. Treat., 2010, 122(1), 35–43 CrossRef CAS PubMed.
- K. Chatterjee, L. Sun, L. C. Chow, M. F. Young and C. G. Simon, Combinatorial screening of osteoblast response to 3D calcium phosphate/poly(ε-caprolactone) scaffolds using gradients and arrays, Biomaterials, 2011, 32(5), 1361–1369 CrossRef CAS.
- G. M. Balachander, S. A. Balaji, A. Rangarajan and K. Chatterjee, Enhanced Metastatic Potential in a 3D Tissue Scaffold toward a Comprehensive in Vitro Model for Breast Cancer Metastasis, ACS Appl. Mater. Interfaces, 2015, 7(50), 27810–27822 CrossRef CAS.
- B. A. Baker, P. S. Pine, K. Chatterjee, G. Kumar, N. J. Lin, J. H. McDaniel, M. L. Salit and C. G. Simon, Jr., Ontology analysis of global gene expression differences of human bone marrow stromal cells cultured on 3D scaffolds or 2D films, Biomaterials, 2014, 35(25), 6716–6726 CrossRef CAS.
- G. M. Balachander, B. Rajashekar, P. M. Sarashetti, A. Rangarajan and K. Chatterjee, MiRNomics Reveals Breast Cancer Cells Cultured on 3D Scaffolds Better Mimic Tumors in Vivo than Conventional 2D Culture, ACS Biomater. Sci. Eng., 2018, 4(1), 116–127 CrossRef CAS PubMed.
- J. Winter, S. Jung, S. Keller, R. I. Gregory and S. Diederichs, Many roads to maturity: microRNA biogenesis pathways and their regulation, Nat. Cell Biol., 2009, 11(3), 228–234 CrossRef CAS PubMed.
- J. Hayes, P. P. Peruzzi and S. Lawler, MicroRNAs in cancer: Biomarkers, functions and therapy, Trends Mol. Med., 2014, 20(8), 460–469 CrossRef CAS PubMed.
- M. S. Ebert and P. A. Sharp, Roles for MicroRNAs in Conferring Robustness to Biological Processes, Cell, 2012, 149(3), 515–524 CrossRef CAS PubMed.
- S. R. K. Meka, L. A. Chacko, A. Ravi, K. Chatterjee and V. Ananthanarayanan, Role of Microtubules in Osteogenic Differentiation of Mesenchymal Stem Cells on 3D Nanofibrous Scaffolds, ACS Biomater. Sci. Eng., 2017, 3(4), 551–559 CrossRef CAS PubMed.
- FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- C. Trapnell, L. Pachter and S. L. Salzberg, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics, 2009, 25(9), 1105–1111 CrossRef CAS PubMed.
- C. Trapnell, B. A. Williams, G. Pertea, A. Mortazavi, G. Kwan, M. J. van Baren, S. L. Salzberg, B. J. Wold and L. Pachter, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol., 2010, 28(5), 511–515 CrossRef CAS.
- H. Mi and P. Thomas, PANTHER pathway: an ontology-based pathway database coupled with data analysis tools, Methods Mol. Biol., 2009, 563, 123–140 CrossRef CAS.
- M. B. Stocks, S. Moxon, D. Mapleson, H. C. Woolfenden, I. Mohorianu, L. Folkes, F. Schwach, T. Dalmay and V. Moulton, The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets, Bioinformatics, 2012, 28(15), 2059–2061 CrossRef CAS PubMed.
- B. Langmead, C. Trapnell, M. Pop and S. L. Salzberg, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 2009, 10(3), R25 CrossRef PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, Basic local alignment search tool, J. Mol. Biol., 1990, 215(3), 403–410 CrossRef CAS PubMed.
- L. Qibin and W. Jiang, MIREAP: microRNA discovery by deep sequencing, 2008 Search PubMed.
- S. Anders and W. Huber, Differential expression analysis for sequence count data, Genome Biol., 2010, 11(10), R106 CrossRef CAS.
- A. J. Enright, B. John, U. Gaul, T. Tuschl, C. Sander and D. S. Marks, MicroRNA targets in Drosophila, Genome Biol., 2003, 5(1), R1 CrossRef PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, 13(11), 2498–2504 CrossRef CAS PubMed.
- K. M. Kennedy, A. Bhaw-Luximon and D. Jhurry, Cell-matrix mechanical interaction in electrospun polymeric scaffolds for tissue engineering: implications for scaffold design and performance, Acta Biomater., 2016, 50, 41–55 CrossRef PubMed.
- O. Adachi, H. Sugii, T. Itoyama, S. Fujino, H. Kaneko, A. Tomokiyo, S. Hamano, D. Hasegawa, J. Obata, S. Yoshida, M. Kadowaki, R. Sugiura, M. S. Albougha and H. Maeda, Decorin Promotes Osteoblastic Differentiation of Human Periodontal Ligament Stem Cells, Molecules, 2022, 27(23), 8224 CrossRef CAS PubMed.
- B. Zhu, F. Xue, G. Li and C. Zhang, CRYAB promotes osteogenic differentiation of human bone marrow stem cells via stabilizing beta-catenin and promoting the Wnt signalling, Cell Proliferation, 2020, 53(1), e12709 CrossRef PubMed.
- K. S. Tsai, S. Y. Kao, C. Y. Wang, Y. J. Wang, J. P. Wang and S. C. Hung, Type I collagen promotes proliferation and osteogenesis of human mesenchymal stem cells via activation of ERK and Akt pathways, J. Biomed. Mater. Res., Part A, 2010, 94(3), 673–682 CrossRef PubMed.
- K. Ishida, C. Acharya, B. A. Christiansen, J. H. Yik, P. E. DiCesare and D. R. Haudenschild, Cartilage oligomeric matrix protein enhances osteogenesis by directly binding and activating bone morphogenetic protein-2, Bone, 2013, 55(1), 23–35 CrossRef CAS PubMed.
- O. Mizrahi, D. Sheyn, W. Tawackoli, I. Kallai, A. Oh, S. Su, X. Da, P. Zarrini, G. Cook-Wiens, D. Gazit and Z. Gazit, BMP-6 is more efficient in bone formation than BMP-2 when overexpressed in mesenchymal stem cells, Gene Ther., 2013, 20(4), 370–377 CrossRef CAS PubMed.
- R. Gruber, W. Graninger, K. Bobacz, G. Watzek and L. Erlacher, BMP-6-induced osteogenic differentiation of mesenchymal cell lines is not modulated by sex steroids and resveratrol, Cytokine, 2003, 23(4–5), 133–137 CrossRef CAS PubMed.
- F. Tian, Y. Wang and D. D. Bikle, IGF-1 signaling mediated cell-specific skeletal mechano-transduction, J. Orthop. Res., 2018, 36(2), 576–583 CrossRef CAS PubMed.
- J. Zhang, Z. Ma, K. Yan, Y. Wang, Y. Yang and X. Wu, Matrix Gla Protein Promotes the Bone Formation by Up-Regulating Wnt/beta-Catenin Signaling Pathway, Front. Endocrinol., 2019, 10, 891 CrossRef PubMed.
- B. Peruzzi, A. Cappariello, A. Del Fattore, N. Rucci, F. De Benedetti and A. Teti, c-Src and IL-6 inhibit osteoblast differentiation and integrate IGFBP5 signalling, Nat. Commun., 2012, 3, 630 CrossRef PubMed.
- Y. Chen, H. C. Whetstone, A. Youn, P. Nadesan, E. C. Chow, A. C. Lin and B. A. Alman, Beta-catenin signaling pathway is crucial for bone morphogenetic protein 2 to induce new bone formation, J. Biol. Chem., 2007, 282(1), 526–533 CrossRef CAS PubMed.
- J. H. Kim, X. Liu, J. Wang, X. Chen, H. Zhang, S. H. Kim, J. Cui, R. Li, W. Zhang, Y. Kong, J. Zhang, W. Shui, J. Lamplot, M. R. Rogers, C. Zhao, N. Wang, P. Rajan, J. Tomal, J. Statz, N. Wu, H. H. Luu, R. C. Haydon and T. C. He, Wnt signaling in bone formation and its therapeutic potential for bone diseases, Ther. Adv. Musculoskeletal Dis., 2013, 5(1), 13–31 CrossRef CAS PubMed.
- P. J. Marie, E. Haÿ and Z. Saidak, Integrin and cadherin signaling in bone: role and potential therapeutic targets, Trends Endocrinol. Metab., 2014, 25(11), 567–575 CrossRef CAS.
- A. Grosso, M. G. Burger, A. Lunger, D. J. Schaefer, A. Banfi and N. Di Maggio, It Takes Two to Tango: Coupling of Angiogenesis and Osteogenesis for Bone Regeneration, Front. Bioeng. Biotechnol., 2017, 5, 68 CrossRef PubMed.
- P. J. Marie and E. Hay, Cadherins and Wnt signalling: a functional link controlling bone formation, BoneKEy Rep., 2013, 2, 330 Search PubMed.
- F. Khodabandehloo, S. Taleahmad, R. Aflatoonian, F. Rajaei, Z. Zandieh, M. Nassiri-Asl and M. B. Eslaminejad, Microarray analysis identification of key pathways and interaction network of differential gene expressions during osteogenic differentiation, Hum. Genomics, 2020, 14(1), 43 CrossRef CAS PubMed.
- A. W. James, Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation, Scientifica, 2013, 2013, 1–17 CrossRef PubMed.
- S. Thomas and B. G. Jaganathan, Signaling network regulating osteogenesis in mesenchymal stem cells, J. Cell Commun. Signaling, 2022, 16(1), 47–61 CrossRef CAS PubMed.
- Y. Lin, H. Dai, G. Yu, C. Song, J. Liu and J. Xu, Inhibiting KCNMA1-AS1 promotes osteogenic differentiation of HBMSCs via miR-1303/cochlin axis, J. Orthop. Surg. Res., 2023, 18(1), 73 CrossRef PubMed.
- Y. Wu, H. Ai, Y. Zou, Q. Yang, C. Dou and J. Xu, Osteoclast-derived extracellular miR-106a-5p promotes osteogenic differentiation and facilitates bone defect healing, Cell. Signalling, 2023, 102, 110549 CrossRef CAS PubMed.
- Y. Li, C. Feng, M. Gao, M. Jin, T. Liu, Y. Yuan, G. Yan, R. Gong, Y. Sun, M. He, Y. Fu, L. Zhang, Q. Huang, F. Ding, W. Ma, Z. Bi, C. Xu, N. Sukhareva, D. Bamba, R. Reiters, F. Yang, B. Cai and L. Yang, MicroRNA-92b-5p modulates melatonin-mediated osteogenic differentiation of bone marrow mesenchymal stem cells by targeting ICAM-1, J. Cell. Mol. Med., 2019, 23(9), 6140–6153 CrossRef CAS PubMed.
- B. Zhou, K. Peng, G. Wang, W. Chen, P. Liu, F. Chen and Y. Kang, miR-483-3p promotes the osteogenesis of human osteoblasts by targeting Dikkopf 2 (DKK2) and the Wnt signaling pathway, Int. J. Mol. Med., 2020, 46(4), 1571–1581 CAS.
- M. J. Kuang, K. H. Zhang, J. Qiu, A. B. Wang, W. W. Che, X. M. Li, D. L. Shi and D. C. Wang, Exosomal miR-365a-5p derived from HUC-MSCs regulates osteogenesis in GIONFH through the Hippo signaling pathway, Mol. Ther. – Nucleic Acids, 2021, 23, 565–576 CrossRef CAS PubMed.
- P. Liu, Y. Zhuang, B. Zhang, H. Huang, P. Wang, H. Wang, Y. Cong, S. Qu, K. Zhang and X. Wei, miR-140-3p regulates the osteogenic differentiation ability of bone marrow mesenchymal stem cells by targeting spred2-mediated autophagy, Mol. Cell. Biochem., 2021, 476(12), 4277–4285 CrossRef CAS PubMed.
- F. Xue, J. Wu, W. Feng, T. Hao, Y. Liu and W. Wang, MicroRNA-141 inhibits the differentiation of bone marrow-derived mesenchymal stem cells in steroid-induced osteonecrosis via E2F3, Mol. Med. Rep., 2022, 26(1), 1–8 Search PubMed.
- M. Zheng, J. Tan, X. Liu, F. Jin, R. Lai and X. Wang, miR-146a-5p targets Sirt1 to regulate bone mass, Bone Rep., 2021, 14, 101013 CrossRef CAS PubMed.
- D. Liang, G. Song and Z. Zhang, miR-216a-3p inhibits osteogenic differentiation of human adipose-derived stem cells via Wnt3a in the Wnt/beta-catenin signaling pathway, Exp. Ther. Med., 2022, 23(4), 309 CrossRef CAS PubMed.
- F. Ji, L. Zhu, J. Pan, Z. Shen, Z. Yang, J. Wang, X. Bai, Y. Lin and J. Tao, hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p, Front. Cell. Dev. Biol., 2020, 8, 470 CrossRef PubMed.
- R. Xu, X. Shen, Y. Si, Y. Fu, W. Zhu, T. Xiao, Z. Fu, P. Zhang, J. Cheng and H. Jiang, MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment, Aging Cell, 2018, 17(4), e12794 CrossRef PubMed.
- W. Li, Z. Chen, C. Cai, G. Li, X. Wang and Z. Shi, MicroRNA-505 is involved in the regulation of osteogenic differentiation of MC3T3-E1 cells partially by targeting RUNX2, J. Orthop. Surg. Res., 2020, 15(1), 143 CrossRef.
- M. Bruderer, R. G. Richards, M. Alini and M. J. Stoddart, Role and regulation of RUNX2 in osteogenesis, Eur. Cells Mater., 2014, 28, 269–286 CrossRef CAS PubMed.
- N. Su, M. Jin and L. Chen, Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models, Bone Res., 2014, 2, 14003 CrossRef PubMed.
- Y. Zhang, L. Ling, D. O. A. A. Ajay, Y. M. Eio, A. J. van Wijnen, V. Nurcombe and S. M. Cool, FGFR2 accommodates osteogenic cell fate determination in human mesenchymal stem cells, Gene, 2022, 818, 146199 CrossRef CAS PubMed.
- W. Chen, P. Wang, Y. Lu, T. Jin, X. Lei, M. Liu, P. Zhuang, J. Liao, Z. Lin, B. Li, Y. Peng, G. Pan, X. Lv, H. Zhang, Z. Ou, S. Xie, X. Lin, S. Sun, S. Ferrone, B. A. Tannous, Y. Ruan, J. Li and S. Fan, Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation, Theranostics, 2019, 9(20), 5739–5754 CrossRef CAS PubMed.
- N. Hoang Dong, L. Audrey, M. N. Leopold, M. Javier, A. C. Hugues, B. Luigi, B. Gilles, M. S. Scott and R. Sophie, Osteoclast microRNA Profiling in Rheumatoid Arthritis to Capture the Erosive Factor, JBMR Plus, 2023, e10776 CrossRef CAS PubMed.
- X. Zhu, Q. Huang, J. Luo, D. Kong and Y. Zhang, Mini-review: Gene regulatory network benefits from three-dimensional chromatin conformation and structural biology, Comput. Struct. Biotechnol. J., 2023, 21, 1728–1737 CrossRef CAS PubMed.
- T. E. Chan, M. P. H. Stumpf and A. C. Babtie, Gene Regulatory Network Inference from Single-Cell Data Using Multivariate Information Measures, Cell Syst., 2017, 5(3), 251–267 CrossRef CAS PubMed .e3.
- W. W. Tang, S. Dietmann, N. Irie, H. G. Leitch, V. I. Floros, C. R. Bradshaw, J. A. Hackett, P. F. Chinnery and M. A. Surani, A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development, Cell, 2015, 161(6), 1453–1467 CrossRef CAS PubMed.
- X. Lai, O. Wolkenhauer and J. Vera, Understanding microRNA-mediated gene regulatory networks through mathematical modelling, Nucleic Acids Res., 2016, 44(13), 6019–6035 CrossRef CAS PubMed.
- X. S. Ke, C. M. Liu, D. P. Liu and C. C. Liang, MicroRNAs: key participants in gene regulatory networks, Curr. Opin. Chem. Biol., 2003, 7(4), 516–523 CrossRef CAS PubMed.
- J. B. Lian, G. S. Stein, A. J. van Wijnen, J. L. Stein, M. Q. Hassan, T. Gaur and Y. Zhang, MicroRNA control of bone formation and homeostasis, Nat. Rev. Endocrinol., 2012, 8(4), 212–227 CrossRef CAS PubMed.
- X. Han and Z. Fan, MicroRNAs Regulation in Osteogenic Differentiation of Mesenchymal Stem Cells, Front. Dent. Med., 2021, 2, 747068 CrossRef.
- J. Wang, S. Liu, J. Li, S. Zhao and Z. Yi, Roles for miRNAs in osteogenic differentiation of bone marrow mesenchymal stem cells, Stem Cell Res. Ther., 2019, 10(1), 197 CrossRef PubMed.
- G. Kumar, C. K. Tison, K. Chatterjee, P. S. Pine, J. H. McDaniel, M. L. Salit, M. F. Young and C. G. Simon, The determination of stem cell fate by 3D scaffold structures through the control of cell shape, Biomaterials, 2011, 32(35), 9188–9196 CrossRef CAS PubMed.
- G. Kumar, M. S. Waters, T. M. Farooque, M. F. Young and C. G. Simon, Jr., Freeform fabricated scaffolds with roughened struts that enhance both stem cell proliferation and differentiation by controlling cell shape, Biomaterials, 2012, 33(16), 4022–4030 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. Wilkinson and R. O. Oreffo, The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6(12), 997–1003 CrossRef CAS PubMed.
- P. Chandorkar, W. Posch, V. Zaderer, M. Blatzer, M. Steger, C. G. Ammann, U. Binder, M. Hermann, P. Hortnagl, C. Lass-Florl and D. Wilflingseder, Fast-track development of an in vitro 3D lung/immune cell model to study Aspergillus infections, Sci. Rep., 2017, 7(1), 11644 CrossRef CAS PubMed.
- N. Celik, M. H. Kim, M. Yeo, F. Kamal, D. J. Hayes and I. T. Ozbolat, miRNA induced 3D bioprinted-heterotypic osteochondral interface, Biofabrication, 2022, 14(4), 044104 CrossRef.
- P. D. Mariner, E. Johannesen and K. S. Anseth, Manipulation of miRNA activity accelerates osteogenic differentiation of hMSCs in engineered 3D scaffolds, J. Tissue Eng. Regener. Med., 2012, 6(4), 314–324 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm01597a |
| ‡ These authors contributed equally to this work. |
| § Current affiliation: Department of Orthopedic Surgery, Rush University Medical Center, Chicago, IL, USA. |
| ¶ Current affiliation: Department of Biomedical Engineering, Texas A&M University, Texas 77843, USA. |
| This journal is © The Royal Society of Chemistry 2024 |