Mesenchymal stromal cells modulate YAP by verteporfin to mimic cartilage development and construct cartilage organoids based on decellularized matrix scaffolds
Jiayi
Zhu
ab,
Wanqing
Lun
ab,
Qi
Feng
bcde,
Xiaodong
Cao
 *bcde and
Qingtao
Li
*bcde and
Qingtao
Li
 *ab
*ab
aSchool of Medicine, South China University of Technology, Guangzhou 510006, P. R. China. E-mail: mcqtli@scut.edu.cn
bNational Engineering Research Centre for Tissue Restoration and Reconstruction, Guangzhou 510006, P. R. China. E-mail: caoxd@scut.edu.cn
cSchool of Materials Science and Engineering, South China University of Technology, Guangzhou 510641, P. R. China
dKey Laboratory of Biomedical Engineering of Guangdong Province, South China University of Technology, Guangzhou 510006, P. R. China
eKey Laboratory of Biomedical Materials and Engineering of the Ministry of Education, South China University of Technology, Guangzhou 510006, P. R. China
First published on 30th June 2023
Abstract
The mechanical elasticity or stiffness of the ECM modulates YAP activity to regulate the differentiation of stem cells during the development and defect regeneration of cartilage tissue. However, the understanding of the scaffold-associated mechanobiology during the initiation of chondrogenesis and hyaline cartilaginous phenotype maintenance remains unclear. In order to elucidate such mechanisms to promote articular cartilage repair by producing more hyaline cartilage, we identify the relationship between YAP subcellular localization and variation of the cartilage structure and organization during the early postnatal explosive growth in incipient articular cartilage. Next, we prepared a decellularized cartilage scaffold with different stiffness (2–33 kPa) to investigate the effect of scaffold stiffness on the formation of hyaline cartilage by mesenchymal stem cells and the change of YAP activity. Furthermore, we simulated the decrease of cellular YAP activity during postnatal cartilage development by inhibiting YAP activity with verteporfin, and realized that the timing of drug incorporation was critical to regulate the differentiation of MSCs to hyaline chondrocytes and inhibit their hypertrophy and fibrosis. On this basis, we constructed hyaline cartilage organoids by decellularized matrix scaffolds. Collectively, the results herein demonstrate that YAP plays a critical role during in vitro chondrogenic differentiation which is tightly regulated by biochemical and mechanical regulation.
1. Introduction
The structure of articular cartilage is mostly made up of chondrocytes embedded in a thick extracellular matrix, which allows for low friction mobility and weight bearing.1 Meanwhile, the absence of vascularization and low proliferation of chondrocytes lead to articular cartilage with poor self-healing capacity.2 Despite the fact that there are several therapies for articular cartilage degeneration, they cannot repair the deficiency and eliminate symptoms. Unfortunately, fibrous cartilage and/or hypertrophic cartilage are the most frequent repair outcomes following conventional microfracture or autologous chondrocyte transplantation therapy; highly organized hyaline cartilage was very often observed.3 There are vast differences among the structural compositions of the ECM and chondrocyte phenotypes.4 It is well established that the mechanical properties of extracellular matrices control many cell functions such as cell shape, adhesion, migration, differentiation, and proliferation.5,6 The optimal mechanical conditions for in vitro cartilage tissue engineering likely depend on the desired outcome (e.g., homeostasis, maturation, or mineralization).7–9 Recent studies have discovered that the mechanical microenvironment has a significant impact on the chondrogenic differentiation behavior of BMSCs, particularly when it comes to differentiation into hyaline cartilage or fibrocartilage.10–12 However, due to the complex process of chondrogenic differentiation, the more detailed mechanism for the interaction of chondrocyte phenotypes, the maturation state and the stiffness of the scaffold is poorly understood.In recent years, several studies have been conducted to recreate the tissue engineering cartilage applying a new scaffold containing a decellularized extracellular matrix (dECM) derived from cartilage.13,14 The dECM scaffold exhibits natural adhesion to the cells and growth factors, and can provide a biochemical microenvironment conducive to cell growth.15 A new method for examining the mechanobiology of in vitro chondrogenesis is provided by scaffolds based on the dECM, which also have mechanically and chemically stable qualities while maintaining biocompatibility and biodegradability. In Drosophila, the YAP/TAZ complex was found to be a major regulator of organ growth during development and is a downstream effector of the Hippo signaling system.16 More recently, YAP/TAZ has proven to be an important mechanosignalling complex and is involved in regulating chondrogenesis, chondrocyte maturation and hypertrophy.17,18 YAP directly regulates SOX6 expression by binding to TEAD, promotes early chondrocyte proliferation, and inhibits COL10A1 expression by interacting with RUNX2 during chondrocyte maturation.5 However, in neural crest cells, the interaction of YAP with the Wnt-β-catenin pathway promotes osteogenesis and inhibits chondrogenesis.19 Additionally, another study found that the differentiation of human induced pluripotent stem cells into chondrocytes is associated with inhibiting the activity of YAP in them.20 Understanding how the ECM regulates MSCs to control chondrogenesis and preserve a stable articular cartilage phenotype will therefore contribute to both fundamental understanding and as-yet undiscovered methods of controlling hyaline/fibrocartilage regeneration.
In the current study, we analyze first the correlation between the activity of mechanical signaling factor YAP and cartilage development in rat articular chondrocytes. Then, using a decellularization technique and dECM sponge scaffolds with solid contents of 1%, 2%, and 4%, we created a decellularized matrix of porcine articular cartilage. We then characterized the scaffolds' physicochemical properties and biocompatibility, and we looked at the nuclear localization of the YAP transcriptional regulator in MSCs. Tests were performed on the expression levels of the genes RUNX2, PPAR, SOX9, COL2A1, ACAN, COL1A1, COL3A1, COL10A1, MMP13, and YAP at 7 and 14 days, respectively. In order to control YAP activity and the chondrogenic differentiation, which was confirmed by RT-PCR and histological staining, verteporfin, a YAP-specific inhibitor, was introduced. The mechanobiological parameters that can influence chondrocytes and their matrix formation to enhance articular cartilage regeneration are confirmed by this investigation.
2. Materials and methods
2.1 Preparation of decellularized cartilage matrix
Fresh porcine articular cartilage was from the knee joints of 6-month-old pigs obtained from a local abattoir 4–8 h after slaughter. The decellularization was carried out according to a modified method from the literature. Briefly, cartilage was cut into small pieces, washed with deionization, and freeze-thawed three times at −80 °C versus room temperature in hypotonic buffer (10 mM Tris-HCl, pH 8.0). The tissue was rapidly freeze-thawed 3 times in liquid nitrogen, and then incubated in hypotonic buffer at 45 °C for 24 hours, and then stirred with ionic detergent (containing 0.1% (w/v) SDS, peptidase 10 KIU mL−1, 0.1% (w/v) EDTA in hypotonic buffer) at 45 °C for 24 hours. The samples were washed twice in PBS containing peptidase 10 KIU mL−1 for 30 minutes, and then washed at 45 °C for 24 hours. The samples were placed in nuclease solution (50 mM Tris-HCl solution, pH 7.5, containing 10 mM MgCl2 and 50 μg mL−1 bovine serum Albumin), then incubated at 37 °C for 3 hours. The cartilage tissue was washed with 1% Triton X-100 for 24 hours, with hypotonic buffer for more than 20 hours, and then washed with deionized water for 3 days. The samples were lyophilized and stored at −20 °C.The decellularized tissue was broken up in cold water with a stirrer to produce a decellularized matrix slurry and lyophilized. The flakes of decellularized cartilage tissue were dissolved in a dilute acid solution containing 1 mg mL−1 pepsin at 1% to obtain the decellularized matrix solution. The insoluble material in the decellularized matrix solution was removed by centrifugation at 3000 rpm for 20 minutes. The pH of the decellularized matrix solution was adjusted to 6–7 with a 1 M solution of NaOH, and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes to allow the decellularized matrix to settle, and the supernatant was discarded. The washing/centrifugation steps were repeated twice to remove the pepsin. Finally, the decellularized extracellular matrix (dECM) powder product was obtained by lyophilization and stored at −20 °C.
000 rpm for 30 minutes to allow the decellularized matrix to settle, and the supernatant was discarded. The washing/centrifugation steps were repeated twice to remove the pepsin. Finally, the decellularized extracellular matrix (dECM) powder product was obtained by lyophilization and stored at −20 °C.
2.2 Residual DNA and collagen quantification of dECM
DNA in natural cartilage and decellularized cartilage matrix was extracted by using a Genomic DNA Purification Kit (K0512, Thermo). And the DNA concentration was quantified using a NanoDrop ND-1000 ultramicro-UV spectrophotometer. The collagen content in natural cartilage and decellularized cartilage matrix was quantified using a Hydroxyproline Assay Kit (BC0255, Solarbio) according to the manufacturers protocol.2.3 Preparation of the dECM scaffolds through EDC/NHS cross-linking
The 1%, 2%, and 4% dECM solutions were prepared by dissolving a given amount of dECM powder in water. The dECM solution was loaded into PDMS molds and lyophilized. The lyophilized sponges were immersed in 95% ethanol solution containing 50 mM 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride (EDC) and 20 mM N-hydroxysuccinimide (NHS) for 12 hours. After cross-linking treatment, the dECM scaffolds were washed with distilled water and lyophilized.2.4 Characterization of dECM scaffolds
The mechanical properties of equilibrium swollen dECM scaffolds were characterized by uniaxial compression measurements on a dynamic thermomechanical analyzer (DMA Q800). The dimensions of the sample were 10 mm × 10 mm × 10 mm. The following tests were performed on each sample: ramp strain −50.00% to −100.00%, 1.222 μm N−1. The apparent morphology of cartilage tissue after decellularization was observed by a scanning electron microscope (Q25, FEI; Merlin, ZEISS), as well as the characteristic structure of collagen after solubilization treatment of the decellularized matrix, and the porous structure of the dECM scaffolds. These samples were placed in conductive adhesives and sprayed gold at 30 seconds in the process of SEM sample preparation.2.5 Biocompatibility study dECM scaffolds
Rat BMSCs (RASMX-01001, OriCell) were cultured with low-sugar DMEM medium (Gibco) containing 10% FBS (EVERY GREEN), 1% penicillin/streptomycin (Gibco) at 37 °C under a humidified atmosphere of 5% CO2. For cell proliferation assays, BMSCs (5 × 104) were resuspended in 100 μL of complete medium and seed onto dECM scaffolds (disk-shaped, 4 mm diameter and 2 mm thick). The rate of cell proliferation was detected using a CCK8 assay kit (Beyotime, Shanghai, China). After 1, 3 and 7 days of co-culture, CCK8 reagents (10 μL) were added, and the absorbance at 450![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm was measured. The Calcein/PI Live/Dead Viability/Cytotoxicity Assay Kit (Beyotime, China) was applied to stain live and dead cells. After 1 day of co-culture, the status of the cells was observed by measuring the fluorescence signal using laser scanning confocal microscopy (Leica TCS SP8, Leica Microsystems, Wetzlar, Germany).
nm was measured. The Calcein/PI Live/Dead Viability/Cytotoxicity Assay Kit (Beyotime, China) was applied to stain live and dead cells. After 1 day of co-culture, the status of the cells was observed by measuring the fluorescence signal using laser scanning confocal microscopy (Leica TCS SP8, Leica Microsystems, Wetzlar, Germany).
2.6 In vitro chondrogenic differentiation test
3 × 105 BMSCs were co-cultured with different dECM scaffolds in low sugar DMEM medium with 10% FBS, 1% penicillin/streptomycin for 48 hours, and then inducing differentiation by chondrogenic induction medium (high-sugar DMEM (Gibco), 100 nM dexamethasone (Sigma), 50 μg mL−1 ascorbic acid (Sigma), 1 mM sodium pyruvate (Sigma), 50 μg mL−1L-proline (Sigma), 1% ITS (Sigma), 10 ng mL−1 TGF-β3 (PeproTech), 1% FBS). The medium was changed on alternate days. The samples were harvested at day 7 and 14, and fixed in 4% paraformaldehyde or RNA extraction.For verteporfin (MCE, CL318952) treatment, the final concentration of verteporfin was 5 μM in chondrogenic induction medium. The chondrogenic induction experiments were divided into three groups: Control group (without verteporfin), D0 + verteporfin group (verteporfin was added from the beginning of induced differentiation, and D7 + verteporfin group (verteporfin was added after 7 days of induced differentiation). The medium was changed on alternate days. The samples were harvested at day 14 and 28, and fixed in 4% paraformaldehyde or RNA extraction.
2.7 Immunofluorescence staining
The experiments involving animals were performed according to the stipulated rules for experimental usage of laboratory animals (the regulation of the administration of affairs concerning experimental animals of P. R. China). All animal experiments were approved by the Committee of Laboratory Animal Experimentation of South China University of Technology. The knee joints were prepared from 5–28 day old male Sprague-Dawley rats, fixed by 4% paraformaldehyde for 1 day and then decalcified in 10% EDTA for one or four weeks.Frozen sections (20 μm) of rat knee joints or dECM scaffolds with rat BMSCs were obtained by cryosection (Leica). Slides were immersed in PBS to remove any residual OCT. Cells were permeabilized and washed with PBS 3 times for 5–10 minutes each time; 0.1%Triton X-100 for 15 minutes, washed with PBS 3 times for 5–10 minutes each time; and blocked with 3% BSA/PBS solution at room temperature for 1 hour. The primary antibody against YAP (Santa Cruz Biotechnology sc-101199, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) was incubated overnight at 4 °C. Alexa 488 Goat-anti-mouse (Invitrogen A11001, 1
200 dilution) was incubated overnight at 4 °C. Alexa 488 Goat-anti-mouse (Invitrogen A11001, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dilution) was incubated for 2 hours at room temperature, and then washed three times with PBS (10 min each). The actin cytoskeletion was stained with rhodamine phalloidin (Bryotime C2207S, 1
400 dilution) was incubated for 2 hours at room temperature, and then washed three times with PBS (10 min each). The actin cytoskeletion was stained with rhodamine phalloidin (Bryotime C2207S, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) for 30 minutes at room temperature. After washing three times with PBS, the nuclei were stained with DAPI for 5 minutes. Confocal images were captured using a Leica TCS-SP8 laser scanning confocal microscope. According to previous studies, the localization of YAP can be divided into 3 types: preferential localization of YAP in the nucleus (N, almost overlapping with the nucleus); uniform or homogeneous distribution of YAP in the nucleus and cytoplasm (N/C); and preferential cytoplasmic YAP distribution (C), which is mainly localized in the cytoplasm, with little YAP seen in the nucleus. The data are expressed as percentage values. Values are mean ± standard deviation (n = 3). *p < 0.05, **p < 0.01.
200 dilution) for 30 minutes at room temperature. After washing three times with PBS, the nuclei were stained with DAPI for 5 minutes. Confocal images were captured using a Leica TCS-SP8 laser scanning confocal microscope. According to previous studies, the localization of YAP can be divided into 3 types: preferential localization of YAP in the nucleus (N, almost overlapping with the nucleus); uniform or homogeneous distribution of YAP in the nucleus and cytoplasm (N/C); and preferential cytoplasmic YAP distribution (C), which is mainly localized in the cytoplasm, with little YAP seen in the nucleus. The data are expressed as percentage values. Values are mean ± standard deviation (n = 3). *p < 0.05, **p < 0.01.
2.8 Quantitative real-time PCR
BMSCs in rats were cultured under specific conditions for a certain period of time. The expression level of their mRNA was detected by qRT-PCR. Total RNA was isolated using the Hipure total RNA kit (Magen, Shanghai, China) according to the manufacturer's instructions. qRT-PCR was performed using the Brilliant SYBR Green QPCR Master Mix (TakaRa) with PCR Amplifier (Life technologies, AB qPCR Q6 Flex). The relative expression levels of the target gene were calculated following the 2−ΔΔCT method (Table 1).| Target genes | Forward primer 5′ → 3′ | Reverse primer 5′ → 3′ |
|---|---|---|
| GAPDH | GGCTGCCTTCTCTTGTGACA | TTGAACTTGCCGTGGGTAGA |
| RUNX2 | GGGAACCAAGAAGGCACAGA | GGATGAGGAATGCGCCCTAA |
| PPARγ | GCCGAGTCTGTGGGGATAAA | CCCAAACCTGATGGCATTGTG |
| SOX9 | AGCACTCCGGGCAATCT | CTGCTCAGCTCACCGATGTC |
| COL2A1 | GCCAGGATGCCCGAAAATTA | TCACCTCTGGGTCCTTGTTC |
| ACAN | GAAATCCAGAACCTTCGCTCC | AAGTCCAGTGTGTAGCGTGT |
| COL1A1 | TGACTGGAAGAGCGGAGAGT | GATAGCGACATCGGCAGGAT |
| COL3A1 | AAAGGATGGGCCGAGAGGTC | GGGCCTCCTTCACCTTTCTC |
| COL10A1 | TGCTAGTGTCCTTGACGCTG | TCCTCTTACTGAAATCTCTTTACCC |
| MMP13 | CAAGCAGCTCCAAAGGCTAC | TGGCTTTTGCCAGTGTAGGT |
| YAP | TGACCCTCGTTTTGCCATGA | TCCGTATTGCCTGCCGAAAT |
2.9 Histology and immunohistochemistry
Tissue sections of rat knee joints and dECM scaffolds with rat BMSCs were prepared by dehydration with gradient alcohol, paraffin embedding, and pathological section. Hematoxylin and eosin (HE) staining, safranin-O staining, alcian blue staining and immunohistochemical staining (COL1/COL2/COL10) were conducted according to routine protocols. Digital images were acquired using a digital scanner (3DHISTECH, P250 FLASH).2.10 Statistical analysis
Quantitative data are expressed as mean ± standard deviation and statistical analysis was performed using an unpaired two-tailed t-test. Differences regarding gene expression are given as the mean of 95% confidence intervals and were assessed using two-way ANOVA and Tukey's HSD post-processing test. All analyses were performed using SPSS 17.0 software. p < 0.05 was considered statistically significant.3. Results
3.1 YAP expression in articular chondrocytes gradually reduces during rat postnatal development
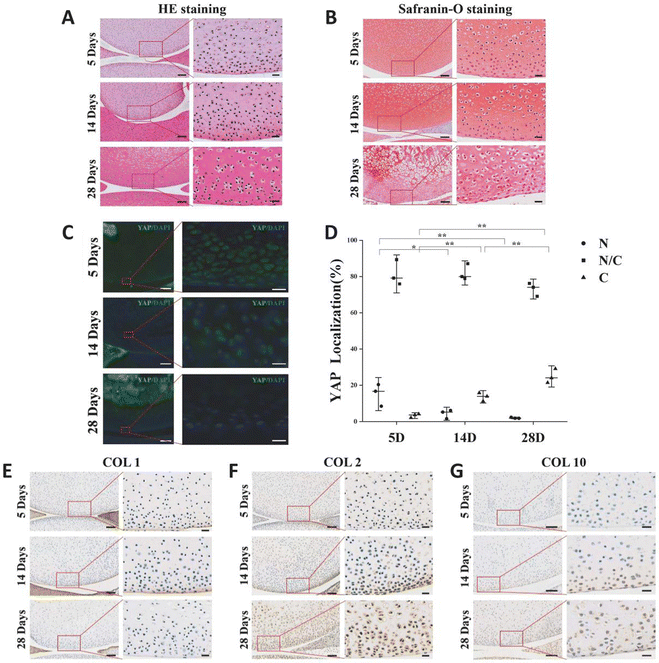
The variation of extracellular matrix and the pattern of YAP expression in articular cartilage during rat postnatal development will be explored. Rats that were 5 days old, 14 days old, and 28 days old had their knee joints inspected. The chondrocytes in the surface layer of cartilage grow and mature as the extracellular matrix of rat articular cartilage accumulates with age. In comparison to cells located deeper in the matrix, chondrocytes in the superficial zone are flatter, smaller, and typically denser (Fig. 1A). The secondary ossification centers occurred, the chondrocyte count decreased overall, and the cartilage layer gradually got thinner (Fig. 1A and B). The number of YAP protein-expressing cells in the chondrocytes of the superficial layer of cartilage gradually decreased during the early stages of rat articular cartilage development, while the number of cells expressing the protein gradually increased (Fig. 1C and D). The results of 5-day-old sections showed that the number of cells expressing YAP protein only in the nucleus accounted for 15% of the total, and this percentage decreased to 4% at 14-day-old, and this value was only 2% at 28-day-old; while the percentage of cells expressing YAP protein only in the cytoplasm increased from the initial 3% to 14%, and reached 35% at 28-day-old.COL1 was present in the articular cartilage's outermost layer, but as the rats developed, its expression gradually decreased (Fig. 1E). As the rats mature and expand, more COL2 accumulates in the articular cartilage matrix as hyaline cartilage (Fig. 1F). In the rat articular cartilage's superficial layer, we similarly saw a rise in COL10 as the cartilage matured (Fig. 1G). These findings imply that the YAP-related signaling pathway is a regulatory target for cartilage growth and development and is connected to cartilage development.
3.2 The dECM scaffold characterization
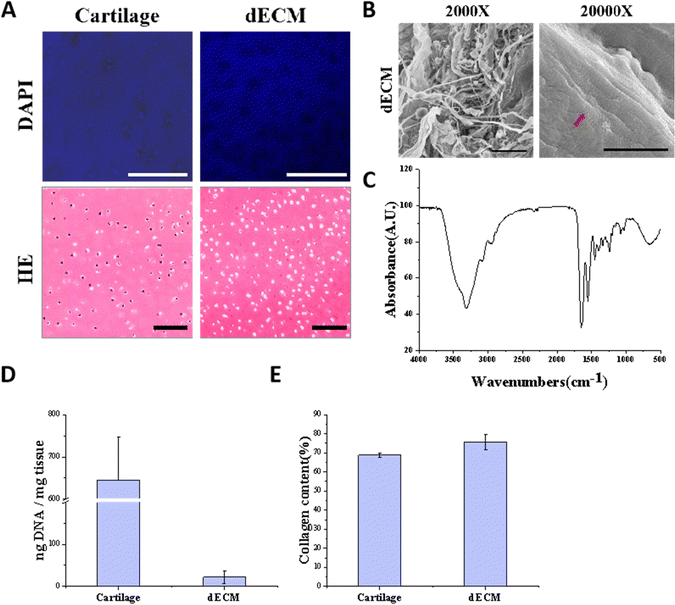
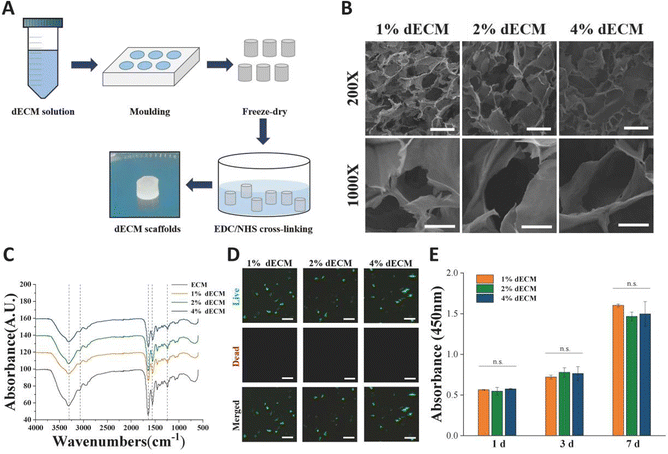
To mimic the chondrogenic microenvironment, we prepared a soluble porcine decellularized articular cartilage matrix. HE staining and DAPI staining demonstrated that the DNA residue in decellularized articular cartilage was basically free compared with normal articular cartilage tissue (Fig. 2A). Quantitative analysis of residual DNA within dECM was carried out and the residual DNA was less than 50 ng mg−1 (Fig. 2D). Moreover, collagen is one of crucial ECM components. By using SEM, the apparent morphology of the decellularized matrix could be seen, and the collagen fibers still had their distinctive threaded structure (Fig. 2B). The characteristic peaks of collagen were detectable by FTIR (Fig. 2C). The hydroxyproline kit demonstrated a good retention of the collagen component in the articular cartilage tissue after decellularization (Fig. 2E) indicating that the preparation and dissolution process of the decellularized matrix does not disrupt the secondary structure of the collagen fraction in the cartilage matrix.The scaffolds in each group were found to have a porous structure by using SEM (Fig. 3B), and there was no significant difference in pore size between the scaffolds in each group, pore size are around 100 to 200 μm, and the porous structure was favorable for cell growth. Furthermore, the secondary structure of the collagen fraction in each group of scaffolds remained unaltered after cross-linking by EDC/NHS as detected by FTIR (Fig. 3C). We cultured rat BMSCs on different groups of decellularized matrix sponge scaffolds. We demonstrated that the scaffold materials were not significantly toxic to the cells using a live-dead staining assay (Fig. 3D), and we demonstrated that there was no statistical difference in the effect of different groups of materials on cell proliferation using a CCK8 kit assay (Fig. 3E), which indicated that each group of decellularized matrix sponge scaffolds possessed good biocompatibility.
3.3 YAP localization of BMSCs in dECM scaffolds
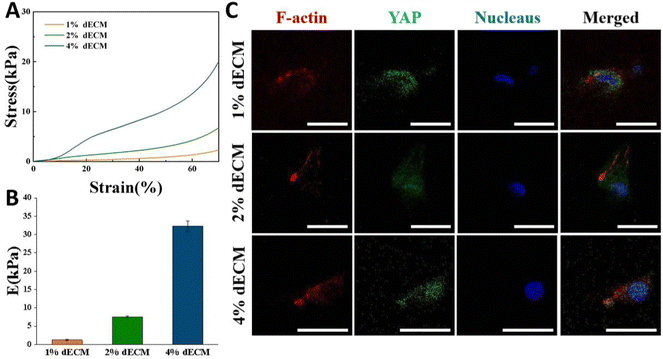
By varying the dECM density, the initial elastic modulus of dECM scaffolds was tuned as follows, a soft matrix in which E = ∼2 kPa, and a stiff matrix in which E = ∼35 kPa (Fig. 4A and B). In all groups, scaffold stiffness is comparable to the stiffness of mature cartilage.21BMSCs seeded into the dECM scaffold for 24 h so that they could be exposed to a three-dimensional environment. On scaffolds with low stiffness, YAP in cells was expressed more in the cytoplasm, i.e., in a non-activated state. As the scaffold stiffness increased, YAP in cells was gradually activated, and the greater the stiffness, the more it tended to be expressed in the nucleus (Fig. 4C). The distinctions in the cellular sublocalization of these YAP proteins imply that the three scaffold groups we prepared are capable of offering distinct mechanical microenvironments for cells. This variation can influence intracellular mechanical signal transduction, which in turn affects cell proliferation and differentiation or phenotypic maintenance.
3.4 Changes of YAP localization and differentiation of BMSCs
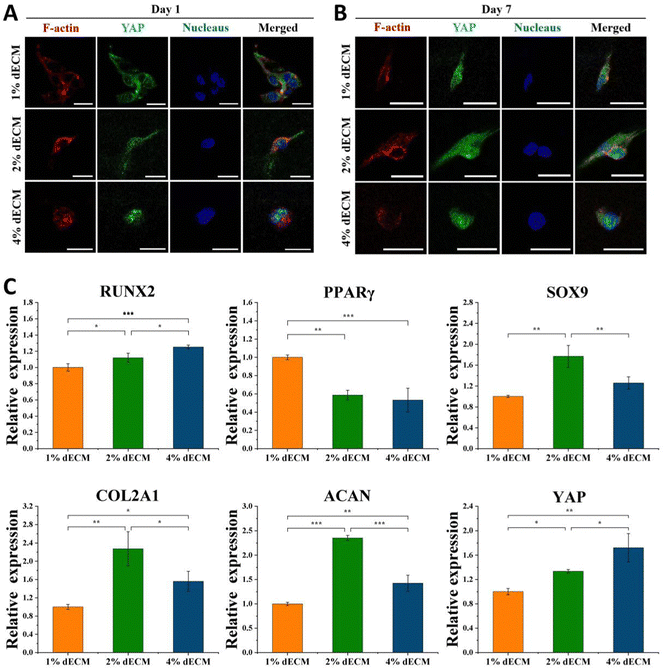
To further verify the effect of mechanical microenvironmental differences on cell proliferation and differentiation or phenotype maintenance during BMSC cell differentiation, we induced differentiation of BMSC cells co-cultured with different groups of decellularized matrix scaffolds excluding exogenous growth factors. The experimental results showed that the greater the scaffold stiffness, the more obvious the nuclear localization of YAP protein in the first day of induced differentiation without exogenous TGF (Fig. 5A). In contrast, as induction of differentiation proceeded, at 7 days of induction of differentiation, an increase in YAP expression level was seen in all groups of cells (Fig. 5B). Similarly, the timeframe of YAP/TAZ nuclear localization may be significant, increasing YAP/TAZ nuclear localization. After 7 days of forced differentiation, the relative expression of the YAP gene, as well as the osteogenesis-related gene RUNX2, was higher in cells from the stiffer scaffold group, while the relative expression of the lipogenesis-related gene PPAR was lower (Fig. 5C). Among the groups, the relative expression of hyaline chondrogenic genes SOX9, COL2A1, and ACAN was the highest in the 2% ECM group with moderate material stiffness (Fig. 5C), indicating that the mechanical microenvironment provided by this group was most suitable for the differentiation of BMSC cells into chondrocytes.YAP localization changed in all groups of cells after 14 days of differentiation induction without exogenous TGF, with YAP expressed in both nucleus and cytoplasm in the 1% ECM group of cells, with minor changes; YAP was both cytoplasm and nucleus in the 2% ECM group of cells. In cells from the 2% ECM group, YAP was expressed in both the cytoplasm and the nucleus, but the expression in the nucleus was more apparent; YAP in cells from the 4% ECM group definitely tended to be expressed in the nucleus (Fig. 6A).
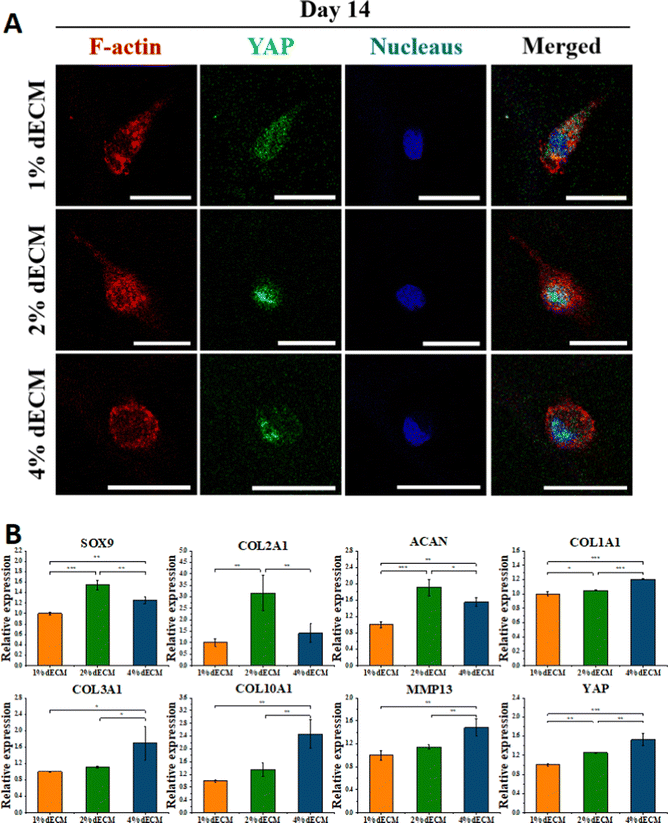
After 14 days of induced differentiation, the relative expression of the YAP gene was higher in cells of the group with a stiffer scaffold material, while the relative expression of fibrocartilage-related genes COL1A1, COL3A1 and hypertrophic cartilage-related genes COL10A1, MMP13 was higher (Fig. 6B). And the relative expression of hyaline cartilage-related genes SOX9, COL2A1, and ACAN was highest in the 2% ECM group cells with moderate material stiffness (Fig. 6B). These results illustrate that the differentiation of BMSCs to hyaline chondrocytes and maintenance of their cell phenotype are not favorable when the stiffness is too low or too high, and the provision of appropriate mechanical signals at different periods of cell life has an important role in cell differentiation and phenotype maintenance.
The 2% ECM sponge scaffold was chosen to co-culture with BMSC cells to provide a mechanistic microenvironment suitable for differentiation of stem cells into chondrocytes and further modulate YAP signaling to promote chondrocyte differentiation and phenotype maintenance.
3.5 Verteporfin regulate the differentiation of BMSCs to hyaline chondrocytes
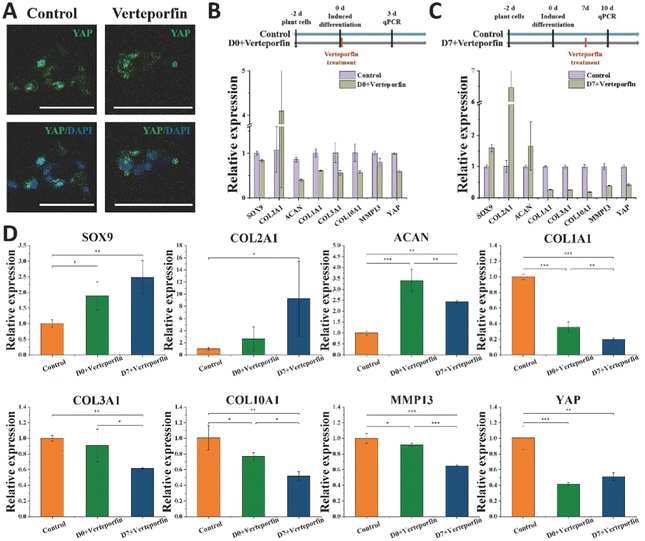
Vereporfin, an inhibitor of YAP activity, was experimentally used at a dose of 5 μM for the inhibitor cell assay (Fig. 7A) to confirm the geographical and temporal variability of YAP during chondrocyte development. Therefore, we tested whether matrix stiffness regulates the YAP under continuous treatment for 3 days. In the Day 0 + verteporfin group, the relative expression of hyaline cartilage-related gene COL2A1 was significantly increased and that of fibrocartilage-related gene COL1A1 was significantly decreased in the cells, but the relative expression of cartilage-related genes SOX9 and ACAN was also decreased (Fig. 7B). In the Day 7 + verteporfin group, the relative expression of hyaline cartilage-related genes COL2A1, SOX9 and ACAN could be found to be significantly increased, while the relative expression of fibrocartilage-related gene COL1A1 was significantly decreased (Fig. 7C).We further experimented that the relative expression of hyaline cartilage-related genes SOX9 and COL2A1 was highest in the experimental group of cells treated with drug after 14 days of induction, and the relative expression of fibrocartilage-related genes COL1A1, COL3A1 and COL1A1, COL10A1, MMP13 were the lowest in the experimental group (Fig. 7D). This indicates that after inducing differentiation for a period of time, then inhibiting the expression of YAP in cells can better promote the differentiation of BMSC to hyaline chondrocytes and maintain their cell phenotype. The activation of YAP in cells at the initial stage was beneficial for tissue regeneration, while the inhibition of YAP protein activity after a period of time could better differentiate the cells toward hyaline cartilage and maintain the phenotype for cartilage tissue regeneration.
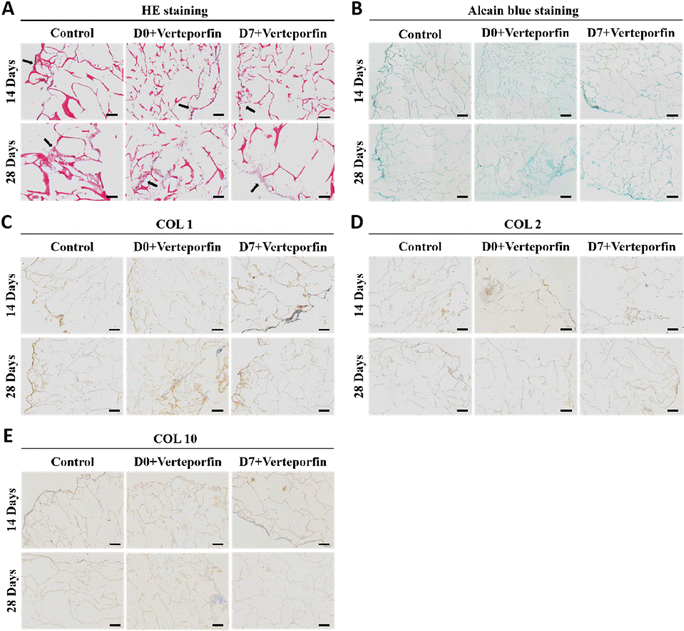
After 14 and 28 days of induced differentiation, sections and staining were performed. The results of HE staining showed that the extracellular matrix was accumulating on the scaffold material, which indicated that the cells were growing well on the material and the accumulation of extracellular matrix was conducive to the regenerative repair of the tissue (Fig. 8A). The accumulation of newborn cartilage-like matrix was also seen on Alcian Blue staining, where the accumulation of cartilage-like matrix was most obvious in the experimental group that added Verteporfin (D7). The accumulation of cartilage-like matrix was most evident in the experimental group with verteporfin (D7-Verteporfin) (Fig. 8B). The immunohistochemical results of COL1, COL2, COL10 showed that COL2 accumulated continuously as the induction of differentiation proceeded, with a better accumulation of COL2 in the D7-Verteporfin group (Fig. 8D); and with time, the expression of COL1 and COL10 in the group with verteporfin inhibiting YAP activity was decreased compared with the Control group (Fig. 8C and E). This indicates that inhibition of YAP activity at the late stages of differentiation of BMSCs better promotes the differentiation of BMSCs to hyaline chondrocytes and is able to generate hyaline cartilage organoids in long-term induction of differentiation experiments.
4. Discussion
In this study, we induced differentiation of BMSCs co-cultured with dECM sponge scaffolds in vitro to mimic the process of cartilage tissue regeneration and repair, and regulated YAP activity by verteporfin to propose a more optimal idea of cartilage regeneration and repair. We examined knee samples from 5-, 14-, and 28-day-old rats and discovered that YAP signaling is altered concurrently with changes in the kinds of collagen in the articular cartilage tissue. These results confirm YAP-related signaling pathways are associated with cartilage development and are regulatory targets for cartilage growth and development.We prepared dECM scaffolds with different mechanical properties. dECM sponge scaffolds of each group have porous structures to facilitate cell growth. With the increase of scaffold stiffness, YAP was gradually activated in the cells, leading to the tendency for nuclear expression. Without exogenous growth factor stimulation, induced differentiation was carried out on BMSCs co-cultured with several groups of dECM sponge scaffolds. After 7 days of induction differentiation, the relative expression of the YAP gene was higher in the group with a higher scaffold stiffness, while the relative expression of osteogenesis-related gene RUNX2 was higher and the relative expression of lipogenesis-related gene PPARγ was lower. After 14 days of induction differentiation, the relative expression of YAP genes was higher in the group of cells with greater scaffold material stiffness, while the relative expression of fibrocartilage-related genes COL1A1 and COL3A1 and hypertrophic cartilage-related genes COL1A1 and COL3A1 was higher. And the relative expression of hyaline cartilage-related genes SOX9, COL2A1, and ACAN was highest in the cells of 2% dECM sponge scaffolds group with moderate material stiffness. It has been shown that normal cartilage stiffness is around 6 kPa, and the 2% ECM stent is close to this stiffness.21 These results indicate that the differentiation of BMSC cells into hyaline chondrocytes and the maintenance of their cell phenotype are not favorable when the stiffness is too low or too high, and that the provision of appropriate mechanical signals at different periods of cell life has an important role in cell differentiation and phenotype maintenance.
Cell surface mechanosensitive receptors such as integrins and channel proteins such as Piezo1 sense the mechanical environment they are in, and these receptors and channels activate complex downstream signaling pathways that in turn regulate the physiopathological processes of articular cartilage.22 The most critical of the upstream signals of YAP are changes in the mechanical microenvironment that trigger mechanical signaling, such as changes in ECM stiffness, which can modulate YAP activity and thus participate in the regulation of important biological functions such as cell proliferation and differentiation. Related content has been added to the discussion.23 YAP is regulated by mechanical signals that modulate cell fate in response to the cellular microenvironment, and cartilage and bone are two particular tissues regulated by mechanical signals, as tissue adaptation and remodeling in response to load is essential to maintain their integrity.5 When mechanical adaptation mechanisms are dysregulated, bone and joint lesions such as osteoporosis or osteoarthritis can occur. Therefore, YAP signaling is an important signaling molecule that acts like a central mediator to maintain continuous adaptation of bone and cartilage tissues in response to alterations in the mechanical environment.9 Chondrocyte proliferation is regulated by YAP, which induces the expression of Sox6, required for proliferation, while inhibiting the expression of COL10, a marker of hypertrophic chondrocytes in vitro and in vivo.5 Since YAP inhibits the BMP response, which is essential for chondrocyte differentiation in vitro, inhibition of YAP activity is necessary to regulate the differentiation of stem cells to chondrocytes.24 Furthermore, YAP inhibits chondrocyte differentiation in vitro through downregulation of Wnt/β-catenin signaling, leading to the finding that chondrocyte dedifferentiation is associated with elevated YAP/TAZ levels induced by RhoA signaling.25 These studies demonstrate the inhibition of chondrocyte differentiation by YAP/TAZ, while acting as a compensatory mechanism to prevent eventual hypertrophic differentiation.
We found that after a period of differentiation induction, inhibition of YAP expression in the cells could better promote the differentiation of BMSCs to hyaline chondrocytes and maintain their cell phenotype. This indicates the spatial and temporal variability of YAP requirement during chondrocyte differentiation, with the activation of YAP in cells at the initial stage facilitating cell proliferation and growth, and the inhibition of YAP protein activity after a period of time to better differentiate the cells toward hyaline chondrocytes and maintain the phenotype. Other studies have previously used verteporfin to modulate YAP activity in cells for the treatment of osteoarthritis, and have achieved better results. Combined with our results, we can verify that there is a positive impact in cartilage regeneration and repair through YAP inhibitors. Based on the results of the induced differentiation and verteporfin treatment, we constructed cartilage organoids in vitro with decellularized matrix scaffolds and further validated our conclusions by histological staining. The construction of cartilage organoids is one of the hot spots in regenerative medicine, and decellular matrix material has more advantages than matrix gum in the construction of cartilage organoids, and the construction of cartilage organoids based on cartilage decellular matrix material induced cell aggregation is a new idea of cartilage regeneration and repair.26 We discovered a relationship between the collagen content and the YAP activity of the chondrocytes in the rat articular cartilage at various developmental stages. Based on these findings, we can develop hyaline cartilage-like tissues for cartilage injury repair using tissue engineering techniques. However, we also need to take into account the risks associated with excessive activation or inhibition of YAP, and we need to carefully control YAP activity at various stages of BMSC differentiation in order to regenerate hyaline cartilage tissues.
5. Conclusions
In conclusion, our results reveal a different requirement for YAP activity at different stages of cartilage development or regeneration. We now have novel methods for creating cartilage-like tissues for cartilage regeneration and repair in vitro by modifying cellular behavior by focusing on YAP activity. Additionally, these results imply that biomaterials with adequate mechanical properties and strong biocompatibility, such as decellularized matrix, may find useful applications in the regeneration of cartilage defects.Author contributions
Jiayi Zhu: methodology, formal analysis, writing – original draft, review & editing. Wanqing Lun: methodology, resources/reagents. Qi Feng: writing – review & editing. Xiaodong Cao: conceptualization, technical supports, writing – review & editing. Qingtao Li: conceptualization, writing – review & editing, funding acquisition, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021A1515010905), and the National Natural Science Foundation of China (Grant No. 52073103).References
- C. B. Carballo, Y. Nakagawa, I. Sekiya and S. A. Rodeo, Clin. Sports Med., 2017, 36, 413–425 CrossRef PubMed.
- E. A. Makris, A. H. Gomoll, K. N. Malizos, J. C. Hu and K. A. Athanasiou, Nat. Rev. Rheumatol., 2015, 11, 21–34 CrossRef CAS.
- H. Kwon, W. E. Brown, C. A. Lee, D. Wang, N. Paschos, J. C. Hu and K. A. Athanasiou, Nat. Rev. Rheumatol., 2019, 15, 550–570 CrossRef.
- A. R. Armiento, M. Alini and M. J. Stoddart, Adv. Drug Delivery Rev., 2019, 146, 289–305 CrossRef CAS.
- Y. Deng, A. Wu, P. Li, G. Li, L. Qin, H. Song and K. K. Mak, Cell Rep., 2016, 14, 2224–2237 CrossRef CAS.
- I. M. Moya and G. Halder, Nat. Rev. Mol. Cell Biol., 2019, 20, 211–226 CrossRef CAS PubMed.
- S. Dupont, L. Morsut, M. Aragona, E. Enzo, S. Giulitti, M. Cordenonsi, F. Zanconato, J. Le Digabel, M. Forcato, S. Bicciato, N. Elvassore and S. Piccolo, Nature, 2011, 474, 179–183 CrossRef CAS PubMed.
- S. G. Szeto, M. Narimatsu, M. Lu, X. He, A. M. Sidiqi, M. F. Tolosa, L. Chan, K. De Freitas, J. F. Bialik, S. Majumder, S. Boo, B. Hinz, Q. Dan, A. Advani, R. John, J. L. Wrana, A. Kapus and D. A. Yuen, J. Am. Soc. Nephrol., 2016, 27, 3117–3128 CrossRef CAS PubMed.
- M. Zarka, E. Hay and M. Cohen-Solal, Front. Cell Dev. Biol., 2021, 9, 788773 CrossRef.
- M. Bhattacharjee, J. Coburn, M. Centola, S. Murab, A. Barbero, D. L. Kaplan, I. Martin and S. Ghosh, Adv. Drug Delivery Rev., 2015, 84, 107–122 CrossRef CAS.
- B. J. Huang, J. C. Hu and K. A. Athanasiou, Biomaterials, 2016, 98, 1–22 CrossRef CAS PubMed.
- Q. Feng, H. C. Gao, H. J. Wen, H. H. Huang, Q. T. Li, M. H. Liang, Y. Liu, H. Dong and X. D. Cao, Acta Biomater., 2020, 113, 393–406 CrossRef CAS.
- M. Chen, Z. Feng, W. Guo, D. Yang, S. Gao, Y. Li, S. Shen, Z. Yuan, B. Huang, Y. Zhang, M. Wang, X. Li, L. Hao, J. Peng, S. Liu, Y. Zhou and Q. Guo, ACS Appl. Mater. Interfaces, 2019, 11, 41626–41639 CrossRef CAS.
- Y. Lu, Y. Wang, H. Zhang, Z. Tang, X. Cui, X. Li, J. Liang, Q. Wang, Y. Fan and X. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 24553–24564 CrossRef CAS PubMed.
- A. J. Vernengo, S. Grad, D. Eglin, M. Alini and Z. Li, Adv. Funct. Mater., 2020, 30, 1909044 CrossRef CAS.
- A. Elosegui-Artola, I. Andreu, A. E. M. Beedle, A. Lezamiz, M. Uroz, A. J. Kosmalska, R. Oria, J. Z. Kechagia, P. Rico-Lastres, A. L. Le Roux, C. M. Shanahan, X. Trepat, D. Navajas, S. Garcia-Manyes and P. Roca-Cusachs, Cell, 2017, 171(1397–1410), e1314 Search PubMed.
- Y. Deng, J. Lu, W. Li, A. Wu, X. Zhang, W. Tong, K. K. Ho, L. Qin, H. Song and K. K. Mak, Nat. Commun., 2018, 9, 4564 CrossRef.
- X. Zhang, D. Cai, F. Zhou, J. Yu, X. Wu, D. Yu, Y. Zou, Y. Hong, C. Yuan, Y. Chen, Z. Pan, V. Bunpetch, H. Sun, C. An, T. Yi-Chin, H. Ouyang and S. Zhang, Biomaterials, 2020, 232, 119724 CrossRef CAS.
- X. Zhao, L. Tang, T. P. Le, B. H. Nguyen, W. Chen, M. Zheng, H. Yamaguchi, B. Dawson, S. You, I. M. Martinez-Traverso, S. Erhardt, J. Wang, M. Li, J. F. Martin, B. H. Lee, Y. Komatsu and J. Wang, Sci. Signaling, 2022, 15, eabn9009 CrossRef CAS.
- A. Yamashita, H. Yoshitomi, S. Kihara, J. Toguchida and N. Tsumaki, Stem Cells Transl. Med., 2021, 10, 115–127 CrossRef CAS PubMed.
- H. Iijima, G. Gilmer, K. Wang, A. C. Bean, Y. He, H. Lin, W. Y. Tang, D. Lamont, C. Tai, A. Ito, J. J. Jones, C. Evans and F. Ambrosio, Nat. Commun., 2023, 14, 18 CrossRef CAS.
- T. Hodgkinson, D. C. Kelly, C. M. Curtin and F. J. O’Brien, Nat. Rev. Rheumatol., 2022, 18, 67–84 CrossRef.
- J. Liu, J. Wang, Y. Liu, S. A. Xie, J. Zhang, C. Zhao, Y. Zhou, W. Pang, W. Yao, Q. Peng, X. Wang and J. Zhou, Circ. Res., 2023, 132, 87–105 CrossRef CAS.
- A. Karystinou, A. J. Roelofs, A. Neve, F. P. Cantatore, H. Wackerhage and C. De Bari, Arthritis Res. Ther., 2015, 17, 1–14 CrossRef CAS.
- B. Yang, H. Sun, F. Song, M. Yu, Y. Wu and J. Wang, Int. J. Biochem. Cell Biol., 2017, 87, 104–113 CrossRef CAS.
- R. Simsa, T. Rothenbucher, H. Gurbuz, N. Ghosheh, J. Emneus, L. Jenndahl, D. L. Kaplan, N. Bergh, A. M. Serrano and P. Fogelstrand, PLoS One, 2021, 16, e0245685 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |