Open Access Article
Open Access Article3-Hydroxyflavone derivatives: promising scaffolds for fluorescent imaging in cells
Xueke Zhao†
a,
Xiang Li†bc,
Shuyu Liangb,
Xiongwei Dong *a and
Zhe Zhang*d
*a and
Zhe Zhang*d
aNational Local Joint Engineering Laboratory for Advanced Textile Processing and Clean Production, Wuhan Textile University, Wuhan, Hubei 430073, P. R. China. E-mail: zxk2686903512@gmail.com; xwdong@wtu.edu.cn
bCollege of Chemistry and Molecular Engineering, Peking University, Beijing, 100871, P. R. China. E-mail: 199284lixiang@pku.edu.cn; liangshuyu@stu.pku.edu.cn
cSchool of Chemistry, Central China Normal University, Wuhan, Hubei 430079, P. R. China
dInstitute of Environmental Research at Greater Bay Area, Key Laboratory for Water Quality and Conservation of the Pearl River Delta, Ministry of Education, Guangzhou Key Laboratory for Clean Energy and Materials, Guangzhou University, Guangzhou 510006, China. E-mail: zhezhang2018@gzhu.edu.cn
First published on 27th August 2021
Abstract
As a typical class of excited-state intramolecular proton transfer (ESIPT) molecules, 3-hydroxyflavone derivatives (3HF, also known as flavonols) have received much attention recently. Thereinto, the role of hydrophobic microenvironment is significant importance in promoting the process and effects of ESIPT, which can be regulated by the solvents, the existence of metal ions and proteins rich with α-helix structures or the advanced DNA structures. Considering that plenty of biological macromolecules offer cellular hydrophobic microenvironment, enhancing the ESIPT effects and resulting in dual emission, 3HF could be a promising scaffold for the development of fluorescent imaging in cells. Furthermore, as the widespread occurance of compounds with biological activity in plants, 3HF derivatives are much more secure to be cellular diagnosis and treatment integrated fluorescent probes. In this review, multiple regulatory strategies for the fluorescence emission of 3HF derivatives have been collectively and comprehensively analyzed, including the solvent effects, metal chelation, interaction with proteins or DNAs, which would be beneficial for ESIPT-promoting or ESIPT-blocking processes and then enhance or control the fluorescence emission of 3HF effectively. We expect that this review would provide a new perspective to develop novel 3HF-based fluorescent sensors for imaging in cells and plants.
1. Introduction
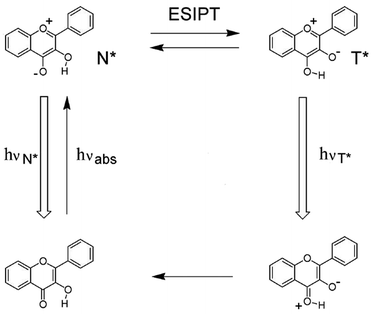
Proton transfer is one of the most fundamental processes in biology and chemistry such as the acid–base neutralization reactions1–3 and proton transfer induced fluorescence effects.4,5 Depending on the reaction in the ground- or excited-states, whether they are adiabatic or non-adiabatic, various types of proton transfer reactions have been classified.6 Intermolecular proton transfer usually involves the transfer of protons from the donor to the acceptor in biological system, which requires the formation of a dimer or complex between the proton donor and acceptor.7–10 On the contrary, intramolecular proton transfer involves the transfer of proton within the same molecule, among which excited-state intramolecular proton transfer (ESIPT) has attracted widespread attention, because of its large Stokes shift without self-reabsorption and its easy inversion in the proton transferred keto tautomer.11–13Properties of the ESIPT molecules are mainly due to its intrinsic four-level photocycle of the enol (E)–keto (K) phototautomerization (E → E* → K* → K →…) process.12 As shown in Fig. 1, the typical ESIPT molecule (2-(2-hydroxyphenyl)-benzothiazole) exists in the enol form at the electronic ground state (E), and forms an intramolecular hydrogen bond upon photoexcitation (E*). As a result, an ultrafast ESIPT process occurs, and the proton transfers from the donor to the acceptor through an intramolecular hydrogen bond, making the excited enol form (E*) transfer into the excited keto form (K*). Eventually, reverse proton transfer occurs after radiative decay of K* to the ground state, and the initial E form is obtained. Here, the geometry relaxation in ESIPT process and the differences between enol absorption and keto emission give rise to the large Stokes shift and complete elimination of self-absorption.12,14
 | ||
| Fig. 1 Schematic representation of ESIPT photocycle. Illustrated by 2-(2-hydroxyphenyl)-benzothiazole (HBT). | ||
Up to now, a large number of ESIPT molecules have been developed, including flavonols,15–17 quinolones,18,19 anthraquinones,20 pyridyls,21–23 azoles,24 salicylidene anilines25 and so on, among which, the widely distributed subgroup of flavonoids flavonols (in this review also regarded as 3-hydroxyflavone, 3HF) have attracted much attention recently, due to unique and beneficial photophysical characteristics, and biocompatibility.26–33
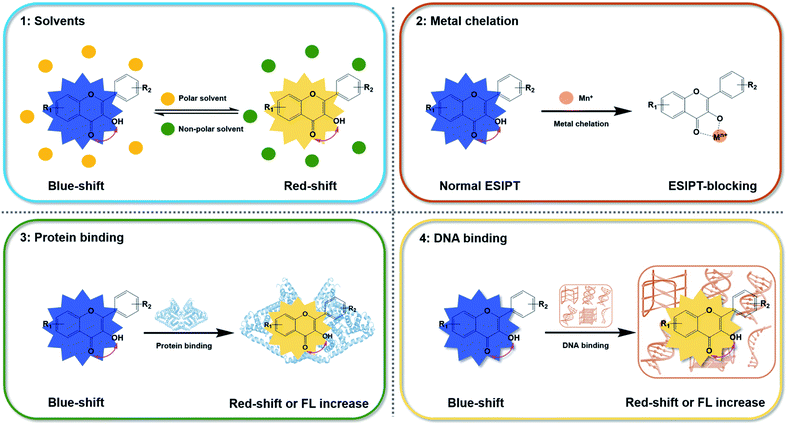
Hydrophobic microenvironment is significant importance in promoting the ESIPT process of 3HF derivatives, which can be regulated by the solvents, the existence of metal ions, proteins rich with α-helix structures or the advanced DNA structures. Considering plenty of biological macromolecules within cells offer hydrophobic microenvironment, enhancing the ESIPT effects of 3HF derivatives and then resulting in a dual emission, with normal fluorescence emission around 400 nm and tautomeric emission at longer wavelengths,34 3HF could be a promising scaffold for the development of fluorescent imaging in cells.
Similar to most ESIPT molecules, 3HF derivatives also have defects in practical applications, such as low fluorescence emission efficiency and environment-sensitive spectral responses. For the former, fluorescence emission decay of the enol form of 3HF is partly caused by intersystem crossing (ISC), which leads to triplet excited state of the enol form.35 For the latter, the normal (N*) and tautomer (T*) emissions of 3HF derivatives are complicated in protic and aprotic solvents, because the solute–solvent interactions have effects on the structure of 3HF derivatives, via the formation of intermolecular hydrogen bond or the break of intramolecular hydrogen bond.36,37 In this regard, modulating four-level photocycle of the phototautomerization process can effectively control or stabilize the fluorescence emission of 3HF derivatives, which plays an important role in the design of 3HF based fluorescence probes and optical sensors.
In this review, multiple regulatory strategies for the fluorescence emission of 3HF have been collectively and comprehensively analyzed, such as solvent effects, chelating with metal ions, and binding with proteins or DNAs (Fig. 2). As the widespread occurrence of compounds in plants, 3HF are much more secure than other small molecule fluorescent probes. We expect that the study of the fluorescent emission of 3HF would provide a new perspective to develop novel 3HF fluorescence sensors for imaging in cells or plants, and further as cellular diagnosis and treatment integrated fluorescent probes, based on the regulatory of ESIPT process.
2. Emission mechanism and solvent effects
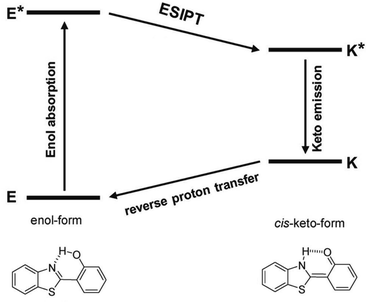
As shown in Fig. 3, 3HF derivatives exhibit dual emission due to their ESIPT process that results in normal (N*) and tautomer (T*) excited forms (kESIPT > 1012 s−1),38,39 and the emissions of N* and T* forms exhibit well-separated bands, whose positions and intensities provide much information about the microenvironment.40 The T* band is predominantly observed in nonpolar solvents, which is the large Stokes-shifted band.41,42 Furthermore, the N* band is typically observed in polar or protic solvents with normal Stokes' shift.41,42 The intensity ratio of the N* and T* bands, IN*/IT*, can always be used into various ratiometric detections.43As mentioned above, the formation of intramolecular hydrogen bond is essential for the ESIPT process, because the proton transfer takes place along the locus of intramolecular hydrogen bond.44 Therefore, the formation of intermolecular hydrogen bond with solvents can effectively hamper the ESIPT process.45,46 In polar or protic solvents, such as water, dimethylsulfoxide (DMSO) and methanol (MeOH), the proton donor and acceptor of ESIPT molecules can form intermolecular hydrogen bond with the solvent molecules, preventing the formation of intramolecular hydrogen bond in the ESIPT molecules (Fig. 4a). Therefore, in polar or protic solvents, the normal enol form transforms into the solvated enol form, which leads to the rise of the N* band and the suppression of the T* band.45,46
 | ||
| Fig. 4 (a) The enol and solvated forms of 3HF; (b) solvents classification by physicochemical properties. | ||
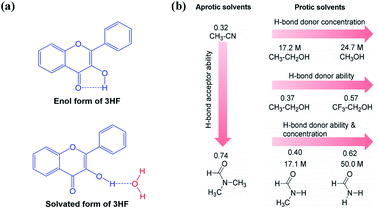
The dual emission behaviour is observed in a plenty of ESIPT molecules, and a five-membered ring structure containing intramolecular hydrogen bond is formed in this process (Fig. 4b).47 Next, we use 3HF as an example to analyse the effects of solvents on the fluorescence emission of 3HF derivatives. 3HF forms weak intramolecular hydrogen bond between 4-carbonyl and 3-hydroxyl groups (Fig. 4a). Here, we have summarized the effects of various organic solvents (aprotic and protic solvents) on 3HF fluorescence emission. As shown in Fig. 4b, the aprotic solvents, acetonitrile (MeCN) and N,N-dimethylformamide (DMF) have similar polarities, but their abilities acting as hydrogen bond acceptors are different.44 The protic solvents can be classified into the following three categories: (1) ethanol (EtOH) ∼ methanol (MeOH) which differ by the hydrogen bond donor (OH-proton) concentrations, (2) ethanol ∼ 2,2,2-trifluoroethanol (TFE) which differ by their hydrogen bond donor abilities and (3) N-methylformamide (MF) ∼ formamide (F) which differ by their hydrogen bond donor (NH-proton) concentrations and hydrogen bond donor abilities (Fig. 4b).48 It is worth noting that there are significant differences between amides as hydrogen bond donors and alcohols.
Position of the absorption maximum of 3HF is hardly affected by the solvent properties, which varies in the range of 339 nm to 345 nm in the above-mentioned solvents (Fig. 5a). Just as previous studies,49–51 two emission bands of 3HF are also observed in all studied solvents (Fig. 5a), which indicates that ESIPT process has occurred in all systems. The fluorescence quantum yield (φ) of 3HF does not change regularly with all studied solvents. However, the lowest fluorescence quantum yield is observed in DMF, which should be caused by the larger hydrogen bond acceptor ability of DMF (Fig. 5a). The low fluorescence quantum yield value in hydrogen bond basic solvent (DMF) might be caused by disrupting the intramolecular hydrogen bond of 3HF and forming other non-emissive species.49
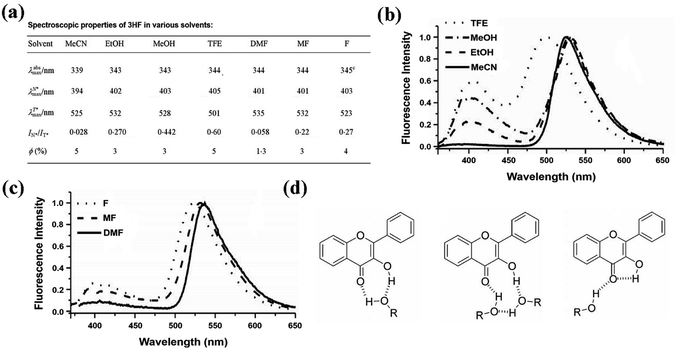 | ||
| Fig. 5 (a) Spectroscopic properties of 3HF in different polar solvents; (b) normalized fluorescence spectra of 3HF in MeCN and different alcohols: TFE, MEOH and EtOH. Excitation wavelength = 350 nm; (c) normalized fluorescence spectra of 3HF in different amide-based solvents: F, MF and DMF. Excitation wavelength = 350 nm; (d) possible hydrogen bonded forms of 3HF with alcohols.48 | ||
In aprotic solvents (MeCN and DMF), the relative intensity of the T* band is much larger than the N* band for 3HF (Fig. 5a), indicating that an extremely fast ESIPT process leads to nearly completely transforms the N* state into T* state. However, the different hydrogen bond acceptor abilities of MeCN and DMF hardly affect the IN*/IT* ratio for 3HF, suggesting that 3HF and its derivatives are not sensitive to this solvent property.40 A significant increase of intensity ratio IN*/IT* is observed in all analyzed protic solvents comparing with that in aprotic solvents (Fig. 5a), indicating that hydrogen bond donors may inhibit the ESIPT processes of 3HF and its derivatives.40,52 With the increase in the concentration of hydrogen bond donors from EtOH to MeOH, the intensity ratio IN*/IT* of 3HF is strongly increased (Fig. 5a and b). With the increase of hydrogen bond donor ability from EtOH to TFE, the intensity ratio IN*/IT* of 3HF is also significantly enhanced, which is accompanied by the slight red shift of the N* band, and the large blue shift (ca. 31 nm) and significant broadening of the T* band (Fig. 5a and b). When the solvent is changed from MF to F, both the hydrogen bond donor ability and the donor concentration are increased at the same time, resulting in a blue shift (ca. 9 nm) of T* band and a substantial increase in the intensity ratio IN*/IT* of 3HF (Fig. 5a and c), all of which is in line with the results in alcohols.
Next, we systematically summarized the effects of hydrogen bond donor ability and donor concentration on the intensity ratio IN*/IT* of 3HF. Since the formation of intermolecular hydrogen bond with solvent can interrupt the ESIPT process,52 the intermolecular hydrogen bond between 3HF and alcohols may also interfere the formation of intramolecular hydrogen bond of 3HF and its ESIPT process (Fig. 5d).37 Therefore, the relative intensity of N* band increases in protic solvents, and also increases as the hydrogen bond donor concentration. In the highly hydrogen bond acidic TFE, the interference of intramolecular hydrogen bond is not as well as other alcohols, because the far less basic hydroxyl group of TFE forms much weaker intermolecular hydrogen bond with the 3-hydroxyl group of 3HF. The more separated between hydrogen bond donor (NH hydrogen) and acceptor (carbonyl oxygen) in amide solvents than that in alcohols, leading to differences in their hydrogen bond network. In these cases, intermolecular hydrogen bond may form between the acidic hydrogen of the solvent and the 4-carbonyl group, and this hydrogen bond is directly affected by the hydrogen bond acidcity of solvents. Overall, the dual emission and ESIPT kinetics of 3HF are significantly affected by such a hydrogen bond.53,54
The dual emission and intensity ratio IN*/IT* of 3HF derivatives are sensitive to the solvent polarity, and the ratio of them ranges from unity up to the complete disappearance of one of the forms. Actually, this uniquely fluorescent characteristic of 3HF derivatives has already be used as probes for polarity, electrostatic fields, and the structural variations in micelles and biomembrane models.55,56 Thus, 3HF derivatives can also be further used as other environment-sensitive probes and labels for biological applications.
3. Metal chelation
In addition to interaction with solvent molecules, coordination with metal ions (such as Cu2+, Zn2+, Ru2+, Al3+ and so on) can also modulate the ESIPT process of various organic molecules (such as flavones, aminosalicylimines, quinolines, azoles and so on).57–65 For the hydroxyflavones, metal chelation usually occurs at the 3-hydroxyl, 5-hydroxyl, and 2-carbonyl groups, which can be used for Cu2+, Zn2+, Al3+, Hg2+, Pb2+ and Fe3+ Sening.66–75 The complexation of metal ions with 3HF derivatives at the α-hydroxy-carbonyl site can interrupt the ESIPT process, and then change the fluorescence emission.57,58,65,76 A comprehensive summary of the effects of metal chelation on the ESIPT process and fluorescence emission of 3HF derivatives is important for the development of novel 3HF based metal fluorescence probes and sensors. Here, we first take several 3HF derivatives as examples to illustrate the effects of metal chelation on their ESIPT process.Typically, flavonoids have two strong UV absorption bands: the band in 320–380 nm and the band in 240–270 nm.77 The UV-visible spectra of 3HF titrated by Zn2+ showed that the band at 344 nm decreases with the increasing concentrations of Zn2+, and a new band appears at 408 nm (Fig. 6a).76 The stoichiometry for this complex is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and this complex has a stability constant: log
1, and this complex has a stability constant: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β3HF–Zn = 6.60 ± 0.10.76 The chelation of Zn2+ occurs at the α-hydroxy-carbonyl group and makes pronounced variations of the ligand geometry, which also leads to complete deprotonation of the hydroxyl group and mainly affects the geometry of γ-pyrone ring.76 The torsion angle between the chromone and the B ring is 15.7°, and the Zn2+ is outside the chromone plane.76
β3HF–Zn = 6.60 ± 0.10.76 The chelation of Zn2+ occurs at the α-hydroxy-carbonyl group and makes pronounced variations of the ligand geometry, which also leads to complete deprotonation of the hydroxyl group and mainly affects the geometry of γ-pyrone ring.76 The torsion angle between the chromone and the B ring is 15.7°, and the Zn2+ is outside the chromone plane.76
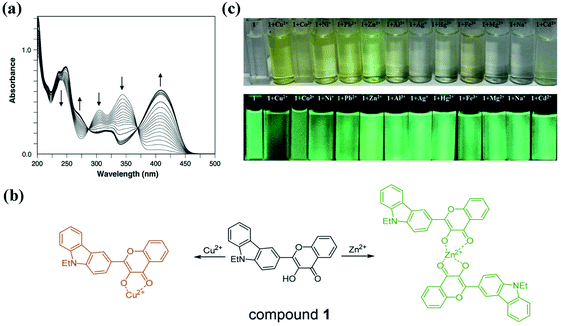 | ||
Fig. 6 (a) The absorption spectra of 3HF (40 μM) in titrations with Zn2+ (0–120 μM) in methanol.76 (b) The binding models of compound 1 with Cu2+ and Zn2+; (c) change of colour in natural light and fluorescence under 365 nm of compound 1 in methanol–HEPES buffer solution (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, pH 7.4) upon the addition of cations.57 1, pH 7.4) upon the addition of cations.57 | ||
Next, 2-(9-ethyl-9H-carbazol-3-yl)-3-hydroxy-4H-chromen-4-one (compound 1) is used as an example to illustrate the fluorescent changes caused by metal chelation.57 This compound can chelate with Cu2+ and Zn2+ in different modes, and the complex ratios of compound 1 with Cu2+ and Zn2+ are 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, respectively (Fig. 6b). As shown in Fig. 6c, the chelation with Zn2+ makes compound 1 change from colourless to green, and increases the fluorescence intensity. However, the chelation with Cu2+ makes compound 1 change from colourless to yellow, and reduces the fluorescence intensity. The hydroxyl group on the chromone of compound 1 is involved in the coordination with Cu2+ and Zn2+. Two oxygen atoms of hydroxyl and carbonyl groups in compound 1 bind with Cu2+ to form the 1
2, respectively (Fig. 6b). As shown in Fig. 6c, the chelation with Zn2+ makes compound 1 change from colourless to green, and increases the fluorescence intensity. However, the chelation with Cu2+ makes compound 1 change from colourless to yellow, and reduces the fluorescence intensity. The hydroxyl group on the chromone of compound 1 is involved in the coordination with Cu2+ and Zn2+. Two oxygen atoms of hydroxyl and carbonyl groups in compound 1 bind with Cu2+ to form the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex. Then, because the d10 orbital of Cu2+ is not filled with electrons, the intramolecular photo-induced electron transfer (PeT) from the excited compound 1 to Cu2+ can quench the fluorescence of compound 1.78–80 For the Zn2+ complex, coordination of two compound 1 molecules with Zn2+ is also at the oxygen atoms in hydroxyl and carbonyl groups.81 However, the filled d10 orbital of Zn2+ cannot accept electrons from the excited compound 1. Conversely, the chelation with Zn2+ increases the electron accepting ability of the 3-hydroxyflavone group of compound 1, which enhances the intramolecular charge transfer from carbazole to 3-hydroxyflavone moiety and then increases the fluorescence intensity.80 Overall, coordination with metal ions can modulate the intramolecular charge transfer process of the excited 3HF derivatives, thereby quenching or enhancing the fluorescence. On the other hand, chelation should also mask the ESIPT process of 3HF derivatives, resulting in the single peak of fluorescence emission.
1 complex. Then, because the d10 orbital of Cu2+ is not filled with electrons, the intramolecular photo-induced electron transfer (PeT) from the excited compound 1 to Cu2+ can quench the fluorescence of compound 1.78–80 For the Zn2+ complex, coordination of two compound 1 molecules with Zn2+ is also at the oxygen atoms in hydroxyl and carbonyl groups.81 However, the filled d10 orbital of Zn2+ cannot accept electrons from the excited compound 1. Conversely, the chelation with Zn2+ increases the electron accepting ability of the 3-hydroxyflavone group of compound 1, which enhances the intramolecular charge transfer from carbazole to 3-hydroxyflavone moiety and then increases the fluorescence intensity.80 Overall, coordination with metal ions can modulate the intramolecular charge transfer process of the excited 3HF derivatives, thereby quenching or enhancing the fluorescence. On the other hand, chelation should also mask the ESIPT process of 3HF derivatives, resulting in the single peak of fluorescence emission.
Since chelation with metal ions can block the ESIPT process of 3HF derivatives, we developed a highly sensitive and selective Zn2+ probe (compound 2) (Fig. 7a), which contains a 3-hydroxychromone (3HC) chromophore and a zinc-selective metal chelator di(2-picolyl)amine (DPA).65 This 3HF derivative can form a Zn2+ complex with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Zn2+-to-ligand ratio and function as a turn-on fluorescent probe for Zn2+. One Zn2+ coordinates to three DPA nitrogen atoms in one compound 2 molecule and to two oxygen atoms of hydroxyl and carbonyl groups in another compound 2 molecule, thereby blocking the ESIPT process through displacing the hydroxyl group in compound 2. In the Zn2+ free form, DPA can transfer its electron to 3-hydroxyflavone moiety in the excited state, and then quench the fluorescent emission of compound 2. After adding Zn2+, both the three nitrogen atoms of DPA and the two oxygen atoms of hydroxyl and carbonyl groups are involved in the coordination with Zn2+, which could suppress the intramolecular electron transfer process of the excited compound 2 and result in the strong fluorescence emission at 466 nm. The fluorescence response mechanism of Zn2+ is attributed to the suppression of photo-induced electron transfer (PeT) exerted by deprotonation of 3-hydroxyflavone moiety and occupation of the lone electron pairs of DPA. As shown in Fig. 7b, the presences of Cu2+ (1 equiv.), Co2+ (1 equiv.), Cd2+ (10 equiv.) and Hg2+ (100 equiv.) has effects on the fluorescence response of compound 2 to Zn2+, which is line with Lewis acid–Lewis base interactions and the principle of hard and soft acids and bases (HASB). Zn2+ is hard metal ion, both Cu2+ and Co2+ are intermediate metal ion, and both Hg2+ and Cd2+ are soft metal ions, therefore, when reacting with the hard nitrogen and oxygen atoms, the order of stabilities for the complexes between compound 2 and these metal ions is as following: Zn2+ > Cu2+ and Co2+ > Hg2+ and Cd2+. Since the Zn2+ content in human normal prostate cell line (RWPE-1) is much higher than that of cancerous cell line (DU145), we have successfully applied compound 2 to identify the prostate cancer cells (Fig. 7c). Thus, by simultaneously coordinating with the electron donor moiety and the 3-hydroxyflavone moiety, we can suppress the ESIPT process and enhance the fluorescence emission of these 3HF derivatives containing electron donor moiety. According to this fluorescence emission mechanism and HASB, we can also design various 3HF based fluorescent probes for detecting other metals, which may have broad application prospects in intracellular detection for metal ions.
2 Zn2+-to-ligand ratio and function as a turn-on fluorescent probe for Zn2+. One Zn2+ coordinates to three DPA nitrogen atoms in one compound 2 molecule and to two oxygen atoms of hydroxyl and carbonyl groups in another compound 2 molecule, thereby blocking the ESIPT process through displacing the hydroxyl group in compound 2. In the Zn2+ free form, DPA can transfer its electron to 3-hydroxyflavone moiety in the excited state, and then quench the fluorescent emission of compound 2. After adding Zn2+, both the three nitrogen atoms of DPA and the two oxygen atoms of hydroxyl and carbonyl groups are involved in the coordination with Zn2+, which could suppress the intramolecular electron transfer process of the excited compound 2 and result in the strong fluorescence emission at 466 nm. The fluorescence response mechanism of Zn2+ is attributed to the suppression of photo-induced electron transfer (PeT) exerted by deprotonation of 3-hydroxyflavone moiety and occupation of the lone electron pairs of DPA. As shown in Fig. 7b, the presences of Cu2+ (1 equiv.), Co2+ (1 equiv.), Cd2+ (10 equiv.) and Hg2+ (100 equiv.) has effects on the fluorescence response of compound 2 to Zn2+, which is line with Lewis acid–Lewis base interactions and the principle of hard and soft acids and bases (HASB). Zn2+ is hard metal ion, both Cu2+ and Co2+ are intermediate metal ion, and both Hg2+ and Cd2+ are soft metal ions, therefore, when reacting with the hard nitrogen and oxygen atoms, the order of stabilities for the complexes between compound 2 and these metal ions is as following: Zn2+ > Cu2+ and Co2+ > Hg2+ and Cd2+. Since the Zn2+ content in human normal prostate cell line (RWPE-1) is much higher than that of cancerous cell line (DU145), we have successfully applied compound 2 to identify the prostate cancer cells (Fig. 7c). Thus, by simultaneously coordinating with the electron donor moiety and the 3-hydroxyflavone moiety, we can suppress the ESIPT process and enhance the fluorescence emission of these 3HF derivatives containing electron donor moiety. According to this fluorescence emission mechanism and HASB, we can also design various 3HF based fluorescent probes for detecting other metals, which may have broad application prospects in intracellular detection for metal ions.
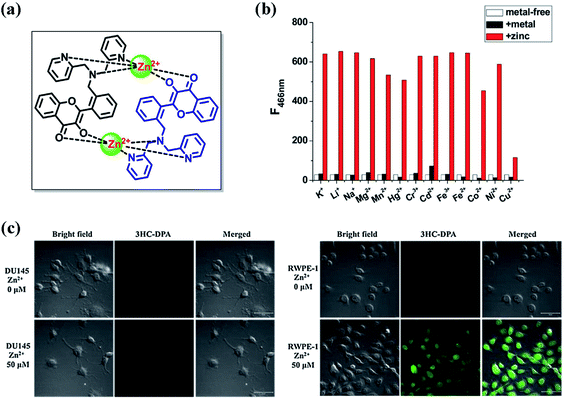 | ||
| Fig. 7 (a) The complex structure of compound 2 with zinc; (b) metal ion selectivity of compound 2: white bar, fluorescence intensity of compound 2; black bar, fluorescence intensity in the presence of metal ion (K+, Li+, Na+, Mg2+, Mn2+, Hg2+ ions, 100 equiv.; Cr3+, Cd2+,10 equiv.; Fe3+, Fe2+, Co2+, Ni2+, Cu2+, 1 equiv. to compound 2); red bar, fluorescence intensity after subsequent addition of Zn2+ (1 equiv.) to the mixture. Excitation wavelength = 382 nm; (c) fluorescence images of human cancerous prostate cell line (DU145) and human normal prostate cell line (RWPE-1) with compound 2.65 | ||
Two oxygen atoms of the hydroxyl and carbonyl groups can participate in the coordination with metal ions, leading to the ESIPT process suppression of 3HF derivatives. In addition, coordination with different metal ions has different effects on the excited state charge transfer process of the 3HF derivatives, thereby enhancing or quenching the fluorescence emission of the 3HF derivatives. Designing suitable electron donor groups to participate in the coordination with metal ions can regulate the PeT process and the fluorescence emission of 3HF derivatives, which has certain guiding significance for designing fluorescence probes for metal ions.
4. Interaction with protein
Proteins are biological macromolecules having specific three-dimensional structure, whose distribution of charges and hydrophobic sites are heterogeneous. Protein-small molecule interaction can regulate the activity of protein or the physicochemical property of small molecule.82–85 At present, some literatures have shown that proteins are often used as targets for therapeutically active flavonoids.86–88 Furthermore, several spectroscopic studies on flavonoid–protein interactions also reveal that interaction with protein can alter the fluorescent emission of various flavonoid derivatives.89–95 Thus, as a typical class of flavonoids, 3HF derivatives should also have the potential to interact with protein, which may be a potential approach to regulate the fluorescence emission of 3HF derivatives.Liu et al. had found that 5-amino-2-benzooxazol-2-yl-phenol (compound 3) can interact with α-helical proteins (such as bovine serum albumins, BSA) to enhance its fluorescence emission intensity and increase its ESIPT efficiency in vitro and in vivo.97 Interaction with protein might also be a way for 3HF derivatives to address the above-mentioned problems in fluorescence emission. 2-(6-Diethylaminobenzo[b]furan-2-yl)-3-hydroxychromone (compound 4, also named as FA, Fig. 8a), belonging to 3-hydroxychromone derivatives, is primarily used as a polarity calibrator in the hydrophobic solvents.98,99 This probe should also bind to BSA with a high binding constant (107 M−1), and this binding is in equimolar ratios. The fluorescence spectra of compound 4 bound to different serum albumins showed that there are two groups of fluorescent emissions after binding to BSA at equimolar concentration ratios (Fig. 8b).96 One group shows two well-resolved bands of comparable intensities, one in 530–560 nm, and the other in 600–630 nm range of spectrum. The two bands are the emissions of N* and T* forms, whose relative intensities vary dramatically in different species. In the second group, they all have the strong emission of N* band, but the observation of T* band is difficult. The fluorescence emission of compound 4 in the aqueous medium is negligibly small, but the binding of compound 4 to these serum albumins can dramatically enhance its fluorescence emission.96 The serum albumins can provide strongly hydrophobic and highly electronically polarizable environment for compound 4.96 Although the binding sites in all mentioned serum albumins are similar, different serum albumins have different ability to form intermolecular hydrogen bonds, which leads to differences in the promotion of ESIPT process of compound 4.96 Since 3HC and 3HF derivatives have similar chemical structures, interaction with BSA might also be a method of amplifying the fluorescence emission or enhancing the ESIPT process of 3HF derivatives.
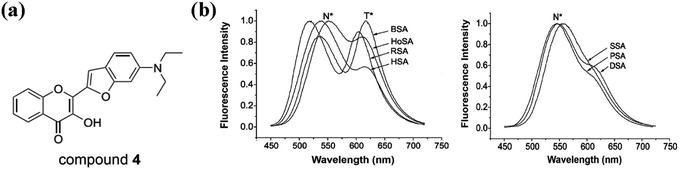 | ||
| Fig. 8 (a) The structure of compound 4; (b) fluorescence emission spectra of bovine (BSA), dog (DSA), horse (HoSA), human (HSA), pig (PSA), rabbit (RSA) and sheep (SSA) serum albumins in complex with compound 4. Excitation wavelength = 443 nm.96 | ||
Considering the high binding constant of compound 4 with BSA, we can also use BSA to regulate the fluorescence emission of 3HF derivatives. We prepared two 3HF derivatives (compound 5 and 6, Fig. 9a), and analyzed the effects of BSA on their fluorescence emission.100 Compound 5 emits fluorescence at 450 nm (N* band) in Tris-buffer, which reduces gradually with the increase of a new emission band at 543 nm (T* band) upon titration with BSA (Fig. 9b). Therefore, BSA can induce the proton transfer from the hydroxylic proton of 3HF group to adjacent carbonyl oxygen atom through intramolecular hydrogen bond, and then enhance the T* form fluorescence emission of compound 5. It is obvious that compound 5 is a typical ESIPT molecule and capable to work as a ratiometric fluorophore in hydrophobic microenvironment. For compound 6, the background fluorescence emission is very weak, and the fluorescence emission increases dramatically at 497 nm (N* band) along with a weak shoulder peak around 550 nm (T* band) upon titration with BSA (Fig. 9c). The non-typical ESIPT spectroscopic properties of compound 6 should be caused by the strong electron-donating amino group in the 4′-position of 3HF. Under normal conditions, the bond vibrations of N–H and the resulting energy-consuming may quench the fluorescence emission of compound 6 in Tris-buffer. Upon addition of BSA, the intermolecular hydrogen bonds between the amino group and BSA would weaken the vibrations of N–H, restore the fluorescence emission of N* band, and finally emit a very weak T* band in the presence of high concentrations of BSA. Molecular docking simulations further support the interaction between protein and these 3HF derivatives.100 The hydroxylic proton of compound 5 would participate in the formation of intermolecular hydrogen bond with serum albumin, and this hydrogen bond can make the proton easy to lose and facilitate the ESIPT process of compound 5 (Fig. 9d). As expected, the amino group in the 4′-position of compound 6 would participate in the formation of two intermolecular hydrogen bonds with acid residues of serum albumin, which might promote the fluorescence recovery of compound 6 (Fig. 9d). Based on the promotion of ESIPT process by the interaction between compound 5 and protein, we have successfully designed a derivative of compound 5 as a ratiometric fluorescent probe for intracellular detection of hydrogen peroxide, and the intracellular microenvironment can facilitate the ESIPT process of compound 5.100 Besides, we have also successfully designed a derivative of compound 6 as a turn-on fluorescent probe for intracellular detection of glutathione, and the intracellular microenvironment can enhance the fluorescence emission of compound 6.100 For compound 4, it can bind to different types of serum albumins to promote its ESIPT process or enhance the fluorescence emission of its N* band. However, through changing the substituents of 3HF derivatives, we can also regulate the fluorescence emission of 3HF derivatives in the same solution environment.
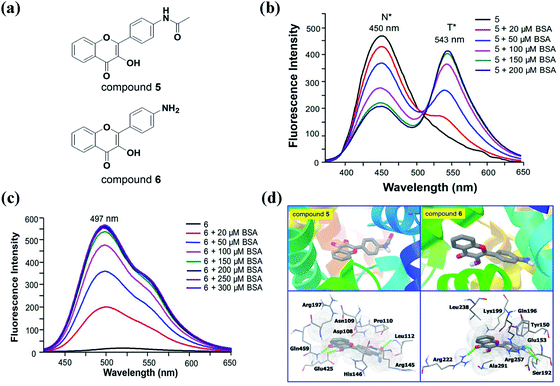 | ||
| Fig. 9 (a) The structure of compound 5 and 6; (b, c) fluorescence spectra of 20 μM compound 5 (b) and 6 (c) upon addition of different concentrations of BSA in 50 mM Tris-buffer (pH 7.4). Excitation wavelength for compound 5 = 365 nm. Excitation wavelength for compound 6 = 395 nm; (d) the interactions between compound 5 and 6 and BSA visualized by molecular docking simulations.100 | ||
Overall, we can modulate the interaction between proteins and 3HF derivatives by utilizing different proteins or designing different substituents, thereby regulating the fluorescence emission or ESIPT process of 3HF derivatives. Complex microenvironment in the cell can also act as a medium for regulating the fluorescence emission of 3HF derivatives, since many intracellular macromolecules may interact with 3HF derivatives. Based on both the intracellular microenvironment and the interaction with protein to regulate the fluorescence emission of 3HF derivatives, we can develop a variety of 3HF based ratiometric and turn-on fluorescent probes for intracellular detection.
5. Interaction with DNA
In addition to protein, DNA is another important biological macromolecule in organisms whose base sequence controls the heredity of life. The interaction of small molecules with DNA has been utilized to explore the structural and functional features of DNA.101 Small molecules bind to DNA by four modes referred to as intercalative binding within the base pairs, groove binding through van der Waal's interaction, electrostatic binding at the negatively charged DNA phosphate backbone, and bases binding through π–π stacking.102,103 At present, it has been reported that some 3HF derivatives (such as 3HF, fisetin, and quercetin) can bind to ss-DNA, ds-DNA, triplex DNA and G-quadruplex, thereby modulating their fluorescence emission and ESIPT process.103–108 Therefore, summarizing the effects of DNA binding on the fluorescence emission of 3HF derivatives has guiding significance for designing novel DNA based sensors and probes.Chattopadhyay and the co-workers found that 3HF can bind to the groove of calf thymus DNA (ctDNA) helix with a binding constant of 1.6 × 103 L mol−1, thereby facilitating the ESIPT process of 3HF.104 Increase in the tautomer fluorescence intensity (T* band) at the cost of the normal species (N* band) reveals that the combination between 3HF and ctDNA in aqueous buffer solution increases the efficiency of ESIPT. Based on the interaction with ctDNA, we can effectively improve the ESIPT efficiency of 3HF.
Unlike the groove binding mode of 3HF and ctDNA, fisetin (Fis), a 3HF derivative containing four hydroxyl groups, can utilize these hydroxyl groups to form hydrogen bonds with the base pairs, thereby intercalating into dsDNA.104,109 Interestingly, the abasic site (AP site) embedded in dsDNA can be used as a binding pocket favorable for the ESIPT process of Fis.106,107 Besides, compared to other flavonoids, Fis also selectively binds with triplex DNA to emit intense green fluorescence in contrast to binding with ssDNA, dsDNA, i-motif, and G-quadruplexes.105 Overall, 3HF derivatives with multiple hydroxyl groups, such as Fis, may also have potential as novel AP site or triplex DNA based probes.
In addition to binding to BSA, we found that binding to G-quadruplex also greatly enhances the fluorescence emission (N* band) of compound 6 (Fig. 10a).103 A variety of G-quadruplexes (TBA is the only exception) can selectively amplify the fluorescent signals of compound 6, whereas the presence of ssDNA (G4TTA-C) and ctDNA have moderate effects on the emission.103 Through molecular docking simulations, we found that compound 6 forms strong interactions with parallel G-quadruplex (such as c-MYC) through multiple π–π stacks and one hydrogen bond, and only forms weak interactions with ssDNA and TBA through hydrogen bonds (Fig. 10b). The strong π–π stacking interactions are formed between the guanines in G-quadruplex and the 3HF group, which dramatically enhances the fluorescence emission (N* band) of compound 6. Based on the enhancement of the fluorescence emission of compound 6 by G-quadruplex, we designed a fluorescent probe (compound 7) with high sensitivity and high selectivity for hydrogen sulfide in aqueous solution (Fig. 10c). The azide group of compound 7 can be reduced to an amino group by hydrogen sulfide to form compound 6. Then, using G4 to amplify the fluorescent signal of compound 6, we can detect the ultra-low level of hydrogen sulfide (limit of detection as low as 2.64 nM) in the aqueous solution. Therefore, the strong π–π stacking interaction between the 3HF group and the guanines in G-quadruplex can amplify the fluorescent signal of 3HF derivatives, which has broad application prospects in designing 3HF derivatives as G-quadruplex based fluorescent probes and sensors.
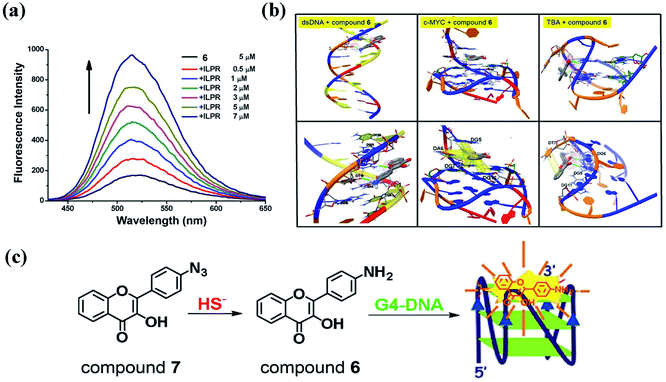 | ||
| Fig. 10 (a) Fluorescence spectra of compound 6 in titrations with G-quadruplexe ILPR. Excitation wavelength = 395 nm; (b) the interactions between compound 6 and ds-DNA, the parallel G-quadruplex (c-MYC) and the anti-parallel quadruplex (TBA); (c) compound 7 recognizing hydrogen sulfide based on G-quadruplex.103 | ||
3HF derivatives with a small number of hydroxyl groups cannot form strong hydrogen bonds with DNA and cannot insert into the dsDNA, but they can bind to the groove of dsDNA to increase the efficiency of ESIPT. For the 3HF derivatives with more hydroxyl groups (such as Fis), they can form multiple hydrogen bonds with DNA to insert into the AP site of dsDNA or bind to the two terminals of the triplex DNA, which would facilitate the ESIPT process of 3HF derivatives. Moreover, the 3HF group can also form a strong π–π stacking interaction with G-quadruplexes to enhance the fluorescence emission (N* band) of 3HF derivatives (such as compound 6). Through binding with various types of DNA, we can modulate the ESIPT process and N* band emission of 3HF derivatives, all of which can help us design a plenty of DNA-based 3HF derivatives for fluorescence detection.
6. Conclusions
This review summarizes various regulating strategies for the fluorescence emission of 3HF derivatives based on chelation with metal ions and interaction with solvents, proteins, and DNAs. Analysis of these regulatory strategies can help us design new 3HF based fluorescence sensors and probes for the detection of microenvironment, metal ions, structures of proteins and DNA, and other molecules.Complexation of metal ions at α-hydroxy-carbonyl site of the 3HF group can suppress the ESIPT process of with 3HF derivatives, and enhance or quench the fluorescence emission of the N* band. Utilizing different electron donor groups to participate in the coordination with metal ions, we can also regulate the PeT process and the fluorescence emission of 3HF derivatives. Therefore, we can design various novel 3HF based fluorescent probes for the detection of metal ions in solution or in the biological system. Based on the intracellular microenvironment and the interaction between protein and 3HF derivatives, we can also modulate the fluorescence emission and ESIPT process of 3HF derivatives, and develop a variety of ratiometric and turn-on fluorescent probes for intracellular detection or cell imaging. Interaction with DNA is another effective method to regulate the fluorescence emission of 3HF derivatives, so we can also design a plenty of DNA based 3HF derivatives for fluorescence detection.
For the development of 3HF derivatives as novel fluorescent probes and sensors, we still need to make efforts in the following areas. First, suitable 3HF derivatives for the fluorescence detection in different microenvironment need to be designed. Second, when the detection object is metal ion, we need to design suitable coordination groups and consider the differences in the electronic arrangement of different metal ions, and then speculate the effects of coordination on the fluorescence emission of 3HF derivatives. Third, if the interaction with protein is used to regulate the fluorescence emission of 3HF derivatives, we should consider the influences of the type and source of proteins and the type of substituents on the interaction between the 3HF derivatives and protein. Moreover, when designing novel DNA based 3HF derivatives for fluorescence detection, we need to consider the effects of hydroxyl substituents and DNA conformation on the binding patterns of 3HF derivatives and DNA, and analyze how their binding regulates the fluorescence emission and ESIPT process of 3HF derivatives. In order to eliminate the interferences by microenvironment and false signals in cell imaging application, advanced probe design should combine the ESIPT process of 3HF derivatives with other photophysical process, such as photo-induced electron transfer, intramolecular charge transfer, aggregation-induced emission, to produce probes with dual/triple interplaying sensing mechanisms.110–112 Besides, designing novel 3HF-based fluorophores with a longer emission wavelength will facilitate the realization of fluorescence imaging in vivo.113,114
3HF derivatives are a class of highly promising ESIPT molecules for cell imaging, based on the existence of metal ions and hydrophobic microenvironment within cells. The use of multiple regulatory strategies to modulate the fluorescence emission and ESIPT process of 3HF derivatives encourages us to develop various 3HF based fluorescence probes and sensors. We thus hope that, from reading this review, the reader can gain fundamental knowledge of these regulatory strategies, and also gain inspirations for designing 3HF derivatives as novel fluorescence probes and sensors for imaging.
Author contributions
Xueke Zhao: writing – review and editing. Xiang Li: writing – original draft. Shuyu Liang: writing – review and editing. Xiongwei Dong: supervision, funding acquisition, writing – review and editing. Zhe Zhang: funding acquisition, writing, – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Dan Zhang at Central China Normal University for help in checking this paper. This work was supported by the National Natural Science Foundation of China [21701128]; Hubei Provincial Natural Science Foundation of China [2017CFB206]; Educational Commission of Hubei Province of China [Q20171601]; Guangdong Natural Science Foundation [2019A1515011358]; Science and Technology Research Project of Guangzhou [202002030257].Notes and references
- A. Douhal, S. K. Kim and A. H. Zewail, Nature, 1995, 378, 260–263 CrossRef CAS PubMed
.
- A. Douhal, F. Lahmani and A. H. Zewail, Chem. Phys., 1996, 207, 477–498 CrossRef CAS
.
- M. Rini, B. Z. Magnes, E. Pines and E. T. Nibbering, Science, 2003, 301, 349–352 CrossRef CAS PubMed
.
- X. Zhang, L. Guo, F. Y. Wu and Y. B. Jiang, Org. Lett., 2003, 5, 2667–2670 CrossRef CAS PubMed
.
- X. Peng, Y. Wu, J. Fan, M. Tian and K. Han, J. Org. Chem., 2005, 70, 10524–10531 CrossRef CAS PubMed
.
- A. Weller, Zeitschrift für Elektrochemie, 1952, 56, 662–668 CAS
.
- P. T. Chou, J. H. Liao, C. Y. Wei, C. Y. Yang, W. S. Yu and Y. H. Chou, J. Am. Chem. Soc., 2000, 122, 986–987 CrossRef CAS
.
- W. T. Hsieh, C. C. Hsieh, C. H. Lai, Y. M. Cheng, M. L. Ho, K. K. Wang, G. H. Lee and P. T. Chou, ChemPhysChem, 2008, 9, 293–299 CrossRef CAS PubMed
.
- W. P. Hu, J. L. Chen, C. C. Hsieh and P. T. Chou, Chem. Phys. Lett., 2010, 485, 226–230 CrossRef CAS
.
- P. T. Chou, W. S. Yu, C. Y. Wei, Y. M. Cheng and C. Y. Yang, J. Am. Chem. Soc., 2001, 123, 3599–3600 CrossRef CAS
.
- T. Iijima, A. Momotake, Y. Shinohara, T. Sato, Y. Nishimura and T. Arai, J. Phys. Chem. A, 2010, 114, 1603–1609 CrossRef CAS
.
- J. Goodman and L. E. Brus, J. Am. Chem. Soc., 1978, 100, 7472–7474 CrossRef CAS
.
- C. Azarias, S. Budzák, A. D. Laurent, G. Ulrich and D. Jacquemin, Chem. Sci., 2016, 7, 3763–3774 RSC
.
- B. Dick and N. P. Ernsting, J. Phys. Chem., 1987, 91, 4261–4265 CrossRef CAS
.
- P. T. Chou, Y. C. Chen, W. S. Yu and Y. M. Cheng, Chem. Phys. Lett., 2001, 340, 89–97 CrossRef CAS
.
- Y. Liu, J. Zhao, Y. Wang, J. Tian, X. Fei and H. Wang, J. Mol. Liq., 2017, 233, 303–309 CrossRef CAS
.
- A. C. Sedgwick, L. Wu, H. H. Han, S. D. Bull, X. P. He, T. D. James, J. L. Sessler, B. Z. Tang, H. Tian and J. Yoon, Chem. Soc. Rev., 2018, 47, 8842–8880 RSC
.
- M. W. Chung, T. Y. Lin, C. C. Hsieh, K. C. Tang, H. Fu, P. T. Chou, S. H. Yang and Y. Chi, J. Phys. Chem. A, 2010, 114, 7886–7891 CrossRef CAS PubMed
.
- T. C. Chien, L. G. Dias, G. M. Arantes, L. G. C. Santos, E. R. Triboni, E. L. Bastos and M. J. Politi, J. Photochem. Photobiol., A, 2008, 194, 37–48 CrossRef CAS
.
- S. J. Schmidtke, D. F. Underwood and D. A. Blank, J. Am. Chem. Soc., 2004, 126, 8620–8621 CrossRef CAS PubMed
.
- M. Kijak, Y. Nosenko, A. Singh, R. P. Thummel and J. Waluk, J. Am. Chem. Soc., 2007, 129, 2738–2739 CrossRef CAS
.
- Y. Nosenko, G. Wiosna-Sałyga, M. Kunitski, I. Petkova, A. Singh, W. J. Buma, B. Brutchy and J. Waluk, Angew. Chem., Int. Ed., 2008, 120, 6126–6129 CrossRef
.
- C. L. Chen, C. W. Lin, C. C. Hsieh, C. H. Lai, G. H. Lee, C. C. Wang and P. T. Chou, J. Phys. Chem. A, 2008, 113, 205–214 CrossRef
.
- D. LeGourriérec, V. A. Kharlanov, R. G. Brown and W. Rettig, J. Photochem. Photobiol., A, 2000, 130, 101–111 CrossRef
.
- E. Hadjoudis and I. M. Mavridis, Chem. Soc. Rev., 2004, 33, 579–588 CAS
.
- A. S. Klymchenko, Y. Mély, A. P. Demchenko and G. Duportail, Biochim. Biophys. Acta, Biomembr., 2004, 1665, 6–19 CrossRef CAS PubMed
.
- A. S. Klymchenko, S. Oncul, P. Didier, E. Schaub, L. Bagatolli, G. Duportail and Y. Mély, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 495–499 CrossRef CAS PubMed
.
- D. Treutter, Environ. Chem. Lett., 2006, 4, 147–157 CrossRef CAS
.
- P. H. Kim, K. H. Son, H. W. Chang and S. S. Kang, J. Pharmacol. Sci., 2004, 96, 229–245 CrossRef
.
- K. H. Lu, P. N. Chen, Y. H. Hsieh, C. Y. Lin, F. Y. Cheng, P. C. Chiu, S. C. Chu and Y. S. Hsieh, Food Chem. Toxicol., 2016, 97, 177–186 CrossRef CAS PubMed
.
- C. G. M. Heijnen, G. R. M. M. Haenen, F. A. A. Van Acker, W. J. F. Van der Vijgh and A. Bast, In Vitro, 2001, 15, 3–6 CrossRef CAS
.
- Q. Lang, H. Zhang, J. Li, F. Xie, Y. Zhang, B. Wan and L. Yu, Mol. Biol. Rep., 2010, 37, 1577–1583 CrossRef CAS
.
- F. I. Baptista, A. G. Henriques, A. Silva, J. Wiltfang and O. A. B. da Cruze Silva, ACS Chem. Neurosci., 2014, 5, 83–92 CrossRef CAS PubMed
.
- B. Dereka, R. Letrun, D. Svechkarev, A. Rosspeintner and E. Vauthey, J. Phys. Chem. B, 2014, 119, 2434–2443 CrossRef PubMed
.
- J. Zhao, S. Ji, Y. Chen, H. Guo and P. Yang, Phys. Chem. Chem. Phys., 2012, 14, 8803–8817 RSC
.
- B. J. Schwartz, L. A. Peteanu and C. B. Harris, J. Phys. Chem., 1992, 96, 3591–3598 CrossRef CAS
.
- A. Sytnik, D. Gormin and M. Kasha, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 11968–11972 CrossRef CAS PubMed
.
- P. K. Sengupta and M. Kasha, Chem. Phys. Lett., 1979, 68, 382–385 CrossRef CAS
.
- V. I. Tomin, A. P. Demchenko and P. T. Chou, J. Photochem. Photobiol., C, 2015, 22, 1–18 CrossRef CAS
.
- A. S. Klymchenko and A. P. Demchenko, Phys. Chem. Chem. Phys., 2003, 5, 461–468 RSC
.
- D. D. Pant, H. C. Joshi, P. B. Bisht and H. B. Tripathi, Chem. Phys., 1994, 185, 137–144 CrossRef CAS
.
- P. B. Bisht, H. B. Tripathi and D. D. Pant, J. Photochem. Photobiol., A, 1995, 90, 103–108 CrossRef CAS
.
- P. T. Chou, M. L. Martinez and J. H. Clements, J. Phys. Chem., 1993, 97, 2618–2622 CrossRef CAS
.
- G. J. Woolfe and P. J. Thistlethwaite, J. Am. Chem. Soc., 1981, 103, 6916–6923 CrossRef CAS
.
- D. McMorrow and M. Kasha, J. Am. Chem. Soc., 1983, 105, 5133–5134 CrossRef CAS
.
- A. J. Strandjord and P. F. Barbara, J. Phys. Chem., 1985, 89, 2355–2361 CrossRef CAS
.
- M. Kasha, J. Chem. Soc., Faraday Trans. 2, 1986, 82, 2379–2392 RSC
.
- A. S. Klymchenko, C. Kenfack, G. Duportail and Y. Mély, J. Chem. Sci., 2007, 119, 83–89 CrossRef CAS
.
- A. S. Klymchenko, V. G. Pivovarenko and A. P. Demchenko, Spectrochim. Acta, Part A, 2003, 59, 787–792 CrossRef
.
- A. S. Klymchenko, V. G. Pivovarenko, T. Ozturk and A. P. Demchenko, New J. Chem., 2003, 27, 1336–1343 RSC
.
- A. S. Klymchenko and A. P. Demchenko, New J. Chem., 2004, 28, 687–692 RSC
.
- D. McMorrow and M. Kasha, J. Phys. Chem., 1984, 88, 2235–2243 CrossRef CAS
.
- A. S. Klymchenko, V. G. Pivovarenko and A. P. Demchenko, J. Phys. Chem. A, 2003, 107, 4211–4216 CrossRef CAS
.
- V. V. Shynkar, A. S. Klymchenko, E. Piemont, A. P. Demchenko and Y. Mely, J. Phys. Chem. A, 2004, 108, 8151–8159 CrossRef CAS
.
- A. S. Klymchenko, G. Duportail, T. Ozturk, V. G. Pivovarenko, Y. Mély and A. P. Demchenko, Chem. Biol., 2002, 9, 1199–1208 CrossRef CAS
.
- J. E. Kwon and S. Y. Park, Adv. Mater., 2011, 23, 3615–3642 CrossRef CAS PubMed
.
- H. Gao and X. Wu, Chem. Heterocycl. Compd., 2018, 54, 125–129 CrossRef CAS
.
- C. Lapouge, L. Dangleterre and J. P. Cornard, J. Phys. Chem. A, 2006, 110, 12494–12500 CrossRef CAS PubMed
.
- G. J. Park, M. M. Lee, G. R. You, Y. W. Choi and C. Kim, Tetrahedron Lett., 2014, 55, 2517–2522 CrossRef CAS
.
- X. Han, M. R. Kumar, A. Hoogerbrugge, K. K. Klausmeyer, M. M. Ghimire, L. M. Harris, M. A. Omary and P. J. Farmer, Inorg. Chem., 2018, 57, 2416–2424 CrossRef CAS PubMed
.
- S. Sinha, B. Chowdhury and P. Ghosh, Inorg. Chem., 2016, 55, 9212–9220 CrossRef CAS
.
- L. Xue, G. Li, D. Zhu, Q. Liu and H. Jiang, Inorg. Chem., 2012, 51, 10842–10849 CrossRef CAS PubMed
.
- Y. Jin, S. Wang, Y. Zhang and B. Song, Sens. Actuators, B, 2016, 225, 167–173 CrossRef CAS
.
- Y. Xu and Y. Pang, Chem. Commun., 2010, 46, 4070–4072 RSC
.
- X. Li, J. Li, X. Dong, X. Gao, D. Zhang and C. Liu, Sens. Actuators, B, 2017, 245, 129–136 CrossRef CAS
.
- G. T. Castro and S. E. Blanco, Spectrochim. Acta, Part A, 2004, 60, 2235–2241 CrossRef CAS PubMed
.
- É. Balogh-Hergovich, J. Kaizer and G. Speier, J. Mol. Catal. A: Chem., 2000, 159, 215–224 CrossRef
.
- R. F. De Souza and W. F. De Giovani, Spectrochim. Acta, Part A, 2005, 61, 1985–1990 CrossRef
.
- P. Viswanathan, V. Sriram and G. Yogeeswaran, J. Agric. Food Chem., 2000, 48, 2802–2806 CrossRef CAS
.
- D. Wang, X. Fan, S. Sun, S. Du, H. Li, J. Zhu, Y. Tang, M. Chang and Y. Xu, Sens. Actuators, B, 2018, 264, 304–311 CrossRef CAS
.
- J.-L. Fillaut, J. Andriès, R. D. Marwaha, P.-H. Lanoë, L. Olivier, L. Toupet and J. A. Gareth Williams, J. Organomet. Chem., 2008, 693, 228–234 CrossRef CAS
.
- D. Svechkarev, B. Dereka and A. Doroshenko, J. Phys. Chem. A, 2011, 115, 4223–4230 CrossRef CAS PubMed
.
- V. K. Gupta, N. Mergu and A. K. Singh, Sens. Actuators, B, 2014, 202, 674–682 CrossRef CAS
.
- Y. Su, H. Dong, M. Li, C. Lai, C. Huang and Q. Yong, Polymers, 2019, 11, 1377 CrossRef CAS
.
- M. Aurélien, F. Cécilia and C. Jean-Paul, J. Phys. Chem. B, 2018, 122, 8943–8951 CrossRef PubMed
.
- J. P. Cornard, L. Dangleterre and C. Lapouge, Chem. Phys. Lett., 2006, 419, 304–308 CrossRef CAS
.
- C. A. Rice-Evans, N. J. Miller and G. Paganga, Free Radical Biol. Med., 1996, 20, 933–956 CrossRef CAS PubMed
.
- F. Wang, L. Wang, X. Chen and J. Yoon, Chem. Soc. Rev., 2014, 43, 4312–4324 RSC
.
- Z. A. Li, X. Lou, H. Yu, Z. Li and J. Qin, Macromolecules, 2008, 41, 7433–7439 CrossRef CAS
.
- S. Anbu, R. Ravishankaran, M. F. C. Guedes da Silva, A. A. Karande and A. J. Pombeiro, Inorg. Chem., 2014, 53, 6655–6664 CrossRef CAS PubMed
.
- M. Grazul and E. Budzisz, Coord. Chem. Rev., 2009, 253, 2588–2598 CrossRef CAS
.
- A. L. Hopkins and C. R. Groom, Nat. Rev. Drug Discovery, 2002, 1, 727–730 CrossRef CAS PubMed
.
- W. J. Jusko and M. Gretch, Drug Metab. Rev., 1976, 5, 43–140 CrossRef CAS
.
- D. C. Turner and L. Brand, Biochemistry, 1968, 7, 3381–3390 CrossRef CAS PubMed
.
- T. Yuan, A. M. Weljie and H. J. Vogel, Biochemistry, 1998, 37, 3187–3195 CrossRef CAS
.
- P. C. Ferriola and V. Cody, Biochem. Pharmacol., 1989, 38, 1617–1624 CrossRef CAS PubMed
.
- R. I. Brinkworth, M. J. Stoermer and D. P. Fairlie, Biochem. Biophys. Res. Commun., 1992, 188, 631–637 CrossRef CAS
.
- M. J. Arts, G. R. Haenen, L. C. Wilms, S. A. Beetstra, C. G. Heijnen, H. P. Voss and A. Bast, Food Chem., 2002, 50, 1184–1187 CrossRef CAS
.
- B. Sengupta and P. K. Sengupta, Biochem. Biophys. Res. Commun., 2002, 299, 400–403 CrossRef CAS
.
- F. Zsila, Z. Bikádi and M. Simonyi, Biochem. Pharmacol., 2003, 65, 447–456 CrossRef CAS
.
- J. Tian, J. Liu, J. Xie, X. Yao, Z. Hu and X. Chen, J. Photochem. Photobiol., B, 2004, 74, 39–45 CrossRef CAS PubMed
.
- C. Dufour and O. Dangles, Biochim. Biophys. Acta, Gen. Subj., 2005, 1721, 164–173 CrossRef CAS
.
- V. V. Sentchouk and E. V. Bondaryuk, J. Appl. Spectrosc., 2007, 74, 731–737 CrossRef CAS
.
- A. Banerjee, K. Basu and P. K. Sengupta, J. Photochem. Photobiol., B, 2008, 90, 33–40 CrossRef CAS PubMed
.
- X. Hu, S. Cui and J. Qin Liu, Spectrochim. Acta, Part A, 2010, 77, 548–553 CrossRef
.
- S. Ercelen, A. S. Klymchenko, Y. Mély and A. P. Demchenko, Int. J. Biol. Macromol., 2005, 35, 231–242 CrossRef CAS
.
- N. Jiang, C. Yang, X. Dong, X. Sun, D. Zhang and C. Liu, Org. Biomol. Chem., 2014, 12, 5250–5259 RSC
.
- A. S. Klymchenko, T. Ozturk, V. G. Pivovarenko and A. P. Demchenko, Tetrahedron Lett., 2001, 42, 7967–7970 CrossRef CAS
.
- S. Ercelen, A. S. Klymchenko and A. P. Demchenko, Anal. Chim. Acta, 2002, 464, 273–287 CrossRef CAS
.
- Y. Hu, X. Gao, X. Li, H. Liang, D. Zhang and C. Liu, Sens. Actuators, B, 2018, 262, 144–152 CrossRef CAS
.
- S. E. Osborne and A. D. Ellington, Chem. Rev., 1997, 97, 349–370 CrossRef CAS PubMed
.
- D. Sarkar, P. Das, S. Basak and N. Chattopadhyay, J. Phys. Chem. B, 2008, 112, 9243–9249 CrossRef CAS PubMed
.
- Y. Hu, X. Li, Z. Zhang, G. Chen, H. Liang, D. Zhang and C. Liu, Sens. Actuators, B, 2018, 272, 308–313 CrossRef CAS
.
- B. Jana, S. Senapati, D. Ghosh, D. Bose and N. Chattopadhyay, J. Phys. Chem. B, 2011, 116, 639–645 CrossRef
.
- Y. Wang, Y. Hu, T. Wu, X. Zhou and Y. Shao, Anal. Chem., 2015, 87, 11620–11624 CrossRef CAS
.
- S. Xu, Y. Shao, K. Ma, Q. Cui, G. Liu and F. Wu, Sens. Actuators, B, 2012, 171, 666–671 CrossRef
.
- S. Xu, Y. Shao, K. Ma, Q. Cui, G. Liu, F. Wu and M. Li, Analyst, 2011, 136, 4480–4485 RSC
.
- A. Capan, M. S. Bostan, E. Mozioglu, M. Akoz, A. C. Goren, M. S. Eroglu and T. Ozturk, J. Photochem. Photobiol., B, 2015, 153, 391–396 CrossRef CAS
.
- A. M. Pyle, J. P. Rehmann, R. Meshoyrer, C. V. Kumar, N. J. Turro and J. K. Barton, J. Am. Chem. Soc., 1989, 111, 3051–3058 CrossRef CAS
.
- S. Chen, P. Hou, B. Zhou, X. Song and J. W. Foley, RSC Adv., 2013, 3, 11543–11546 RSC
.
- C. S. Abeywickrama, K. A. Bertman and Y. Pang, Chem. Commun., 2019, 55, 7041–7044 RSC
.
- R. Long, C. Tang, J. Xu, T. Li, C. Tong, Y. Guo, S. Shi and D. Wang, Chem. Commun., 2019, 55, 10912–10915 RSC
.
- P. Hou, H. Li and S. Chen, Tetrahedron, 2016, 72, 3531–3534 CrossRef CAS
.
- C. Song, H. Peng and X. Song, Sens. Actuators, B, 2015, 221, 951–955 CrossRef
.
Footnote |
| † X. K. Zhao and X. Li contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |