Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multivalent glycan arrays
Marco
Mende
a,
Vittorio
Bordoni
 a,
Alexandra
Tsouka
ab,
Felix F.
Loeffler
a,
Alexandra
Tsouka
ab,
Felix F.
Loeffler
 a,
Martina
Delbianco
a,
Martina
Delbianco
 a and
Peter H.
Seeberger
a and
Peter H.
Seeberger
 *ab
*ab
aDepartment of Biomolecular Systems, Max Planck Institute of Colloids and Interfaces, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: peter.seeberger@mpikg.mpg.de
bDepartment of Chemistry and Biochemistry, Freie Universität Berlin, Arnimallee 22, 14195 Berlin, Germany
First published on 12th July 2019
Abstract
Glycan microarrays have become a powerful technology to study biological processes, such as cell–cell interaction, inflammation, and infections. Yet, several challenges, especially in multivalent display, remain. In this introductory lecture we discuss the state-of-the-art glycan microarray technology, with emphasis on novel approaches to access collections of pure glycans and their immobilization on surfaces. Future directions to mimic the natural glycan presentation on an array format, as well as in situ generation of combinatorial glycan collections, are discussed.
1. Introduction
Glycans decorate the surface of many cells, forming a thick layer (glycocalyx) that mediates a variety of important cellular processes.1 This 100 nm–1 mm thick glycan layer comprises highly diverse structures, including glycoproteins, glycolipids, and glycopolymers. Several complex biological processes, such as protein folding, cell–cell interaction, cell adhesion, and signaling, are the result of the interactions of glycans with themselves (carbohydrate–carbohydrate interactions, CCIs) or with glycan binding proteins (carbohydrate–protein interactions, CPIs).2–4 In addition, pathogens use these glycans as receptors for the attachment to host cells and subsequent invasion.5,6 At the same time, pathogenic glycans are recognized by the immune system, which initiate the immune response.7,8 Pathological events, such as tumor metastasis, inflammation, and infections, are all mediated by glycan–protein interactions.A better understanding of these CPIs is of fundamental importance. Yet, in comparison to polynucleotides and proteins, the study of glycans and CPIs has been slower for multiple reasons: the complexity of carbohydrate synthesis and their difficult isolation from natural sources has hampered a detailed analysis of such compounds. The limited access to collections of pure materials precluded high-throughput screening formats. Even though glycan arrays have become extremely popular and primary analytical tools for the study of CPIs,9–11 they are limited due to glycan availability. Additionally, CPIs are very weak (typically in the micromolar range) and glycan binding proteins can often interact with many substrates with low specificity. Nature’s strategy to enhance binding strength and specificity is multivalency, where multiple carbohydrate units bind to one protein to gain stronger affinity than the sum of the single contributions.12–14 Chemists have aimed to reproduce nature, developing several synthetic multivalent systems that mimic natural supramolecular interactions.15 Nevertheless, recreating the binding thermodynamics of natural interfaces in a microarray format, is extremely challenging.16 Different approaches aimed to mimic the natural glycan presentation on an array surface. The most common approach involves the direct printing of glycans, controlling the density by varying the concentration or by surface functionalization.17–19 Alternatively, prearranged multivalent systems, based on natural or unnatural scaffolds, can be immobilized on surfaces, aiming at more defined glycan presentation.9,20
A challenging approach is the direct synthesis of glycans or multivalent glycan systems on the array. However, in comparison to other biomolecules, chemical carbohydrate synthesis on surfaces is far more difficult, due to the demanding reaction parameters. Only the synthesis of disaccharides has been achieved to date.21 Enzymatic synthesis on surfaces is more common, e.g., for the synthesis of N-glycans or the discovery of glycosyltransferases.22–25 Yet, whether glycans can also be synthesized in situ in a molecularly defined and multivalent fashion, remains to be shown.
We review the state of the art of glycan microarrays, from access to glycan collections, to surface immobilization, and analysis. We will focus on current approaches to mimic natural interfaces and new directions in surface functionalization. Moreover, we will describe how simple glycans and more complex multivalent scaffolds are printed or grown from surfaces to elucidate important cellular processes.
2. Access to glycan collections
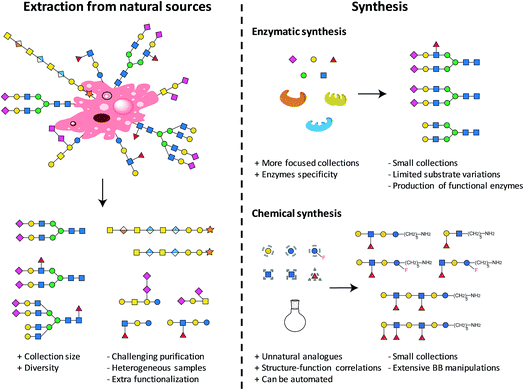
The first step towards the production of a glycan microarray is the identification of suitable glycans. Two approaches are available and currently used to access glycans (see Fig. 1): isolation from natural sources and/or synthesis (enzymatic or chemical). Natural glycans can be readily obtained from animal tissues, plant material, or from cultured pathogens.26 Large collections in terms of size and diversity could be accessed, when completely uncharacterized binders need to be identified.10 Nevertheless, the isolation procedures and characterization of the final carbohydrates could be extremely challenging, often resulting in mixtures of compounds. Heterogeneous samples, often containing minor impurities, could culminate in non-reproducible results. Moreover, extracted glycans generally require an extra functionalization step for immobilization on surfaces. Compound collections obtained from chemical synthesis are generally smaller, more focused, and less diverse. Generating a set of related glycans, with the possibility of including non-natural glycans, is of great interest for the elucidation of structure–activity relationships. Chemically obtained compounds are highly pure, reducing the possibility of false results. A reactive linker can be easily installed during synthesis, facilitating subsequent immobilization. Using these two approaches, many glycans were prepared and printed on arrays.9,10 The microarray with the currently largest diversity is represented by the mammalian array (version 5.3) of the Consortium for Functional Glycomics (CFG), which includes more than 600 synthetic and isolated compounds.26 A microbial glycan microarray is also available, including more than 300 carbohydrates. However, covering the huge diversity of microbial glycans, often containing rare sugars, remains a major challenge.27 Efforts to access such glycans in a well-defined manner are still needed.2.1. Automated glycan assembly
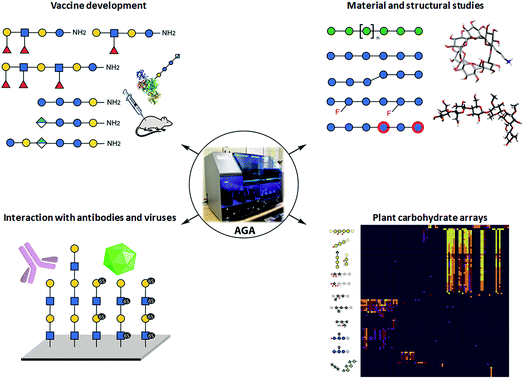
Collections of well-defined glycans are fundamental tools to elucidate glycan interactions. With the aim to explore the natural and unnatural diversity of glycans, systematic strategies for the chemical, enzymatic, and chemo-enzymatic synthesis of glycans were developed. Nevertheless, the challenging installation of the glycosidic linkage, that requires regio- and stereo-control, poses a bottleneck. Enzymatic synthesis relies on the specificity of the enzymes to form the desired glycosidic linkage, but to date, it is limited by the availability of suitable glycosyltransferases.28 Such enzymes are extremely efficient with natural substrates, but often tolerate only limited substrate variations, hindering access to chemically modified glycans and unnatural structures. In addition, the in vitro production of functional enzymes is sometimes troublesome.29 Despite several challenges, enzymatic synthesis remains a powerful option, when poorly reactive monosaccharides such as sialic acid or particularly challenging linkages such as β-mannosides need to be installed.30 Efforts to standardize this process resulted in two fully automated systems.31,32Chemical synthesis offers the unique opportunity to access well-defined natural and unnatural structures. Collections of complex synthetic glycans, including heparin sulfate glycans, GPI-anchors, and high-mannose oligosaccharides, were used to create custom arrays to characterize lectin and antibody specificity and to study the human response to infections and allergies.33 The biggest drawback of this approach is the enormous synthetic effort required. Automated Glycan Assembly (AGA) speeds up the process, allowing for quick and reliable access to glycans.34,35 The sequential addition of sugar building blocks (BBs) on a solid support replaces the purification steps with simple washing cycles. The coupling cycle, consisting of glycosylation, capping, and deprotection, has been optimized to achieve nearly quantitative conversion in around 1.5 h.36,37 Moreover, the glycan is attached to the solid support through a linker that, upon UV irradiation, liberates the target glycan already equipped with an amino-linker for subsequent surface functionalization.38 Collections of natural and unnatural glycans found applications in vaccine development,39,40 materials science,36,41,42 and structural studies37 (see Fig. 2). Well-defined linear β(1,3) and branched β(1,3)β(1,6) glucans permitted to conclude that most individuals form antibodies that bind to both linear (protective) and branched (non-protective) epitope.43 Synthetic keratan sulfate (KS) analogues, with different sulfation patterns, helped to identify the specific interaction between the disulfated KS tetrasaccharide and the adeno-associated virus AAVrh10 gene-therapy vector (see Fig. 2).44 Frameshifts of the S. pneumoniae serotype 8 (ST8) capsular polysaccharides were used to identify the glycotopes recognized by antibodies against ST8. The insights were essential for the preparation of a semisynthetic Streptococcus pneumoniae serotype 8 glycoconjugate vaccine candidate.40 AGA was exploited to determine the binding epitopes of many plant cell-wall glycan-directed mAbs.45,46 A total of 88 synthetic oligosaccharides, including arabinogalactan-, rhamnogalacturonan-, xylan-, and xyloglucan were printed on a microarray aiming to comprehensively map the epitopes of plant cell-wall glycan-directed antibodies (see Fig. 2).47
3. Printing on surfaces
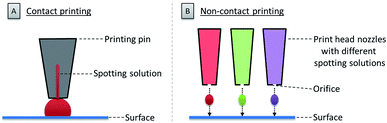
Once the glycan collection has been produced, the glycans are printed onto the array surface. High accuracy and reproducibility are essential for a reliable microarray. Two technologies are mainly used to deposit bioactive molecules, such as carbohydrates, on a reactive surface. These rely on contact and non-contact printing (see Fig. 3).48Contact technologies (see Fig. 3A) are naturally more precise, mainly relying on pin printing and microstamping of arrays. A pin printing setup consists of a robotically controlled print head, equipped with one to dozens of differently shaped solid pins. The pins soak a certain volume of a spotting solution (dissolved biomolecule or building blocks) from wells of a microtiter plate upon dipping. Nanoliters of the solution can then be deposited as a droplet on the reactive surface by bringing the pins in contact with the surface. The transfer process relies on favorable surface energies between the spotting solution, the surface, and the pin. An alternative to pin printing is microcontact printing, where crosslinked polydimethylsiloxane (PDMS) microstamps with micro-features are used.49 Spray-on or robotic feature–feature ink transfer is applied to coat the stamp with the spotting solution. The substance is then transferred to the surface upon contact between the stamp and the surface. This technique is mainly used to array one compound on a surface, while pin printing allows for the deposition of different molecules at the same time. In both cases, extensive washing steps and refilling after iterative cycles are necessary.
Non-contact printing technologies (see Fig. 3B) rely on the ejection of spotting solution from a reservoir through an orifice as a droplet or stream onto the microarray surface. Common inkjet printing technology uses a solution of a dissolved biomolecule or building block, serving as the “ink”. The solutions are ejected from a cartridge by a print head nozzle at a distance of 1–5 mm from the surface. The ejection process can be triggered by mainly three different methods: piezo actuation, valve-jet, or thermal inkjet. All three methods are based on a reversible and rapid change of pressure within the cartridge to release small droplets of the spotting solution. Non-contact printing approaches are highly flexible, since they allow for a fast switching between various cartridges and frequent refilling is avoided. Furthermore, because the method is contact-free, there is no risk of surface disruption. Possible clogging of the nozzle and process-related contaminations are the main drawback of this technique.
The combinatorial laser-induced transfer method (cLIFT) helps to circumvent contamination and clogging issues.50 Currently the method is restricted to the chemical synthesis of peptide and peptoid51 arrays, but may be expanded to glycan array synthesis. Novel micro- and nanoprinting technologies exploit cantilevers from atomic force microscopy to pattern surfaces, such as the well-known dip-pen nanolithography.52 Meanwhile, the technique evolved to sophisticated microfluidic and lithographic setups, enabling photochemical patterning of surfaces with different monosaccharides in high resolution.53 Another recent scanning probe approach shows the layer-by-layer printing and synthesis of peptides with a resolution of 50 μm.54 After deposition of the compounds onto the surface, immobilization can be achieved in many different approaches.
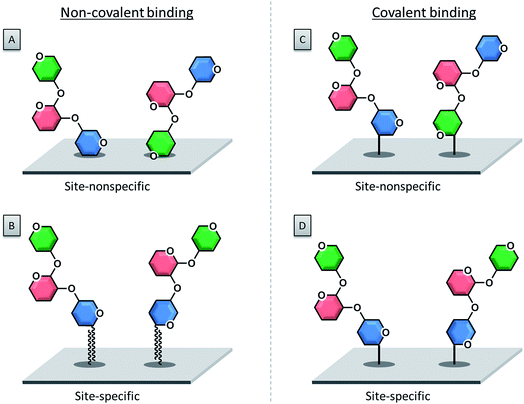
3.1. Non-covalent immobilization
The first immobilization of glycans onto a surface was reported in 2002, following a non-covalent adsorption approach.55 Non-covalent attachment of either modified or unmodified glycans to a surface is mediated by electrostatic interactions, hydrogen bonds or van der Waals forces. Today the selective covalent attachment of sugar molecules to a microarray is preferred, because it results in more stable and well-defined binding sites, enabling more precise biomolecular interactions.With this approach, unmodified polysaccharides were spotted onto a nitrocellulose-coated glass slide.55,56 Charged polysaccharides like heparin are particularly suitable for this approach, since the negatively charged sulfate groups can be efficiently attached to positively charged poly-L-lysine coated glass slides via electrostatic interactions.57,58
Nitrocellulose or PVDF (polyvinylidene difluoride) membranes were used to immobilize lipid-conjugated glycans (neoglycolipids) via hydrophobic interactions (van der Waals forces). The neoglycolipids were prepared by reductive amination of the sugar compound and an amino-conjugated lipid.59–68 A similar attachment strategy used fluorous tagged glycans for immobilization on a Teflon/epoxy coated glass slide.69–71 The perfluorinated alkyl chain allows for easy purification and permits strong binding to the surface that survives extensive washing steps. Another fluorous approach was carried out on aluminum oxide coated glass slides, which were covalently functionalized with a phosphonate, tagged with a perfluoroalkyl chain.72,73 On-spot analysis via mass spectrometry was also possible. Importantly, when hydrophobic surfaces are used, a blocking step before biomolecular screenings is required.
The strong biotin–streptavidin interaction (Kd ∼ 10−15 M) was exploited to manufacture glycan arrays. Streptavidin-coated surfaces in combination with biotinylated glycans were utilized.74–77 Similarly, DNA hybridization was employed to prepare glycan microarrays. The glycans were functionalized with an oligonucleotide that was hybridized with the complementary oligonucleotide attached to a surface.78
3.2. Covalent immobilization
The covalent attachment of a glycan to a surface is usually preferred, because it minimizes the risk of compound leaching during the washing steps. Glass slides coated with a silane or thin polymer film are employed, which are functionalized with various functional groups for the coupling reaction.Photo-labile groups such as aryl(trifluoromethyl)diazirine79 or 4-azido-2,3,5,6-tetrafluorophenyl80 are common functionalities that, upon UV irradiation, turn into reactive carbene or nitrene species. These reactive compounds are able to react easily with the spotted unprotected sugar compounds via simple insertion reactions to form stable covalent bonds. Another approach makes use of phthalimide-modified surfaces.81,82 Upon UV irradiation, the carbonyl groups of phthalimide can readily undergo a photochemical hydrogen abstraction reaction with the desired sugar which ends in stable covalent bonds between the compound and the surface. The reaction between boronic acid functionalized surfaces and diols of the sugars was also exploited to produce carbohydrate microarrays.83
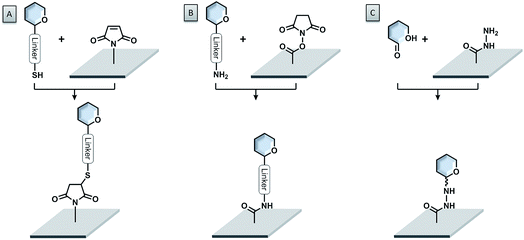
A very powerful strategy exploits the thiol–maleimide chemistry. The reaction is very fast under mild conditions and highly selective. Glycans are either functionalized with maleimide groups and coupled to thiol-coated surfaces,84–86 or thio-sugars are attached to maleimide-coated surfaces (see Fig. 5A).87–94 With this approach, even challenging glycans like glycosylphosphatidylinositols (GPIs) could be easily printed onto microarrays.95,96 The formation of disulfide bonds, either between thiosulfonate-conjugated glycans and thiol-functionalized surfaces or between thiol-conjugated glycans and pyridyl disulfide-modified surfaces, was also successful.97,98 Nevertheless, the possible oxidation of the thiol due to exposure to air can create problems.
The most widely used immobilization strategy employs amino-functionalized glycans and N-hydroxysuccinimide (NHS) ester-coated surfaces (see Fig. 5B). The reaction between these two compounds under slightly basic conditions (pH ∼ 8.5) leads to the formation of a very stable amide bond with very good selectivity.11,99–102 The NHS ester-coated glass slides are commercially available and the synthesis of amine-functionalized glycans follows standard protocols.103 Amine-modified glycans can be readily accessed also from automated strategies.35 Alternatively, the amino-modified glycans can be attached to cyanuric chloride-functionalized surfaces via nucleophilic aromatic substitution.104,105
Unprotected linker-free glycans can be immobilized in a site-specific way, using hydrazide- (see Fig. 5C) or oxyamine-modified surfaces.106,107 These functional groups are highly nucleophilic and able to react easily with the reducing ends of the glycans to form stable adducts. Similarly, an aldehyde-functionalized surface and oxyamine-modified sugars were used to prepare glycosaminoglycan microarrays.108 Another glycosaminoglycan microarray was produced on an amine-coated surface, using a deaminated heparin, bearing an aldehyde functionality.109
Epoxide-coated surfaces in combination with hydrazide-functionalized sugars offer a valuable alternative to form stable covalent bonds (see Fig. 6C).110–115 Moreover, epoxide-coated surfaces are very versatile and can be used in combination with many different nucleophiles such as amine- or thiol-conjugated carbohydrates (see Fig. 6A and B).116,117 Cycloadditions with dienophile-conjugated carbohydrates also show good selectivity and can be carried out under mild conditions. Diels–Alder reaction between a cyclopentadiene-linked sugar and a benzoquinone-coated surface,118 as well as the very fast tetrazine–norbornene inverse electron demand Diels–Alder reaction were applied.119 Nevertheless, the lack of long-term stability of some of these compounds limits their applications.
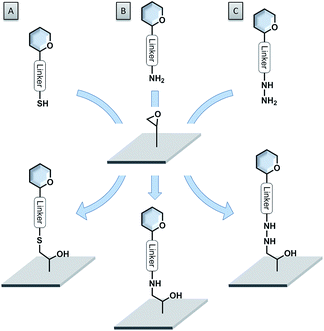 | ||
| Fig. 6 Versatility of epoxide-coated surfaces. (A) Thiol-functionalized sugar; (B) amine-functionalized sugar; (C) hydrazide-functionalized sugar. | ||
The copper-catalyzed azide–alkyne click reaction (CuAAC) was applied in glycan microarray production, because of its high selectivity and compatibility with a broad range of functional groups. Glycans functionalized with azide groups are coupled to alkyne-functionalized surfaces (or inverted functionalization).120–124 In addition, azide-modified glycans were used to prepare microarrays through chemoselective Staudinger ligation.125 Photochemical attachment of a 4-azido-2,3,5,6-tetrafluorophenyl-conjugated sugar to a polymer monolayer offered a very mild alternative.80,126 After spotting the compound onto the surface, irradiation with UV light converts the azide functionality to a reactive nitrene species, which is able to react with the polymer monolayer to from a stable covalent bond.
4. Multivalent presentation
Carbohydrate–protein interactions are very weak. However, usually multiple simultaneous interactions between several carbohydrate ligands and one receptor occur, which increases the binding strength. This concept is called multivalency. For a multivalent interaction to take place, the spatial distribution and orientation of the sugar groups are crucial.127–129 To translate this to carbohydrate microarrays, the glycan presentation and, especially the density and orientation, need to be considered in detail.In conventional array platforms, single monovalent glycans are randomly attached to the array surface via a linker, yielding a certain – but uncontrolled – multivalent display, which may be sufficient to elicit a high-avidity binding event. These systems usually rely on two-dimensional arrangements of monovalent glycans, with very little control over spatial organization. However, carbohydrate–protein interactions vary quite significantly and high glycan density may either enhance it via multivalency or suppress it via steric hindrance. Therefore, several studies were conducted to identify the optimal presentation of carbohydrates by varying the glycan concentration during printing and the flexibility of the attachment point.130,131 To date, full control on spatial organization is still a big challenge and clustering effects can cause unreproducible results.
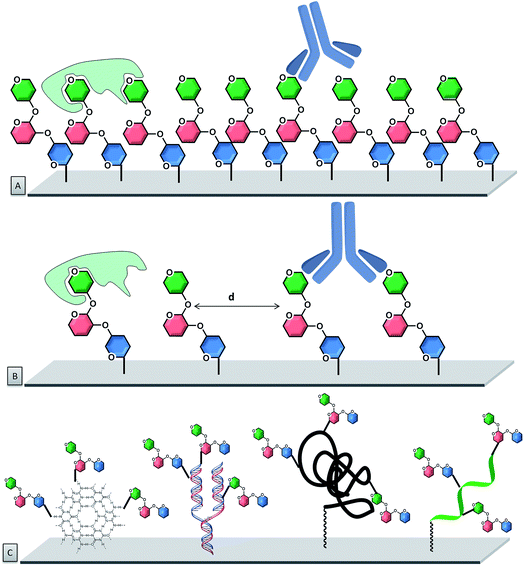
To improve control over glycan presentation, multivalent glycoconjugates with various valencies and spatial arrangements have been designed and immobilized on arrays. Scaffolds, based on natural glycoproteins, neoglycoproteins/neoglycopeptides, glycodendrimers, multivalent display on DNA, glycoclusters, and glycopolymers, have been used (Fig. 7).13,132–144
Natural glycoproteins, such as the heavily glycosylated mucins, were used as multivalent glycan systems for the production of arrays. This microarray retained the three-dimensional presentation of mucin oligosaccharides, without modifications of the protein backbone and permitted the discovery of biologically important motifs for bacterial–host interactions.145 A similar approach uses natural proteins, such as bovine serum albumin (BSA) or human serum albumin (HSA), for the production of neoglycoproteins/neoglycopeptides (proteins or peptides with glycans covalently attached via non-native linkage), which display multiple copies of each glycan. Presynthesized glycoconjugates can be immobilized on epoxide slides. This strategy permits immobilization of both synthetic carbohydrates as well as natural carbohydrates, presented on glycoproteins. Important factors that affect the binding are the number of glycans on a neoglycopeptide, the linker length between the individual sugars, the distance between neoglycopeptide probes on the surface, and the type of protein.113,146–148 These parameters can be tuned to affect the recognition process. Variations in the neoglycoprotein density revealed differences in specificity for antibodies that were not apparent at low density.149
Oligonucleotide hybridization permits to tailor spatial geometry. The rigidity of the double strand nucleic acid with well-defined nucleotide spacing permits to adjust the ligand presentation on this supramolecular scaffold.150–152 Similarly, peptide nucleic acids (PNAs) have been used to tag glycans and evaluate their multivalent interactions with lectins. From an assembly stand-point, stable PNA–DNA duplexes can be achieved with shorter sequences than the corresponding DNA homoduplexes (10–14-mer PNA typically provides sufficient duplex stability).153
Chemical ligation of sugars at different positions within a PNA oligomer has been achieved154 by using thiol moieties embedded in the backbone of the PNA, chemoselectively conjugated to a maleimide-glycan. DNA microarrays permitted the combinatorial pairing of diverse PNA-tagged glycan conjugates. The use of adjacent hybridization sites produced assemblies, emulating the diversity of di-, tri- and tetra-antennary glycans, mimicking the geometry of the HIV gp120 glycan epitope. The combinatorial synthesis of an extended library of PNA-encoded glycoconjugates represents the largest array of heteroglycan conjugates reported to date.155–157
Unnatural scaffolds, like dendrimers, were used for microarray analyses of CPIs.158–161 Well-defined 3D saccharide arrangements on microarrays were constructed upon covalent binding of the dendrimers to the chip surface. Carbohydrates were attached to the dendrimer arm via “click” chemistry, prior or following the attachment of the whole construct to the chip. The multivalency can be precisely controlled with the structure of the glycodendrimer, with valencies ranging from one to eight sugars. Other unnatural alternatives used for multivalent presentation are glycoclusters. Calix[4]arenes are a suitable platform that can be easily derivatized at the upper and lower rims, resulting in well-organized three-dimensional architectures.162,163 With such systems, the primary importance of the spatial arrangement, compared to the number of carbohydrate residues, was highlighted.164
The importance of spatial orientation was observed by 16 different fucosylated glycomimetics, bearing one to eight fucose moieties, synthesized with antenna-like, linear (or comb-like), or crown-like arrangements.165 Binding properties using DNA directed immobilization (DDI)-based glycan microarrays showed that no chelate effect was present, with a one to one interaction between fucose and the lectin. Synthetic glycopolymers have been used to generate mucin-like structures which, as do natural ones, possess rigid extended structures.124,166 Polymers of low polydispersity, displaying α-GalNAc residues, were produced by reversible addition-fragmentation chain transfer (RAFT) polymerization. This new class of orthogonally end-functionalized mucin-mimetics was printed on a microarray, where GalNAc valency and interligand spacing could be controlled. This system again proved that glycan valency and organization are critical parameters that determine the modes through which these interactions occur.
5. Characterization and binding measurements
The readout of a glycan microarray is an important step to obtain precise and convincing data. To detect binding events of glycan binding proteins (GBPs) or successful enzymatic glycosylations on the array, different methods are available. The most frequently applied method is the detection of fluorescently-labeled binders, which directly or indirectly bind to the glycans on the microarray (see Fig. 8). The binding event can be visualized with a fluorescence scanner in several ways: either the GBP is fluorescently labelled or a fluorescently tagged secondary reagent (e.g. antibody) is used to bind to the GBP or to a tag (e.g. biotin, His tag) on the GBP. As discussed, multivalency plays a crucial role for CPIs and the glycan density on the microarray is essential to achieve differential binding. If the density is too low, the GBPs are sometimes unable to properly bind to the glycans, which results in a loss of signals and thus to misleading results.167 Additional problems can be caused by the label, which can reduce the activity or influence the selectivity of the GBPs.168 Unfortunately, indirect labeling of GBPs is often not possible, because fluorescently-labeled secondary reagents are not available.9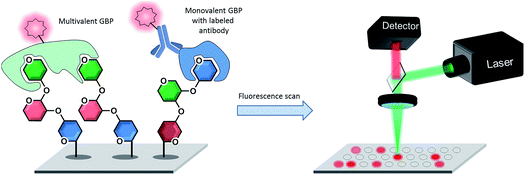 | ||
| Fig. 8 Detection of directly or indirectly fluorescently labeled glycan binding proteins (GBPs) binding to specific glycans on a microarray by fluorescence scanning. | ||
Mass spectrometry is a label-free method to monitor chemical or enzymatic glycosylations directly on an array. Thiol-linked sugars were deposited on a gold surface, whereby self-assembled monolayers are formed. Elongation reactions were then monitored by an on-slide mass spectrometry technique named SAMDI-TOF-MS.21,24,169 With a similar non covalent approach, glycosylation of carbohydrates immobilized on modified gold surfaces using van der Waals forces between aliphatic170,171 or perfluorinated172 carbon chains was monitored. Multiple detection techniques could be used as proven by a multifunctional microarray platform consisting of a glass surface coated with an indium-tin oxide layer. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, fluorescence spectroscopy, and optical microscopy can be employed on the same surface.25
Surface plasmon resonance (SPR) imaging is an alternative label-free method for the analysis of glycan microarrays that allows for determination of the thickness of layers on a metal surface in the nanometer range. SPR has the advantage of real-time monitoring of GBP binding events, which allows for measuring of kinetic and thermodynamic parameters. Metal surfaces (e.g. gold) are mandatory for this approach to excite surface plasmons within the metal by irradiation with polarized light. SPR was used to screen interactions between GBPs and glycans of the pathogen Schistosoma mansoni.146 BSA–mannose-conjugates with different mannose substituents were attached to a gold surface and incubated with ConA to measure KD values and relate it to multivalency.173 Additionally, SPR permitted to identify ligand specificity of plant lectins174 and to better understand siglec-8 (ref. 76) or ConA175 binding specificities.
The above mentioned analysis technologies are the most common, but many others, such as evanescent-field fluorescence,176–178 ellipsometry,179 electrochemoluminescence,180 detection of radioactivity,181,182 oblique-incidence reflectivity (OI-RD) microscopy,183 frontal affinity chromatography,184,185 isothermal calorimetry,186 and cantilever-based detection187 exist. Nevertheless, multivalency cannot be detected directly with one of the analytical techniques. Experiments using multivalent scaffolds have to be compared to those with the monovalent analogue. New technologies to systematically vary the glycan density directly on the microarray are required, to understand multivalent events.
6. Conclusions and outlook
The direct in situ synthesis on surfaces is already well-established and commercialized for oligonucleotides (e.g., Affymetrix, Agilent, Illumina) and peptides (e.g., Intavis, JPT, PEPperPRINT). A big part of this success is based on the enabling technologies, that had a major impact on high-throughput analysis and screening. To translate these technologies to in situ glycan synthesis will be far more challenging: to date, only the chemical synthesis of disaccharides on a “macroarray” surface has been shown,21 whereas enzymatic synthesis is more promising.22–25 Furthermore, multivalency is usually neglected in oligonucleotide and peptide synthesis.In contrast, multivalency is essential for GBPs, because of the naturally weak (KD ∼ μm) protein–glycan affinity, compensated by multiple binding sites.188 Since the advent of glycan microarrays, the main focus has been on the analysis of glycans on surfaces, with less interest in the control of molecular density and spacing. Yet, a defined way of presenting glycans on microarrays is the key step to strong GBP binding. Therefore, strategies are required to display glycans in a molecularly defined spatial order.
Different scaffolds or density variations have been proposed and quite successfully applied for multivalent glycan display.166,167 Especially, the density variation presentation on surfaces leads to random and non-homogeneous systems, which lack reproducibility. Most scaffolds offer defined spacing, but lack flexibility, because the spacing cannot be changed easily. An elegant solution is DNA technology to display glycans. Only recently, this was shown for the display different molecules.189,190 Using DNA-origami structures as scaffolds, multiple glycan structures can be placed in a wide variety of 2D and 3D configurations at exact positions in a controlled and reproducible way. Moreover, it may be used to exactly space the glycans to generate a perfectly matching template for the binding sites of multivalent GBPs. Thereby, control in the screening processes for pathogen interaction with a large variety of structures is possible.
The defined generation of many diverse scaffolds with defined glycan spacing will be one of the future research goals in glycan array technology. Progress in the field of in situ synthesis of scaffolds has been made. By growing brush-like glycopolymers directly on the surface via in situ photo-polymerization, glycan microarrays with multivalent display were generated.20 Different polymer lengths were produced with different amounts of sugar units on the polymer scaffold, by changing the irradiation time.
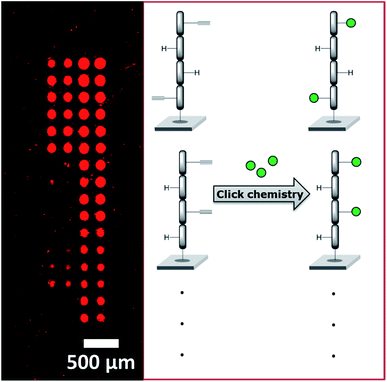
We recently employed a novel laser-based transfer setup50 to generate peptide scaffolds for multivalent display. We synthesized arrays of peptide tetramers, containing all 16 possible sequences of L-glycine and L-propargylglycine. The propargylglycine offers an alkyne group for copper catalyzed click chemistry to attach up to four glycan azides to the peptide backbone. Depending on the amount and position of the α-D-mannose, we obtained differential binding of the lectin concanavalin A (see Fig. 9). This approach may serve as a basis to generate large and complex compound collections for the multivalent display of many different glycans in an orthogonal synthesis strategy. With our laser-based approach, molecules can be synthesized directly on surfaces step-by-step, by “printing” and stacking solid polymer nanolayers,51,191 which embed all kinds of different chemicals and building blocks. Especially for peptide synthesis and applications in disease research, this offers a rapid strategy to generate diverse microarrays.192–197 In the future, this technology may be exploited for the in situ synthesis of glycopeptides, glycans, and DNA in a microarray format.
A large gap remains in the multivalent display and analysis of complex glycans that needs to be filled. Advances accessing glycans and their synthesis and immobilization on surfaces show promising directions for future glycan microarray research. Precisely defined multivalent arrangements on DNA or other structural scaffolds will enable the identification of cooperative effects between identical or diverse collections of glycans. Simultaneously, novel tools based on the presentation of single or multiple glycan molecules in specific arrangements and stoichiometry on the surfaces will be developed.
In the future, newly developed platforms will enable highly parallelized screenings, testing tens of thousands of combinations simultaneously in a microarray-based assay format. The field of protein–glycan interactions will benefit as researchers will be able to uncover the conformation of glycans in a biological environment and open new roads to develop efficient vaccines.
Conflicts of interest
P. H. S. declares a significant financial interest in GlycoUniverse GmbH & Co KGaA, the company that commercializes the synthesis instrument, building blocks and other reagents. F. F. L. is named on a pending patent application related to laser-based microarray synthesis.Acknowledgements
We thank the Max-Planck Society, the Minerva Fast Track Program, and the MPG-FhG Cooperation Project Glyco3Dysplay, and the German Federal Ministry of Education and Research (BMBF, grant no. 13XP5050A) for generous financial support.References
- A. Varki, R. Cummings, J. Esko, H. Freeze, P. Stanley, G. Hart and P. H. Seeberger, Essentials of glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 2017 Search PubMed.
- Y. C. Lee and R. T. Lee, Acc. Chem. Res., 1995, 28, 321–327 CrossRef CAS.
- J. Rojo, J. C. Morales and S. Penadés, in Host-Guest Chemistry: Mimetic Approaches to Study Carbohydrate Recognition, ed. S. Penadés, Springer Berlin Heidelberg, Berlin, Heidelberg, 2002, vol. 218, pp. 45–92 Search PubMed.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS.
- D. F. Smith and R. D. Cummings, Curr. Opin. Virol., 2014, 7, 79–87 CrossRef CAS.
- K. A. Kline, S. Fälker, S. Dahlberg, S. Normark and B. Henriques-Normark, Cell Host Microbe, 2009, 5, 580–592 CrossRef CAS.
- Y. van Kooyk and G. A. Rabinovich, Nat. Immunol., 2008, 9, 593–601 CrossRef CAS.
- P. R. Crocker, J. C. Paulson and A. Varki, Nat. Rev. Immunol., 2007, 7, 255–266 CrossRef CAS.
- S. Park, J. C. Gildersleeve, O. Blixt and I. Shin, Chem. Soc. Rev., 2013, 42, 4310–4326 RSC.
- C. D. Rillahan and J. C. Paulson, Annu. Rev. Biochem., 2011, 80, 797–823 CrossRef CAS.
- M. D. Disney and P. H. Seeberger, Chem. Biol., 2004, 11, 1701–1707 CrossRef CAS.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 CrossRef CAS.
- J. L. Jiménez Blanco, C. Ortiz Mellet and J. M. García Fernández, Chem. Soc. Rev., 2013, 42, 4518–4531 RSC.
- C. Fasting, C. A. Schalley, M. Weber, O. Seitz, S. Hecht, B. Koksch, J. Dernedde, C. Graf, E.-W. Knapp and R. Haag, Angew. Chem., Int. Ed., 2012, 51, 10472–10498 CrossRef CAS.
- M. Delbianco, P. Bharate, S. Varela-Aramburu and P. H. Seeberger, Chem. Rev., 2016, 116, 1693–1752 CrossRef CAS.
- X. Han, Y. Zheng, C. J. Munro, Y. Ji and A. B. Braunschweig, Curr. Opin. Biotechnol., 2015, 34, 41–47 CrossRef CAS.
- L. Wang, R. D. Cummings, D. F. Smith, M. Huflejt, C. T. Campbell, J. C. Gildersleeve, J. Q. Gerlach, M. Kilcoyne, L. Joshi, S. Serna, N.-C. Reichardt, N. Parera Pera, R. J. Pieters, W. Eng and L. K. Mahal, Glycobiology, 2014, 24, 507–517 CrossRef CAS.
- P.-H. Liang, S.-K. Wang and C.-H. Wong, J. Am. Chem. Soc., 2007, 129, 11177–11184 CrossRef CAS.
- F. Broecker and P. H. Seeberger, in Small Molecule Microarrays: Methods and Protocols, ed. M. Uttamchandani and S. Q. Yao, Springer New York, New York, NY, 2017, vol. 1518, pp. 227–240 Search PubMed.
- S. Bian, S. B. Zieba, W. Morris, X. Han, D. C. Richter, K. A. Brown, C. A. Mirkin and A. B. Braunschweig, Chem. Sci., 2014, 5, 2023–2030 RSC.
- L. Ban and M. Mrksich, Angew. Chem., Int. Ed., 2008, 47, 3396–3399 CrossRef CAS.
- S. Serna, J. Etxebarria, N. Ruiz, M. Martin-Lomas and N.-C. Reichardt, Chem.–Eur. J., 2010, 16, 13163–13175 CrossRef CAS.
- O. Blixt, K. Allin, O. Bohorov, X. Liu, H. Andersson-Sand, J. Hoffmann and N. Razi, Glycoconjugate J., 2008, 25, 59–68 CrossRef CAS PubMed.
- L. Ban, N. Pettit, L. Li, A. D. Stuparu, L. Cai, W. L. Chen, W. Y. Guan, W. Q. Han, P. G. Wang and M. Mrksich, Nat. Chem. Biol., 2012, 8, 769–773 CrossRef CAS.
- A. Beloqui, J. Calvo, S. Serna, S. Yan, I. B. Wilson, M. Martin-Lomas and N. C. Reichardt, Angew. Chem., Int. Ed., 2013, 52, 7477–7481 CrossRef CAS.
- X. Song, J. Heimburg-Molinaro, R. D. Cummings and D. F. Smith, Curr. Opin. Chem. Biol., 2014, 18, 70–77 CrossRef CAS.
- R. B. Zheng, S. A. F. Jégouzo, M. Joe, Y. Bai, H.-A. Tran, K. Shen, J. Saupe, L. Xia, M. F. Ahmed, Y.-H. Liu, P. S. Patil, A. Tripathi, S.-C. Hung, M. E. Taylor, T. L. Lowary and K. Drickamer, ACS Chem. Biol., 2017, 12, 2990–3002 CrossRef CAS.
- J.-i. Kadokawa, Chem. Rev., 2011, 111, 4308–4345 CrossRef CAS.
- F. Pfrengle, Curr. Opin. Chem. Biol., 2017, 40, 145–151 CrossRef CAS.
- L. Wen, G. Edmunds, C. Gibbons, J. Zhang, M. R. Gadi, H. Zhu, J. Fang, X. Liu, Y. Kong and P. G. Wang, Chem. Rev., 2018, 118, 8151–8187 CrossRef CAS.
- T. Li, L. Liu, N. Wei, J.-Y. Yang, D. G. Chapla, K. W. Moremen and G.-J. Boons, Nat. Chem., 2019, 11, 229–236 CrossRef CAS.
- J. Zhang, C. Chen, M. R. Gadi, C. Gibbons, Y. Guo, X. Cao, G. Edmunds, S. Wang, D. Liu, J. Yu, L. Wen and P. G. Wang, Angew. Chem., Int. Ed., 2018, 57, 16638–16642 CrossRef CAS.
- P. H. Seeberger, Perspect. Sci., 2017, 11, 11–17 CrossRef.
- M. Guberman and P. H. Seeberger, J. Am. Chem. Soc., 2019, 141, 5581–5592 CrossRef CAS.
- A. Pardo-Vargas, M. Delbianco and P. H. Seeberger, Curr. Opin. Chem. Biol., 2018, 46, 48–55 CrossRef CAS.
- Y. Yu, A. Kononov, M. Delbianco and P. H. Seeberger, Chem.–Eur. J., 2018, 24, 6075–6078 CrossRef CAS.
- M. Delbianco, A. Kononov, A. Poveda, Y. Yu, T. Diercks, J. Jiménez-Barbero and P. H. Seeberger, J. Am. Chem. Soc., 2018, 140, 5421–5426 CrossRef CAS.
- L. Krock, D. Esposito, B. Castagner, C.-C. Wang, P. Bindschadler and P. H. Seeberger, Chem. Sci., 2012, 3, 1617–1622 RSC.
- M. Guberman, M. Bräutigam and P. Seeberger, Chem. Sci., 2019, 10, 5634 RSC.
- B. Schumann, H. S. Hahm, S. G. Parameswarappa, K. Reppe, A. Wahlbrink, S. Govindan, P. Kaplonek, L. A. Pirofski, M. Witzenrath, C. Anish, C. L. Pereira and P. H. Seeberger, Sci. Transl. Med., 2017, 9, eaaf5347 CrossRef.
- K. Naresh, F. Schumacher, H. S. Hahm and P. H. Seeberger, Chem. Commun., 2017, 53, 9085–9088 RSC.
- Y. Yu, S. Gim, D. Kim, Z. A. Arnon, E. Gazit, P. H. Seeberger and M. Delbianco, J. Am. Chem. Soc., 2019, 141, 4833–4838 CrossRef CAS.
- M. W. Weishaupt, H. S. Hahm, A. Geissner and P. H. Seeberger, Chem. Commun., 2017, 53, 3591–3594 RSC.
- H. S. Hahm, F. Broecker, F. Kawasaki, M. Mietzsch, R. Heilbronn, M. Fukuda and P. H. Seeberger, Chem, 2017, 2, 114–124 CAS.
- C. Ruprecht, P. Dallabernardina, P. J. Smith, B. R. Urbanowicz and F. Pfrengle, ChemBioChem, 2018, 19, 793–798 CrossRef CAS.
- D. Schmidt, F. Schuhmacher, A. Geissner, P. H. Seeberger and F. Pfrengle, Chem.–Eur. J., 2015, 21, 5709–5713 CrossRef CAS.
- C. Ruprecht, M. P. Bartetzko, D. Senf, P. Dallabernadina, I. Boos, M. C. F. Andersen, T. Kotake, J. P. Knox, M. G. Hahn, M. H. Clausen and F. Pfrengle, Plant Physiol., 2017, 175, 1094–1104 CrossRef CAS.
- V. Romanov, S. N. Davidoff, A. R. Miles, D. W. Grainger, B. K. Gale and B. D. Brooks, Analyst, 2014, 139, 1303–1326 RSC.
- J. L. Wilbur, A. Kumar, H. A. Biebuyck, E. Kim and G. M. Whitesides, Nanotechnology, 1996, 7, 452–457 CrossRef CAS.
- F. F. Loeffler, T. C. Foertsch, R. Popov, D. S. Mattes, M. Schlageter, M. Sedlmayr, B. Ridder, F. X. Dang, C. von Bojnicic-Kninski, L. K. Weber, A. Fischer, J. Greifenstein, V. Bykovskaya, I. Buliev, F. R. Bischoff, L. Hahn, M. A. Meier, S. Brase, A. K. Powell, T. S. Balaban, F. Breitling and A. Nesterov-Mueller, Nat. Commun., 2016, 7, 11844 CrossRef CAS.
- D. S. Mattes, B. Streit, D. R. Bhandari, J. Greifenstein, T. C. Foertsch, S. W. Munch, B. Ridder, C. Bojničić-Kninski, A. Nesterov-Mueller, B. Spengler, U. Schepers, S. Brase, F. F. Loeffler and F. Breitling, Macromol. Rapid Commun., 2019, 40, 1800533 CrossRef.
- R. D. Piner, J. Zhu, F. Xu, S. Hong and C. A. Mirkin, Science, 1999, 283, 661–663 CrossRef CAS.
- D. J. Valles, Y. Naeem, C. Carbonell, A. M. Wong, D. R. Mootoo and A. B. Braunschweig, ACS Biomater. Sci. Eng., 2019, 5, 3131 CrossRef CAS.
- J. Atwater, D. S. Mattes, B. Streit, C. von Bojnicic-Kninski, F. F. Loeffler, F. Breitling, H. Fuchs and M. Hirtz, Adv. Mater., 2018, 30, 1801632 CrossRef.
- D. Wang, S. Liu, B. J. Trummer, C. Deng and A. Wang, Nat. Biotechnol., 2002, 20, 275–281 CrossRef CAS.
- V. I. Dyukova, N. V. Shilova, O. E. Galanina, A. Y. Rubina and N. V. Bovin, Biochim. Biophys. Acta, Gen. Subj., 2006, 1760, 603–609 CrossRef CAS.
- E. L. Shipp and L. C. Hsieh-Wilson, Chem. Biol., 2007, 14, 195–208 CrossRef CAS.
- C. J. Rogers, P. M. Clark, S. E. Tully, R. Abrol, K. C. Garcia, W. A. Goddard and L. C. Hsieh-Wilson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9747–9752 CrossRef CAS.
- S. Fukui, T. Feizi, C. Galustian, A. M. Lawson and W. Chai, Nat. Biotechnol., 2002, 20, 1011–1017 CrossRef CAS.
- T. Feizi and W. Chai, Nat. Rev. Mol. Cell Biol., 2004, 5, 582–588 CrossRef CAS.
- A. S. Palma, T. Feizi, Y. Zhang, M. S. Stoll, A. M. Lawson, E. Díaz-Rodríguez, M. A. Campanero-Rhodes, J. Costa, S. Gordon, G. D. Brown and W. Chai, J. Biol. Chem., 2006, 281, 5771–5779 CrossRef CAS.
- A. S. Palma, Y. Liu, H. Zhang, Y. Zhang, B. V. McCleary, G. Yu, Q. Huang, L. S. Guidolin, A. E. Ciocchini, A. Torosantucci, D. Wang, A. L. Carvalho, C. M. G. A. Fontes, B. Mulloy, R. A. Childs, T. Feizi and W. Chai, Mol. Cell. Proteomics, 2015, 14, 974–988 CrossRef CAS.
- A. S. Palma, T. Feizi, R. A. Childs, W. Chai and Y. Liu, Curr. Opin. Chem. Biol., 2014, 18, 87–94 CrossRef CAS.
- Z. M. Khan, Y. Liu, U. Neu, M. Gilbert, B. Ehlers, T. Feizi and T. Stehle, J. Virol., 2014, 88, 6100–6111 CrossRef CAS.
- F. Klein, C. Gaebler, H. Mouquet, D. N. Sather, C. Lehmann, J. F. Scheid, Z. Kraft, Y. Liu, J. Pietzsch, A. Hurley, P. Poignard, T. Feizi, L. Morris, B. D. Walker, G. Fätkenheuer, M. S. Seaman, L. Stamatatos and M. C. Nussenzweig, J. Exp. Med., 2012, 209, 1469–1479 CrossRef CAS.
- C. Gao, Y. Liu, H. Zhang, Y. Zhang, M. N. Fukuda, A. S. Palma, R. P. Kozak, R. A. Childs, M. Nonaka, Z. Li, D. L. Siegel, P. Hanfland, D. M. Peehl, W. Chai, M. I. Greene and T. Feizi, J. Biol. Chem., 2014, 289, 16462–16477 CrossRef CAS.
- S. Hanashima, S. Götze, Y. Liu, A. Ikeda, K. Kojima-Aikawa, N. Taniguchi, D. Varón Silva, T. Feizi, P. H. Seeberger and Y. Yamaguchi, ChemBioChem, 2015, 16, 1502–1511 CrossRef CAS.
- Y. Liu, R. A. Childs, A. S. Palma, M. A. Campanero-Rhodes, M. S. Stoll, W. Chai and T. Feizi, in Carbohydrate Microarrays: Methods and Protocols, ed. Y. Chevolot, Humana Press, Totowa, NJ, 2012, vol. 808, pp. 117–136 Search PubMed.
- K.-S. Ko, F. A. Jaipuri and N. L. Pohl, J. Am. Chem. Soc., 2005, 127, 13162–13163 CrossRef CAS.
- S. K. Mamidyala, K.-S. Ko, F. A. Jaipuri, G. Park and N. L. Pohl, J. Fluorine Chem., 2006, 127, 571–579 CrossRef CAS.
- G.-S. Chen and N. L. Pohl, Org. Lett., 2008, 10, 785–788 CrossRef CAS.
- S.-H. Chang, J.-L. Han, S. Y. Tseng, H.-Y. Lee, C.-W. Lin, Y.-C. Lin, W.-Y. Jeng, A. H. J. Wang, C.-Y. Wu and C.-H. Wong, J. Am. Chem. Soc., 2010, 132, 13371–13380 CrossRef CAS.
- Y. Li, E. Arigi, H. Eichert and S. B. Levery, J. Mass Spectrom., 2010, 45, 504–519 CrossRef CAS.
- O. E. Galanina, M. Mecklenburg, N. E. Nifantiev, G. V. Pazynina and N. V. Bovin, Lab Chip, 2003, 3, 260–265 RSC.
- Y. Guo, H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis and K. Drickamer, Nat. Struct. Mol. Biol., 2004, 11, 591–598 CrossRef CAS.
- B. S. Bochner, R. A. Alvarez, P. Mehta, N. V. Bovin, O. Blixt, J. R. White and R. L. Schnaar, J. Biol. Chem., 2005, 280, 4307–4312 CrossRef CAS.
- K. Godula and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 9963–9965 CrossRef CAS.
- Y. Chevolot, C. Bouillon, S. Vidal, F. Morvan, A. Meyer, J.-P. Cloarec, A. Jochum, J.-P. Praly, J.-J. Vasseur and E. Souteyrand, Angew. Chem., Int. Ed., 2007, 46, 2398–2402 CrossRef CAS.
- F. Crevoisier, H. Gao, H. Sigrist, J. L. Ridet, N. Kusy, N. Sprenger, S. Angeloni, S. Guinchard and S. Kochhar, Glycobiology, 2004, 15, 31–41 CrossRef.
- Z. Pei, H. Yu, M. Theurer, A. Waldén, P. Nilsson, M. Yan and O. Ramström, ChemBioChem, 2007, 8, 166–168 CrossRef CAS.
- G. T. Carroll, D. Wang, N. J. Turro and J. T. Koberstein, Langmuir, 2006, 22, 2899–2905 CrossRef CAS PubMed.
- D. Wang, G. T. Carroll, N. J. Turro, J. T. Koberstein, P. Kováč, R. Saksena, R. Adamo, L. A. Herzenberg, L. A. Herzenberg and L. Steinman, Proteomics, 2007, 7, 180–184 CrossRef CAS.
- H.-Y. Hsiao, M.-L. Chen, H.-T. Wu, L.-D. Huang, W.-T. Chien, C.-C. Yu, F.-D. Jan, S. Sahabuddin, T.-C. Chang and C.-C. Lin, Chem. Commun., 2011, 47, 1187–1189 RSC.
- S. Park and I. Shin, Angew. Chem., Int. Ed., 2002, 41, 3180–3182 CrossRef CAS.
- S. Park, M.-r. Lee, S.-J. Pyo and I. Shin, J. Am. Chem. Soc., 2004, 126, 4812–4819 CrossRef CAS PubMed.
- I. Shin, A. D. Zamfir and B. Ye, Methods Mol. Biol., 2008, 441, 19–39 CrossRef CAS.
- B. T. Houseman, E. S. Gawalt and M. Mrksich, Langmuir, 2003, 19, 1522–1531 CrossRef CAS.
- E. W. Adams, D. M. Ratner, H. R. Bokesch, J. B. McMahon, B. R. O’Keefe and P. H. Seeberger, Chem. Biol., 2004, 11, 875–881 CrossRef CAS.
- D. M. Ratner, E. W. Adams, J. Su, B. R. O’Keefe, M. Mrksich and P. H. Seeberger, ChemBioChem, 2004, 5, 379–383 CrossRef CAS PubMed.
- M. A. Brun, M. D. Disney and P. H. Seeberger, ChemBioChem, 2006, 7, 421–424 CrossRef CAS.
- D. M. Ratner and P. H. Seeberger, Curr. Pharm. Des., 2007, 13, 173–183 CrossRef CAS.
- J. H. Seo, K. Adachi, B. K. Lee, D. G. Kang, Y. K. Kim, K. R. Kim, H. Y. Lee, T. Kawai and H. J. Cha, Bioconjugate Chem., 2007, 18, 2197–2201 CrossRef CAS.
- S. Matthies, P. Stallforth and P. H. Seeberger, J. Am. Chem. Soc., 2015, 137, 2848–2851 CrossRef CAS PubMed.
- B. Schumann, R. Pragani, C. Anish, C. L. Pereira and P. H. Seeberger, Chem. Sci., 2014, 5, 1992–2002 RSC.
- F. Kamena, M. Tamborrini, X. Liu, Y.-U. Kwon, F. Thompson, G. Pluschke and P. H. Seeberger, Nat. Chem. Biol., 2008, 4, 238–240 CrossRef CAS.
- M. Tamborrini, X. Liu, J. P. Mugasa, Y.-U. Kwon, F. Kamena, P. H. Seeberger and G. Pluschke, Bioorg. Med. Chem., 2010, 18, 3747–3752 CrossRef CAS.
- L. G. Harris, W. C. E. Schofield, K. J. Doores, B. G. Davis and J. P. S. Badyal, J. Am. Chem. Soc., 2009, 131, 7755–7761 CrossRef CAS.
- E. A. Smith, W. D. Thomas, L. L. Kiessling and R. M. Corn, J. Am. Chem. Soc., 2003, 125, 6140–6148 CrossRef CAS.
- O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C.-H. Wong and J. C. Paulson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17033–17038 CrossRef CAS.
- X. Song, Y. Lasanajak, B. Xia, J. Heimburg-Molinaro, J. M. Rhea, H. Ju, C. Zhao, R. J. Molinaro, R. D. Cummings and D. F. Smith, Nat. Methods, 2011, 8, 85–90 CrossRef CAS.
- C. L. Pereira, A. Geissner, C. Anish and P. H. Seeberger, Angew. Chem., Int. Ed., 2015, 54, 10016–10019 CrossRef CAS.
- H.-Y. Lee, C.-Y. Chen, T.-I. Tsai, S.-T. Li, K.-H. Lin, Y.-Y. Cheng, C.-T. Ren, T.-J. R. Cheng, C.-Y. Wu and C.-H. Wong, J. Am. Chem. Soc., 2014, 136, 16844–16853 CrossRef CAS.
- O. Bohorov, H. Andersson-Sand, J. Hoffmann and O. Blixt, Glycobiology, 2006, 16, 21C–27C CrossRef CAS.
- M. Schwarz, L. Spector, A. Gargir, A. Shtevi, M. Gortler, R. T. Altstock, A. A. Dukler and N. Dotan, Glycobiology, 2003, 13, 749–754 CrossRef CAS.
- A. Gargir, A. Shtevi, E. Fire, L. Nimrichter, M. Gortler, N. Dotan, O. Weisshaus, R. T. Altstock and R. L. Schnaar, Glycobiology, 2004, 14, 197–203 Search PubMed.
- M.-r. Lee and I. Shin, Org. Lett., 2005, 7, 4269–4272 CrossRef CAS.
- A. Reinhardt, Y. Yang, H. Claus, C. L. Pereira, A. D. Cox, U. Vogel, C. Anish and P. H. Seeberger, Chem. Biol., 2015, 22, 38–49 CrossRef CAS.
- S. E. Tully, M. Rawat and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2006, 128, 7740–7741 CrossRef CAS.
- J. L. de Paz, D. Spillmann and P. H. Seeberger, Chem. Commun., 2006, 3116–3118 RSC.
- M.-r. Lee and I. Shin, Angew. Chem., Int. Ed., 2005, 44, 2881–2884 CrossRef CAS.
- S. Park and I. Shin, Org. Lett., 2007, 9, 1675–1678 CrossRef CAS.
- S. Park, M.-R. Lee and I. Shin, Nat. Protoc., 2007, 2, 2747–2758 CrossRef CAS.
- X. Tian, J. Pai and I. Shin, Chem.–Asian J., 2012, 7, 2052–2060 CrossRef CAS.
- S. Park, M.-R. Lee and I. Shin, in Small Molecule Microarrays: Methods and Protocols, ed. M. Uttamchandani and S. Q. Yao, Humana Press, Totowa, NJ, 2010, vol. 669, pp. 195–208 Search PubMed.
- M.-R. Lee, S. Park and I. Shin, in Carbohydrate Microarrays: Methods and Protocols, ed. Y. Chevolot, Humana Press, Totowa, NJ, 2012, vol. 808, pp. 103–116 Search PubMed.
- S. Götze, N. Azzouz, Y.-H. Tsai, U. Groß, A. Reinhardt, C. Anish, P. H. Seeberger and D. Varón Silva, Angew. Chem., Int. Ed., 2014, 53, 13701–13705 CrossRef PubMed.
- S. Götze, A. Reinhardt, A. Geissner, N. Azzouz, Y.-H. Tsai, R. Kurucz, D. Varón Silva and P. H. Seeberger, Glycobiology, 2015, 25, 984–991 CrossRef.
- B. T. Houseman and M. Mrksich, Chem. Biol., 2002, 9, 443–454 CrossRef CAS.
- H. S. G. Beckmann, A. Niederwieser, M. Wiessler and V. Wittmann, Chem.–Eur. J., 2012, 18, 6548–6554 CrossRef CAS.
- X.-L. Sun, C. L. Stabler, C. S. Cazalis and E. L. Chaikof, Bioconjugate Chem., 2006, 17, 52–57 CrossRef CAS.
- C.-Y. Huang, D. A. Thayer, A. Y. Chang, M. D. Best, J. Hoffmann, S. Head and C.-H. Wong, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15–20 CrossRef CAS.
- O. Michel and B. J. Ravoo, Langmuir, 2008, 24, 12116–12118 CrossRef CAS.
- O. J. Barrett, A. Pushechnikov, M. Wu and M. D. Disney, Carbohydr. Res., 2008, 343, 2924–2931 CrossRef CAS.
- K. Godula, D. Rabuka, K. T. Nam and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 4973–4976 CrossRef CAS.
- M. Köhn, R. Wacker, C. Peters, H. Schröder, L. Soulère, R. Breinbauer, C. M. Niemeyer and H. Waldmann, Angew. Chem., Int. Ed., 2003, 42, 5830–5834 CrossRef.
- A. Tyagi, X. Wang, L. Deng, O. Ramström and M. Yan, Biosens. Bioelectron., 2010, 26, 344–350 CrossRef CAS.
- K. J. Doores, D. P. Gamblin and B. G. Davis, Chem.–Eur. J., 2006, 12, 656–665 CrossRef CAS.
- A. Imberty, Y. M. Chabre and R. Roy, Chem.–Eur. J., 2008, 14, 7490–7499 CrossRef CAS.
- P. I. Kitov, J. M. Sadowska, G. Mulvey, G. D. Armstrong, H. Ling, N. S. Pannu, R. J. Read and D. R. Bundle, Nature, 2000, 403, 669–672 CrossRef CAS PubMed.
- C.-H. Liang, S.-K. Wang, C.-W. Lin, C.-C. Wang, C.-H. Wong and C.-Y. Wu, Angew. Chem., 2011, 123, 1646–1650 CrossRef.
- D. Valles, Y. Naeem, A. Rozenfeld, R. Aldasooky, A. Wong, C. Carbonell, D. R. Mootoo and A. Braunschweig, Faraday Discuss., 2019 10.1039/c9fd00028c.
- R. J. Payne and C.-H. Wong, Chem. Commun., 2010, 46, 21–43 RSC.
- P. M. Rendle, A. Seger, J. Rodrigues, N. J. Oldham, R. R. Bott, J. B. Jones, M. M. Cowan and B. G. Davis, J. Am. Chem. Soc., 2004, 126, 4750–4751 CrossRef CAS.
- I. Otsuka, B. Blanchard, R. Borsali, A. Imberty and T. Kakuchi, ChemBioChem, 2010, 11, 2399–2408 CrossRef CAS.
- D. Ponader, F. Wojcik, F. Beceren-Braun, J. Dernedde and L. Hartmann, Biomacromolecules, 2012, 13, 1845–1852 CrossRef CAS.
- J. Rieger, F. Stoffelbach, D. Cui, A. Imberty, E. Lameignere, J.-L. Putaux, R. Jérôme, C. Jérôme and R. Auzély-Velty, Biomacromolecules, 2007, 8, 2717–2725 CrossRef CAS.
- L. Baldini, A. Casnati, F. Sansone and R. Ungaro, Chem. Soc. Rev., 2007, 36, 254–266 RSC.
- S. Cecioni, R. Lalor, B. Blanchard, J.-P. Praly, A. Imberty, S. E. Matthews and S. Vidal, Chem.–Eur. J., 2009, 15, 13232–13240 CrossRef CAS PubMed.
- A. Dondoni and A. Marra, Chem. Rev., 2010, 110, 4949–4977 CrossRef CAS.
- S. André, R. J. Pieters, I. Vrasidas, H. Kaltner, I. Kuwabara, F.-T. Liu, R. M. J. Liskamp and H.-J. Gabius, ChemBioChem, 2001, 2, 822–830 CrossRef.
- M. A. Mintzer, E. L. Dane, G. A. O’Toole and M. W. Grinstaff, Mol. Pharmaceutics, 2012, 9, 342–354 CrossRef CAS.
- C. D. Heidecke and T. K. Lindhorst, Chem.–Eur. J., 2007, 13, 9056–9067 CrossRef CAS.
- D. A. Fulton and J. F. Stoddart, Bioconjugate Chem., 2001, 12, 655–672 CrossRef CAS.
- A. Bernardi, J. Jiménez-Barbero, A. Casnati, C. De Castro, T. Darbre, F. Fieschi, J. Finne, H. Funken, K.-E. Jaeger, M. Lahmann, T. K. Lindhorst, M. Marradi, P. Messner, A. Molinaro, P. V. Murphy, C. Nativi, S. Oscarson, S. Penadés, F. Peri, R. J. Pieters, O. Renaudet, J.-L. Reymond, B. Richichi, J. Rojo, F. Sansone, C. Schäffer, W. B. Turnbull, T. Velasco-Torrijos, S. Vidal, S. Vincent, T. Wennekes, H. Zuilhof and A. Imberty, Chem. Soc. Rev., 2013, 42, 4709–4727 RSC.
- M. Kilcoyne, J. Q. Gerlach, R. Gough, M. E. Gallagher, M. Kane, S. D. Carrington and L. Joshi, Anal. Chem., 2012, 84, 3330–3338 CrossRef CAS.
- A. R. de Boer, C. H. Hokke, A. M. Deelder and M. Wuhrer, Glycoconjugate J., 2008, 25, 75–84 CrossRef CAS.
- E. W. Adams, J. Ueberfeld, D. M. Ratner, B. R. O’Keefe, D. R. Walt and P. H. Seeberger, Angew. Chem., Int. Ed., 2003, 42, 5317–5320 CrossRef CAS.
- J. C. Manimala, Z. Li, A. Jain, S. VedBrat and J. C. Gildersleeve, ChemBioChem, 2005, 6, 2229–2241 CrossRef CAS.
- Y. Zhang, C. Campbell, Q. Li and J. C. Gildersleeve, Mol. BioSyst., 2010, 6, 1583–1591 RSC.
- F. Morvan, S. Vidal, E. Souteyrand, Y. Chevolot and J.-J. Vasseur, RSC Adv., 2012, 2, 12043–12068 RSC.
- N. Spinelli, E. Defrancq and F. Morvan, Chem. Soc. Rev., 2013, 42, 4557–4573 RSC.
- V. Wittmann and R. J. Pieters, Chem. Soc. Rev., 2013, 42, 4492–4503 RSC.
- Z. L. Pianowski and N. Winssinger, Chem. Soc. Rev., 2008, 37, 1330–1336 RSC.
- C. Scheibe, A. Bujotzek, J. Dernedde, M. Weber and O. Seitz, Chem. Sci., 2011, 2, 770–775 RSC.
- K.-T. Huang, K. Gorska, S. Alvarez, S. Barluenga and N. Winssinger, ChemBioChem, 2011, 12, 56–60 CrossRef CAS.
- K. Gorska, K.-T. Huang, O. Chaloin and N. Winssinger, Angew. Chem., Int. Ed., 2009, 48, 7695–7700 CrossRef CAS.
- A. Novoa, T. Machida, S. Barluenga, A. Imberty and N. Winssinger, ChemBioChem, 2014, 15, 2058–2065 CrossRef CAS.
- X. Zhou, C. Turchi and D. Wang, J. Proteome Res., 2009, 8, 5031–5040 CrossRef CAS.
- H. M. Branderhorst, R. Ruijtenbeek, R. M. J. Liskamp and R. J. Pieters, ChemBioChem, 2008, 9, 1836–1844 CrossRef CAS.
- T. Fukuda, S. Onogi and Y. Miura, Thin Solid Films, 2009, 518, 880–888 CrossRef CAS.
- N. Parera Pera, H. M. Branderhorst, R. Kooij, C. Maierhofer, M. van der Kaaden, R. M. J. Liskamp, V. Wittmann, R. Ruijtenbeek and R. J. Pieters, ChemBioChem, 2010, 11, 1896–1904 CrossRef CAS.
- V. Böhmer, Angew. Chem., Int. Ed. Engl., 1995, 34, 713–745 CrossRef.
- A. Ikeda and S. Shinkai, Chem. Rev., 1997, 97, 1713–1734 CrossRef CAS.
- L. Moni, G. Pourceau, J. Zhang, A. Meyer, S. Vidal, E. Souteyrand, A. Dondoni, F. Morvan, Y. Chevolot, J.-J. Vasseur and A. Marra, ChemBioChem, 2009, 10, 1369–1378 CrossRef CAS.
- B. Gerland, A. Goudot, G. Pourceau, A. Meyer, V. Dugas, S. Cecioni, S. Vidal, E. Souteyrand, J.-J. Vasseur, Y. Chevolot and F. Morvan, Bioconjugate Chem., 2012, 23, 1534–1547 CrossRef CAS.
- K. Godula and C. R. Bertozzi, J. Am. Chem. Soc., 2012, 134, 15732–15742 CrossRef CAS.
- H. S. Kim, J. Y. Hyun, S.-H. Park and I. Shin, RSC Adv., 2018, 8, 14898–14905 RSC.
- Y. Fei, Y.-S. Sun, Y. Li, K. Lau, H. Yu, H. A. Chokhawala, S. Huang, J. P. Landry, X. Chen and X. Zhu, Mol. BioSyst., 2011, 7, 3343–3352 RSC.
- W. Guan, L. Ban, L. Cai, L. Li, W. Chen, X. Liu, M. Mrksich and P. G. Wang, Bioorg. Med. Chem. Lett., 2011, 21, 5025–5028 CrossRef CAS.
- A. Sanchez-Ruiz, S. Serna, N. Ruiz, M. Martin-Lomas and N.-C. Reichardt, Angew. Chem., Int. Ed., 2011, 50, 1801–1804 CrossRef CAS.
- A. Beloqui, A. Sanchez-Ruiz, M. Martin-Lomas and N.-C. Reichardt, Chem. Commun., 2012, 48, 1701–1703 RSC.
- T. R. Northen, J.-C. Lee, L. Hoang, J. Raymond, D.-R. Hwang, S. M. Yannone, C.-H. Wong and G. Siuzdak, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3678–3683 CrossRef CAS.
- S. Tao, T.-W. Jia, Y. Yang and L.-Q. Chu, ACS Sens., 2017, 2, 57–60 CrossRef CAS.
- M. Fais, R. Karamanska, S. Allman, S. A. Fairhurst, P. Innocenti, A. J. Fairbanks, T. J. Donohoe, B. G. Davis, D. A. Russell and R. A. Field, Chem. Sci., 2011, 2, 1952–1959 RSC.
- M. Dhayal and D. M. Ratner, Langmuir, 2009, 25, 2181–2187 CrossRef CAS.
- H. Tateno, A. Mori, N. Uchiyama, R. Yabe, J. Iwaki, T. Shikanai, T. Angata, H. Narimatsu and J. Hirabayashi, Glycobiology, 2008, 18, 789–798 CrossRef CAS.
- K. Takahara, T. Arita, S. Tokieda, N. Shibata, Y. Okawa, H. Tateno, J. Hirabayashi and K. Inaba, Infect. Immun., 2012, 80, 1699–1706 CrossRef CAS.
- A. Kuno, N. Uchiyama, S. Koseki-Kuno, Y. Ebe, S. Takashima, M. Yamada and J. Hirabayashi, Nat. Methods, 2005, 2, 851–856 CrossRef CAS.
- Y. Fei, Y.-S. Sun, Y. Li, H. Yu, K. Lau, J. Landry, Z. Luo, N. Baumgarth, X. Chen and X. Zhu, Biomolecules, 2015, 5, 1480–1498 CrossRef CAS.
- E. Han, L. Ding, S. Jin and H. Ju, Biosens. Bioelectron., 2011, 26, 2500–2505 CrossRef CAS.
- M. Shipp, R. Nadella, H. Gao, V. Farkas, H. Sigrist and A. Faik, Glycoconjugate J., 2008, 25, 49–58 CrossRef CAS.
- H. L. Pedersen, J. U. Fangel, B. McCleary, C. Ruzanski, M. G. Rydahl, M.-C. Ralet, V. Farkas, L. von Schantz, S. E. Marcus, M. C. F. Andersen, R. Field, M. Ohlin, J. P. Knox, M. H. Clausen and W. G. T. Willats, J. Biol. Chem., 2012, 287, 39429–39438 CrossRef CAS.
- Y. Y. Fei, A. Schmidt, G. Bylund, D. X. Johansson, S. Henriksson, C. Lebrilla, J. V. Solnick, T. Borén and X. D. Zhu, Anal. Chem., 2011, 83, 6336–6341 CrossRef CAS.
- H. Tateno, S. Nakamura-Tsuruta and J. Hirabayashi, Nat. Protoc., 2007, 2, 2529–2537 CrossRef CAS.
- J. Iwaki and J. Hirabayashi, Trends Glycosci. Glycotechnol., 2018, 30, SE137–SE153 CrossRef.
- Y. Takeda and I. Matsuo, in Lectins: Methods and Protocols, ed. J. Hirabayashi, Springer New York, New York, NY, 2014, vol. 1200, pp. 207–214 Search PubMed.
- K. Gruber, T. Horlacher, R. Castelli, A. Mader, P. H. Seeberger and B. A. Hermann, ACS Nano, 2011, 5, 3670–3678 CrossRef CAS.
- A. Geissner and P. H. Seeberger, Annu. Rev. Anal. Chem., 2016, 9, 223–247 CrossRef CAS.
- C. Kielar, F. V. Reddavide, S. Tubbenhauer, M. Cui, X. Xu, G. Grundmeier, Y. Zhang and A. Keller, Angew. Chem., Int. Ed., 2018, 57, 14873–14877 CrossRef CAS.
- W. Hawkes, D. Huang, P. Reynolds, L. Hammond, M. Ward, N. Gadegaard, J. F. Marshall, T. Iskratch and M. Palma, Faraday Discuss., 2019 10.1039/c9fd00023b.
- G. Paris, J. Heidepriem, A. Tsouka, M. Mende, S. Eickelmann and F. F. Loeffler, Proc. SPIE 10875, Microfluidics, BioMEMS, and Medical Microsystems XVII, 2019, vol. 10875, p. 108750C Search PubMed.
- A. Palermo, L. K. Weber, S. Rentschler, A. Isse, M. Sedlmayr, K. Herbster, V. List, J. Hubbuch, F. F. Loffler, A. Nesterov-Muller and F. Breitling, Biotechnol. J., 2017, 12, 1700197 CrossRef.
- L. K. Weber, A. Isse, S. Rentschler, R. E. Kneusel, A. Palermo, J. Hubbuch, A. Nesterov-Mueller, F. Breitling and F. F. Loeffler, Eng. Life Sci., 2017, 17, 1078–1087 CrossRef CAS.
- L. K. Weber, A. Palermo, J. Kugler, O. Armant, A. Isse, S. Rentschler, T. Jaenisch, J. Hubbuch, S. Dubel, A. Nesterov-Mueller, F. Breitling and F. F. Loeffler, J. Immunol. Methods, 2017, 443, 45–54 CrossRef CAS.
- M. C. L. C. Freire, L. Pol-Fachin, D. F. Coelho, I. F. T. Viana, T. Magalhaes, M. T. Cordeiro, N. Fischer, F. F. Loeffler, T. Jaenisch, R. F. Franca, E. T. A. Marques and R. D. Lins, ACS Omega, 2017, 2, 3913–3920 CrossRef CAS PubMed.
- F. F. Loeffler, J. Pfeil and K. Heiss, Methods Mol. Biol., 2016, 1403, 569–582 CrossRef PubMed.
- T. Jaenisch, K. Heiss, N. Fischer, C. Geiger, F. R. Bischoff, G. Moldenhauer, L. Rychlewski, A. Sie, B. Coulibaly, P. H. Seeberger, L. S. Wyrwicz, F. Breitling and F. F. Loeffler, Mol. Cell. Proteomics, 2019, 18, 642 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |