Self-assembled DNA nanomaterials with highly programmed structures and functions
Zhihao
Li
,
Jie
Wang
,
Yingxue
Li
,
Xinwen
Liu
and
Quan
Yuan
 *
*
Key Laboratory of Analytical Chemistry for Biology and Medicine (Ministry of Education), College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, 430072, People's Republic of China. E-mail: yuanquan@whu.edu.cn
First published on 18th December 2017
Abstract
Long admired as the repository of genetic information, DNA has more recently been exploited as a powerful material for self-assembly. Taking advantage of the specificity and programmability of the Watson–Crick base-pairing principle, DNA is able to form diversiform nanostructures and nanomaterials. More importantly, the assembled DNA nanomaterials can be modulated at nanometer spatial resolution with designable structures and functions. Such nanoscale precision endows DNA nanomaterials with well-defined orientations, spacings, stereo-relationships, and even optimized interactions with biomolecules, contributing to a revolution in the development of materials sophistication. Benefiting from these structural and functional properties, DNA nanomaterials have found a wide range of applications in materials science, analytical science, and biomedicine. Here, we summarize the recent progress in the design and applications of DNA nanomaterials which possess intriguing structures and functions, with a highlight on three kinds of important DNA nanomaterials including DNA tetrahedrons, DNA hydrogels and functional DNA nanomaterials. Finally, some of the current challenges are proposed in the design and applications of DNA nanomaterials, and we also provide our understandings of the prospects of several DNA nanomaterials, aiming to motivate future multidisciplinary and interdisciplinary research.
1. Introduction
Not merely recognized as the repository of genetic codes, DNA has recently been exploited as a powerful molecule for molecular nanotechnology. The field of DNA nanotechnology can be traced back to Seeman's innovative idea of constructing immobile junctions from DNA in 1982.1 Since then, DNA nanotechnology has developed with an astounding speed over the past few decades. From Holliday junctions, to DNA tiles2 and two-dimensional lattices,3,4 to higher-order DNA origami5–7 and DNA nanocomposites,8,9 accumulation of a large number of DNA nanostructures has spurred this field. Taking advantage of the precise and predictable nature of the Watson–Crick base-pairing principle, DNA can be highly programmed to form diversiform nanostructures and nanomaterials by self-assembly. More importantly, the assembled DNA nanomaterials can be modulated at nanometer spatial resolution with unique structures and functions. For example, the shapes, orientations and spacings of DNA nanomaterials can be precisely modulated by altering the number of base pairs, DNA chain length and DNA molecule number.10 The precise modulation of DNA nanomaterials provides an efficient strategy to construct functional materials with complex, hierarchical and ordered structures.11 Furthermore, the DNA configuration can be reversibly controlled by external stimuli such as temperature, light, and biomolecules, leading to responsive changes in the shapes and structures of DNA nanomaterials.12,13 In addition, integration of functional nucleic acids can help in optimizing the interaction between DNA nanomaterials and biomolecules by endowing DNA nanomaterials with functions such as molecular recognition, catalytic activity and therapeutic effects, extensively expanding the application of DNA nanomaterials.14,15 Owing to these structural and functional properties, DNA nanomaterials have contributed to a revolution in the development of materials sophistication for applications in materials synthesis,16 analytical science,17,18 and medical diagnosis and therapy.19–21Fuelled by the advances in design and preparation techniques over the past few decades, an explosion of DNA nanomaterials have been synthesized successfully. These DNA nanomaterials exhibit myriad geometries such as stars,6 polyhedrons22 and complex curvatures,23 and they also display various functions ranging from materials organization,16 molecular recognition24 to catalysis.25 Among them, several nanomaterials with intriguing structures and functions have been attracting considerable attention and have gradually become hot topics.26,27 DNA tetrahedrons, as the simplest three-dimensional DNA nanomaterials, are rigid nanostructures composed of regular edges and acmes. Owing to their unique structure, DNA tetrahedrons exhibit lots of merits such as precise addressability, chirality, high stability and excellent enzyme tolerance, providing powerful scaffolds for the rapidly developing area of materials assembly, bioimaging and therapy.28,29 For specific applications, DNA nanomaterials equipped with complex structures and special properties are usually required to perform complicated tasks. In this regard, it's of great importance to construct DNA nanomaterials with complex structures and functions. DNA hydrogels are a class of porous nanomaterials composed of highly crosslinked DNA networks.30 Their networks and porous architecture endow DNA hydrogels with large surface area, high loading capacity, excellent mechanical stability and cell adhesion ability.30 Moreover, since the configuration of DNA molecules can be reversibly controlled, DNA hydrogels are easily designed with stimuli-responsive properties for the construction of smart materials and shape-memory devices.31 Based on these structural and functional properties, DNA hydrogels have attracted substantial interest in smart materials, controlled drug release and target-responsive detection.32 Apart from these nanomaterials with unique structures, functional nucleic acid-based DNA nanomaterials, which are designed with intriguing functions, have been gaining increased attention in recent years. In addition to self-assembly, functional nucleic acids can impart extra functions such as molecular recognition, catalytic activities and therapeutic effects.33,34 For instance, aptamers can bind to targets selectively by folding themselves into specific secondary and tertiary structures, exhibiting high selectivity and high target binding affinity for applications in biosensors, target imaging and therapy.35 DNAzymes, which are able to catalyze a series of biochemical reactions such as hydrolysis of phosphodiester and degradation of peroxide, offer a highly promising tool to build responsive nanomaterials.36 Leveraging these advances, we envision that such attractive DNA nanomaterials will play an irreplaceable role in the development of evolvable functional materials.
In this Review, we describe some of the most significant progresses made in the design and applications of DNA nanomaterials over the past few years. Firstly, we briefly introduce the principle of DNA self-assembly. Then, we highlight three kinds of DNA nanomaterials with highly programmed structures and functions, including DNA tetrahedrons, DNA hydrogels and functional DNA nanomaterials. Taking these nanomaterials as examples, the design and applications of DNA nanomaterials with different structures and functions are addressed. Finally, we present some of the current challenges faced by DNA nanomaterials and attempt to evaluate some of the critical steps in exploring future opportunities in smart therapeutics, diagnostic nanoplatforms, and even interdisciplinary research.
2. Assembly of DNA nanomaterials
DNA is composed of a phosphate–deoxyribose backbone and natural bases including adenine (A), thymine (T), cytosine (C), and guanine (G). According to Watson–Crick base-pairing rules, two single-stranded DNA molecules (ssDNAs) can hybrid with each other by the hydrogen bonding formed between A and T and C and G. In nature, these two strands are generally fully complementary with the form of double helix DNA (dsDNA),37 as shown in Fig. 1a. If they are only partially complementary, a single stranded overhang will form at the end of DNA molecules, called ‘sticky ends’.37 When the two sticky ends are complementary, they can cohere by hydrogen bonding and form a longer double-stranded DNA molecule (Fig. 1b). As a result, DNA molecules can be assembled to produce new arrangements of sequences. Due to their polymer nature, the flexibility and rigidity of DNA assemblies can be easily tailored by changing the number of base pairs and by modulating the combination of ssDNA and dsDNA.38 For example, a DNA double helix with more base pairs is usually more tolerant to heat and pH. Besides, a relatively flexible ssDNA can be linked with a rigid dsDNA to form stable motifs with desired structures. These features are very important factors in the design of DNA self-assemblies.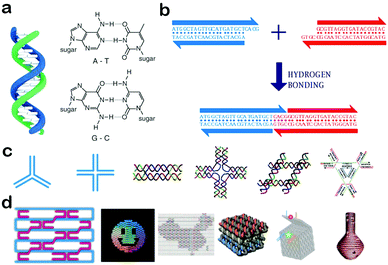 | ||
| Fig. 1 Self-assembly of DNA nanostructures and nanomaterials. (a) Structure of a DNA double helix (left) and Watson–Crick base-pairing rules (right). (b) Affinity in sticky-ended cohesion. (c) Junction structures and multi-crossover tiles. (d) DNA origami nanostructures. (Figures reprinted with permission from: (a) and (b) ref. 38. Copyright 2010, Wiley-VCH; (c) and (d) ref. 20. Copyright 2013, Wiley-VCH.) | ||
Based on the self-recognition properties of DNA, various DNA motifs can be designed with predefined sequences by obeying Watson–Crick rules. In the very beginning, Holliday junctions were used as building blocks to construct larger arrangements. However, these branched DNA junctions are comparably flexible and the assembly of Holliday junctions usually didn’t form stable nanostructures. To solve this problem, a group of DNA structures called crossover tiles were designed with greater rigidity, including double-crossover (DX) tiles,39 triple-crossover (TX) tiles40 and paranemic-crossover (PX) tiles41 (Fig. 1c). In crossover structures, an ssDNA usually starts from one helix, and then crosses over to another adjacent helix, thus connecting the two helixes together. Based on these crossover tiles, a lot of well-defined two-dimensional architectures with programmable periodicities and designable geometries were successfully fabricated, and have greatly paved the way for the rapid development of DNA nanotechnology.4 In an advanced development, a multitude of 3D polyhedral structures have been designed and synthesized, e.g., tetrahedrons, cubes, octahedrons and dodecahedrons.28 In addition, DNA tiles can be connected by sticky-ends, and used to assemble high-ordered superstructures and lattices which include DNA dendrimers, DNA hydrogels, and DNA crystals.42,43 In 2006, Ruthemund adopted a simple ‘scaffolded DNA origami’ method to fold DNA into arbitrary 2D shapes, creating a new century of DNA nanotechnology.5 This method involved folding long single-stranded DNA into a desired shape by using hundreds of short staple DNA strands. The origami approach can significantly improve the structural versatility and complexity of DNA structures, and was used to generate various shapes including a smiley face and a cube (Fig. 1d). In the subsequent development, large DNA structures with predefined dimensions and functionalities have been created, including hollow spheres, twisted and curved shapes, 3D curved surfaces and so on (Fig. 1d).23
The high programmability of DNA endows assembled DNA nanostructures and nanomaterials with nanometer precision, and further offers new opportunities to arrange functional components for various applications. Functional nucleic acids are some of the most important functional components, as they can bind to a series of target molecules with high selectivity and exhibit diverse properties.44,45 For instance, aptamers can fold into 3D structures to offer a highly selective cavity with which targets can snugly bind.46,47 Another example is DNAzymes which possess the ability to catalyze a lot of chemical and biological reactions.48,49 In this regard, introduction of aptamers and DNAzymes into DNA nanomaterials can endow them with special properties of molecular recognition and catalysts. Besides, since DNA can be functionalized with other biomolecules and nanoparticles, DNA nanomaterials can be modified with dyes, quantum dots, gold nanoparticles and other nanomaterials, to access fluorescence and plasmonic properties.50,51 These functionalized DNA nanomaterials greatly enrich the field of DNA nanotechnology and open up new doors for extending DNA nanomaterials to the applications in bioanalysis and biomedicine.
3. DNA tetrahedrons
Because of their simple structure and analogy to natural viral capsids, DNA hollow polyhedrons have attracted particular attention and hold great promise for applications in scaffolding and assembling functional materials.52,53 To date, diverse polyhedrons such as cubes, tetrahedrons, octahedrons, bipyramids, dodecahedrons and icosahedrons have been constructed.53,54 Among them, a tetrahedron is the most simple and intriguing pattern. As the simplest three-dimensional DNA nanomaterial, a DNA tetrahedron is composed of regular edges and acmes to form a rigid nanostructure. Owing to its unique structure, a DNA tetrahedron displays a series of attractive properties and functions. For example, the edges and acmes can provide well-defined positions to build sophisticated nanomaterials, and even enable precise assembly of chiral nanocomplexes.55,56 Moreover, the edges and acmes are conjugated to form a rigid structure which offers a DNA tetrahedron with high thermal stability and excellent enzyme tolerance.57,58 In addition, since a DNA tetrahedron shares a similarity with natural viral capsids, it can be easily taken up by cells through a caveolin-dependent pathway.59 Benefiting from these fascinating properties, DNA tetrahedrons have started to become a powerful and irreplaceable unit in materials assembly, bioimaging and therapy.The DNA tetrahedron architecture was first constructed by Turberfield and colleagues.28 To achieve this, four predesigned oligonucleotides with equimolar quantities were combined in buffer to form a stable tetrahedron structure after a thermal annealing. Then, an enzymatic ligation was performed to covalently connect the four nicks in the DNA backbone (Fig. 2a). As such, the synthesis of a DNA tetrahedron is very simple and highly efficient with tunable edge length. Apart from the employment of short oligonucleotides as building blocks to assemble a DNA tetrahedron, DNA tiles can also be used to achieve a more rigid 3D DNA tetrahedron. Linuma and co-workers reported the assembly of tetrahedrons through a hierarchical approach which utilized identically symmetrical three-point DNA tiles as templates.23 All the tiles were designed with a short overhang in order to converge into a closed structure which is called a DNA framewire tetrahedron. In other works, Mao et al. created a group of DNA tetrahedrons with more complex and sophisticated architectures. For example, in order to regulate the opening and closing processes of the surface pores, they introduced extra single-stranded DNA tails into the tetrahedron structure (Fig. 2b).60 Such tails can be modulated to cap or uncap the surface pores by interacting with other DNA nanostructures, providing prospects for applications in controllable nanoswitches. Besides, inspired by the layer-by-layer structure of multi-wall carbon nanotubes, Mao et al. fabricated a Russian-doll-like DNA tetrahedron in a layer-by-layer assembly method (Fig. 2c).61 Moreover, the layers can be dissociated in response to several stimuli, providing a new strategy for the design of dynamic smart materials. Based on the well-defined self-assembly of DNA strands, DNA tetrahedrons can be programmed with elaborate architectures and special functions, demonstrating their potential in materials assembly, biosensors and biomedicine.
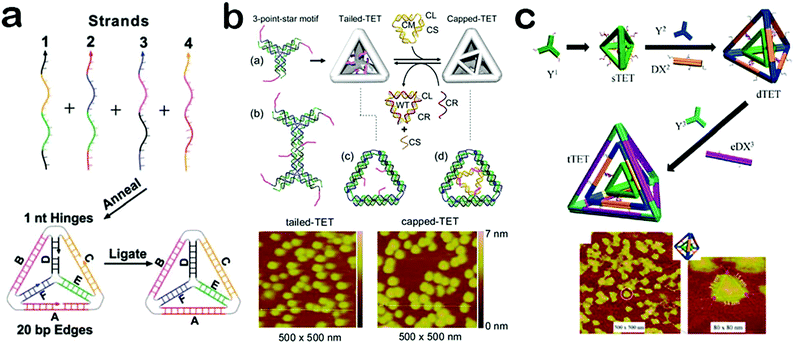 | ||
| Fig. 2 Construction of DNA tetrahedrons. (a) DNA tetrahedron assembled from oligonucleotides. (b) DNA tetrahedron with controlled surface pores. (c) DNA tetrahedron with a layer-by-layer structure. (Figures reprinted with permission from: (a) ref. 28. Copyright 2005, American Association for the Advancement of Science; (b) ref. 60. Copyright 2012, American Chemical Society; (c) ref. 61. Copyright 2015, American Chemical Society.) | ||
In general, a DNA tetrahedron displays a rigid nanostructure formed with six edges and four acmes. By covalent interactions, the edges and acmes can be modified with different materials such as proteins, drug molecules and nanoparticles.62–64 Moreover, based on the highly controllable features of DNA nanostructures, precise positioning of functional materials on the DNA tetrahedron is generally feasible. Mao et al. have utilized DNA tetrahedrons for the assembly of molecule-like gold clusters which reveals defined compositions and specific 3D directional bonds (Fig. 3a).65 In this molecule-like architecture, a symmetric DNA tetrahedron is designed with two unpaired ssDNAs distributed at each edge. When ssDNA functionalized Au nanoparticles (DNA–Au) are encapsulated into the tetrahedron structure, the unpaired ssDNAs can interact with other DNA–Au species via hybridization through the face of the tetrahedron, greatly enhancing structural rigidity. The size or number of nanoparticles can be adjusted via rational design of tetrahedron structures. Moreover, it's possible to replace the Au nanoparticles with other materials (such as magnetic nanoparticles and quantum dots) to construct new functional nanomaterials. Aside from nanoparticles, DNA tetrahedrons can be used to co-assemble with biomolecules such as proteins. For example, Alexander and co-workers have reported that a tetrahedron can be utilized as a skeleton to guide the assembly of protein fragments (Fig. 3b).66 This method realizes the construction of DNA/protein-based hybrid assemblies, greatly expanding the range of self-assembled DNA nanomaterials. Remarkably, DNA tetrahedrons can be modified at specific locations for conjugation with nanoparticles and biomolecules to create delicate nanomaterials such as clusters, crystals and even more sophisticated nanostructures. Such delicate nanomaterials can be equipped with different properties ranging from materials assembly, catalytic activities to biomimetic functions, greatly expanding the repertoire of evolvable functional materials.67,68
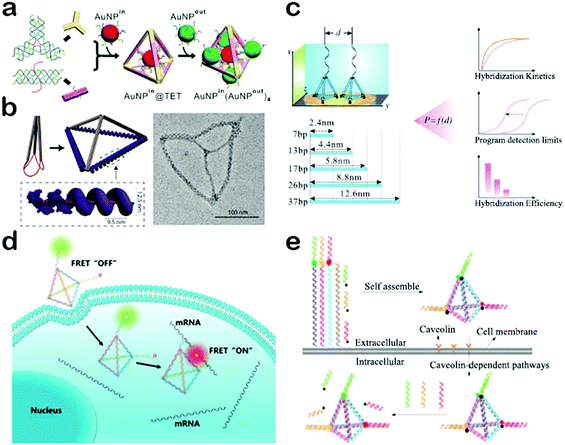 | ||
| Fig. 3 Applications of DNA tetrahedrons. (a) DNA tetrahedron for the assembly of molecule-like gold clusters. (b) DNA tetrahedron utilized as a skeleton to guide the assembly of proteins. (c) DNA tetrahedron-based biosensing interface for ultrasensitive detection of DNA. (d) DNA tetrahedron nanotweezer for the detection of tumor-related mRNA. (e) Multicolor nanoprobe-based DNA tetrahedron for simultaneous detection of three tumor-related mRNAs. (Figures reprinted with permission from: (a) ref. 65. Copyright 2015, American Chemical Society; (b) ref. 66. Copyright 2017, American Chemical Society; (c) ref. 69. Copyright 2015, Wiley-VCH; (d) ref. 75. Copyright 2017, American Chemical Society; (e) ref. 78. Copyright 2017, American Chemical Society.) | ||
If the edges and acmes of DNA tetrahedrons are conjugated with biological probes, DNA tetrahedrons can be used to construct biosensors for detection of biomolecules such as proteins, RNA and even cells. Moreover, DNA tetrahedron-based probes can be well designed at nanoscale spatial resolution and thus it's highly possible to optimize the interaction between DNA and targets in order to enhance the detection selectivity and sensitivity. For example, Fan et al. engineered a biosensing interface with DNA tetrahedrons and gold electrodes for ultrasensitive detection of DNA (Fig. 3c).69 They synthesized a series of DNA tetrahedrons with different sizes and fixed them onto the surface of the gold electrodes. On the acme of each individual tetrahedron, a complementary DNA is conjugated as the capture probe. As such, the spacing and interactions between the capture probe and target can be precisely regulated by the size of the tetrahedron. Consequently, both the thermodynamics and kinetics of DNA hybridization were then found to be significantly affected by a strong dependence on the tetrahedron size. They further optimized the hybridization efficiency and hybridization time by simply adjusting the size of the tetrahedron. Notably, the detection sensitivity can be remarkably improved by over four orders of magnitude, achieving attomolar sensitivity with the help of enzyme amplification. Based on this principle, DNA tetrahedrons have also been used to construct biosensors for sensitive and selective detection of a lot of targets including miRNA, proteins and small molecules.70–72 These biosensors may play significant roles in medical diagnostics, particularly in cancer biomarker detection.
The aforementioned DNA tetrahedron biosensors all work in cell-free settings, exhibiting excellent detection performance. Compared to cell-free settings, intracellular conditions are more complex.73 Hence, biosensors for intracellular and in vivo applications are usually required to possess additional properties, such as high stability and a good transmembrane ability.73,74 Because of its rigid structure, a DNA tetrahedron reveals high tolerance to enzymatic degradation and heating. In addition, since a DNA tetrahedron can be rapidly internalized by cells via a caveolin-dependent pathway, tetrahedron-based biosensors exhibit an excellent transmembrane ability.59 Benefiting from such salient properties, DNA tetrahedron-based biosensors have found widespread application in intracellular imaging and detection. The Tan group has constructed a DNA tetrahedron nanotweezer based on fluorescence resonance energy transfer (FRET) for the detection of tumor-related mRNA (Fig. 3d).75 mRNA has been reported to present important information related to cancer progression, and thus the detection of miRNA has potential utility in the early detection of cancer.76 Tan et al. utilized four dye-modified ssDNAs to self-assemble a tetrahedron probe.75 In the absence of target mRNA, FRET cannot occur because the donor and acceptor fluorophores are kept at a long distance. In contrast, when target mRNA is present, the structure of the tetrahedron is altered, and at the same time, donor and acceptor fluorophores are linked in close proximity, resulting in high FRET efficiency. Such a tetrahedron nanotweezer can be used to distinguish cancer cells from normal cells with high efficiency. However, this method focuses on only one kind of miRNA and may bring about false positive(s), because some tumor-related mRNAs are expressed in normal cells.77,78 To solve this problem, Zhang et al. developed a multicolor nanoprobe based DNA tetrahedron to simultaneously detect three tumor-related mRNAs (Fig. 3e).78 To be specific, the tetrahedron was designed with three mRNA probes anchored at each acme. In the off state, the fluorescence of the probe was quenched by the complementary sequence conjugated with the quencher via DNA hybridization. In the presence of targets, the target mRNA hybridized with the probe to form a longer duplex and the quencher sequence was off from the DNA tetrahedron, leading to the restoration of fluorescence. These DNA tetrahedron-based biosensors provide efficient strategies for imaging of cancer-related biomarkers, offering excellent paradigms to rationally construct self-assembled in vivo nanodiagnostic platforms. Moreover, owing to their hollow nanostructure, DNA tetrahedrons can also be used as valuable carriers for drug delivery to build high-efficiency nanotherapeutic systems.
4. DNA hydrogels
The properties of DNA nanomaterials depend on their structures and architectures. For specific applications, it necessary for DNA nanomaterials to be designed with complex structures so as to possess more properties.79 Therefore, it's of critical importance to equip DNA nanomaterials with elaborate structures. DNA origami and DNA hydrogels represent two important classes within this field. For DNA origami, a number of excellent reviews have been published with an emphasis on their design and application.80–83 Thus, DNA hydrogels will be introduced in this section as the typical example of DNA nanomaterials with complex structures. DNA hydrogels, which are composed of highly crosslinked DNA molecules, have started to become a powerful part of DNA nanomaterials because of their special structural and functional properties.84–86 Crosslinked DNA networks provide an efficient strategy to enhance the mechanical strength of DNA nanomaterials, and at the same time endow DNA hydrogels with porous structure and large surface area, demonstrating their advantages including high loading capacity and strong cell adhesion.87,88 In addition, DNA hydrogels are capable of undergoing shape changes and stiffness changes under specific external stimuli, exhibiting switchable physical, chemical and structural properties.89,90 These intriguing properties have resulted in the implementation of DNA hydrogels as important functional nanomaterials for applications in nanoswitches, controlled release systems and target-responsive detection.91–93In general, DNA hydrogels can be synthesized through two approaches, self-assembly and chemical crosslinking.94 In the self-assembly approach, X tiles, Y tiles and T-shaped tiles with sticky ends were usually used as building blocks and enzymatic catalysis-induced ligation served as the crosslinker. For example, using Y DNA tiles and linkers as building blocks, Liu et al. constructed a kind of DNA hydrogel through sticky-end hybridization and enzymatic catalysis (Fig. 4a).95 The mechanical strength of the hydrogel can be finely tuned by simply changing the ratio between Y tiles and linkers. Moreover, the hydrogel can reversely respond to stimuli including heat and enzymes. These properties allow DNA hydrogels to be excellent candidates for bio-responsive vehicles. In another study by Luo et al., single-stranded DNA was used as the building block to produce a hydrogel (Fig. 4b).96 They used a special polymerase enzyme to elongate the DNA sequences combined with a rolling circle amplification and a multi-primed chain amplification. The resulting DNA hydrogel exhibits liquid-like properties and can be used to create an electric circuit with water as a switch. This work expanded the applications of DNA hydrogels due to their great potential in electric switches and flexible circuits. Instead of employing enzymatic ligation to connect DNA together, chemical crosslinking can be exploited as an efficient approach to build DNA-based hybrid networks.97,98 In this way, DNA mainly serves as a crosslinker and a functional switch in response to specific stimuli. For instance, the Willner group used pH- or Ag+-induced crosslinking to construct a DNA–polymer hybrid hydrogel (Fig. 4c).98 They successfully synthesized a kind of DNA-functionalized polymer chain which can be crosslinked to form hydrogels under the stimulus of pH or Ag+. Furthermore, the resulting hydrogels display designable properties of switchable phase transition in response to stimuli of pH and Ag+ ions. This dual stimuli-responsive hydrogel synthesized by chemical crosslinking could find significant applications in biosensing and controlled drug delivery. In a word, DNA hydrogels assembled via the two approaches have well-defined structures and diverse functions, highly promising for future applications.
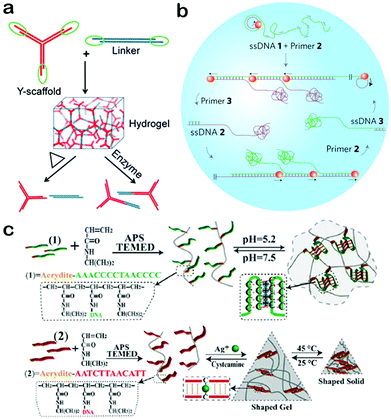 | ||
| Fig. 4 Construction of DNA hydrogels. (a) DNA hydrogel assembled from Y tiles and linkers. (b) DNA hydrogel formed through rolling circle amplification by using single-stranded DNA as the building block. (c) DNA hydrogel synthesized through chemical crosslinking. (Figures reprinted with permission from: (a) ref. 95. Copyright 2011, Wiley-VCH; (b) ref. 96. Copyright 2012, Nature Publishing Group; (c) ref. 98. Copyright 2014, Wiley-VCH.) | ||
When assembled by target-specificity DNA sequences, DNA hydrogels can respond to a lot of biological stimuli because of the target-responsive conformations of DNA molecules.99,100 More significantly, DNA hydrogels can be precisely controlled at nanometer resolution to optimize the interaction between the hydrogel and targets, further improving the detection performance.101 In this regard, DNA hydrogels have attracted much attention in biosensors, especially for biomedical use. Ju and co-workers reported the assembled DNA hydrogel as a switchable material for the detection of thrombin (Fig. 5a).102 Thrombin is a critical serine protease and plays a significant role in hemolysis and hemostasis.103 It's of great importance to detect thrombin for both biomedicine and research applications. Ju et al. employed Y DNA tiles and aptamer linkers to assemble pure DNA hydrogels with rigid space which can encapsulate small-sized gold nanoparticles.102 In the presence of thrombin, aptamer linkers preferentially bind with thrombin rather than the complementary sequences of DNA tiles, leading to the dissolution of DNA hydrogels. Thus, the gold nanoparticles can be released from the hydrogel and diffuse into the solution where FRET occurs between the negatively charged gold nanoparticles and positively charged quantum dots. This strategy shows excellent feasibility in a complex serum system. Yang et al. developed a target-responsive hydrogel for the colorimetric detection of multiple targets (Fig. 5b).104 They predesigned two ssDNA sequences which can bind with part of the aptamer sequence, and grafted them onto polymer chains to form monomers. Once aptamers and monomers are mixed, they can hybrid with each other and generate a tightly crosslinked hydrogel. When the hydrogel is formed in the microfluidic paper, the flow can be stopped and this can be exploited as a readout signal by carrying the indicator. In contrast, in the presence of targets, no hydrogel is formed and the flow would produce a colorimetric readout signal. Moreover, multiple targets including cocaine, adenosine, and Pb2+ can be detected simultaneously. This strategy can be further used to set up a versatile point-of-care testing device. Lee et al. combined the DNA hydrogel with microfluidics to construct a point-of-care testing platform for the visual detection of pathogens.105 Compared to traditional diagnostic platforms, the platform developed using DNA hydrogels is simple enough to prepare and operate with a low cost and a short detection time, promising great potential for biomedical diagnosis, especially the tests for infectious diseases.
 | ||
| Fig. 5 Applications of DNA hydrogels. (a) Target-responsive DNA hydrogel for the detection of thrombin. (b) DNA hydrogel combined with microfluidic paper for the colorimetric detection of multiple targets. (c) Stimuli-responsive DNA hydrogel as an efficient vehicle for targeted gene regulation. (Figures reprinted with permission from: (a) ref. 102. Copyright 2013, American Chemical Society; (b) ref. 104. Copyright 2015, American Chemical Society; (c) ref. 106. Copyright 2015, American Chemical Society.) | ||
As mentioned above, DNA hydrogels are capable of loading a mass of drugs. Moreover, it can be programmed to respond to specific stimuli, leading to a collapse of the network structure and release of the drug. DNA hydrogels exhibit several advantages such high biocompatibility, robust stability, and easy synthesis and modification. Such properties make DNA hydrogels ideal carriers for stimuli-responsive drug release. Tan and co-workers designed a stimuli-responsive DNA hydrogel as an efficient vehicle for targeted gene regulation (Fig. 5c).106 Gene regulation is a highly promising tool for the treatment of several diseases such as cancer and viral infections.107,108 However, the lack of useful gene delivery carriers has greatly limited the applications of gene therapy.109 To solve this problem, Tan et al. designed three kinds of DNA monomers to create a DNA hydrogel through the hybridization at the sticky ends of the monomers.106 When an aptamer was introduced into the hydrogel, it exhibited a target-responsive behavior and can lead to the dissolution of the hydrogel. Furthermore, the dissolved DNA monomers can strongly inhibit the proliferation and growth of the target cancer cells, exhibiting an excellent therapeutic effect. This study demonstrates that DNA hydrogels can be used as useful carriers for targeted cancer therapy.
No matter self-assembled DNA hydrogels or chemical crosslinking DNA hydrogels, the most intriguing property is their stimuli responsiveness. In the presence of the target, DNA hydrogels usually undergo structural changes, leading to the generation of detection signals or release of drug and materials, highlighting their value in biosensing and controlled drug delivery. Besides, DNA hydrogels exhibit switchable mechanical and electronic properties, standing a good chance of information storage and processing.110
5. Functional nucleic acids
Beyond the structural properties of self-assembly, DNA also displays unique sequence-dictated functions. Contemporaneous with the advances in DNA nanostructures, DNA nanomaterials with intriguing functions have promoted the development of DNA nanotechnology. In the past few decades, several nucleic acids have been demonstrated to reveal additional functions such as molecular recognition, catalysis and therapeutic effects.111,112 These special nucleic acids are called ‘functional nucleic acids’, which include ribozymes, aptamers, DNAzymes, i-motif structures, and so on. Among them, aptamers and DNAzymes are two kinds of the most widely known functional nucleic acids, and have been widely utilized as functional components for the construction of functional nucleic acid-based DNA nanomaterials. To date, functional nucleic acid-based DNA nanomaterials have been demonstrated to be of significant practical utility in biosensing, environmental monitoring, drug delivery and cancer therapy.113,114 In this section, we focus on the design and applications of functional DNA nanomaterials including aptamer-based and DNAzyme-based nanomaterials.5.1. Aptamer-based DNA nanomaterials
Aptamers are single-stranded functional oligonucleotides which are able to bind with specific target molecules by folding into distinct secondary or tertiary structures.115,116 They are generally selected from large random DNA or RNA pools through Systematic Evolution of Ligands by Exponential Enrichment (SELEX).117,118 During the selection, only the target-binding molecules are isolated as aptamers so that they have an excellent binding ability with targets. Owing to their high specificity to targets, aptamers are evaluated as strong rivals to antibodies, while displaying additional advantages such as a wide range of targets, small sizes, easy modification, nontoxicity and low immunogenicity.119 Therefore, the integration of aptamers would impart DNA nanomaterials with intriguing functions including high binding affinity for target molecules, holding great promise for biomolecule detection, targeted imaging and therapy, which will be specifically discussed in the following.Because of their high specificity to a variety of targets including metal ions, small molecules, nucleic acids, proteins, cells and even tissues, aptamer-based DNA nanomaterials have been widely applied in detection and diagnosis.120,121 For example, Tan et al. constructed an aptamer-integrated DNA tetrahedron-based electrochemical biosensor for the detection of cancerous exosomes (Fig. 6a).122 Exosomes are cell-secreted nanoscale extracellular vesicles (50–100 nm) which naturally carry a lot of bioinformation from their parental cells, including nucleic acids, transmembrane proteins, and cytosolic proteins.123 Therefore, exosomes can be exploited as biomarkers to aid diagnosis, treatment and prognosis.124 Tan et al. selected a single-stranded aptamer, LZH8, which can bind to hepatocarcinoma cell exosomes with high specificity.122 Utilizing this aptamer as a building block, they constructed an aptamer-integrated DNA tetrahedron and immobilized the tetrahedron to the surface of gold electrodes. In the presence of hepatocarcinoma cell exosomes, the binding of aptamers and exosomes on the gold electrodes can hinder the electron transfer between the gold surface and the ferricyanide–ferrocyanide redox couple in covered solution, leading to a decrease of the generated signal in square wave voltammetry analysis. Based on the biosensor, a detection limit of 2.09 × 104 mL−1 was achieved. Aside from exosomes, proteins and DNA, aptamer-based DNA nanomaterials have been extensively used in the detection of metal ions, DNA, RNA and cells, playing a key role in environmental monitoring and biomedical diagnostics.125–128 Besides their applications in detection, aptamer-based DNA nanomaterials have also attracted increasing attention in intracellular imaging owing to their controllable internalization, outstanding stability, and resistance to nuclease degradation.129 Recently, Tan and co-workers reported a switchable aptamer micelle flare (SAMF) for the imaging of intracellular ATP (Fig. 6b).130 The imaging probe, named a switchable aptamer, is constructed from three elements: an anti-ATP aptamer, a short DNA sequence complementary to part of the aptamer, and a PEG linker for connection. A fluorophore and a quencher are attached to the 3′ end and 5′ end, respectively, for signal creation. To form a DNA micelle, a diacyllipid tail is added at the 5′ terminus with the quencher. Because of the hydrophobic interaction between lipophilic moieties, these switchable aptamers can spontaneously assemble into micelle structures once their concentration exceeds the critical micelle concentration. When transported into cells, aptamers in the SAMF will combine with ATP targets and thus undergo conformational change, which separates the fluorophore from its quencher and thus generates a fluorescent signal. Successful imaging of the distribution and concentration of ATP in living cells is realized. Apart from DNA micelles, the Tan group constructed another nanostructure of aptamers, referred to as a circular bivalent aptamer (cb-aptamer), which exhibits significantly improved efficiency for imaging of tumors.131 In addition, aptamer-based DNA nanomaterials are recognized as ideal carriers of specific drugs and therapeutic agents.132,133 For instance, Tan et al. designed and produced an aptamer-tethered DNA nanotrain (aptNTr) for the delivery of chemotherapyeutic drugs (Fig. 6c).134 AptNTr is constructed through hybridization chain reaction with two short DNA building blocks (M1 and M2) self-assembling into a long linear DNA structure. Driven by the π–π stacking interaction, DOX, a chemotherapeutic drug, can intercalate into the double-stranded DNA structure with quenching of the fluorescence. Since the sequences of M1 and M2 were designed fully with intercalation sites, the drug payload of aptNTr can be as high as nearly 160 loading sites per molecule, which greatly enhanced the therapeutic effect.
 | ||
| Fig. 6 Applications of aptamer-based DNA nanomaterials. (a) Aptamer-integrated DNA tetrahedron-based electrochemical biosensor for the detection of cancerous exosomes. (b) Aptamer-based DNA micelle for targeted imaging. (c) Aptamer-tethered DNA nanotrains for targeted transport of drugs. (Figures reprinted with permission from: (a) ref. 122. Copyright 2017, American Chemical Society; (b) ref. 130. Copyright 2013, American Chemical Society; (c) ref. 134. Copyright 2013, National Academy of Sciences.) | ||
In addition, aptNTr exhibits fewer side effects and higher biocompatibility than free DOX. Such results suggest that aptNTr can be a promising platform of targeted drug delivery for cancer therapy. Aside from DNA nanotrains, the Tan group also constructed another material, an aptamer-based DNA nanoflower as a highly efficient carrier of chemotherapeutic drugs.135,136 With high specificity to targets, aptamers can impart the function of molecular recognition to DNA nanomaterials with great potential in detection, targeted imaging and therapy.
5.2. DNAzyme-based DNA nanomaterials
DNAzymes, another class of functional nucleic acids obtained from SELEX, are able to function as catalysts for a great many biological and chemical reactions.137–139 A DNAzyme generally consists of a substrate strand and an enzyme strand, and its catalytic function is highly dependent on the cofactor which is usually a kind of metal ion or amino acid.140–142 When a specific cofactor exists, the enzyme strand can cleave the substrate strand with high efficiency and separate it into two parts. Based on their specific binding ability and high catalytic activity, DNAzymes can be integrated into DNA nanomaterials to provide a new function of molecular recognition and catalysis, which makes DNAzyme-based DNA nanomaterials potential candidates to construct cofactor-responsive switches. This section focuses on the design and applications of DNAzyme-based DNA nanomaterials.Taking advantage of the cofactor-responsive properties of DNAzymes, DNAzyme-based nanomaterials can be utilized as sensing platforms for the detection of metal ions and amino acids. Tan et al. reported a DNAzyme-based DNA dendrimer as an efficient system for intracellular molecular sensing (Fig. 7a).143 First, they mixed a histidine-dependent DNAzyme and three extra oligonucleotides with equal moles to form a Y-DNA which then self-assemble to produce a DNA dendrimer from the hybridization of the sticky ends. When the dendrimer forms, a quencher and a fluorophore both conjugated with two individual oligonucleotides are kept in close proximity, leading to the quenching of fluorescence. In the presence of histidine, the substrate strand of the DNAzyme is cleaved and the produced shorter DNA with a fluorophore is then released from the dendrimer. Thus, the fluorescent signal is significantly improved. This DNAzyme-based dendrimer can be further applied to in vivo imaging of histidine. Moreover, when the DNAzyme is replaced with an aptamer, the DNA dendrimer can be utilized to detect more targets. Such a sensing platform also reveals other advantages, such as excellent biocompatibility, high stability, easy preparation and high delivery capability. In another work, Xiang and co-workers constructed a dual-colored DNAzyme-based DNA tetrahedron for the simultaneous detection of Pb2+ and UO22+ in living cells (Fig. 7b).144 They designed four oligonucleotides which contain two DNAzyme sequences of Pb2+ and UO22+, and then mixed them to assemble a tetrahedron architecture. In such a tetrahedron, the two DNAzyme sequences were attached with two kinds of fluorophores, respectively, but the fluorescence was quenched by adjacent quenchers. In the presence of target metal ions, DNAzymes catalyzed the cleavage of the substrate strand and short DNA sequences with fluorophores would be separated from the tetrahedron, leading to a restoration of fluorescence. This DNAzyme-based tetrahedron probe can be delivered into cells, and can reflect the intracellular distribution and concentration of Pb2+ and UO22+. In addition to the cofactors, DNAzyme-based nanomaterials can also be implemented to detect DNA targets. For example, the Willner group utilized the Zn2+-dependent ligation DNAzyme as a biocatalyst for the detection of DNA mutants (Fig. 7c).145 The Zn2+-dependent DNAzyme can act as a versatile amplification unit which greatly improves the detection sensitivity. The results reviewed in this section indicate that DNAzyme-based DNA nanomaterials may make a critical impact in bioanalysis, bioimaging and bio-responsive drug delivery.
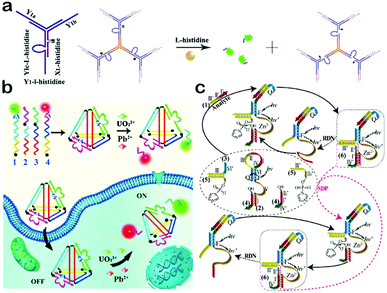 | ||
| Fig. 7 Applications of DNAzyme-based DNA nanomaterials. (a) DNAzyme-based DNA dendrimers for intracellular imaging of histidine. (b) DNAzyme-based tetrahedron for the simultaneous imaging of intracellular Pb2+ and UO22+. (c) Zn2+-dependent DNAzyme machinery for the detection of DNA mutants. (Figures reprinted with permission from: (a) ref. 143. Copyright 2014, American Chemical Society; (b) ref. 144. Copyright 2016, Elsevier; (c) ref. 145. Copyright 2012, American Chemical Society.) | ||
The development of DNA nanomaterials reveals a current trend of seeing the integration of functional nucleic acids into DNA nanoarchitectures to access a lot of special functions. Owing to these special functions, DNA nanomaterials have attracted increasing attention in the fields of biosensing, bioimaging and drug delivery. It is likely that functional nucleic acid-based DNA nanomaterials are many-fold greater in terms of the complexity of structures and functions, providing desirable and critical prerequisites for the construction of diagnostic nanodevices and therapeutic agents.
6. Conclusion and perspectives
DNA nanomaterials, with high programmability in structure and functions, have found a wide range of applications in biosensing, bioimaging and biomedicine. In this Review, we have summarized the major progress in the design and applications of DNA nanomaterials over the past five years. The specificity and programmability of the Watson–Crick base-pairing principle make DNA a powerful building block for engineering DNA nanomaterials at the nanoscale, indicating its great advantages in building sophisticated architectures with specific properties and functions. In addition, at the nanometer precision, interaction between DNA and biomolecules can be precisely designed and significantly optimized, contributing to a remarkable improvement in bio-responsive sensitivity and selectivity. Benefiting from these properties, DNA nanomaterials have made important strides in a variety of applications in nanomaterials assembly, biosensors, diagnosis, controlled drug delivery and therapy.Although great progress has been made, the field of DNA nanomaterials is still in its infancy and much work lies ahead to offer the scientific community. For example, as DNA nanostructures get larger, the error rate of self-assembly increases. Hence, it's of great importance to investigate the kinetic and thermodynamic aspects of assembly, so as to develop a highly efficient strategy with accurate controllability over the self-assembly process. Another example of prospect is the development of modified nucleotides, which can largely expand the library of DNA sequences to access more properties. Recently, the Tan group has synthesized an artificial functional base by incorporating an azobenzene into a natural T base.146 This new molecular base is named zT which can form stable base pairs with a natural A base. Moreover, it exhibits photo-responsive behavior and can be utilized to exogenously regulate the self-assembly of a DNA double helix, indicating that it can be exploited as a new functional element of DNA molecules. It's highly possible that functional DNA elements with distinct properties can contribute to the construction of multifarious functional DNA nanomachines and nanodevices. With further developments and improvements, DNA nanomaterials will have a more significant impact in materials science and therapeutic and diagnostic platforms, and may open up new doors for interdisciplinary research.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFA0208000), the National Natural Science Foundation of China (21675210 and 21422105), the National Natural Science Foundation of Hubei Province (2015CFA032), and the Ten Thousand Talents Program for Young Talents.Notes and references
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- X. J. Li, X. P. Yang, J. Qi and N. C. Seeman, J. Am. Chem. Soc., 1996, 118, 6131–6140 CrossRef CAS.
- Y. He, Y. Tian, Y. Chen, Z. Deng, A. E. Ribbe and C. Mao, Angew. Chem., Int. Ed., 2005, 44, 6694–6696 CrossRef CAS PubMed.
- Y. Liu, Y. G. Ke and H. Yan, J. Am. Chem. Soc., 2005, 127, 17140–17141 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Hogberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS PubMed.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- M. Pumera, Chem. Soc. Rev., 2010, 39, 4146–4157 RSC.
- L. A. Lanier and H. Bermudez, Curr. Opin. Chem. Eng., 2015, 7, 93–100 CrossRef PubMed.
- Z. G. Wang and B. Q. Ding, Acc. Chem. Res., 2014, 47, 1654–1662 CrossRef CAS PubMed.
- J. Li, A. A. Green, H. Yan and C. Fan, Nat. Chem., 2017, 9, 1056–1067 CrossRef PubMed.
- A. Cangialosi, C. Yoon, J. Liu, Q. Huang, J. Guo, T. D. Nguyen, D. H. Gracias and R. Schulman, Science, 2017, 357, 1126–1130 CrossRef CAS PubMed.
- J. P. Goertz and I. M. White, Angew. Chem., Int. Ed., 2017, 56, 13411–13415 CrossRef CAS PubMed.
- L. Gong, Z. Zhao, Y. F. Lv, S. Y. Huan, T. Fu, X. B. Zhang, G. L. Shen and R. Q. Yu, Chem. Commun., 2015, 51, 979–995 RSC.
- W. Sun, E. Boulais, Y. Hakobyan, W. L. Wang, A. Guan, M. Bathe and P. Yin, Science, 2014, 346, 1258361 CrossRef PubMed.
- D. Li, S. P. Song and C. H. Fan, Acc. Chem. Res., 2010, 43, 631–641 CrossRef CAS PubMed.
- Y. Du and S. J. Dong, Anal. Chem., 2017, 89, 189–215 CrossRef CAS PubMed.
- B. Zhu, L. H. Wang, J. Li and C. H. Fan, Chem. Rev., 2017, 17, 1–19 Search PubMed.
- J. Li, C. H. Fan, H. Pei, J. Y. Shi and Q. Huang, Adv. Mater., 2013, 25, 4386–4396 CrossRef CAS PubMed.
- C. Angell, S. B. Xie, L. F. Zhang and Y. Chen, Small, 2016, 12, 1117–1132 CrossRef CAS PubMed.
- R. Iinuma, Y. G. Ke, R. Jungmann, T. Schlichthaerle, J. B. Woehrstein and P. Yin, Science, 2014, 344, 65–69 CrossRef CAS PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed.
- H. M. Meng, H. Liu, H. Kuai, R. Peng, L. Moa and X. B. Zhang, Chem. Soc. Rev., 2016, 45, 2583–2602 RSC.
- R. Orbach, F. Remacle, R. D. Levinea and I. Willner, Chem. Sci., 2014, 5, 1074–1081 RSC.
- Z. G. Wang, Q. Liu and B. Ding, Chem. Mater., 2014, 26, 3364–3367 CrossRef CAS.
- Z. G. Wang, P. Zhan and B. Ding, ACS Nano, 2013, 7, 1591–1598 CrossRef CAS PubMed.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- X. W. Liu, Y. Xu, T. Yu, C. Clifford, Y. Liu, H. Yan and Y. Chang, Nano Lett., 2012, 12, 4254–4259 CrossRef CAS PubMed.
- D. Wang, Y. Hu, P. F. Liu and D. Luo, Acc. Chem. Res., 2017, 50, 733–739 CrossRef CAS PubMed.
- J. Liu, Soft Matter, 2011, 7, 6757–6767 RSC.
- W. Lu, X. Le, J. Zhang, Y. Huang and T. Chen, Chem. Soc. Rev., 2017, 46, 1284–1294 RSC.
- J. Li, L. T. Mo, C. H. Lu, T. Fu, H. H. Yang and W. H. Tan, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- Y. S. Kim, N. H. A. Raston and M. B. Gu, Biosens. Bioelectron., 2016, 76, 2–19 CrossRef PubMed.
- J. Zheng, R. Yang, M. Shi, C. Wu, X. Fang, Y. Li, J. Li and W. Tan, Chem. Soc. Rev., 2015, 44, 3036–3055 RSC.
- N. C. Seeman and P. S. Lukeman, Rep. Prog. Phys., 2005, 68, 237–270 CrossRef CAS PubMed.
- D. Y. Yang, M. J. Campolongo, T. N. N. Tran, R. C. H. Ruiz, J. S. Kahn and D. Luo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 648–669 CrossRef CAS PubMed.
- E. Winfree, F. R. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- T. H. LaBean, H. Yan, J. Kopatsch, F. R. Liu, E. Winfree, J. Reif and N. C. Seeman, J. Am. Chem. Soc., 2000, 122, 1848–1860 CrossRef CAS.
- H. Yan, X. P. Zhang, Z. Y. Shen and N. C. Seeman, Nature, 2002, 415, 62–65 CrossRef CAS PubMed.
- Y. G. Li, Y. D. Tseng, S. Y. Kwon, L. D’Espaux, J. S. Bunch, P. L. McEuen and D. Luo, Nat. Mater., 2004, 3, 38–42 CrossRef CAS PubMed.
- M. G. Warner and J. E. Hutchison, Nat. Mater., 2003, 2, 272–277 CrossRef CAS PubMed.
- S. Modi, D. Bhatia, F. C. Simmel and Y. Krishnan, J. Phys. Chem. Lett., 2010, 1, 1994–2005 CrossRef CAS.
- H. Shen, J. Wang, H. Liu, Z. Li, F. Jiang, F. B. Wang and Q. Yuan, ACS Appl. Mater. Interfaces, 2016, 8, 19371–19378 CAS.
- X. H. Fang and W. H. Tan, Acc. Chem. Res., 2010, 43, 48–57 CrossRef CAS PubMed.
- Y. Yang, X. Yang, Z. Xu, S. Wu, D. Wan, A. Cao, L. Liao, Q. Yuan and X. Duan, Adv. Funct. Mater., 2017, 1604096 CrossRef.
- J. W. Liu, Z. H. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- K. Hwang, P. Wu, T. Kim, L. Lei, S. Tian, Y. Wang and Y. Lu, Angew. Chem., Int. Ed., 2014, 53, 13798–13802 CrossRef CAS PubMed.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef PubMed.
- X. Lim, Nature, 2017, 546, 687–689 CrossRef CAS PubMed.
- Y. L. Li, C. D. Mao and Z. X. Deng, Chin. J. Chem., 2017, 35, 801–810 CrossRef CAS.
- C. Zhang, M. Su, Y. He, Y. J. Leng, A. E. Ribbe, G. S. Wang, W. Jiang and C. D. Mao, Chem. Commun., 2010, 46, 6792–6794 RSC.
- N. Xie, S. Liu, X. Yang, X. He, J. Huang and K. Wang, Analyst, 2017, 142, 3322–3332 RSC.
- D. Zhu, H. Pei, G. Yao, L. Wang, S. Su, J. Chao, L. Wang, A. Aldalbahi, S. Song, J. Shi, J. Hu, C. Fan and X. Zuo, Adv. Mater., 2016, 28, 6860–6865 CrossRef CAS PubMed.
- W. Yan, L. Xu, C. Xu, W. Ma, H. Kuang, L. Wang and N. A. Kotov, J. Am. Chem. Soc., 2012, 134, 15114–15121 CrossRef CAS PubMed.
- K. J. Huang, Y. J. Liu and Q. F. Zhai, J. Mater. Chem. B, 2015, 3, 8180–8187 RSC.
- Z. Xia, P. Wang, X. Liu, T. Liu, Y. Yan, J. Yan, J. Zhong, G. Sun and D. He, Biochemistry, 2016, 55, 1326–1331 CrossRef CAS PubMed.
- L. Liang, J. Li, Q. Li, Q. Huang, J. Shi, H. Yan and C. Fan, Angew. Chem., Int. Ed., 2014, 53, 7745–7750 CrossRef CAS PubMed.
- C. Zhang, C. Tian, X. Li, H. Qian, C. H. Hao, W. Jiang and C. D. Mao, J. Am. Chem. Soc., 2012, 134, 11998–12001 CrossRef CAS PubMed.
- Z. Y. Liu, C. Tian, J. W. Yu, Y. L. Li, W. Jiang and C. D. Mao, J. Am. Chem. Soc., 2015, 137, 1730–1733 CrossRef CAS PubMed.
- J. Li, C. Y. Hong, S. X. Wu, H. Liang, L. P. Wang, G. M. Huang, X. Chen, H. H. Yang, D. H. Shangguan and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 11210–11213 CrossRef CAS PubMed.
- Q. M. Feng, Y. H. Guo, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 17637–17644 CAS.
- N. Li, M. Wang, X. Gao, Z. Yu, W. Pan, H. Wang and B. Tang, Anal. Chem., 2017, 89, 6670–6677 CrossRef CAS PubMed.
- Y. L. Li, Z. Y. Liu, G. M. Yu, W. Jiang and C. D. Mao, J. Am. Chem. Soc., 2015, 137, 4320–4323 CrossRef CAS PubMed.
- D. Schiffels, V. A. Szalai and J. A. Liddle, ACS Nano, 2017, 11, 6623–6629 CrossRef CAS PubMed.
- H. Lee, A. K. R. Lytton-Jean, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis1, A. Sehgal, W. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed.
- N. Xie, J. Huang, X. Yang, J. Yang, K. Quan, H. Wang, L. Ying, M. Ou and K. Wang, Chem. Commun., 2016, 52, 2346–2349 RSC.
- M. H. Lin, J. J. Wang, G. B. Zhou, J. B. Wang, N. Wu, J. X. Lu, J. M. Gao, X. Q. Chen, J. Y. Shi, X. L. Zuo and C. H. Fan, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou and J. Shi, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
- D. Zhu, H. Pei, G. Yao, L. Wang, S. Su, J. Chao, L. Wang, A. Aldalbahi, S. Song, J. Shi, J. Hu, C. Fan and X. Zuo, Adv. Mater., 2016, 28, 6860–6865 CrossRef CAS PubMed.
- Y. Aiba, J. Sumaoka and M. Komiyama, Chem. Soc. Rev., 2011, 40, 5657–5668 RSC.
- Y. J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS PubMed.
- D. S. Lee, H. Qian, C. Y. Tay and D. T. Leong, Chem. Soc. Rev., 2016, 45, 4199–4225 RSC.
- L. He, D. Q. Lu, H. Liang, S. T. Xie, C. Luo, M. M. Hu, L. J. Xu, X. B. Zhang and W. H. Tan, ACS Nano, 2017, 11, 4060–4066 CrossRef CAS PubMed.
- X. Hu, Y. Wang, H. Liu, J. Wang, Y. Tan, F. Wang, Q. Yuan and W. Tan, Chem. Sci., 2017, 8, 466–472 RSC.
- D. Sidransky, Science, 1997, 278, 1054–1059 CrossRef CAS PubMed.
- S. Wang, M. C. Xia, J. Liu, S. C. Zhang and X. R. Zhang, ACS Sens., 2017, 2, 735–739 CrossRef CAS PubMed.
- K. Ariga, Q. Ji, J. P. Hill, Y. Bando and M. Aono, NPG Asia Mater., 2012, 4, e17 CrossRef.
- T. Torring, N. V. Voigt, J. Nangreave, H. Yan and K. V. Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Nat. Nanotechnol., 2011, 6, 763–772 CrossRef CAS PubMed.
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- N. Oliva, J. Conde, K. Wang and N. Artzi, Acc. Chem. Res., 2017, 50, 669–679 CrossRef CAS PubMed.
- C. Li, Z. Xu, Y. Shao, P. Chen, Y. Xing, Z. Yang, Z. Li and D. Liu, Mater. Chem. Front., 2017, 1, 654–659 RSC.
- C. K. McLaughlin, G. D. Hamblin and H. F. Slaiman, Chem. Soc. Rev., 2011, 40, 5647–5656 RSC.
- Z. G. Wang and B. Q. Ding, Adv. Mater., 2013, 25, 3905–3914 CrossRef CAS PubMed.
- X. He, B. Wei and Y. Mi, Chem. Commun., 2010, 46, 6308–6310 RSC.
- Y. Eguchi, T. Kato, T. Tanaka and T. Maruyama, Chem. Commun., 2017, 58, 5802–5805 RSC.
- X. Zhou, C. Li, Y. Shao, C. Chen, Z. Yang and D. Liu, Chem. Commun., 2016, 57, 10668–10671 RSC.
- J. S. Kahn, A. Trifonov, A. Cecconello, W. Guo, C. Fan and I. Willner, Nano Lett., 2015, 15, 7773–7778 CrossRef CAS PubMed.
- P. Song, D. Ye, X. Zuo, J. Li, J. Wang, H. Liu, M. T. Hwang, J. Chao, S. Su, L. Wang, J. Shi, L. Wang, W. Huang, R. Lal and C. Fan, Nano Lett., 2017, 17, 5193–5198 CrossRef CAS PubMed.
- J. Wang, J. Chao, H. Liu, S. Su, L. Wang, W. Huang, I. Willner and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 2171–2175 CrossRef CAS PubMed.
- Y. H. Roh, R. C. H. Ruiz, S. M. Peng, J. B. Lee and D. Luo, Chem. Soc. Rev., 2011, 40, 5730–5744 RSC.
- Y. Z. Xing, E. Cheng, Y. Yang, P. Chen, T. Zhang, Y. W. Sun, Z. Q. Yang and D. S. Liu, Adv. Mater., 2011, 23, 1117–1121 CrossRef CAS PubMed.
- J. B. Lee, S. M. Peng, D. Y. Yang, Y. H. Roh, H. Funabashi, N. Park, E. J. Rice, L. W. Chen, R. Long, M. M. Wu and D. Luo, Nat. Nanotechnol., 2012, 7, 816–820 CrossRef CAS PubMed.
- J. S. Kahn, Y. W. Hu and I. Willner, Acc. Chem. Res., 2017, 50, 680–690 CrossRef CAS PubMed.
- W. W. Guo, C. H. Lu, X. J. Qi, R. Orbach, M. Fadeev, H. H. Yang and I. Willner, Angew. Chem., Int. Ed., 2014, 53, 10134–10138 CrossRef CAS PubMed.
- V. Tjong, L. Tang, S. Zauscher and A. Chilkoti, Chem. Soc. Rev., 2014, 43, 1612–1626 RSC.
- C. E. Castro, H. J. Su, A. E. Marras, L. Zhou and J. Johnson, Nanoscale, 2015, 7, 5913–5921 RSC.
- Z. Zhu, C. Wu, H. Liu, Y. Zhou, X. Liu, K. Huang, C. J. Yang and W. Tan, Angew. Chem., Int. Ed., 2010, 49, 1052–1056 CrossRef CAS PubMed.
- L. Zhang, J. P. Lei, L. Liu, C. F. Li and H. X. Ju, Anal. Chem., 2013, 85, 11077–11082 CrossRef CAS PubMed.
- J. Wang, Y. R. Wei, X. X. Hu, Y. Y. Fang, X. Y. Li, J. Liu, S. F. Wang and Q. Yuan, J. Am. Chem. Soc., 2015, 137, 10576–10584 CrossRef CAS PubMed.
- X. F. Wei, T. Tian, S. S. Jia, Z. Zhu, Y. Ma, J. J. Sun, Z. Y. Lin and C. J. Yang, Anal. Chem., 2015, 87, 4275–4282 CrossRef CAS PubMed.
- H. Y. Lee, H. Jeong, Y. Jung, B. Jang, Y. C. Seo, H. Lee and H. Lee, Adv. Mater., 2015, 27, 3513–3517 CrossRef CAS PubMed.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. H. Xu, C. Cui, Y. Liu, W. J. Hou, Y. Y. Wang, L. Q. Zhang, I. T. Teng, H. H. Yang and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS PubMed.
- I. M. Verma and N. Somia, Nature, 1997, 389, 239–242 CrossRef CAS PubMed.
- J. Chen, Z. Guo, L. Lin, Y. Hu, H. Tian, M. Chen and X. Chen, Mater. Chem. Front., 2017, 1, 937–946 RSC.
- S. Li and L. Huang, Gene Ther., 2000, 7, 31–34 CrossRef CAS PubMed.
- M. Ebara, K. Uto, N. Idota, J. M. Hoffman and T. Aoyagi, Soft Mater., 2013, 9, 3071–3080 Search PubMed.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS PubMed.
- F. Pu, J. Ren and X. Qu, Adv. Mater., 2014, 26, 5742–5757 CrossRef CAS PubMed.
- S. F. Torabi and Y. Lu, Curr. Opin. Biotechnol., 2014, 28, 88–95 CrossRef CAS PubMed.
- Y. T. Tang, B. X. Ge, D. Sen and H. Z. Yu, Chem. Soc. Rev., 2014, 43, 518–529 RSC.
- H. M. Meng, T. Fu, X. B. Zhang and W. Tan, Natl. Sci. Rev., 2015, 2, 71–84 CrossRef.
- Q. Yuan, Y. Wu, J. Wang, D. Q. Lu, Z. L. Zhao, T. Liu, X. B. Zhang and W. H. Tan, Angew. Chem., 2013, 125, 14215–14219 CrossRef.
- G. Zhu, M. Ye, M. J. Donovan, E. Song, Z. Zhao and W. Tan, Chem. Commun., 2012, 48, 10472–10480 RSC.
- R. Wang, D. Lu, H. Bai, C. Jin, G. Yan, M. Ye, L. Qiu, R. Chang, C. Cui, H. Liang and W. Tan, Chem. Sci., 2016, 7, 2157–2162 RSC.
- J. Wang, T. Wei, X. Y. Li, B. H. Zhang, J. X. Wang, C. Huang and Q. Yuan, Angew. Chem., Int. Ed., 2014, 53, 1616–1620 CrossRef CAS PubMed.
- L. Wang and G. Arrabito, Analyst, 2015, 140, 5821–5848 RSC.
- Y. Tan, X. Hu, M. Liu, X. Liu, X. Lv, Z. Li, J. Wang and Q. Yuan, Chem. – Eur. J., 2017, 23, 10683–10689 CrossRef CAS PubMed.
- S. Wang, L. Q. Zhang, S. Wan, S. Cansiz, C. Cui, Y. Liu, R. Cai, C. Y. Hong, I. T. Teng, M. L. Shi, Y. Wu, Y. Y. Dong and W. H. Tan, ACS Nano, 2017, 11, 3943–3949 CrossRef CAS PubMed.
- H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J. J. Lee and J. O. Lötvall, Nat. Cell Biol., 2007, 9, 654–659 CrossRef CAS PubMed.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. Lebleu, E. A. Mittendorf, J. Weitz and N. Rahbari, Nature, 2015, 523, 177–182 CrossRef CAS PubMed.
- D. Koirala, P. Shrestha, T. Emura, K. Hidaka, S. Mandal, M. Endo, H. Sugiyama and H. B. Mao, Angew. Chem., 2014, 126, 8275–8279 CrossRef.
- B. Kim, I. H. Jung, M. Kang, H. K. Shim and H. Y. Woo, J. Am. Chem. Soc., 2012, 134, 3133–3138 CrossRef CAS PubMed.
- L. Qiu, C. Wu, M. You, D. Han, T. Chen, G. Zhu, J. Jiang, R. Yu and W. Tan, J. Am. Chem. Soc., 2013, 133, 12952–12955 CrossRef PubMed.
- C. D. Medley, S. Bamrungsap, W. Tan and J. E. Smith, Anal. Chem., 2011, 83, 727–734 CrossRef CAS PubMed.
- J. Wang, H. J. Shen, X. X. Hu, Y. Li, Z. H. Li, J. F. Xu, X. F. Song, H. B. Zeng and Q. Yuan, Adv. Sci., 2016, 3, 1500289 CrossRef PubMed.
- C. C. Wu, T. Chen, D. Han, M. X. You, L. Peng, S. Cansiz, G. Z. Zhu, C. M. Li, X. L. Xiong, E. Jimenez, C. J. Yang and W. H. Tan, ACS Nano, 2013, 7, 5724–5731 CrossRef CAS PubMed.
- H. L. Kuai, Z. L. Zhao, L. T. Mo, H. Liu, X. X. Hu, T. Fu, X. B. Zhang and W. H. Tan, J. Am. Chem. Soc., 2017, 139, 9128–9131 CrossRef CAS PubMed.
- H. Xing, K. Hwang, J. Li, S. F. Torabi and Y. Lu, Curr. Opin. Chem. Eng., 2014, 4, 79–87 CrossRef PubMed.
- X. Hu, Y. Wang, Y. Tan, J. Wang, H. Liu, Y. Wang, S. Yang, M. Shi, S. Zhao, Y. Zhang and Q. Yuan, Adv. Mater., 2017, 1605235 CrossRef PubMed.
- G. Z. Zhu, J. Zheng, E. Song, M. Donovan, K. J. Zhang, C. Liu and W. H. Tan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7998–8003 CrossRef CAS PubMed.
- R. Hu, X. B. Zhang, Z. Zhao, G. Zhu, T. Chen, T. Fu and W. H. Tan, Angew. Chem., Int. Ed., 2014, 53, 5821–5826 CrossRef CAS PubMed.
- L. Mei, G. Zhu, L. Qiu, C. Wu, H. Chen, H. Liang, S. Cansiz, Y. Lv, X. B. Zhang and W. H. Tan, Nano Res., 2015, 8, 3447–3460 CrossRef CAS PubMed.
- Y. Yuan, Y. Chai, R. Yuan, Y. Zhuo and X. Gan, Chem. Commun., 2013, 49, 7328–7330 RSC.
- Y. Chen and R. M. Corn, J. Am. Chem. Soc., 2013, 135, 2072–2075 CrossRef CAS PubMed.
- X. H. Mao, A. J. Simon, H. Pei, J. Y. Shi, J. Li, Q. Huang, K. W. Plaxco and C. H. Fan, Chem. Sci., 2016, 7, 1200–1204 RSC.
- P. J. J. Huang, M. Vazin and J. Liu, Anal. Chem., 2014, 86, 9993–9999 CrossRef CAS PubMed.
- Z. Wu, H. Fan, N. S. R. Satyavolu, W. J. Wang, R. Lake, J. H. Jiang and Y. Lu, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef CAS PubMed.
- R. Orbach, F. Remacle, R. D. Levine and I. Willner, Chem. Sci., 2014, 5, 1074–1081 RSC.
- H. M. Meng, X. B. Zhang, Y. F. Lv, Z. L. Zhao, N. N. Wang, T. Fu, H. H. Fan, H. Liang, L. P. Qiu, G. Z. Zhu and W. H. Tan, ACS Nano, 2014, 8, 6171–6181 CrossRef CAS PubMed.
- W. J. Zhou, W. B. Liang, D. X. Li, R. Yuan and Y. Xiang, Biosens. Bioelectron., 2016, 85, 573–579 CrossRef CAS PubMed.
- F. Wang, J. Elbaz and I. Willner, J. Am. Chem. Soc., 2012, 134, 5504–5507 CrossRef CAS PubMed.
- R. Wang, J. Cheng, X. Zhu, L. Zhou, W. Xuan, Y. Liu, Q. Liu and W. Tan, J. Am. Chem. Soc., 2017, 139, 9104–9107 CrossRef CAS PubMed.
| This journal is © the Partner Organisations 2018 |





