Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multiplexed photoluminescent sensors: towards improved disease diagnostics
Xiao-Peng
He
*a,
Xi-Le
Hu
a,
Tony D.
James
 *b,
Juyoung
Yoon
*b,
Juyoung
Yoon
 *c and
He
Tian
*a
*c and
He
Tian
*a
aKey Laboratory for Advanced Materials & Institute of Fine Chemicals, East China University of Science and Technology (ECUST), 130 Meilong Rd., Shanghai 200237, P. R. China. E-mail: xphe@ecust.edu.cn; tianhe@ecust.edu.cn
bDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
cDepartment of Chemistry and Nano Science, Ewha Womans University, Seoul 03760, Korea. E-mail: jyoon@ewha.ac.kr
First published on 25th August 2017
Abstract
The ability to simultaneously monitor multiple analytes in, for example, a single microplate well, is important for both basic research and clinical applications. In particular, for disease diagnosis there is a growing awareness that determination of a single disease biomarker is insufficient to pathologically confirm a disease state. Consequently, much recent literature has been directed towards the development of multiplexed photoluminescent sensors which can simultaneously detect multiple and diverse biomarkers that exist in a homogenous solution or a single cell, accelerating the progress towards precise disease diagnosis. This tutorial review highlights a selection of recent contributions towards this emerging interdisciplinary field that incorporates chemistry, chemical biology, materials sciences and medical sciences.
Key learning points(1) Why there is a need for multiplexed, photoluminescent sensing of disease biomarkers?(2) The diverse materials used for constructing multiplexed photoluminescent sensors. (3) Photoluminescence detection of multiple disease biomarkers in a homogenous solution or within a single cell. (4) Large-scale, multicentre clinical-specimen analysis is needed to demonstrate the clinical value of multiplexed photoluminescent sensors. |
1. Introduction
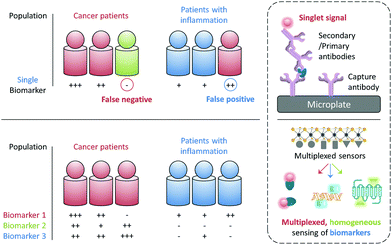
The early and precise diagnosis of fatal human diseases such as cancer can help significantly improve the overall therapeutic efficacy and patient prognosis. However, it has been difficult to pathologically confirm a cancer by referring to the serological test of just a single biomarker, which may either be overexpressed during other abnormal states (false positive) or, more problematically absent in a number of individuals with cancer (false negative). For example, the alpha-fetoprotein (AFP), a biomarker used clinically for the serological test of liver cancer, is also overexpressed in some patients with chronic liver diseases with a false-positive rate of up to 58%. In contrast, it is suggested that 35–65% of patients diagnosed with hepatocellular carcinoma did not show an elevated AFP expression at a cutoff of 20 ng mL−1.1 Therefore, the use of multiple biomarkers to synergistically confirm a disease state has become increasingly important for the more precise and earlier identification of disease states (Fig. 1).2While the confirmation of a set of biomarkers for a given disease requires multicentre, large-scale clinical trials, it would be ideal if these biomarkers can be simultaneously reported using a single sensing system and working with a simple blood or cell sample. The most popular analytical technique for protein biomarker detection is currently the enzyme-linked immunosorbent assay (ELISA). However, besides its high cost, laborious manipulation and long detection time, this biochemical assay has not been proven for the simultaneous detection of multiple biomarkers in a single test (Fig. 1). Therefore, the development of advanced sensing systems that can quickly unveil the existence of more than one specific biomarker has become an intriguing area of research for both chemists and biologists.
With the rapid development of modern materials science and nanotechnology, a number of functional materials have been developed for biomedical applications and, especially for disease diagnosis.3 In addition to the single detection of an analyte, a number of multiplexed sensing systems have been developed over the past few years. These systems can display multiplexed photoluminescent signals for multiple biomarkers in a homogenous solution (Fig. 1). Unlike solid-state techniques, solution-based homogeneous determination of biomarkers is much simpler in manipulation and faster in signal readout since it just relies on the selective interaction of the sensor with the targets in solution to produce a sensing signal. Here we highlight the diverse tactics and materials used in developing multiplexed photoluminescent sensors for homogeneous, multiple biomarker detection.
Multiplexed photoluminescent sensors for homogeneous detection of multiple analytes are constructed by the self-assembly between a range of probe-luminophores (e.g. fluorophores and QDs) and material substrates such as 2D materials and inorganic/organic nanoparticles for the simultaneous detection of different pathologically relevant species in an aqueous environment. Molecularly functionalised nanoparticles (e.g. upconversion materials and conjugated polymers) of different emission colours for multi-colour imaging of cells, pathogens and animals are discussed. While in the perspective section we discuss other optical and electrical multiplexing strategies for clinical diagnoses. Traditional sensing strategies have focused on improving single-analyte specificity; however, this could reduce diagnostic precision (Fig. 1). With this review we suggest that rational design of multi-analyte sensors can significantly advance disease diagnosis.
2. Multiplexed sensors
2.1 Multiplexed photoluminescent sensors based on low-dimensional materials for homogeneous, photoluminogenic detection of multiple analytes
Ever since the discovery of graphene – an atomic-thin carbon material with exceptional optoelectronic properties – there has been ever-increasing interest in the development of two-dimensional (2D) materials for many applications.4 The material substrate used for constructing fluorogenic biosensors mainly relies on water-soluble graphene oxide (GO).5 On the basis of the strong π-stacking between the base pairs of DNA and GO, a multicolour GO probe for the detection of three tumour-suppressor genes (p16, p21 and p53) in homogenous solution was developed.6 Three different single-strand DNA (ssDNA) sequences tagged with fluorophores of different emission wavelengths were used to simultaneously assemble with GO, resulting in the fluorescence quenching of all fluorophores. The quenching was ascribed to the Förster resonance energy transfer (FRET) from the fluorophores to GO (Fig. 2a). The resulting GO/multi-DNA sensor displayed a selectively enhanced fluorescence upon hybridization with a pairing gene sequence because the double-strand DNA (dsDNA) formed resulted in a weakened interaction with the GO quenching platform (Fig. 2b). In particular, the multiplexed system has proven to only show a selective fluorescence enhancement with a given tumour-suppressor gene without interference from other unselective genes (Fig. 2a). This research sets the basis for the multiplexed sensing of tumour-related DNAs.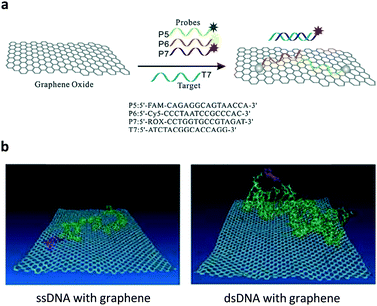 | ||
| Fig. 2 (a) Schematic illustration of the co-assembly of three different ssDNA probes tagged with different fluorophores to graphene oxide and the selective hybridization of an ssDNA probe with a target ssDNA, forming a dsDNA. (b) A molecular dynamic simulation showing the different interaction modes of ssDNA and dsDNA with the graphene surface. Reproduced with permission from ref. 6. Copyright: John Wiley & Sons. | ||
In a subsequent study, it was shown that the GO multiplexed system was also suitable for the simultaneous detection of three virus genes.7 Three looped DNAs tagged with different coloured fluorophores for human immunodeficiency virus (HIV), variola virus and ebola virus were coated onto GO, quenching the fluorescence. The presence of the multiple gene sequences of the viruses in the solution of the multiplexed GO sensor led to the fluorescence enhancement of all three fluorophores, similarly due to the formation of dsDNAs. Meanwhile, this multi-ssDNA-functionalized GO sensor has also been extended to the multiplexed detection of a mixture of DNAs, proteins and ions.8
MicroRNAs (miRNAs) are non-coding RNAs typically with 19–25 nucleotides. A number of these miRNAs have been shown to regulate cellular processes in diseases. Multiple fluorescent ssDNA probes for miRNAs were used to assemble with GO, achieving multiplexed miRNA detection in the presence of a nuclease for signal amplification.9 A Y-shaped DNA scaffold was then developed incorporating the cleavage sequence (each tagged with a different fluorophore) of three nucleases.10 In a homogeneous solution consisting of GO and multiple nucleases, the three different emissions of the Y-shaped DNA were quenched simultaneously, enabling a multiplexed system for nuclease analysis.
The hepatitis C virus (HCV) infection can lead to cirrhosis and ultimately hepatocarcinoma. Over 170 million people have HCV infections worldwide. Based on the DNA/GO assembly, a multifunctional system for the detection of helicase biomarkers that exist in HCV and severe acute respiratory syndrome coronavirus was developed.11 Unwinding the dsDNAs of these viruses by helicases led to the simultaneous fluorescence quenching of the ssDNA probes (Fig. 3). In contrast, the presence of a helicase inhibitor largely prevented the fluorescence quenching due to inhibition of helicase activity. Using this feature, a high-throughput, multiplexed assay for screening helicase inhibitors of multiple viruses was developed. This study highlights the potential of using GO multiplex systems for not only biomarker detection, but more broadly drug discovery.
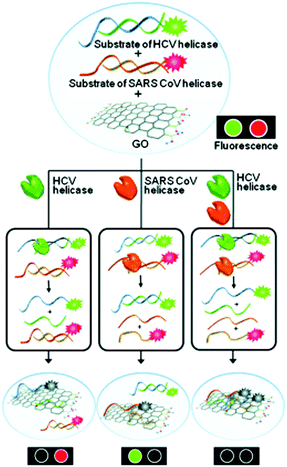 | ||
| Fig. 3 Multiplexed sensing of two different helicases acting on two dsDNA probes in the presence of GO as a quenching platform. HCV and SARS CoV are hepatitis C virus and severe acute respiratory syndrome coronavirus, respectively. Reprinted from ref. 11. Copyright: John Wiley & Sons. | ||
Besides DNA based sensing systems, we have demonstrated that glycoligand–receptor interactions12 can also be determined in a multiplexed manner using GO.13 Carbohydrate–protein interactions are involved in a number of pathological processes including cancer metastasis, virus invasion and bacterial infection. Therefore, the effective analysis of these interactions may aid the development of disease diagnosis and therapy. We assembled two glycoligands with different fluorophores to GO. The quenched fluorescence was recovered simultaneously when the receptors of both glycoligands were available.
In addition to organic dyes, quantum dots (QDs) have also been used as optical reporters for GO multiplexed sensors. A homogenous immunoassay was developed by co-assembly of two antibody-conjugated QDs (green-emitting and red-emitting) to GO, quenching the photoluminescence.14 The presence of human enterovirus 71 and Coxsackievirus simultaneously resulted in the recovery of both green and red photoluminescence as a result of antibody–virus binding. Similarly, using QDs as the reporter, the multiplexed detection of two gene markers for a human infectious pathogen, Listeria monocytogenes, was achieved.15
With the tremendous success of graphene, a diverse range of graphene-like 2D materials such as transition-metal dichalcogenides (TMDs) and oxides have been extensively developed for a number of applications.16 Several TMDs have been proven to possess better biocompatibility than GO, and therefore have been introduced for the development of sensors and therapeutic materials.17–19 It has been shown that TMDs including 2D MoS2, TiS2 and TaS2 are suitable platforms to build fluorogenic multiplexed systems for sensing DNAs (Fig. 4).20 Among several competing low-dimensional materials including single-walled nanotubes, gold nanoparticles and GO, TaS2 based systems exhibited the best sensitivity towards multiplexed DNA detection. We have also shown that 2D MoS2 can be used as a substrate to construct multiplexed sensors for influenza viruses (IVs).21 IVs can cause severe respiratory tract infections, and lead to seasonal epidemics. The hemagglutinin, a main surface protein of IVs, is mainly responsible for viral invasion by binding to sialyl glycans (SA) on the surface of avian or human cells. It is reported that IVs that infect birds and humans have a preference for α2,3-SA and α2,6-SA, respectively.22 Taking note of these facts, we have co-assembled α2,6-SA and α2,3-SA coupled with different fluorophores on 2D MoS2, producing multiplexed sensors (Fig. 5). While the system showed a single emission colour with IVs containing single glycan specificity, in the presence of a virus with dual glycan specificity, both emission colours were produced. This research paves the way for the timely warning of IV mutations that can cause global epidemics.
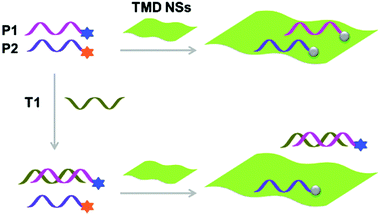 | ||
| Fig. 4 Schematic illustration of the use of single-layer transition-metal dichalcogenide based multiplexed sensors for DNA detection, where P1 and P2 are different fluorophore-tagged ssDNA probes and T1 is a target ssDNA. Reprinted with permission from ref. 20. Copyright: John Wiley & Sons. | ||
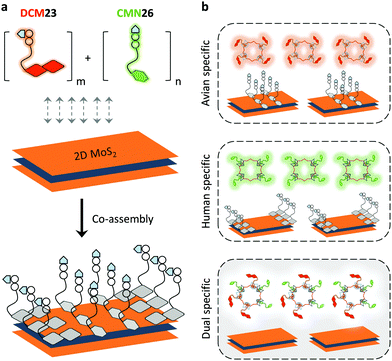 | ||
| Fig. 5 Schematic illustration of (a) the co-assembly of two sialyl glycan probes tagged with different fluorophores to 2D MoS2 and (b) the use of a multiplexed system for the detection of influenza viruses with single or dual glycan specificities, where DCM23 and CMN26 are dicyanomethylene 4H-pyran-tagged Neu5Acα2,3Gal-β1,4Glc and coumarin-tagged Neu5Acα2,6Gal-β1,4Glc probes, respectively. Reprinted with permission from ref. 21. Copyright: The Royal Society of Chemistry. | ||
Proteases are enzymes that catalyze the cleavage of peptide bonds. They play key roles in many diseases such as cancer, viral infection and inflammation. Carbon nanotubes (CNTs), a topical 1D material, have been used as the quenching material for constructing multiplexed sensors for proteases. Three multicolour peptide substrates were conjugated to a CNT platform, leading to fluorescence quenching. The presence of three cancer-related proteases, matrix metalloproteinase-7, matrix metalloproteinase-2 and urokinase-type plasminogen activator, in a homogeneous solution of the sensor produced multicolour fluorescence by specific cleavage of the corresponding peptide substrates.23 In addition, tube-like silicon nanowires have also been proven to be a suitable quenching platform for multiplexed sensing of cancer-related DNA biomarkers.24
2.2 Multiplexed photoluminescent sensors based on nanoparticles for homogeneous, photoluminogenic detection of multiple analytes and multi-colour cell imaging
Inorganic nanoparticles have been the material of choice in advancing modern nanotechnology, owing to their unique optical features at the nanoscale. Of the nanoparticles available, gold nanoparticles (AuNPs) are among the most frequently used for sensing and cell imaging. With the aim of simultaneously probing three tumour-suppressor genes, a molecular beacon (i.e. a dual-labelled stem-loop-structured oligonucleotide with a quenched fluorescence) sensor was developed using the Au–thiol assembly of three multicolour DNA sequences onto a single AuNP core (Fig. 6).25 The universal quenching ability of AuNPs precluded the need to optimise the fluorophore–quencher pair for each DNA probe. This beacon sensor simultaneously displayed a multicolour fluorescence enhancement in the presence of multiple tumour-related DNA markers (p16, p21 and p53).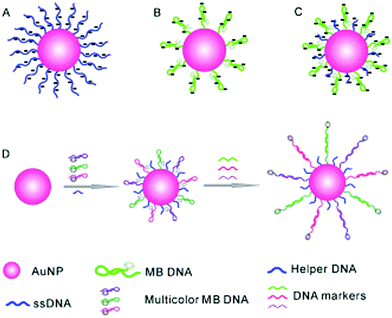 | ||
| Fig. 6 Schematic illustration of a gold nanoparticle based multiplexed sensor for the simultaneous detection of multiple DNA targets. Unlike AuNPs densely assembled with linear ssDNA probes (A, stable but without the ability to form stem–loop structures) and those with sparsely assembled stem–loop structures (B, easy to aggregate), the co-assembly of a helper DNA with ssDNA probes (C) facilitates the formation of stable nanoparticles without affecting the formation of stem-loop structures for multiplexed sensing (D). Reprinted with permission from ref. 25. Copyright: John Wiley & Sons. | ||
Subsequently, this strategy was extended to the multicolour detection of tumour-related mRNA markers.26 A nanoprobe that consists of AuNPs coated with a dense shell of recognition sequences hybridised with different dye-labelled reporter sequences was developed (Fig. 7a). A competitive hybridisation with three tumour-related mRNAs (c-myc mRNA, TK1 mRNA and GalNAc-T mRNA) caused the multicolour fluorescence emission to recover as a result of the formation of more stable duplexes. This nanoprobe has also shown the ability to simultaneously image three mRNAs in a set of cancer cells (Fig. 7b). In addition to AuNPs, silver nanoclusters are also used for multiplexed DNA detection.27 Label-free molecular beacons were developed based on DNA-templated silver nanoclusters with different emissions, achieving simultaneous detection of influenza A subtype H1N1 and H5N1 genes.
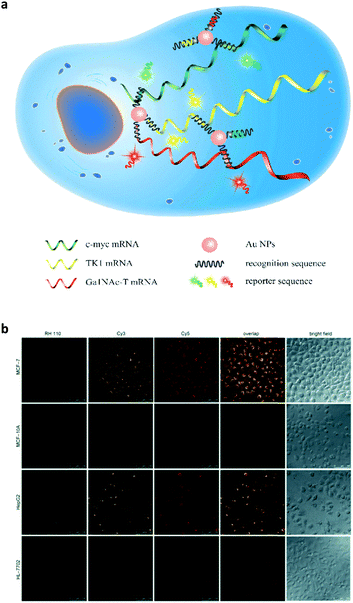 | ||
| Fig. 7 (a) Schematic illustration of a gold nanoparticle based multiplexed sensor for the simultaneous detection of multiple mRNA targets. (b) Multiplexed fluorescence imaging of the multiple mRNA targets in different cells. Reprinted with permission from ref. 26. Copyright: John Wiley & Sons. | ||
QDs represent another class of widely used nanoparticles for bioimaging because of their emission brightness, large Stokes shift and photostability.28 Using sophisticated bioconjugation methods, multicolour immunohistochemistry was achieved in order to simultaneously image a range of different biomarkers on cancer cells and tissues.29 Five QD bioconjugates were synthesised with antibodies against five different biomarkers of breast cancer with multicolour emission.30 These bioconjugates have been used to simultaneously quantify the expression level of the different biomarkers in both cultured breast cancer cells and single paraffin embedded tissue sections. More importantly, the QD–antibody conjugates have been successfully used for endoscopy-guided multicolour cancer diagnosis.31 Conventional diagnosis of colon cancer using endoscopy relies on the clinician's naked eye, which is subjective and may lead to false detection. Multicolour QD bioconjugates coated with matrix metalloproteinase (MMP) 9, MMP 14 and carcinoembryonic antigen were used to help diagnose tumours on sectioned mouse tissue, fresh mouse colons and human colon adenoma. This multiplexed approach imaged using endoscopy provides a rapid and accurate molecular fluorescence imaging technique for colon cancer diagnostics.
In addition to antibody conjugation, peptide-functionalised QDs and QD-based nanocomposite emulsions have been developed for in vivo imaging of therapeutic cells32 and spatiotemporal multicolour labelling of distinct subcellular compartments,33 respectively. DNA-functionalised liposome–QD complex (with a reporter DNA sequence), in combination with magnetic beads (with a capture DNA sequence), has been developed for the multiplexed detection of low-abundance DNA.34 The presence of a target DNA facilitated the formation of a sandwich structure with both the reporter and capture DNA sequences. Subsequently the sandwich ensemble was separated by an external magnetic field and the QDs released an optical signal. This research enables the simultaneous detection of HIV-1 and HIV-2 genes with attomolar sensitivity.
To address the inability of conventional biochemical techniques such as western blotting and single-colour immunohistochemical methods for multiplexed detection of cancer, antibody-conjugated single-band upconversion nanoparticles (UCNPs) with different colours were developed (Fig. 8).35 Compared to QDs, which may suffer from the autofluorescence background of biological samples with blue or ultraviolet emission wavelengths, UCNPs use tissue-transparent near-infrared irradiation to produce photoluminescence. It was shown that the expression of three typical breast cancer biomarkers, estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2, could be simultaneously quantified in cancer cells and tissue specimens, even in triple-negative breast cancer cells that poorly express these biomarkers (Fig. 8).
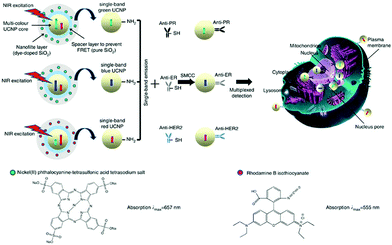 | ||
| Fig. 8 Schematic illustration of the synthesis of multiplexed upconversion nanoparticles for the simultaneous imaging of three biomarkers in breast cancer cells and tissues. Reprinted with permission from ref. 35. Copyright: Nature publishing group. | ||
2.3 Multiplexed photoluminescent sensors based on organic materials for in vitro and in vivo multi-colour imaging
Due to tuneable emission wavelengths (colour) and high quantum yields, emissive conjugated polymers (CPs) have been widely used in biosensing and bioimaging.36,37 An interesting approach based on a simple mixture of bacteria and emissive CPs has been developed.38 Four CPs with different emission colours were synthesised and self-assembled with E. coli cells to produce microparticles where multicolour emissions are achieved by tuning the FRET efficiencies among the CPs (Fig. 9). The use of a FRET mechanism to modulate multicolour emissions of these materials is advantageous since a single wavelength excitation is required and the emission can be modulated using the CPs with different colours. These microparticles have been shown to be well-suited for multiplexed cell detection and identification using flow cytometry.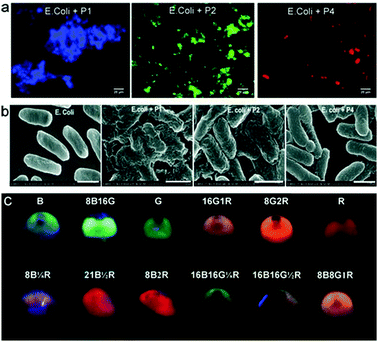 | ||
| Fig. 9 (a) Fluorescence imaging of E. coli based conjugated polymer (CP) microparticles. (b) Scanning electron microscopic images of E. coli alone and the E. coli based CP microparticles. (c) An image of the different multicolour microparticles that can be used for multiplexed cell imaging. Reprinted with permission from ref. 38. Copyright: John Wiley & Sons. | ||
CPs have also been used to construct surface-functionalised nanoparticles for the identification of two different kinds of cancer cells.39 A green-emitting CP grafted with an affibody (i.e. small proteins engineered to strongly bind to target proteins and peptides) for human epidermal growth factor receptor 2 (which is overexpressed on breast cancer cells) and a red-emitting CP grafted with a peptide ligand for integrin (which is overexpressed on colon cancer cells) were produced. These CPs were successfully used to discriminate a breast cancer cell line from a colon cancer cell line in live cell mixtures, using single laser excitation (Fig. 10). Polymer-based nanoparticles can also be loaded with a set of different organic fluorophores using a FRET cascade for multiplexed imaging.40 Multiplexed single excitation in vivo imaging was achieved using the subcutaneous injection of three differently coloured particles in different regions of a mouse (Fig. 11c). The imaging capacities of polymer nanoparticles (RAGE-T3 and R7-M5) were compared with corresponding QDs (QDs-665 and QDs-800). Identical numbers of both particles were injected into different regions of an immunodeficient mouse, and the results indicate that the polymer particles are 10 times brighter than the QDs (Fig. 11a, b and d).
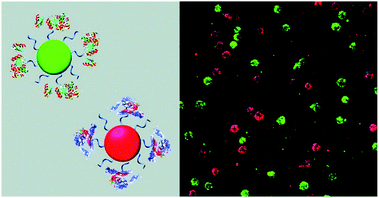 | ||
| Fig. 10 Schematic illustration of affibody-grafted, multicolour conjugated polymer nanoparticles for the discrimination of two different kinds of cancer cells, where the green and red nanoparticles selectively stain breast cancer and colon cancer cells, respectively. Reproduced with permission from ref. 39. Copyright: American Chemical Society. | ||
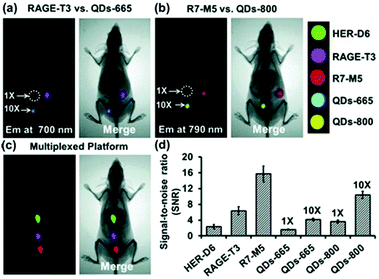 | ||
| Fig. 11 Multiplexed in vivo imaging using multicolour polymer-based nanoparticles in comparison with quantum dots (QDs). Reproduced with permission from ref. 40. Copyright: John Wiley & Sons. | ||
Organic dyes with FRET cascade properties have also been employed to covalently couple with DNA nanoflowers, which were prepared using a straightforward rolling circle replication (Fig. 12).41 The multicolour DNA nanoflowers with an aptamer precursor for cancer cell targeting have proven to be amenable for multiplexed imaging of cancer cells and tracing targeted drug delivery. In addition to the covalent strategy, a supramolecular protocol based on the interaction of a broad-spectrum quencher-type dye, cationic perylene diimide (PDI), with fluorophore-tagged ssDNAs was also developed.42 The fluorescence of the ssDNA probes was quenched by PDI as a result of strong electrostatic contacts, and the presence of multiple complementary DNAs with the assistance of an exonuclease autocatalytic recycling amplification simultaneously showed multicolour fluorescence with picomolar sensitivity. A diverse range of functional materials including metal–organic framework-based molecular beacons43 and lanthanide-based organometallic luminescent complexes44 have also been developed for multiplexed detection of virus genes and enzymatic activity, respectively.
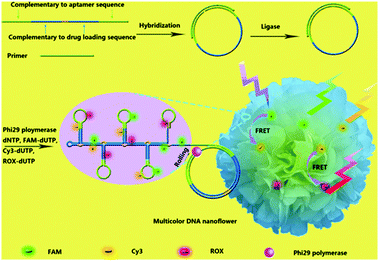 | ||
| Fig. 12 Schematic illustration of the bioassisted, sequence-independent self-assembly of DNA nanoflowers covalently linked with multicolour fluorphores with FRET (Förster resonance energy transfer) cascade properties. Reproduced with permission from ref. 41. Copyright: John Wiley & Sons. | ||
3. Summary and perspective
We have highlighted the recent developments in multiplexed photoluminescent sensors for the homogenous detection of multiple target mixtures and multiplexed imaging of cells and pathogens simultaneously. Based on topical materials including low-dimensional materials such as graphene oxide, transition-metal dichalcogenides and carbon nanotubes, inorganic nanoparticles including AuNPs and QDs, and emissive organic conjugated polymers are all suitable candidates for building such multicolour sensors. With these sensors, multiplexed photoluminescence sensing towards two or more biomolecular markers (including DNA/RNA and proteins) in a homogeneous solution, the multiplexed imaging of multiple markers of a single cell or pathogen and the differentiation of a specific cell in mixed cell cultures are possible.While these systems have displayed significant promise for sensitive multi-target analysis, the specificity of these multiplexed systems can be improved. For example, low-dimensional-material based sensor composites are constructed by the physical adsorption of multiple fluorescent macromolecular ligands with the material substrate in order to acquire sensitivity for the analyte mixtures. However, the recognition and thus the complexation between a ligand probe and a biomolecular analyte lead to the exposure of a certain surface area of the material substrate. This may cause non-specific adsorptions to produce false-positive signals. We anticipate that improved material design will overcome these limitations of low-dimensional materials. Meanwhile, improvements in the quality control of low-dimensional materials will ensure an improved reproducibility of the sensors from batch to batch.
On a second front, the amenability of the multiplexed photoluminescent sensors for clinical samples has been barely demonstrated. In human fluids such as blood, serum and urine, a number of primary and secondary natural metabolites with different structures and charges exist. The conventional way of detecting a single biomarker using ELISA is to first isolate the biomarker of interest by a capture antibody immobilized on a microplate well (rinsing steps are necessary to eliminate non-specific absorbents), and then the biomarker is indicated using a primary antibody in conjugation with a secondary antibody followed by enzymatic reactions (signal production and amplification). This procedure is somewhat laborious though automatic ELISA systems have been developed and already used in hospital environments. However, a set of antibody pairs will be required for the analysis of multiple targets using conventional techniques, which are expensive and sluggish. Therefore, a simplified protocol for the detection of multiple biomarkers in human fluids is challenging yet intriguing. An ideal situation requires that a homogeneous blood/urine sample, when presented with a multiplexed photoluminescent sensor, produces multiple emission colours, which are sensitively readable by conventional microplate readers. These colours correspond to the multiple biomarkers that are produced as a result of the onset of a particular disease. Then, after calibration, the markers are quantified for evaluation of the disease state; however, medical imaging techniques and biopsy analyses may be required for disease confirmation.
A particularly popular technique for developing multiplexed photoluminescent sensors is the use of ligand molecules such as carbohydrates, peptides, aptamers and other small-molecular ligands to replace antibodies, reducing the cost of reagents (obviously these ligands are much cheaper than antibodies in terms of production cost and consumption). However, the specificity of the “smaller” ligand molecules for a target protein must be proven. As an alternative, a particularly elegant approach is the “array sensing” technique pioneered by Anslyn.45 Rather than developing a highly specific receptor for a single analyte, this technique (also known as differential sensing) encourages the use of an array of receptors, which may bind unselectively to multiple analytes, but recognises each of them to varying extents. As a result, a fingerprint response is produced which can give rise to a characteristic pattern for complex mixtures consisting of multiple analytes (Fig. 13). With colorimetric or fluorescence signals, this strategy has been employed for the differential sensing of ions, small neutral molecules, proteins, and cells and bacteria.46 With such a patterning strategy, ligand specificity might be attenuated, leading to flexibility in multiplexed sensor design. Nanomaterials can also be used for pattern analysis.47 An intelligent nanoarray based on molecularly modified AuNPs and a random network of CNTs was prepared for the diagnosis and classification of a number of diseases from exhaled breath. Electrical analyses of 2808 breath samples collected from 1404 subjects having different diseases using the nanoarray verified that each disease had a unique “breathprint”, thus facilitating the development of an easy-to-use and inexpensive personalized diagnostic system. The analogous multiplex sensing of other volatile biomarkers via breath and skin has been demonstrated by the same research group.47
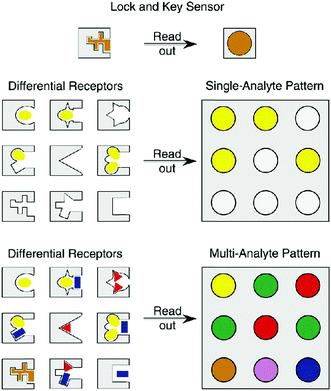 | ||
| Fig. 13 Schematic illustration of the concept of differential sensing (array sensing) for single and multiple analytes. Reproduced with permission from ref. 45. Copyright: John Wiley & Sons. | ||
Furthermore, the direct treatment of a multiplexed photoluminescent sensor with a complex human fluid may cause false-positive signals since the multicolour emissions cover the entire visible-light region. Those with relatively shorter emission wavelengths overlap biological auto-fluorescence. Meanwhile, hydrophobic organic dyes might also bind unselectively to hydrophobic cavities of proteins and DNAs, and can be influenced by changes in the microenvironment such as ionic strength and pH. To tackle this problem, a purification step should be added. We imagine the use of “ligand pairs” in which one ligand (capture ligand) isolates the marker from the complex solution and the other (indicator ligand) identifies the isolated marker. As such, we further envision the development of multiplexed isolating materials incorporating multiplexed sensors for improved disease diagnosis. Besides macromolecules such as nucleosides and proteins, multiplexed sensors for small-molecular cell metabolites and ions are also worthwhile.
Last but not least, it is important to develop improved and diversified multiplexed photoluminescent sensor prototypes. Vibration-induced-emission (VIE) is an intrinsic fluorescence mechanism discovered by Tian and colleagues by serendipity, coined for the subtle alteration in the planar motion of phenazine derivatives.48,49 VIEgens have unique optical properties in that their emission almost spans the visible region (400–750 nm) and have two predominant, well-separated emission peaks. We believe that using VIEgens with tuneable, broad-band emission characteristics, it will be possible to develop single-molecular multiplexed sensors for disease diagnosis.
In addition to the aforementioned perspectives, it should be noted that the most important hurdle for multiplexed photoluminescent sensors to clear before being widely employed in clinics is the ability to be used on a large-scale (typically >1000 specimens) in multicentre clinical trials. We are confident that this goal will be achieved and look forward to the advent of the first FDA-approved multiplexed photoluminescent sensor as a commercially available tool for precise disease diagnosis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Prof. Hong-Yang Wang and Dr Yu-Fei Pan, collaborators of Prof. He Tian and Dr Xiao-Peng He in the development of new molecular probes and sensors for cancer diagnosis, at the Eastern Hepatobiliary Surgery Institute of the Second Military Medical University are warmly thanked for their helpful discussion on the clinical aspects of this review. The authors are financially supported by the 973 project (2013CB733700), the Science and Technology Commission of Shanghai Municipality (15540723800), the National Natural Science Foundation of China (21722801 and 21572058), the Fundamental Research Funds for the Central Universities (222201717003) and the Shanghai Rising-Star Program (16QA1401400) (to X.-P. He). J. Y. thanks the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2012R1A3A2048814). The Catalysis And Sensing for our Environment (CASE) network is thanked for research exchange opportunities. T. D. J. thanks ECUST for a guest professorship.Notes and references
- K. Taketa, Hepatology, 1990, 12, 1420–1432 CrossRef CAS PubMed.
- Q. Shen, J. Fan, X.-R. Yang, Y. Tan, W. Zhao, Y. Xu, N. Wang, Y. Niu, Z. Wu, J. Zhou, S.-J. Qiu, Y.-H. Shi, B. Yu, N. Tang, W. Chu, M. Wang, J. Wu, Z. Zhang, S. Yang, J. Gu, H. Wang and W. Qin, Lancet Oncol., 2012, 13, 817–826 CrossRef CAS PubMed.
- For a recent comprehensive review, see: L. Wu and X. Qu, Chem. Soc. Rev., 2015, 44, 2963–2997 RSC.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS PubMed.
- X.-P. He, Y. Zang, T. D. James, J. Li and G.-R. Chen, Chem. Soc. Rev., 2015, 44, 4239–4248 RSC.
- S. He, B. Song, D. Li, C. Zhu, W. Qi, Y. Wen, L. Wang, S. Song, H. Fang and C. Fan, Adv. Funct. Mater., 2010, 20, 453–459 CrossRef CAS.
- Q. Zhu, D. Xiang, C. Zhang, X. Ji and Z. He, Analyst, 2013, 138, 5194–5196 RSC.
- M. Zhang, B.-C. Yin, W. Tan and B.-C. Ye, Biosens. Bioelectron., 2011, 26, 3260–3265 CrossRef CAS PubMed.
- L. Cui, X. Lin, N. Lin, Y. Song, Z. Zhu, X. Chen and C. J. Yang, Chem. Commun., 2012, 48, 194–196 RSC.
- C.-H. Lu, J. Li, X.-J. Qi, X.-R. Song, H.-H. Yang, X. Chen and G.-N. Chen, J. Mater. Chem., 2011, 21, 10915–10919 RSC.
- H. Jang, S.-R. Ryoo, Y.-K. Kim, S. Yoon, H. Kim, S. W. Han, B.-S. Choo, D.-E. Kim and D.-H. Min, Angew. Chem., Int. Ed., 2013, 52, 2340–2344 CrossRef CAS PubMed.
- H.-L. Zhang, X.-L. Wei, Y. Zang, J.-Y. Cao, S. Liu, X.-P. He, Q. Chen, Y.-T. Long, J. Li, G.-R. Chen and K. Chen, Adv. Mater., 2013, 25, 4097–4101 CrossRef CAS PubMed.
- D.-K. Ji, G.-R. Chen, X.-P. He and H. Tian, Adv. Funct. Mater., 2015, 25, 3483–3487 CrossRef CAS.
- L. Chen, X. Zhang, G. Zhou, X. Xiang, X. Ji, Z. Zheng, Z. He and H. Wang, Anal. Chem., 2012, 84, 3200–3207 CrossRef CAS PubMed.
- Y. Liao, X. Zhou and D. Xing, ACS Appl. Mater. Interfaces, 2014, 6, 9988–9996 CAS.
- For a recent review, see: X.-P. He and H. Tian, Small, 2016, 12, 144–160 CrossRef CAS PubMed.
- J. Hao, G. Song, T. Liu, X. Yi, K. Yang, L. Cheng and Z. Liu, Adv. Sci., 2017, 4, 1600160 CrossRef PubMed.
- D.-K. Ji, Y. Zhang, Y. Zang, J. Li, G.-R. Chen, X.-P. He and H. Tian, Adv. Mater., 2016, 28, 9356–9363 CrossRef CAS PubMed.
- Y.-H. Ma, W.-T. Dou, Y.-F. Pan, L.-W. Dong, Y.-X. Tan, X.-P. He, H. Tian and H.-Y. Wang, Adv. Mater., 2017, 29, 1604253 CrossRef PubMed.
- Y. Zhang, B. Zheng, C. Zhu, X. Zhang, C. Tan, H. Li, B. Chen, J. Yang, J. Chen, Y. Huang, L. Wang and H. Zhang, Adv. Mater., 2015, 27, 935–939 CrossRef CAS PubMed.
- J.-X. Song, X.-Y. Tang, D.-M. Zhou, W. Zhang, T. D. James, X.-P. He and H. Tian, Mater. Horiz., 2017, 4, 431–436 RSC.
- X.-P. He, Y.-L. Zeng, X.-Y. Tang, N. Li, D.-M. Zhou, G.-R. Chen and H. Tian, Angew. Chem., Int. Ed., 2016, 55, 13995–13999 CrossRef CAS PubMed.
- Y. Huang, M. Shi, K. Hu, S. Zhao, X. Lu, Z.-F. Chen, J. Chen and H. Liang, J. Mater. Chem. B, 2013, 1, 3470–3476 RSC.
- S. Su, X. Wei, Y. Zhong, Y. Guo, Y. Su, Q. Huang, S.-T. Lee, C. Fan and Y. He, ACS Nano, 2012, 6, 2582–2590 CrossRef CAS PubMed.
- S. Song, Z. Liang, J. Zhang, L. Wang, G. Li and C. Fan, Angew. Chem., Int. Ed., 2009, 48, 8670–8674 CrossRef CAS PubMed.
- N. Li, C. Chang, W. Pan and B. Tang, Angew. Chem., Int. Ed., 2012, 51, 7426–7430 CrossRef CAS PubMed.
- Y. Zhang, C. Zhu, L. Zhang, C. Tan, J. Yang, B. Chen, L. Wang and H. Zhang, Small, 2015, 11, 1385–1389 CrossRef CAS PubMed.
- M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631–635 CrossRef CAS PubMed.
- Y. Xing, Q. Chaudry, C. Shen, K. Y. Kong, H. E. Zhau, L. W. Chung, J. A. Petros, R. M. O'Regan, M. V. Yezhelyev, J. W. Simons, M. D. Wang and S. Nie, Nat. Protoc., 2007, 2, 1152–1165 CrossRef CAS PubMed.
- M. V. Yezhelyev, A. Al-Hajj, C. Morris, A. I. Marcus, T. Liu, M. Lewis, C. Cohen, P. Zrazhevskiy, J. W. Simons, A. Rogatko, S. Nie, X. Gao and R. M. O'Regan, Adv. Mater., 2007, 19, 3146–3151 CrossRef CAS.
- Y. Park, Y.-M. Ryu, Y. Jung, T. Wang, Y. Baek, Y. Yoon, S. M. Bae, J. Park, S. Hwang, J. Kim, E.-J. Do, S.-Y. Kim, E. Chung, K. H. Kim, S. Kim and S.-J. Myung, ACS Nano, 2014, 8, 8896–8910 CrossRef CAS PubMed.
- J. B. Delehanty, C. E. Bradburne, K. Susumu, K. Boeneman, B. C. Mei, D. Farrell, J. B. Blanco-Canosa, P. E. Dawson, H. Mattoussi and I. L. Medintz, J. Am. Chem. Soc., 2011, 133, 10482–10489 CrossRef CAS PubMed.
- Y. T. Kim, Y.-W. Noh, J.-H. Cho, J. H. Han, B. S. Choi, J. Kwon, K. S. Hong, A. Gokama, Y.-H. Cho and B. H. Chung, J. Am. Chem. Soc., 2009, 131, 17145–17154 CrossRef PubMed.
- J. Zhou, Q.-X. Wang and C.-Y. Zhang, J. Am. Chem. Soc., 2013, 135, 2056–2059 CrossRef CAS PubMed.
- L. Zhou, R. Wang, C. Yao, X. Li, C. Wang, X. Zhang, C. Xu, A. Zeng, D. Zhao and F. Zhang, Nat. Commun., 2015, 6, 6938 CrossRef CAS PubMed.
- L. Feng, C. Zhu, H. Yuan, L. Liu, F. Lv and S. Wang, Chem. Soc. Rev., 2013, 42, 6620–6633 RSC.
- C. Zhu, L. Liu, Q. Yang, F. Lv and S. Wang, Chem. Rev., 2012, 112, 4687–4735 CrossRef CAS PubMed.
- X. Feng, G. Yang, L. Liu, F. Lv, Q. Yang, S. Wang and D. Zhu, Adv. Mater., 2012, 24, 637–641 CrossRef CAS PubMed.
- K. Li, R. Zhan, S.-S. Feng and B. Liu, Anal. Chem., 2011, 83, 2125–2131 CrossRef CAS PubMed.
- A. Wagh, F. Jyoti, S. Mallik, S. Qian, E. Leclerc and B. Law, Small, 2013, 9, 2129–2139 CrossRef CAS PubMed.
- R. Hu, X. Zhang, Z. Zhao, G. Zhu, T. Chen, T. Fu and W. Tan, Angew. Chem., Int. Ed., 2014, 53, 5821–5826 CrossRef CAS PubMed.
- R. Hu, T. Liu, X.-B. Zhang, S.-Y. Huan, C. Wu, T. Fu and W. Tan, Anal. Chem., 2014, 86, 5009–5016 CrossRef CAS PubMed.
- T. Ye, Y. Liu, M. Luo, X. Xiang, X. Ji, G. Zhou and Z. He, Analyst, 2014, 139, 1721–1725 RSC.
- E. Pershagen and K. Eszter Borbas, Angew. Chem., Int. Ed., 2015, 54, 1787–1790 CrossRef CAS PubMed.
- J. J. Lavigne and E. V. Anslyn, Angew. Chem., Int. Ed., 2001, 40, 3118–3130 CrossRef CAS.
- L. You, D. Zha and E. V. Anslyn, Chem. Rev., 2015, 115, 7840–7892 CrossRef CAS PubMed.
- M. K. Nakhleh, H. Amal, R. Jeries, Y. Y. Broza, M. Aboud, A. Gharra, H. Ivgi, S. Khatib, S. Badarneb, L. Har-Shai, L. Glass-Marmor, I. Lejbkowicz, A. Miller, S. Badarny, R. Winer, J. Finberg, S. Cohen-Kaminsky, F. Perros, D. Montani, B. Girerd, G. Garcia, G. Simonneau, F. Nakhoul, S. Baram, R. Salim, M. Hakim, M. Gruber, O. Ronen, T. Marshak, I. Doweck, O. Nativ, Z. Bahouth, D.-Y. Shi, W. Zhang, Q.-L. Hua, Y.-Y. Pan, L. Tan, H. Liu, A. Karban, E. Koifman, T. Rainis, R. Skapars, A. Sivins, G. Ancans, I. Liepniece-Karele, I. Kikuste, I. Lasina, I. Tolmanis, D. Johnson, S. Z. Millstone, J. Fulton, J. W. Wells, L. H. Wilf, M. Humbert, M. Leja, N. Peled and H. Haick, ACS Nano, 2017, 11, 112–125 CrossRef CAS PubMed . For recent elegant studies on the multiplex sensing of other volatile biomarkers via breath and skin, please refer to publications of the Haick group (http://lnbd.technion.ac.il/publications).
- Z. Zhang, Y.-S. Wu, K.-C. Tang, C.-L. Chen, J.-W. Ho, J. Su, H. Tian and P.-T. Chou, J. Am. Chem. Soc., 2015, 137, 8509–8520 CrossRef CAS PubMed.
- W.-T. Dou, W. Chen, X.-P. He, J. Su and H. Tian, Faraday Discuss., 2017, 196, 395–402 RSC.
| This journal is © The Royal Society of Chemistry 2017 |