Cellular modulation by the elasticity of biomaterials
Fengxuan
Han
,
Caihong
Zhu
,
Qianping
Guo
,
Huilin
Yang
and
Bin
Li†
*
Department of Orthopaedics, The First Affiliated Hospital, Orthopaedic Institute, Soochow University, 188 Shizi St, Suzhou, Jiangsu 215006, China
First published on 18th November 2015
Abstract
The behaviors and functions of individual cells, fundamental to the complexity of multicellular organisms, are regulated by their integrated response to a variety of environmental cues such as soluble factors, extracellular matrix (ECM)-mediated signals, and cell–cell interactions. Among these cues, the biomechanical feature of the ECM, represented by its elasticity, has been increasingly recognized as a dominating factor of cell fate. This review article aims to provide an overview of the general principles and recent advances in the field of matrix elasticity-dependent regulation of cellular activities and functions, the underlying biomechanical and molecular mechanisms, as well as pathophysiological implications. A discussion is also provided as to how material design strategies can be used to control the local microenvironment of stem cells to direct their lineage commitment and functions toward tissue development and regeneration.
1. Introduction
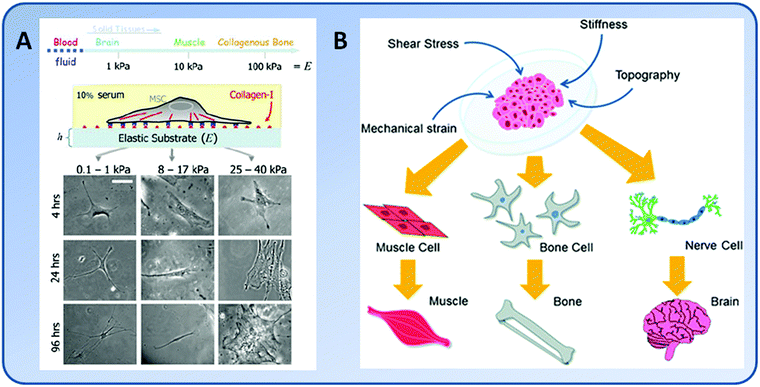
Cells do not live alone in native tissues. Instead, they constantly sense and interact with the surrounding environment, i.e., the extracellular matrix (ECM). Mainly composed of large proteins and polysaccharides, ECM plays a critical role in regulating the majority of cellular activities, including adhesion, migration, proliferation, differentiation, and apoptosis. Under physiological conditions, ECM levels are finely tuned into a status of homeostasis in order to achieve normal growth and differentiation toward morphogenesis and organogenesis. However, under pathological conditions, increased synthesis and/or breakdown of certain ECM components may contribute to cancer progression. Cells respond to the ECM through a complex array of biochemical and biophysical signals which have been extensively studied and reviewed elsewhere.1–3 Recently, growing evidence has suggested that mechanical stimuli, including extrinsic strain, intrinsic stress, substrate elasticity and topography, have a profound impact on the cells.4–8The elasticity of ECM, one of the mechanical stimuli on cells, has been increasingly recognized as an important mediator of cell behaviors. While different terms, including stiffness, rigidity, flexibility, and modulus, have been used to define the elasticity of materials, it generally characterizes the resistance of a material to deformation and is usually represented by the elastic modulus or the Young's modulus of the material. The elasticity of native tissues significantly varies, ranging from soft to rigid environments. For example, the elastic modulus of brain, striated muscle, and osteoid collagen is 0.1–1 kPa, 8–17 kPa, and 25–40 kPa, respectively.9–12 The ECMs that match the elasticity of native tissues preferentially direct the differentiation of stem cells into the lineages of residential tissue cells.8 For instance, human bone marrow mesenchymal stem cells (BMSCs) were effectively differentiated into bone, muscle or neuronal lineages when they were cultured on stiff, medium or soft substrates, respectively.8
Reportedly, the discovery of the effect of ECM elasticity on cell behaviors and functions may date back to the 1970s. Emerman et al. found that mouse epithelial cells (ECs) underwent stronger differentiation on soft collagen gels than on rigid tissue culture plastic dishes.13 Later on, by culturing rat kidney ECs and Swiss 3T3 fibroblastic cells on polyacrylamide (PA) hydrogels that allowed varied flexibility while maintaining a constant chemical environment, Pelham and Wang found that the cells cultured on flexible substrates exhibited reduced spreading and increased motility compared to those on rigid substrates.14 Deroanne et al. also found that tubulogenesis of human umbilical vein ECs (HUVECs) was dependent on the mechanical properties of the underlying substrate. When the cells were cultured on soft matrigels, the expression of actin and focal adhesion (FA) plaques was less than those on rigid matrigels.15 To date, numerous studies have shown that the elasticity of the matrix effectively directs the lineage specification of stem cells in either 2D or 3D environments.8,16–22
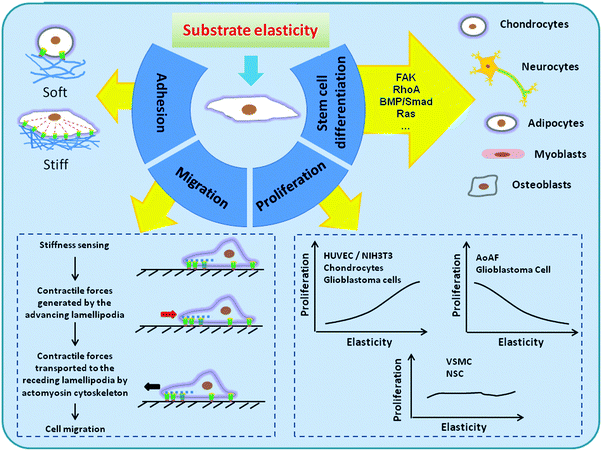
This review article aims to discuss the general principles and recent advances in the field of substrate elasticity-dependent regulation of cellular activities and functions (Fig. 1). We also discuss how material design strategies can be applied to control the local microenvironment, or niche, of stem cells to direct them toward adequate fate and functions.
2. Biomaterials with adjustable elasticity
In vitro, cells are usually cultured on polystyrene-based tissue culture dishes which are intrinsically very rigid surfaces. Considering the fact that in native tissues the majority of cells attach to ECMs of the elastic modulus ranging from 0.01–10 kPa, it is conceivable that most cells in culture are in a highly non-physiologically relevant mechanical environment. Many of the cell behaviors, including cytoskeletal organization, proliferation and differentiation, therefore, may not reflect the real situation in vivo.23 As such, a number of biomaterials that possess adjustable elasticity have been used to study cells in vitro under more physiologically relevant conditions. Fig. 2 summarizes a range of substrate elasticity that have been used in the literature to regulate behaviors of cells from various tissues. To satisfy the needs of different kinds of cells or tissues, biomaterials with different elasticity have been developed. Such materials can be roughly grouped into two major categories, i.e., natural biomaterials and synthetic biomaterials.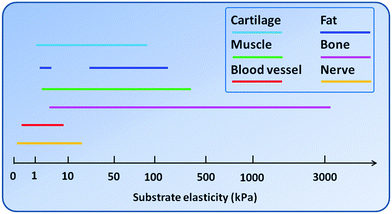 | ||
| Fig. 2 A range of substrate elasticity that have been used in the literature to direct stem cell differentiation toward the cell phenotypes of various tissues. | ||
Natural biomaterials
Derived from the native tissues, many ECM-based natural materials, such as collagen and glycosaminoglycans (GAGs), possess elasticity approximating physiological levels. Examples of natural materials include collagen, hyaluronic acid (HA), gelatin, fibrin, alginate hydrogels, agrose, silk hydrogels, silk–alginate hydrogels, and polyproteins (Table 1). Different strategies have been developed in order to manipulate the elasticity of these materials and scaffolds made from them so that desired native tissue microenvironments can be recapitulated.| Material | Fabrication | Elasticity | Effects on cell behaviors | Cell source | Ref. |
|---|---|---|---|---|---|
| Collagen | 3-D porous scaffolds were synthesized by Col I and hyaluronic acid. The elasticity was tuned by adjusting EDC concentration. | 1–10 kPa | 3-D Col–HA scaffolds can direct hMSCs towards neuronal and glial differentiation via controllable substrate stiffness. | hMSCs | 125 |
| The mechanical properties were tuned using non-enzymatic glycation over various range of ribose. | 175–730 Pa | Increased matrix stiffness resulted in increased sprouting and outgrowth. | ECs | 126 | |
| Gelatin | Gelatin–hydroxyphenylpropionic acid hydrogels (Gtn–HPA) were formed by coupling HPA in the presence of H2O2 and HRP. | 0.6–12.8 kPa | Stiffness of hydrogels strongly affected the cell attachment, FA, migration and proliferation rate of hMSCs. | hMSC | 127 |
| Prepared as above. | 281 Pa, 841 Pa | Proliferation of hMSCs was affected by hydrogel stiffness. Degree of hMSC neurogenesis was tuned by stiffness without biochemical signals. | hMSC | 16 | |
| Gelatin surgical sponge was incubated into the solution of NHS and EDAC by varying the ratio of EDAC/NHS. | 50–1345 Pa | Modest changes in the substrate modulus could have a significant impact on EC function in 3D systems. | ECs | 128 | |
| Alginate | Various elasticities were achieved by tuning the concentration of alginate and calcium ions. | 0.18–19.7 kPa | Greatest enhancement in β-tubulin III expression was seen in hydrogels having elasticity comparable to brain tissues. | NSCs | 129 |
| Gel stiffness can be temporally modulated by light-triggered release of calcium or a chelator from liposomes which is capable of both dynamic stiffening and softening. | 10 Pa–5 kPa | Stiffening inhibited fibroblast spreading. Temporal modulation of stiffness enabled studying the role of dynamic microenvironments. | Fibroblasts | 130 | |
| Silk | Modulated water/methanol annealing were applied to further change the secondary structures for modulating hydrogel stiffness. | 0.6–6 kPa | NSCs grown on the nanofibers expressed preferred neuron differentiation and inhibition of glial differentiation without growth factors. | NSCs | 48 |
| Covalently crosslinking tyrosine residues in silk via HRP and hydrogen peroxide to generate hydrogels with tunable properties. | 0.2–10 kPa | Shows long term survival and exhibits cell–matrix interactions reflective of both silk concentration and gelation conditions. | hMSC | 131 | |
| Agrose | Gel stiffness as a function of agarose gel concentration. | 3–130 Pa | Rate of neurite extension was correlated with the stiffness of agarose gels. | DRGs | 132 |
| Polyprotein | GB1–resilin hydrogels were constructed using photochemical crosslinking and mechanical properties were fine-tuned by adjusting the composition of the elastomeric proteins. | 2 Pa–60 hPa | Mimicked the mechanical properties of muscles. | — | 28 |
| Hyaluronic acid | Methacrylated hyaluronic acid was synthesized to allow for crosslinking via Michael addition and radical polymerization. | 3–100 kPa | Spatially controlling hMSC morphology and proliferation. | hMSC | 133 |
| Silk–alginate | Elasticity is highly dependent on the silk to alginate ratio. | 7–50 kPa | Mechanical and physical properties of alginate hydrogels can be fine-tuned as needed for specific applications. | mESCs, rMSCs | 134 |
The most common approach to achieve variable elasticity is by changing the molecular composition of materials. For example, semi-interpenetrating hydrogels were obtained by mixing HA with different molecular weight (MW) and an atelocollagen solution, followed with inducing collagen fibrillogenesis. The elasticity of composite hydrogels was effectively modulated by simply changing the molecular weight of HA molecules, leading to hydrogels of enhanced stiffness without compromising the biological activity of HA.24 Similarly, highly cross-linked hydrogels can be formed using a blend of high MW and low MW alginates.7
Natural ECM molecules such as collagen are often crosslinked with various crosslinkers (e.g., glutaraldehyde, succinic anhydride, and acyl azide) or enzymes (e.g., lysyl oxidase) to achieve improved elasticity and stability.25,26 While effective, this approach is often accompanied with decrease in gel permeability. By using poly(lactic-co-glycolic acid) (PLGA) microparticles as a filler to bridge interconnected collagen fibrils, the PLGA-filled collagen hydrogels showed a markedly changed storage modulus (4–21 Pa) with minimal change in gel permeability.27
The natural materials often have a porous fibrous structure on the order of cell dimensions which facilitate cell migration and infiltration. However, the potential safety issue and batch-to-batch inconsistency remain to be concerned. In addition, as the natural materials usually have specific biological characteristics, it is sometimes difficult to decouple the effects of matrix elasticity and biochemical signals on cells. Recently, biomimetic polyproteins have been synthesized for both soft and hard tissue regeneration.28 Their mechanical properties can be readily tuned by varying the composition of proteins.
Synthetic biomaterials
Compared to natural biomaterials, synthetic biomaterials are usually bioinert and have well-defined mechanical properties (Table 2). Although a wide gap still exists between these substrates and the physiological environment, they offer the possibility to study cell behaviors under normal and pathological elasticity.| Material | Fabrication | Elasticity | Effects on cell behaviors | Cell source | Ref. |
|---|---|---|---|---|---|
| Polyacrylamide (PA) | Stiffness of PA gels was adjusted by varying the ratios of acrylamide/bis-acrylamide. | 0.1–40 kPa | Soft matrices that mimic brain are neurogenic, stiffer matrices that mimic muscle are myogenic, comparatively rigid matrices that mimic collagenous bone prove osteogenic. | MSCs | 8 |
| (Meth)acrylate-based networks | PEGDMA: networks were photo-polymerized by varying the ratios of PEGDMA/DEGDMA. | 60–850 MPa | Cells exhibited a more differentiated phenotype on the stiffest surface indicated by elevated osteocalcin compared with TCPS. | MG63 | 135 |
| Poly(n-butyl-acrylate) networks (cPnBAs): stiffness was adjusted by the crosslink density of PPGDMA content. | 100 kPa–10 MPa | Promising candidates as soft substrates for passive mechanical stimulation of cells. | L929 | 136 | |
| Poly(dimethylsiloxane) (PDMS) | Using temperature gradient to create a gradient in the crosslinking density of siloxane. | 190 kPa–3.1 MPa | Mineralization is strongly dependent on the stiffness, but is also influenced by the ECM proteins pre-adsorbed on the gradients. | rMSCs | 29 |
| By varying the ratio of crosslinker to oligomer. | 66 kPa–1.1 MPa | MSC proliferation is unaltered but osteogenic differentiation varies with substrate stiffness. | rMSCs | 30 | |
| Polyelectrolyte multilayers (PEMs) | (Poly(L-lysine)/hyaluronan)12 films cross-linked with EDC. | 3–400 kPa | Film stiffness strongly modulates initial myoblast adhesion and proliferation, but also myoblast differentiation into myotubes. | C2C12 | 137 |
| Polyurethane | Polyurethane acrylates reacted with cross-linkers with multi-functionality of acryloxy groups to tune the modulus. | 20–320 MPa | Well defined hierarchical structures showed promising application in ranging from bio-mimetics, microfluidics, to tissue engineering. | — | 35 |
| Poly(ether carbonate urethane) ureas (PECUUs): by varying the soft segment, PEO/PEO–PPO–PEO/TMC | 2–18 MPa | Low moduli polyurethanes may find applications in engineering cardiovascular or other soft tissues. | — | 138 | |
| Polyethylene glycol (PEG)–silica gel | By varying the weight percentage of FS incorporated into the gel. | 7–100 Pa | Effect of matrix stiffness on the differentiation of hMSCs in 3D culture. | hMSCs | 34 |
| Polyvinyl alcohol (PVA) | Gradual freezing–thawing method. | 1–24 kPa | Each soft and stiff hydrogel section promotes effective neurogenesis and osteogenesis, respectively, with the tendency to decrease toward the opposing characteristic's side. | hBMSC | 139 |
| Polyethylene glycol (PEG) | Michael-type addition between thiol- and maleimide-functionalized four-arm-star PEG; varied polymer concentration to modulate hydrogel stiffness. | 0.34–9.1 kPa | The modulus was associated with cell proliferation and function. Gels with low moduli may be useful in stimulating cell engraftment and microvascularization of graft adventitia. | UCBSC | 140 |
| Poly(propylene fumarate)-co-polycaprolactone (PPF-co-PCL) | Different percent compositions of PCL have a wide range of mechanical properties to satisfy diverse requirements in hard and soft tissue replacements. | 2.7 MPa–1.5 GPa | Scaffold surface stiffness correlates with cell attachment, phenotypic expression, proliferation, and differentiation for both bone and nerve cell types. | MC3T3-E1, SPL201, PC12 | 141 |
The most common way to control the elasticity of materials is using monomers and crosslinkers at different concentrations or varying the molecular weight of polymers. Thus far, PA hydrogels have been overwhelmingly used to study the effect of substrate elasticity on cells due to the easy availability and formulation stability. The elasticity of PA gels can be adjusted by varying the crosslinker concentration. Poly(dimethylsiloxane) (PDMS) is also a widely used elastomeric material whose elastic modulus can be adjusted from several tens of kPa to a few MPa simply by changing the ratio of base to curing agent.29–31 PA and PDMS represent two typical substrates of variable elasticity—highly porous hydrogels in which ECM molecules can easily penetrate versus non-penetrating solid surfaces. The cellular responses with respect to elasticity changes may remarkably vary between these two types of substrates.31
Many other synthetic biomaterials have also been used to study the relationship between cell activities and matrix elasticity. A typical hydrogel is poly(ethylene glycol) (PEG)-based hydrogel. In the hydrogels prepared using PEG macromers and poly(lactic acid) (PLA), the elastic modulus increased from 60 to 500 kPa when the initial macromer concentration was doubled from 10%.32 By combining electrospinning and photopolymerization techniques, polyethylene glycol dimethacrylate (PEGDA) hydrogels with tunable elasticity were prepared. Their elastic modulus ranged from 2 to 15 kPa, similar to the elasticity of the intima basement membrane and media layer.33 Since the small mesh size of PEG hydrogels may prevent the deformation and migration of cells, hybrid hydrogels such as PEG–silica nanocomposites have been prepared.34 Polyurethane, with an elasticity of 20–320 MPa, shows promise in mimicking the rigid tissue environment.35 In composite PLA/PLGA porous scaffolds, the elasticity was controlled by changing the ratio of PLA versus PLGA. It was found that PLA-containing scaffolds (100% to 25% PLA) provided stiffness that supported myotube formation of the myoblasts, while pure PLGA scaffolds did not.36
Except the above methods in which the bulk property of the material is changed, there are also other approaches to control the elasticity of biomaterials. For example, discrete substrates such as elastomeric micropost arrays with controlled elasticity have been fabricated by fixing the cross-sectional area of microposts while changing their height. Using this method, the chemistry and bulk mechanics of the material remained constant, while the surface elasticity of it could be changed, making it an ideal tool to decouple the effect of substrate elasticity from other factors.37
3. Effects of matrix elasticity on cell adhesion, spreading, and migration
Cell adhesion
The cells engage with the ECM through focal adhesions or the nearby cells through gap junctions to form a tissue. The FAs, mainly composed of transmembrane adhesion receptors including integrins, vinculins, and paxillins, link the actin–myosin cytoskeleton to the ECM. Cell–matrix interactions largely rely on the mutual interactions between the elasticity of the substrate and intracellular contractility. Therefore, both FAs and the actin–myosin cytoskeleton play an important role in the matrix elasticity sensing of cells.1 When a cell adheres to the substrate and resistance is generated, it senses the resistance through the FA receptors and responds with the organization of actin–myosin cytoskeleton, finally leading to downstream processes such as gene expression and cell differentiation.23As can be imagined, the number and strength of adhesions of cells strongly depend on the elasticity of the underlying matrix. In general, cells have more stable FAs and organized cytoskeleton on relatively rigid substrates and reduced spreading and organization of actin into stress fiber on softer substrates. For example, the vinculin-containing adhesion complexes of ECs and fibroblasts were more diffuse and dynamic on soft gels (E = ∼1 kPa) compared to the stable FAs on stiff gels (E = 30–100 kPa).1 Fibroblasts also exhibited flatter morphology, expressed more as α5-integrin, and generated stronger traction force on stiffer substrates.38,39 Hence, the adhesion strength of the cells is highly matrix elasticity-dependent, which results in differential cell adhesion behaviors and eventually leads to changes in cell morphology, signaling, transcription and consequent functions.
However, the cells' responses to the substrate elasticity are nonlinear—a small elasticity change may result in remarkable change in the cell morphology. Yeung et al. studied the morphology and cytoskeletal structure of fibroblasts and ECs on substrates of varied stiffness (E = 2–55 kPa). When the cells were grown in sparse culture without cell–cell contacts, abrupt changes in actin stress fibers, α5-integrin, and cell spreading occurred at a stiffness of around 3 kPa.38 The matrix elasticity-dependency of the cell shape and cytoskeleton that was evident in single cell culture of fibroblasts or ECs was eliminated when cell–cell contact formed. Therefore, depending on the intercellular communications, the elasticity of matrix affected cells in fundamentally different ways.
Cell spreading
The cell spreading consists of actin-dependent cell membrane extensions and integrin-mediated adhesions, and the actin assembly is affected by the integrin–ECM binding and membrane resistance. As soon as a cell attaches to the substrate, it changes from a rough sphere to a thick disk and receives the signal from integrins. Following that, actin polymerization results in motion and extension of the cell membrane, while myosin contraction and membrane tension forces it to retrograde. Finally, a balance between these processes determines the cell spread area.40In native tissues and in vitro cultures, both ECM composition and elasticity affect the spreading of cells. In general, cells on softer substrates show reduced spreading and reduced organization of actin into stress fibers compared to those on stiffer substrates, as shown in Fig. 1. However, different cell types exhibited different ECM elasticity dependence. For example, the motor neurons develop neurites with extensive branches on softer surfaces instead of rigid ones, while fibroblasts are more spreading on rigid surfaces.9,41 On PEG hydrogels, human mesenchymal stem cells (MSCs) showed maximal spreading at a 13 kPa surface, while HT-1080 fibrosarcoma cells, a tumor cell line, showed a round shape on all PEG surfaces with little elasticity dependence.42 In chondrocytes, the degree of spreading kept improving when the substrate elasticity increased from 4 kPa to 100 kPa. When cultured on nanofibrous gelatin scaffolds (compressive modulus = 0.9–8.2 kPa), dental pulp stem cells (DPSCs) showed round morphology and were separated from each other on low-stiffness surfaces, had few pseudopodia and limited connections on medium-stiffness surfaces, and showed more spread and pseudopodia on high-stiffness surfaces.43
While cell spreading may be dependent on the substrate elasticity, this effect may be masked or even reversed by other compositional or biochemical factors. For example, adenocarcinomic human alveolar basal ECs (A549 cells) showed a positive correlation between spreading and substrate elasticity on PA hydrogels, yet their spreading was not affected on PDMS within the same elasticity range.40 Similarly, vascular smooth muscle cells (VSMCs) showed increased spreading on fibronectin-modified substrates when the stiffness increased from 25 to 135 kPa; however, an exactly opposite trend was seen in VSMCs cultured on laminin-modified substrates.44 On the other hand, there are also conditions that the impact of substrate elasticity exceeds other factors. The adhesion and spreading of rat aortic SMCs, for example, was insensitive to the density of adhesive ligands of soft gels.41
It should be noted that not all cells sense the substrate elasticity. In addition, not all the mechanosensitive cell types show similar responses to the elasticity. The effective responsive range of cells to substrate stiffness varies by cell type. For example, neurons showed best spread on soft substrates (E < 0.5 kPa), while fibroblasts most spread on relatively rigid substrates (E = ∼10 kPa), on which chondrocytes just started to spread.45,46 In another study in which four types of cells were cultured on heparinized PEG-based hydrogels with different elasticity (E = 0.3, 5.2 and 13.7 kPa, respectively), the vascular cells exhibited apparent elasticity-dependent adhesion behaviors, human VSMCs preferred attaching on stiff hydrogels, whereas both adventitial fibroblasts and HUVECs attached to the same degree irrespective of the elasticity of hydrogels.47
Cell migration
Guided cell migration, or directional cell locomotion, is fundamental for many important physiological processes such as tissue morphogenesis, wound healing, and immune responses. Except the many known chemical and biological cues that guide the movement of cells, the elasticity of the substrate also plays a critical role in cell migration, as shown in Fig. 1. For example, a two-fold increase in the migration speed of HT-1080 fibrosarcoma cells was found on stiff gels (13 kPa) compared to those on soft gels (0.34 kPa). Notably, despite the significant changes in cell migration speed over a wide range of elasticity, HT-1080s showed rounded morphology on all surfaces. This implies that while the motility of HT-1080s is strongly influenced by matrix elasticity, they migrate with minimal dependence on cell adhesion – a phenomenon which is distinctively different from the case of human MSCs.42 Similarly, when neural stem cells (NSCs) were cultured on silk hydrogels of various elasticity (E = 0.6–6 kPa), more migrating cells were seen on stiffer gels.48During migration, cells extend lamellipodia and probe the matrix through integrin binding and determine the elasticity of the matrix through traction forces.49 The signal is generated locally by forces applied to a stiff ECM at the tips of lamellipodia and then transported by the actin cytoskeleton to the back of the lamellipodia where it can activate contraction and start a new cycle. Cells precisely sense and respond to the elasticity of the anchoring matrix by localized and proportional strengthening of the integrin–cytoskeleton linkages, allowing stronger force to be exerted on the integrins.50 The stiffer substrate contributes to stronger contractile response. Therefore, rigid substrates can generate contraction and induce movement of cells toward a rigid region, whereas soft substrates may not. Motile cells have been found to align along the direction of highest stiffness and move toward stiffer regions. In a classical study by Lo et al., NIH 3T3 fibroblasts were cultured on PA hydrogels which had a transition in rigidity in the central region. When the cells approached the transition region from the soft side, they easily migrated across the boundary to the stiff side and spread more extensively. In contrast, when the migrating cells came to the boundary from the stiff side, they tended to avoid going to the soft side by turning around or retracting.39 A similar phenomenon was also seen in ECs, which migrated along the direction of greatest stiffness.51 Such a substrate elasticity-guided cell migration, also known as durotaxis or mechanotaxis, implies that changes in tissue elasticity may play a critical role in many pathophysiological processes involving cell migration.50
4. Effects of matrix elasticity on cell proliferation and apoptosis
Cell proliferation is also regulated by ECM elasticity. For example, fibroblasts on flexible substrates exhibit decrease in DNA synthesis and increase in apoptosis.52 Compared to muscle stem cells (MuSCs) cultured on rigid plastic dishes (106 kPa), MuSCs cultured on hydrogels mimicking the elasticity of muscle (12 kPa) showed self-renewal in vitro and extensively contributed to muscle regeneration upon subsequent implantation into mice. Remarkably, GBM tumor cell proliferation was also strongly regulated by ECM elasticity, with cells dividing much more rapidly on rigid than on compliant ECMs.53While it is true that many cells tend to divide much more rapidly on relatively rigid ECMs than on compliant ones,53 the exact impact of ECM elasticity on cell proliferation varies by cell type. Different types of cells may have distinctively different proliferation behaviors on the substrates with different elasticity, as shown in Fig. 1. In native blood vessels, for example, the vascular cells displayed different proliferation behaviors on PEG-based hydrogels within a range of Young's modulus of 0.3–13.7 kPa.47 Proliferation of adventitial fibroblasts increased as the hydrogel elasticity increased, yet proliferation of HUVECs showed a nonlinear elasticity-dependence and proliferated most rapidly on the softest hydrogel. On the other hand, proliferation of human VSMCs was hardly affected by the elasticity of hydrogels.47
It should be noted that not all cells show substrate elasticity-dependent proliferation. For example, normal NIH 3T3 cells undergo less proliferation and more apoptosis on soft substrates than on stiff ones. In contrast, H-ras-transformed cells maintained their growth and apoptotic characteristics regardless of substrate elasticity. The responses in cell spreading area and traction forces to substrate elasticity were similarly diminished, which may explain the unregulated growth behavior of transformed cells.52
In addition, different cell behaviors may respond to substrate elasticity at different elasticity levels. Porcine chondrocytes proliferated less on soft gels (E = 4 kPa) compared with those on stiffer gels (E = 10–100 kPa). However, the differentiated phenotype of chondrocytes was best stabilized when they were grown on 4 kPa gels.54 Such unsynchronized elasticity dependence reminds that for efficient tissue engineering applications, the mechanical properties of scaffolds should be tailored to cater for the different needs of cell expansion and differentiation. For example, pristine chondrocytes are best expanded in vitro on relatively stiff substrates in order to promote cell proliferation. Following that, they are preferably transplanted to soft scaffolds to support the chondrogenic phenotype. Similar situations were also seen in other cell types. For example, proliferation of rat BMSCs (rBMSCs) and rat adipose derived MSCs (rAMSCs) was not apparently affected by substrate stiffness. However, the osteogenic differentiation of them was significantly promoted with stiffness increase, although rBMSCs appeared to express more osteoblast-related markers than rAMSCs at the same stiffness.55 In another example, NSCs showed no difference in proliferation on silk nanofibers (E = 0.6–6 kPa). The cell apoptosis, however, was markedly delayed on the softer substrates which approximated the elasticity of native nerve tissue.48 Therefore, although many cells are impacted by the substrate elasticity, their responsive behaviors are nonlinear. The cell type and phenotypic status, substrate elasticity range, and cell culture conditions all play an important role in deciding the specific cellular responses.
5. Matrix elasticity-mediated differentiation of stem cells
The majority of stem cells are sensitive to tissue-level elasticity. The effect of ECM elasticity on cell differentiation was first observed by Emerman et al., who found that mouse mammary ECs undergo stronger differentiation on soft collagen gels than on stiff tissue culture plastics.13 Recently, numerous studies have revealed that the lineage commitment of stem cells is significantly affected by ECM elasticity (Fig. 1). In general, ECM elasticity that matches native tissue preferentially directs stem cell differentiation into the resident cells of this tissue. For example, human MSCs were found to differentiate into neuron-like cells on soft gels (E = 0.1–1 kPa) which mimicked nerve tissue properties, myoblasts on moderately stiff gels (E = 8–17 kPa) which mimicked muscle tissue, and osteoblasts on stiff gels (E = 25–40 kPa) which mimicked bone tissue, respectively8,56 (Fig. 3).The elasticity of the substrate plays a vital role in stimulating the osteogenic differentiation of stem cells. In general, MSCs preferentially differentiate into osteogenic lineage on stiff substrates. For example, the osteogenic differentiation of hMSCs was significantly enhanced on graphene oxide (GO)-modified collagen scaffolds (E = 38.7 kPa) compared to unmodified ones (E = 14.6 kPa).57 Indeed, within a certain range of elasticity (E = 7–42 kPa), the osteogenic differentiation of hMSCs continued to improve as matrix elasticity increased.58
The adipogenic differentiation of stem cells is also sensitive to substrate elasticity. For example, hADSCs on gels mimicking the native stiffness of adipose tissue (2 kPa) had significantly upregulated adipogenic markers even without the presence of exogenous adipogenic growth factors. As substrate stiffness increased, hADSCs started to lose the rounded morphology and failed to express adipogenic markers. Therefore, a substrate that recapitulates the mechanical properties of adipose tissue can stimulate adipogenesis of hADSCs in the absence of exogenous adipogenic molecules.59
The substrate elasticity is not only critical in directing chondrogenic differentiation of stem cells, but also important in the maintenance of the chondrogenic phenotype. Upon culture on regular plastic dishes, articular chondrocytes tend to lose their chondrogenic phenotype and develop a fibroblast-like phenotype over time. Such a phenotypic change may be reversed by culturing them on soft gels (E = 4 kPa), where they became more round and expressed more type II collagen and aggrecan typical of articular cartilage tissue.54 Similarly, differentiation of NSCs is also regulated by substrate elasticity. NSCs preferred to differentiate to neuronal cells when cultured on 0.1–0.5 kPa hydrogels, whereas differentiate to glial cells on 1–10 kPa hydrogels.60
The elasticity of the substrate also exerts decisive influence on the differentiation of MSCs toward vascular cell types. By using PEGDA hydrogels with tunable elasticity (E = 2–15 kPa) which is similar to the elasticity of the intima basement membrane and media layer, it was found that MSCs seeded on rigid gels (E = 8–15 kPa) were bigger than those on soft gels (2–5 kPa). Depending on the matrix elasticity, cells showed different vascular-specific phenotypes with remarkably high differentiation efficiency. About 95% of MSCs seeded on soft gels (E = 3 kPa) showed expression of Flk-1, an endothelial marker, within 24 hours, while only 20% of cells on rigid gels (E > 8 kPa) had Flk-1 expression. In contrast, 80% of cells seeded on rigid gels demonstrated smooth muscle α-actin markers, while less than 10% of cells on soft gels showed α-actin markers. Such ability to control the differentiation of MSCs into either endothelial or smooth muscle-like cells through the elasticity of the substrate appears to be an effective approach for vascular tissue regeneration.33
The regulation of substrate elasticity on cell differentiation is remarkably strong and mostly dominating and may override that of biochemical signals.61 The substrate elasticity imposes a strictly non-overlapping range of differentiation. When MC3T3-E1 cells were cultured on PA gels, higher alkaline phosphatase (ALP) activity was obtained on stiff gels (E = 9.6–153 kPa), while inhibited ALP expression was seen on soft gels (E = 0.6–4.8 kPa).45 The tubulogenesis of HUVECs, represented by the expression of actin and FA plaques, on soft matrigels was less than on rigid matrigels regardless being coated with collagen or not.15 In addition, the effect of substrate elasticity on stem cell differentiation varies by their origin. For example, while the osteogenic differentiation of both rat BMSCs and rAMSCs was significantly promoted by the substrate stiffness, rBMSCs expressed more osteoblast-related markers than rAMSCs when cultured on substrates of equal elasticity.55
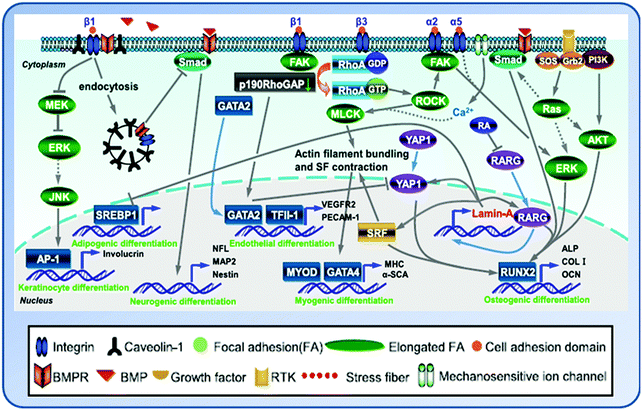
6. Molecular basis of cellular modulation by matrix elasticity
A comprehensive understanding of the cellular responses to the elasticity of the substrate is essential for designing biomaterials that mimic the physiological environment and advancing stem cell-based clinical applications. It is generally believed that cells sense and respond to the microenvironmental elasticity through the dynamics of the actomyosin cytoskeletal network and the mechanosensory proteins in the adhesion complexes that link the cytoskeleton to ECM and the contractile forces that are generated by the cytoskeleton and transmitted to ECM through transcellular structures.6,62,63 However, how the substrate elasticity cue as an external mechanical signal is translated into intracellular signals to trigger changes in gene expression via a cascade of signaling pathways remains to be elucidated.Matrix elasticity regulates integrin binding and reorganization of adhesion ligands on the nanoscale, which are traction dependent and contribute to the commitment of stem cells.7 When a cell is in contact with the substrate, it undergoes the following processes. First, the cell adheres to the substrate by the adhesion plaque proteins (integrins, vinculins and paxillins). The cell applies traction forces on the substrate and produces resistance through the actin–myosin cytoskeletal linkages to the FAs. During this process, the cell senses the restraining force from the substrate and responds with proportional localized strengthening of cytoskeleton linkages, allowing stronger force to be exerted on the integrins. Following that, the cytoskeleton senses and responds to the resistance. Second, the biophysical cue is converted into intracellular signaling cascades. Finally, the gene expression profile of the cell alters, followed with changes at the protein level.63 During the whole process, the mechanotransducing molecules, Rho kinase (ROCK) and FA kinase (FAK), play an important role in transducing the mechanical signal outside in and eventually affect the cell fate and activities.
Sensory receptors of matrix elasticity on the cell membrane
The adhesion receptor integrin which links ECM and the cytoskeleton mediates the response of the stem cell to ECM.64 As the primary cellular mechanosensors for adhesion-dependent mechanical forces, the occupancy and clustering of integrins regulates downstream signaling in response to matrix elasticity. During the osteogenic differentiation of MSCs, integrins on the membrane of MSCs sensed the mechanistic alteration of substrates and dictate the osteogenic differentiation process via ROCK and FAK to subsequent activation of ER1/2.58 When cultured on stiff GO-modified collagen scaffolds, MSCs presented up-regulated molecules involved in cell adhesion, stretched actin filaments and consequently increased cytoskeletal tension and FA formation, and more activated FAK and extracellular-signal-related kinase (ERK) pathways, all of which contributed to the enhanced osteogenic differentiation of the stem cells.57Depending on the elasticity of the substrate, the isoforms of integrin play different roles in stem cell responses. For instance, α2 integrin regulates the osteogenesis of stem cells on stiff substrates, while β3 integrin mediates the myogenesis on medium substrates.65,66 Du et al. found that while the level of cell surface integrin on soft substrates was significantly lower than that on stiff ones, β1 integrin activation in BMSCs was more apparently induced by soft substrates than by stiff ones. The integrin–ligand complexes are more easily ruptured on soft substrates and as a result, the soft substrate markedly enhanced the internalization of integrin, which promoted the neural lineage specification of BMSCs. Moreover, soft substrates suppressed the bone morphogenetic protein (BMP)/Smad pathway at least partially through integrin-regulated BMP receptor endocytosis. Therefore, ECM elasticity affects integrin activity and trafficking to modulate integrin BMP receptor internalization, thus contributing to stem cell lineage specification.67 Except integrins, vinculins and paxillins are also the adhesion plaque proteins.
Mechanotransduction processes
The mechanosensing is a cell's ability to sense and respond to the mechanical properties of its microenvironment. Cells interpret changes in the physical properties of adhesion substrates as changes in adhesion–ligand presentation.7 Following the initial FA formation upon cell adhesion to a substrate, the resulting tension by myosin-dependent traction forces on the substrate leads to the activation of integrin.62 Then the cells respond to the resistance through cytoskeleton organization under different matrix elasticity. The forces generated from the sliding of myosin bundles along actin filaments are transmitted to ECM, causing adhesive protein to assemble together to link the extracellular and intracellular environments. Therefore, the actomyosin contractility is critical for the cells to sense the substrate elasticity. Inhibition of non-muscle myosin II blocks matrix elasticity-directed lineage specification.The initial tension generated by acto–myosin contractility at the beginning of mechanosensing not only allows the adherent cells to exert traction stress on the matrix, but also forces the microtubules to experience resisting compressive forces.68 In addition, the initial tension caused by the acto–myosin contraction and the opposing compressive forces exerted by microtubules may also be transmitted into the nucleus through the cytoskeletal network. These forces can be resisted by the mechano-sensitive nucleoskeletal protein lamin-A on the basis that lamin-A levels in nuclei of stem cells correlate positively with increasing ECM elasticity.69 Recently, proteomics analyses have revealed that tissue elasticity increased the level of lamin-A which stabilized the nucleus and contributed to lineage determination of stem cells. For instance, differentiation of stem cells into fat cells on the soft matrix was enhanced by low lamin-A levels, whereas differentiation of cells into bone cells on the stiff matrix was enhanced by high lamin-A level.70
Molecular pathways
The complex crosstalk network triggered by the substrate elasticity affects the gene expression and fate of cells through a variety of signaling pathways and their interplays (Fig. 4). The mechanical cues embodied by cytoskeletal tension and RhoA signaling are integral to the commitment of stem cell fate.71 The activation of integrin signaling stimulates Rho GTPase and the downstream target protein ROCK to further activate myosin light chain kinase (MLCK), which in turn mediates actin filament polymerization and actomyosin-driven contraction to generate cytoskeleton tension.62 When MSCs were cultured in osteogenic medium on hydrogels with tunable elasticity (E = 7 and 42 kPa), enhanced osteogenic differentiation was seen to accompany with an increase in kinase activities of ROCK, FAK, and ERK1/2 on stiffer matrices (E = 42 kPa). Inhibition of FAK and ROCK resulted in decreased expression of osteogenic markers during osteogenic induction. In addition, FAK affects osteogenic differentiation through ERK1/2, whereas ROCK regulates both FAK and ERK1/2. Therefore, the matrix elasticity influenced MSC osteogenesis through integrin-mediated mechanotransduction.72 Indeed, MSCs on the stiff substrate could recruit β3-integrin to develop more matured FA complexes and subsequently activated RhoA signaling and promoted RhoA-mediated osteogenesis and RhoA/ROCK commitment signals.65The BMP/Smad signaling is also affected by the matrix elasticity. It was found that MSCs on the soft substrate had enhanced β1-integrin internalization and subsequent BMP receptor (BMPR) endocytosis as the BMP/Smad signaling pathway was blocked. As a result, the expression of neuronal genes was up-regulated in the cells. On the other hand, the osteogenesis of MSCs on the stiff substrate may be modulated by the interplay between FAK and RhoA/ROCK, BMP/Smad and Ras-mediated signaling pathways.63,66,73
Other than these, a variety of signaling pathways have also been suggested to be involved in the cellular responses toward substrate elasticity, including β-adrenergic receptor (β-AR) signaling and protein kinase A (PKA) activation through the coordination of microtubules,74 lipoprotein receptor-related protein (LRP) 5/Tie2 signaling,75 the ERK/mitogen-activated protein kinase (MAPK) signaling pathway,31 and the Rho/Rho kinase (ROK)-mediated myosin light chain (MLC) phosphorylation.76 Cells on soft substrates showed reduced phosphotyrosine at adhesion sites, suggesting the possible involvement of both protein tyrosine phosphorylation in the process of cell–matrix interaction.14,50 In addition, the vertebrate transient receptor potential channel vanilloid subfamily 4 (TRPV4) cation channel has been suggested to function as a component of an osmotic/mechanical sensor in vivo.77
7. Interplay between matrix elasticity and other environmental factors
Substrate elasticity alone may promote a certain lineage over another. However, a fundamental fact is that many tissues have similar stiffness, meaning that stimulation from the mechanical properties of the substrate alone is insufficient to decide the cell fate.78 The differentiation of cells into a specific lineage usually involves a spectrum of different factors.Increasing evidence has suggested that substrate elasticity and other physical properties (such as geometry, topography, and roughness) and biochemical signals (such as molecular composition, nutrient supplements, and growth factors) may act in a coordinated fashion to direct stem cell differentiation. For instance, combined use of biomaterials of appropriate elasticity and biochemical treatments led to stronger osteogenic differentiation of rat BMSCs and ADSCs than either treatment alone.55 Using silk–tropoelastin composite matrices which had controlled surface roughness, topological patterns, stiffness, and mechanical strength, it was found that a combination of low roughness and high stiffness promoted myogenic differentiation of C2C12 cells. In contrast, high roughness with micro/nano-scale surface patterns favored hMSC differentiation. Increasing the tropoelastin content promoted osteogenic differentiation of hMSCs.79 To investigate the interplay of multiple environmental factors on cell behaviors, a device which enabled simultaneous control of multiple variables such as the scaffold mechanics and surface chemistry has been developed. Using this device, the local activation of biochemical responses and spatial distribution of FA complexes and transmembrane proteins could be explored to decipher their roles in mechanotransduction.80
Geometric and topographical cues
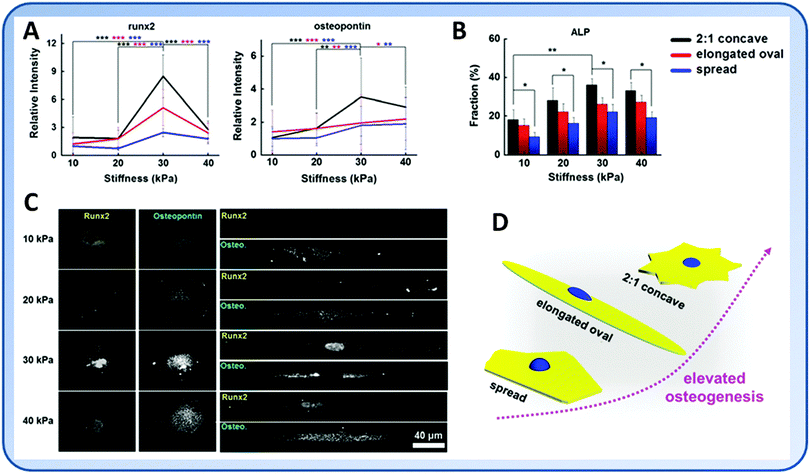
The geometry of cells is an important factor that decides the differentiation of stem cells.81–83 Many studies have shown that well-spread cells are more prone to osteogenic or SMC differentiation instead of adipogenesis or chondrogenesis.71,81 Cells with an increasing aspect ratio and in those having subcellular concave regions had enhanced actomyosin contractility that promoted the osteogenesis.84 Using hydrogels of different stiffness and controlled geometric cues, Lee et al. found that while MSCs tended to undergo osteogenic differentiation on the stiff substrate, patterned cells with increased cytoskeletal tension showed further enhanced osteogenic marker gene expression85 (Fig. 5).The ECM nanotopography alone modulates cell behavior by changing the integrin clustering and FA assembly, leading to changes in cytoskeletal organization and cell mechanical properties. On rigid cell culture plates, hMSCs on gratings exhibited lower instantaneous and equilibrium Young's moduli and apparent viscosity. On the softer PDMS, the effects of nanotopography became insignificant. However, hMSCs on PDMS showed lower mechanical properties than those on culture plates regardless of surface topography. Therefore, both nanotopography and substrate elasticity are important in determining mechanical properties, while nanotopography may be more dominant in determining the organization of the cytoskeleton and FAs.86
Biochemical factors
Substrate elasticity modulates the responsiveness of MSCs to biochemical cues such as growth factors. Using an artificial niche microarray platform, Gobaa et al. found that substrate stiffness imposes a strictly non-overlapping range of adipogenic differentiation of hMSCs, highlighting the dominance of physical factors over biochemical ones. At a given stiffness, a significant protein-dependent effect on adipogenic differentiation was observed. The synergistic interactions between proteins could also be driven by the substrate stiffness.61The matrix stiffness may prime the signaling pathways in stem cells or differentiated cells and synergistically affect the cellular characteristics. When goat articular chondrocytes were cultured on PA gels, the effects of transforming growth factor-β1 (TGF-β1) on chondrocyte mechanics were potent in cells cultured on stiff substrates (E = 90 kPa), while the effects of interleukin 1β (IL-1β) were potent on soft substrates (E = 1 kPa).87 The chondrocytes grown on substrates of adequate stiffness (E = 0.5 MPa, close to the stiffness range of native articular cartilage) had the most prominent proteoglycan deposition and Sox9, Col2α1, and aggrecan gene expression. The combination of ECM stiffness and exogenous TGF-β induced chondrocyte gene expression more robustly than either cue alone through a p38 MAPK-dependent mechanism.88 In fibroblasts, the differentiation into myofibroblast requires both mechanical tension from matrix stiffness and TGF-β.89,90 Combined responses for MSCs to matrix elasticity and BMP-2 cues were also reported, yet with contradictory findings. Zouani et al. discovered that a minimum stiffness (E = 3.5 kPa) existed for MSCs to respond to BMP-2.73 However, they found no synergy between the matrix stiffness and BMP-2 dose, which is in contrast to the findings of Tan et al.91 In another study, the effects of BMP-2, platelet-derived growth factor (PDGF) and substrate elasticity on the differentiation of ADSCs were examined.21 Interestingly, the growth factors affected cell fate only when the cells were cultured on soft substrates, while stiff substrates directed the osteogenic differentiation of ADSCs no matter growth factors were presented or not.21 Clearly, matrix elasticity and growth factors have a synergistic effect on the cellular responses within a certain elasticity range, and in many cases, the impact from one factor overrides that from another.
Matrix composition
The composition of the matrix is important in modulating the cell fate. Substrates with similar elasticity but different molecular compositions may display distinct effects on the same cells. For example, the ECM components, proteoglycans and hyaluronic acid, being able to regulate matrix hydration and therefore resistance to compression, also contribute to the local mechanical environment sensed by cells.92 In a hydrogel system using polymerizable gelatin methacrylate (GelMA), GelMA with osteo-inductive alendronate (Aln) (Aln-GelMA), and PEGDA to achieve various stiffness (E = 4–40 kPa) and Aln density (0–4 μM), it was found that the stiffness and Aln density could synergistically improve the expression of all osteogenesis markers. High Aln density appeared to be more effective than the stiffness.93 Similarly, it was found the combination of high substrate stiffness and α5β1 integrin signaling stimulated by c(RRETAWA), an α5β1 integrin-binding peptide, was sufficient to induce osteogenic differentiation of hMSCs without using any soluble factors.20 Engler et al., on the other hand, found that adhesion and spreading of rat aorta SMCs were dependent on matrix stiffness, but insensitive to the density of adhesive ligands.418. Implications of matrix elasticity in diseases and therapies
Matrix elasticity usually alters during ageing and the progression of diseases, such as cancer,94 liver fibrosis and cirrhosis,23 emphysema,95 scleroderma,96 and cardiovascular diseases.97 The feedback of cells toward local matrix elasticity changes, therefore, has important implications for the ageing, disease development and tissue repair/regeneration.6Matrix elasticity alternation upon tissue development and ageing
The physical properties of tissues and ECM remodeling play a critical role in tissue and organ development. For example, the elasticity of the matrix determines the tubulogenesis of ECs. More ECs switched to a tube-like pattern on the soft matrix. In fact, the reduced tension between ECs and the matrix as a result of decreased matrix elasticity easily triggered intracellular signaling cascade toward tubulogenesis, one of the last steps of angiogenesis.15 During the postnatal development of lung, the tissue elasticity modulated by lysyl oxidase (LOX), an ECM crosslinking enzyme, regulates lung development through lipoprotein receptor-related protein 5 (LRP5)/Tie2 signaling by modulating angiogenesis. The expression of LRP5 and Tie2 was up-regulated in lung microvascular ECs cultured on the stiff matrix compared to those on the soft matrix. Inhibiting LOX disrupted lung ECM structures, softened neonatal lung tissue, significantly down-regulated LRP5 and Tie2 expression, and thereby inhibited postnatal lung development. Therefore, appropriate physical properties of lung tissue are necessary for physiological postnatal lung development, and deregulation of this mechanism contributes to postnatal lung developmental disorders, such as bronchopulmonary dysplasia.75 Other studies also reported the LOX was upregulated in early liver injury and resulted in significant matrix stiffness increase.75,98Cell cycle events regulate cell proliferation during tissue development. Klein et al. found that physiological tissue stiffness inhibited cell cycle in mammary ECs and vascular SMCs. FAK-dependent Rac activation, Rac-dependent cyclin D1 gene induction, and cyclin D1-dependent Rb phosphorylation were strongly inhibited at physiological tissue stiffness and rescued upon matrix stiffening. Most mitogenic events proceed normally when matrix stiffness was altered in the range that controls mitogenesis. Matrix remodeling associated with pathogenesis, therefore, positively regulated cell cycle through a highly selective effect on integrin-dependent signaling to FAK, Rac, and cyclin D1.99
Huynh et al. cultured ECs on hydrogels that match the elasticity of young and aging intima. They found endothelial monolayers exhibit increased permeability and disrupted cell–cell junctions on stiffer matrices, a phenomenon similar to the physiological changes of intima with ageing. The enhanced cell contractility associated with increased matrix stiffness destabilized cell–cell junctions and disrupted cell monolayer integrity, leading to increased leukocyte extravasation and eventually the atherosclerotic plaque formation. Mild inhibition of Rho-dependent cell contractility restored monolayer integrity. Hence, ECM stiffening during aging can lead to substantial endothelial monolayer disruption and atherosclerosis pathogenesis. Therapeutics that target the Rho-dependent cellular contractile response to matrix stiffening instead of the stiffness itself, therefore, may prevent atherosclerosis progression more effectively.97
Pathological implications of matrix elasticity
The progression of diseases often accompanies with alternations in the elasticity of local tissue, which may be monitored using techniques such as magnetic resonance imaging or ultrasound elastography.100 For example, the mechanical properties of normal chondrocytes substantially differed from those of chondrocytes derived from osteoarthritis (OA) tissue. The adhesion forces of normal and OA chondrocytes were 7.06 and 2.97 nN, respectively, and the stiffnesses were 960 and 347 mN m−1, respectively.101 The obesity-associated adipogenesis is also a mechanosensitive process, in which the stiffness of adipocytes increases with the accumulation of lipid droplets.102 The altered ECM elasticity, in turn, drives the resident cells toward a more pathological status.Blood vessels stiffen dramatically during atherosclerosis progression (from E = 40 kPa to E = 110 kPa).103 Such changes may disrupt normal cell–cell contact of ECs and increase vascular permeability and further promote atherosclerosis.97,104 The phenotype of vascular SMCs also changes as a result of the matrix elasticity change.44
There are significant mechanical changes in liver with the fibrosis and non-alcohol fatty liver disease. The elastic modulus of liver tissues varies over several orders of magnitude (from 0.3–0.6 kPa in a normal liver to more than 20 kPa in fibrosis and cirrhosis livers).105,106 As a result of the elasticity change in diseased livers, the behaviors of cells within this tissue greatly alter. The hepatocytes spread, proliferate and dedifferentiate on the stiff matrix, while they remain differentiated and growth arrested on soft ones.107,108 The elasticity change also affects the myofibroblastic differentiation of portal fibroblasts, a key mediator of biliary fibrosis, which requires both TGF-β and a stiff matrix for differentiation.90
Matrix elasticity-mediated tumor progression
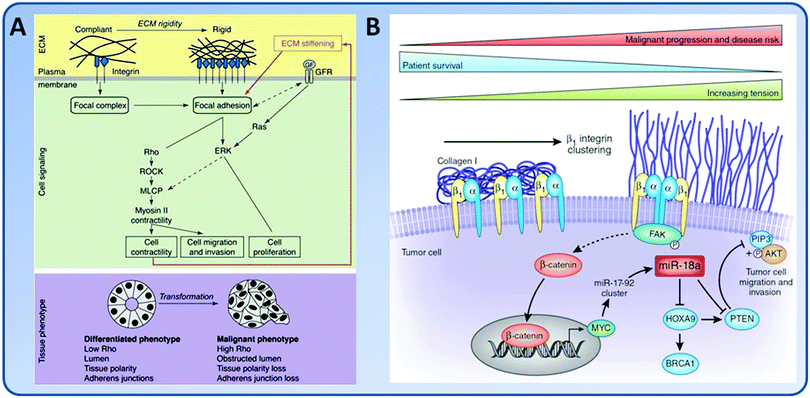
Tumors are stiffer than normal tissues. For example, the healthy mammary gland is very soft (E = ∼200 Pa), while it becomes over one order of magnitude stiffer (E = ∼4 kPa) in the breast cancer.109 Even the stroma around the tumor showed markedly increased stiffness (E = ∼900 Pa).109 Along with the tumor cell growth and invasion, the microenvironment including the biochemical and biomechanical cues also changes. Changes in the matrix elasticity can lead to clustering of integrins and up-regulation of FAs which further increase the contractility and ECM synthesis of tumor cells.98,109,110 In the tumor microenvironment, the increased local elasticity also enhances the branching and permeability of ECs, resulting in a highly disorganized and leaky tumor vascular network.97,111 In addition, alterations in the alternative splicing of proteins are involved in the oncogenic process. Matrix elasticity also regulates alternative splicing through the activation of serine/arginine rich proteins.112Local stiffening of tumor tissue promotes tumor cell proliferation. Glioblastoma multiforme (GBM) is the most common and aggressive form of primary brain tumor in adults. The brain tissue stiffness increases during tumor progression as a result of increased ECM synthesis by GBM cells (from 0.1–1 kPa in a normal brain to 26 kPa in GBM tissue).8,113 The local tissue stiffening promoted GBM proliferation by spatially and biochemically amplifying epidermal growth factor receptor (EGFR) signaling.114 The glioma cells also showed enhanced proliferation along with the increase of matrix stiffness.53 The stiffened ECM promoted FAs, enhanced PI3 kinase (PI3K) activity, and induced the tumor progression.98
The matrix elasticity is an important mediator in the tumor invasion process (Fig. 6). On highly rigid ECMs, GBM tumor cells spread extensively, form prominent stress fibers and mature FAs, and migrate rapidly. However, on ECMs with elasticity comparable to normal brain tissue, tumor cells appear rounded and fail to effectively migrate. Inhibition of nonmuscle myosin II-based contractility blunts this elasticity-sensitivity and rescues cell motility on highly compliant substrates. Therefore, ECM elasticity, by acting through actomyosin contractility, effectively regulates the invasive behaviors of tumor cells (Fig. 6A).53 The invading tumor cells can degrade the underlying ECM through the invadopodia, and then extend large protrusions to invade into the surrounding stroma.115 Increasing the ECM stiffness directly increases the number and activity of invadopodia by FAK and P130Cas signaling pathways.116 Indeed, stiff collagen gels alone were able to induce an invasive phenotype of mammary ECs (MECs) through a FAK/ERK cell signal pathway.117 In the dense region of mammographically dense breast tissue, one of the greatest risk factors in the development of breast carcinoma, the stroma collagen and EC content increased. The increased matrix stiffness promoted proliferation of MECs and the risk of cancer.117 In addition, matrix stiffness can modulate microRNA expression to drive tumor progression (Fig. 6B). For example, increased matrix stiffness caused enhanced expression of miR-18a, as shown by the significantly elevated expression of miR-18a in human breast tumor biopsies. The enhanced expression of miR-18a led to down-regulation of the levels of tumor suppressor phosphatase and tensin homolog (PTEN).118,119
The growth of cancer stem cells (CSCs) also depends on the elasticity of tumor microenvironment. The CSC sub-population of cancer cells resides within a niche with optimum stiffness which relies on the tissue origin of cancer cells. The optimum matrix stiffness for growth and marker expression of CSCs, for example, is 5 kPa for breast MCF7 and MDA231 cells, 25 kPa for colorectal HCT116 cells and gastric AGS cells, and 50 kPa for bone U2OS cells, respectively.120
Implications of matrix elasticity in therapies and tissue regeneration
The impact of matrix elasticity on the diversified array of cell fate and activities has important implications in the therapies based on cells, biomaterial scaffolds, or a combination of them. First, as the lineage commitment of stem cells is largely dependent on the elasticity, introduction of stem cells into diseased tissues which usually have altered biomechanical profiles may lead to unexpected cellular phenotypes unless a mechanically favorable microenvironment is previously created. For example, while the fusion of myoblasts into myotubes occurs independent of substrate elasticity, later myosin/actin striations, which lead to functional sarcomere formation, happened only on the matrix with similar stiffness to the normal muscle (E = ∼12 kPa).10 Another example is chondrocytes, which presented different phenotypes on substrates with different elasticity. The stiffness of chondrocytes significantly decreases in OA patients, accompanied with decreased synthesis yet increased degradation of ECM.101 Therefore, a well controlled microenvironment of adequate stiffness is needed so that therapeutic application of chondrocytes may succeed. This also implies that cell therapies may be most effective at the early stage of disease development when the tissue mechanics do not change much.The scaffold-based tissue engineering strategy is a promising approach to replacing damaged tissues and restoring the biological functions of them. Here, modulating the elasticity of the biomimetic matrix in a way that recapitulates the mechanical heterogeneity of native tissue is critical in achieving complete tissue regeneration. For example, in order to regenerate a complete tooth-like pulpodentin complex, the distinct difference between the soft pulp and rigid dentin should be considered. In a complex scaffold in which the low- and high-stiffness gelatin matrices were integrated, biomineralization took place only in the high-stiffness peripheral area and formed a ring-like structure surrounding the non-mineralized central area. A complete hybrid structure similar to native pulpodentin was successfully regenerated after subcutaneous implantation.43 Recently, we have also prepared a series of biodegradable poly(ether carbonate urethane)urea materials whose elasticity approximated that of native annulus fibrosus (AF) tissue.121 The substrate elasticity-dependent changes in AF-derived stem cells (AFSCs) were similar to the gradual transition in the cells from inner to outer regions of AF tissue.22,122 Such studies, therefore, provide a novel approach to construct tissue replacements that recapitulate native AF tissue, in which the cellular phenotype, biochemical components, and biomechanical characteristics gradually change.123
9. Concluding remarks
The activities and functions of cells are regulated by their integrated response to a variety of microenvironmental cues, including the elasticity of ECM. The interactions between cells and ECM, sensed by the transmembrane adhesion receptors (most notably integrins) and transmitted by the linkage of receptor cytoplasmic domains to the cytoskeleton, are fundamental to the regulation of multiple cellular functions and consequent development of complex tissues. A comprehensive understanding of the responses of cells, especially stem cells, to matrix elasticity as well as its temporal and spatial location is essential for designing biomaterials that approximate the physiological environment to advance tissue engineering endeavors toward clinical applications.Substrate elasticity alone may promote a specific lineage of cells over another. The effect of substrate elasticity may even override that from the biochemical signaling factors. However, the fact that many tissues have similar stiffness implies that the stimulation from the substrate mechanical property alone is insufficient to decide the cell fate.78 The differentiation of cells into a specific lineage usually requires the orchestration of factors from different categories. It should also be noted that different cell behaviors of the same cells may respond to different range of matrix elasticity. Such unsynchronized elasticity dependence reminds that the mechanical properties of the substrate should be specifically tailored to cater for different needs of cell expansion and differentiation in order to achieve efficient tissue engineering applications. In addition, while the majority of studies believe that ECM elasticity plays an important role in regulating cell behaviors, reverse opinions also exist. For instance, Trappmann et al. proposed that it is the pore size of materials, instead of their stiffness, that regulates MSC differentiation.31
Cells reside in a 3D environment. It has been increasingly appreciated that cellular phenotypes are significantly affected by the reduction of dimensionality in which the mechanical and biochemical cues are presented to the cells. The phenotype of stem cells can greatly vary in a 3D environment compared to 2D culture systems. Collective cell behavior differences may be more visible when these physiological mechanical cues are presented to the cells in 3D. In contrast to a 2D situation, the cell fate was not correlated with morphology in a 3D environment. Instead, matrix elasticity regulated integrin binding and reorganization of adhesion ligands on the nanoscale, both of which are contractility dependent and correlated with the osteogenic commitment of MSCs.7 Indeed, the FAs observed in the 3D environment are more mature and consist of more molecules.124 In addition to adhesions, cytoskeletal tension in stem cells differs significantly in 3D where a highly fibrillar ECM transduces unidirectional forces along fibers rather than bidirectionally as in 2D. Ultimately, closer examinations are needed to understand how cells sense mechanical cues in 3D via FA components, and then respond via signaling pathway activation and transcriptional activities to affect lineage commitment.4 Stem cells may also exhibit more tissue-like organizations when grown in 3D microenvironments. It is anticipated that cells may respond to matrix elasticity in a markedly different way in a 3D situation compared to those in 2D. Therefore, future tissue regeneration strategies should create physiologically relevant 3D microenvironments to better mimic the natural niche of cells and recapitulate the intrinsic heterogeneity of native tissue from the cellular, biochemical, and biomechanical aspects. The ability to dynamically regulate the cellular microenvironment as the body does, which is likely a critical requirement for developing differentiated cells from stem cells, may further extend our capability in regenerating tissue substitutes for therapeutic applications.
Acknowledgements
The authors are grateful to the funding support from the National Natural Science Foundation of China (81171479, 81471790, 31530024), Jiangsu Provincial Special Program of Medical Science (BL2012004), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.References
- F. Guilak, D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke and C. S. Chen, Cell Stem Cell, 2009, 5, 17–26 CrossRef CAS PubMed.
- A. Higuchi, Q. Ling, S. S. Kumar, Y. Chang, A. A. Alafaj, M. A. Munusamy, K. Murugan, S. Hsu and A. Umezawa, J. Mater. Chem. B, 2015, 3, 8032–8058 RSC.
- A. Higuchi, Q. D. Ling, Y. Chang, S. T. Hsu and A. Umezawa, Chem. Rev., 2013, 113, 3297–3328 CrossRef CAS PubMed.
- Kshitiz, J. Park, P. Kim, W. Helen, A. J. Engler, A. Levchenko and D. H. Kim, Integr. Biol., 2012, 4, 1008–1018 RSC.
- B. Li, F. Li, K. M. Puskar and J. H. Wang, J. Biomech., 2009, 42, 1622–1627 CrossRef PubMed.
- D. E. Discher, P. Janmey and Y. L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS PubMed.
- N. Huebsch, P. R. Arany, A. S. Mao, D. Shvartsman, O. A. Ali, S. A. Bencherif, J. Rivera-Feliciano and D. J. Mooney, Nat. Mater., 2010, 9, 518–526 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- L. A. Flanagan, Y. E. Ju, B. Marg, M. Osterfield and P. A. Janmey, NeuroReport, 2002, 13, 2411–2415 CrossRef PubMed.
- A. J. Engler, M. A. Griffin, S. Sen, C. G. Bonnemann, H. L. Sweeney and D. E. Discher, J. Cell Biol., 2004, 166, 877–887 CrossRef CAS PubMed.
- A. J. Garcia and C. D. Reyes, J. Dent. Res., 2005, 84, 407–413 CrossRef CAS PubMed.
- H. J. Kong, T. R. Polte, E. Alsberg and D. J. Mooney, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 4300–4305 CrossRef CAS PubMed.
- J. T. Emerman, S. J. Burwen and D. R. Pitelka, Tissue Cell, 1979, 11, 109–119 CrossRef CAS PubMed.
- R. J. Pelham Jr. and Y. Wang, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13661–13665 CrossRef CAS.
- C. F. Deroanne, C. M. Lapiere and B. V. Nusgens, Cardiovasc. Res., 2001, 49, 647–658 CrossRef CAS PubMed.
- L. S. Wang, J. E. Chung, P. P. Chan and M. Kurisawa, Biomaterials, 2010, 31, 1148–1157 CrossRef CAS PubMed.
- J. S. Park, J. S. Chu, A. D. Tsou, R. Diop, Z. Tang, A. Wang and S. Li, Biomaterials, 2011, 32, 3921–3930 CrossRef CAS PubMed.
- Z. Li, Y. Gong, S. Sun, Y. Du, D. Lu, X. Liu and M. Long, Biomaterials, 2013, 34, 7616–7625 CrossRef CAS PubMed.
- Y. Gu, Y. Ji, Y. Zhao, Y. Liu, F. Ding, X. Gu and Y. Yang, Biomaterials, 2012, 33, 6672–6681 CrossRef CAS PubMed.
- N. R. Gandavarapu, D. L. Alge and K. S. Anseth, Biomater. Sci., 2014, 2, 352–361 RSC.
- J. M. Banks, L. C. Mozdzen, B. A. Harley and R. C. Bailey, Biomaterials, 2014, 35, 8951–8959 CrossRef CAS PubMed.
- Q. Guo, C. Liu, J. Li, C. Zhu, H. Yang and B. Li, J. Cell. Mol. Med., 2015, 19, 1582–1592 CrossRef CAS PubMed.
- R. G. Wells, Hepatology, 2008, 47, 1394–1400 CrossRef CAS PubMed.
- X. Xin, A. Borzacchiello, P. A. Netti, L. Ambrosio and L. Nicolais, J. Biomater. Sci., Polym. Ed., 2004, 15, 1223–1236 CrossRef CAS PubMed.
- K. Y. Lee and D. J. Mooney, Chem. Rev., 2001, 101, 1869–1879 CrossRef CAS PubMed.
- W. M. Elbjeirami, E. O. Yonter, B. C. Starcher and J. L. West, J. Biomed. Mater. Res., Part A, 2003, 66, 513–521 CrossRef PubMed.
- R. J. DeVolder, I. W. Kim, E. S. Kim and H. Kong, Tissue Eng., Part A, 2012, 18, 1642–1651 CrossRef CAS PubMed.
- S. Lv, D. M. Dudek, Y. Cao, M. M. Balamurali, J. Gosline and H. Li, Nature, 2010, 465, 69–73 CrossRef CAS PubMed.
- P. Y. Wang, W. B. Tsai and N. H. Voelcker, Acta Biomater., 2012, 8, 519–530 CrossRef CAS PubMed.
- C. M. Cheng, P. R. LeDuc and Y. W. Lin, J. Biomech., 2011, 44, 856–862 CrossRef PubMed.
- B. Trappmann, J. E. Gautrot, J. T. Connelly, D. G. Strange, Y. Li, M. L. Oyen, M. A. Cohen Stuart, H. Boehm, B. Li, V. Vogel, J. P. Spatz, F. M. Watt and W. T. Huck, Nat. Mater., 2012, 11, 642–649 CrossRef CAS PubMed.
- S. J. Bryant, R. J. Bender, K. L. Durand and K. S. Anseth, Biotechnol. Bioeng., 2004, 86, 747–755 CrossRef CAS PubMed.
- K. Wingate, W. Bonani, Y. Tan, S. J. Bryant and W. Tan, Acta Biomater., 2012, 8, 1440–1449 CrossRef CAS PubMed.
- Y. S. Pek, A. C. Wan and J. Y. Ying, Biomaterials, 2010, 31, 385–391 CrossRef CAS PubMed.
- S.-J. Choi, H. N. Kim, W. G. Bae and K.-Y. Suh, J. Mater. Chem., 2011, 21, 14325 RSC.
- M. Levy-Mishali, J. Zoldan and S. Levenberg, Tissue Eng., Part A, 2009, 15, 935–944 CrossRef CAS PubMed.
- M. T. Yang, J. Fu, Y. K. Wang, R. A. Desai and C. S. Chen, Nat. Protoc., 2011, 6, 187–213 CrossRef CAS PubMed.
- T. Yeung, P. C. Georges, L. A. Flanagan, B. Marg, M. Ortiz, M. Funaki, N. Zahir, W. Ming, V. Weaver and P. A. Janmey, Cell Motil. Cytoskeleton, 2005, 60, 24–34 CrossRef PubMed.
- C. M. Lo, H. B. Wang, M. Dembo and Y. L. Wang, Biophys. J., 2000, 79, 144–152 CrossRef CAS PubMed.
- J. Li, D. Han and Y. P. Zhao, Sci. Rep., 2014, 4, 3910 Search PubMed.
- A. Engler, L. Bacakova, C. Newman, A. Hategan, M. Griffin and D. Discher, Biophys. J., 2004, 86, 617–628 CrossRef CAS PubMed.
- T. D. Hansen, J. T. Koepsel, N. N. Le, E. H. Nguyen, S. Zorn, M. Parlato, S. G. Loveland, M. P. Schwartz and W. L. Murphy, Biomater. Sci., 2014, 2, 745–756 RSC.
- T. Qu, J. Jing, Y. Ren, C. Ma, J. Q. Feng, Q. Yu and X. Liu, Acta Biomater., 2015, 16, 60–70 CrossRef CAS PubMed.
- O. V. Sazonova, B. C. Isenberg, J. Herrmann, K. L. Lee, A. Purwada, A. D. Valentine, J. A. Buczek-Thomas, J. Y. Wong and M. A. Nugent, Matrix Biol., 2015, 41, 36–43 CrossRef CAS PubMed.
- C. A. Mullen, T. J. Vaughan, K. L. Billiar and L. M. McNamara, Biophys. J., 2015, 108, 1604–1612 CrossRef CAS PubMed.
- J. Solon, I. Levental, K. Sengupta, P. C. Georges and P. A. Janmey, Biophys. J., 2007, 93, 4453–4461 CrossRef CAS PubMed.
- K. G. Robinson, T. Nie, A. D. Baldwin, E. C. Yang, K. L. Kiick and R. E. Akins, J. Biomed. Mater. Res., Part A, 2012, 100, 1356–1367 CrossRef PubMed.
- S. Bai, W. Zhang, Q. Lu, Q. Ma, D. L. Kaplan and H. Zhu, J. Mater. Chem. B, 2014, 2, 6590–6600 RSC.
- G. Giannone, B. J. Dubin-Thaler, H. G. Dobereiner, N. Kieffer, A. R. Bresnick and M. P. Sheetz, Cell, 2004, 116, 431–443 CrossRef CAS PubMed.
- D. Choquet, D. P. Felsenfeld and M. P. Sheetz, Cell, 1997, 88, 39–48 CrossRef CAS PubMed.
- A. Saez, M. Ghibaudo, A. Buguin, P. Silberzan and B. Ladoux, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8281–8286 CrossRef CAS PubMed.
- H. B. Wang, M. Dembo and Y. L. Wang, Am. J. Physiol.: Cell Physiol., 2000, 279, C1345–C1350 CAS.
- T. A. Ulrich, E. M. de Juan Pardo and S. Kumar, Cancer Res., 2009, 69, 4167–4174 CrossRef CAS PubMed.
- E. Schuh, J. Kramer, J. Rohwedel, H. Notbohm, R. Muller, T. Gutsmann and N. Rotter, Tissue Eng., Part A, 2010, 16, 1281–1290 CrossRef CAS PubMed.
- X. Li, Y. Huang, L. Zheng, H. Liu, X. Niu, J. Huang, F. Zhao and Y. Fan, J. Biomed. Mater. Res., Part A, 2014, 102, 1092–1101 CrossRef PubMed.
- W. L. Murphy, T. C. McDevitt and A. J. Engler, Nat. Mater., 2014, 13, 547–557 CrossRef CAS PubMed.
- S. Kang, J. B. Park, T.-J. Lee, S. Ryu, S. H. Bhang, W.-G. La, M.-K. Noh, B. H. Hong and B.-S. Kim, Carbon, 2015, 83, 162–172 CrossRef CAS.
- Y.-R. V. Shih, K.-F. Tseng, H.-Y. Lai, C.-H. Lin and O. K. Lee, J. Bone Miner. Res., 2011, 26, 730–738 CrossRef CAS PubMed.
- D. A. Young, Y. S. Choi, A. J. Engler and K. L. Christman, Biomaterials, 2013, 34, 8581–8588 CrossRef CAS PubMed.
- K. Saha, A. J. Keung, E. F. Irwin, Y. Li, L. Little, D. V. Schaffer and K. E. Healy, Biophys. J., 2008, 95, 4426–4438 CrossRef CAS PubMed.
- S. Gobaa, S. Hoehnel and M. P. Lutolf, Integr. Biol., 2015, 7, 1135–1142 RSC.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Nat. Rev. Mol. Cell Biol., 2009, 10, 21–33 CrossRef CAS PubMed.
- H. Lv, L. Li, M. Sun, Y. Zhang, L. Chen, Y. Rong and Y. Li, Stem Cell Res. Ther., 2015, 6, 103 CrossRef PubMed.
- R. O. Hynes, Cell, 1992, 69, 11–25 CrossRef CAS PubMed.
- H. Yu, Y. S. Lui, S. Xiong, W. S. Leong, F. Wen, H. Nurkahfianto, S. Rana, D. T. Leong, K. W. Ng and L. P. Tan, Stem Cells Dev., 2013, 22, 136–147 CrossRef CAS PubMed.
- Y. R. Shih, K. F. Tseng, H. Y. Lai, C. H. Lin and O. K. Lee, J. Bone Miner. Res., 2011, 26, 730–738 CrossRef CAS PubMed.
- J. Du, X. Chen, X. Liang, G. Zhang, J. Xu, L. He, Q. Zhan, X. Q. Feng, S. Chien and C. Yang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9466–9471 CrossRef CAS PubMed.
- H. Hirata, H. Tatsumi and M. Sokabe, J. Cell Sci., 2008, 121, 2795–2804 CrossRef CAS PubMed.
- J. D. Pajerowski, K. N. Dahl, F. L. Zhong, P. J. Sammak and D. E. Discher, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 15619–15624 CrossRef CAS PubMed.
- J. Swift, I. L. Ivanovska, A. Buxboim, T. Harada, P. C. Dingal, J. Pinter, J. D. Pajerowski, K. R. Spinler, J. W. Shin, M. Tewari, F. Rehfeldt, D. W. Speicher and D. E. Discher, Science, 2013, 341, 1240104 CrossRef PubMed.
- R. McBeath, D. M. Pirone, C. M. Nelson, K. Bhadriraju and C. S. Chen, Dev. Cell, 2004, 6, 483–495 CrossRef CAS PubMed.
- Y. R. Shih, K. F. Tseng, H. Y. Lai, C. H. Lin and O. K. Lee, J. Bone Miner. Res., 2011, 26, 730–738 CrossRef CAS PubMed.
- O. F. Zouani, J. Kalisky, E. Ibarboure and M. C. Durrieu, Biomaterials, 2013, 34, 2157–2166 CrossRef CAS PubMed.
- T. J. Kim, J. Sun, S. Lu, J. Zhang and Y. Wang, Biomaterials, 2014, 35, 8348–8356 CrossRef CAS PubMed.
- T. Mammoto, E. Jiang, A. Jiang and A. Mammoto, Am. J. Respir. Cell Mol. Biol., 2013, 49, 1009–1018 CrossRef CAS PubMed.
- L. Fan, A. Sebe, Z. Peterfi, A. Masszi, A. C. Thirone, O. D. Rotstein, H. Nakano, C. A. McCulloch, K. Szaszi, I. Mucsi and A. Kapus, Mol. Biol. Cell, 2007, 18, 1083–1097 CrossRef CAS PubMed.
- W. Liedtke, D. M. Tobin, C. I. Bargmann and J. M. Friedman, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(suppl 2), 14531–14536 CrossRef CAS PubMed.
- F. M. Watt and B. L. Hogan, Science, 2000, 287, 1427–1430 CrossRef CAS PubMed.
- X. Hu, S. H. Park, E. S. Gil, X. X. Xia, A. S. Weiss and D. L. Kaplan, Biomaterials, 2011, 32, 8979–8989 CrossRef CAS PubMed.
- J. D. Kubicek, S. Brelsford, P. Ahluwalia and P. R. Leduc, Langmuir, 2004, 20, 11552–11556 CrossRef CAS PubMed.
- L. Gao, R. McBeath and C. S. Chen, Stem Cells, 2010, 28, 564–572 CAS.
- D. Zhang and K. A. Kilian, Biomaterials, 2013, 34, 3962–3969 CrossRef CAS PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS PubMed.
- K. A. Kilian, B. Bugarija, B. T. Lahn and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4872–4877 CrossRef CAS PubMed.
- J. Lee, A. A. Abdeen, T. H. Huang and K. A. Kilian, J. Mech. Behav. Biomed. Mater., 2014, 38, 209–218 CrossRef CAS PubMed.
- E. K. Yim, E. M. Darling, K. Kulangara, F. Guilak and K. W. Leong, Biomaterials, 2010, 31, 1299–1306 CrossRef CAS PubMed.
- C. Chen, J. Xie, L. Deng and L. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 16106–16116 CAS.
- J. L. Allen, M. E. Cooke and T. Alliston, Mol. Biol. Cell, 2012, 23, 3731–3742 CrossRef CAS PubMed.
- J. J. Tomasek, G. Gabbiani, B. Hinz, C. Chaponnier and R. A. Brown, Nat. Rev. Mol. Cell Biol., 2002, 3, 349–363 CrossRef CAS PubMed.
- Z. Li, J. A. Dranoff, E. P. Chan, M. Uemura, J. Sevigny and R. G. Wells, Hepatology, 2007, 46, 1246–1256 CrossRef CAS PubMed.
- S. Tan, J. Y. Fang, Z. Yang, M. E. Nimni and B. Han, Biomaterials, 2014, 35, 5294–5306 CrossRef CAS PubMed.
- L. G. Griffith and M. A. Swartz, Nat. Rev. Mol. Cell Biol., 2006, 7, 211–224 CrossRef CAS PubMed.
- P. Jiang, Z. Mao and C. Gao, Acta Biomater., 2015, 19, 76–84 CrossRef CAS PubMed.
- C. M. Kraning-Rush and C. A. Reinhart-King, Cell Adh. Migr., 2012, 6, 274–279 CrossRef PubMed.
- H. Parameswaran, A. Majumdar and B. Suki, PLoS Comput. Biol., 2011, 7, e1001125 CAS.
- G. E. Pierard, T. Hermanns-Le and C. Pierard-Franchimont, Expert Opin. Med. Diagn., 2013, 7, 119–125 CrossRef CAS PubMed.
- J. Huynh, N. Nishimura, K. Rana, J. M. Peloquin, J. P. Califano, C. R. Montague, M. R. King, C. B. Schaffer and C. A. Reinhart-King, Sci. Transl. Med., 2011, 3, 112–122 Search PubMed.
- K. R. Levental, H. Yu, L. Kass, J. N. Lakins, M. Egeblad, J. T. Erler, S. F. Fong, K. Csiszar, A. Giaccia, W. Weninger, M. Yamauchi, D. L. Gasser and V. M. Weaver, Cell, 2009, 139, 891–906 CrossRef CAS PubMed.
- E. A. Klein, L. Yin, D. Kothapalli, P. Castagnino, F. J. Byfield, T. Xu, I. Levental, E. Hawthorne, P. A. Janmey and R. K. Assoian, Curr. Biol., 2009, 19, 1511–1518 CrossRef CAS PubMed.
- J. Zhou, W. Zhan, Y. Dong, Z. Yang and C. Zhou, Eur. Radiol., 2014, 24, 1659–1667 CrossRef PubMed.
- C. H. Hsieh, Y. H. Lin, S. Lin, J. J. Tsai-Wu, C. H. Herbert Wu and C. C. Jiang, Osteoarthritis Cartilage, 2008, 16, 480–488 CrossRef PubMed.
- N. Shoham, P. Girshovitz, R. Katzengold, N. T. Shaked, D. Benayahu and A. Gefen, Biophys. J., 2014, 106, 1421–1431 CrossRef CAS PubMed.
- Y. Guo, S. Mahony and D. K. Gifford, PLoS Comput. Biol., 2012, 8, e1002638 CAS.
- J. P. Califano and C. A. Reinhart-King, Cell. Mol. Bioeng., 2008, 1, 122–132 CrossRef.
- M. Yin, J. A. Talwalkar, K. J. Glaser, A. Manduca, R. C. Grimm, P. J. Rossman, J. L. Fidler and R. L. Ehman, Clin. Gastroenterol. Hepatol., 2007, 5, 1207–1213 CrossRef PubMed.
- A. M. Gressner, Eur. J. Clin. Chem. Clin. Biochem., 1994, 32, 225–237 CAS.
- L. K. Hansen, J. Wilhelm and J. T. Fassett, in Current Topics in Developmental Biology, ed. P. S. Gerald, Academic Press, 2005, vol. 72, pp. 205–236 Search PubMed.
- J. Fassett, Mol. Biol. Cell, 2005, 17, 345–356 CrossRef PubMed.
- M. J. Paszek, N. Zahir, K. R. Johnson, J. N. Lakins, G. I. Rozenberg, A. Gefen, C. A. Reinhart-King, S. S. Margulies, M. Dembo, D. Boettiger, D. A. Hammer and V. M. Weaver, Cancer Cell, 2005, 8, 241–254 CrossRef CAS PubMed.
- C. M. Kraning-Rush, J. P. Califano and C. A. Reinhart-King, PLoS One, 2012, 7, e32572 CAS.
- D. B. Pink, W. Schulte, M. H. Parseghian, A. Zijlstra and J. D. Lewis, PLoS One, 2012, 7, e33760 CAS.
- F. Bordeleau, J. P. Califano, Y. L. Negron Abril, B. N. Mason, D. J. LaValley, S. J. Shin, R. S. Weiss and C. A. Reinhart-King, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8314–8319 CrossRef CAS PubMed.
- W. J. Polacheck, I. K. Zervantonakis and R. D. Kamm, Cell. Mol. Life Sci., 2013, 70, 1335–1356 CrossRef CAS PubMed.
- V. Umesh, A. D. Rape, T. A. Ulrich and S. Kumar, PLoS One, 2014, 9, e101771 Search PubMed.
- S. P. Carey, T. M. D'Alfonso, S. J. Shin and C. A. Reinhart-King, Crit. Rev. Oncol. Hematol., 2012, 83, 170–183 CrossRef PubMed.
- N. R. Alexander, K. M. Branch, A. Parekh, E. S. Clark, I. C. Iwueke, S. A. Guelcher and A. M. Weaver, Curr. Biol., 2008, 18, 1295–1299 CrossRef CAS PubMed.
- P. P. Provenzano, D. R. Inman, K. W. Eliceiri and P. J. Keely, Oncogene, 2009, 28, 4326–4343 CrossRef CAS PubMed.
- J. K. Mouw, Y. Yui, L. Damiano, R. O. Bainer, J. N. Lakins, I. Acerbi, G. Ou, A. C. Wijekoon, K. R. Levental, P. M. Gilbert, E. S. Hwang, Y. Y. Chen and V. M. Weaver, Nat. Med., 2014, 20, 360–367 CrossRef CAS PubMed.
- V. Seewaldt, Nat. Med., 2014, 20, 332–333 CrossRef CAS PubMed.
- E. Jabbari, S. K. Sarvestani, L. Daneshian and S. Moeinzadeh, PLoS One, 2015, 10, e0132377 Search PubMed.
- C. Zhu, J. Li, C. Liu, P. Zhou, H. Yang and B. Li, Acta Biomater., 2015 DOI:10.1016/j.actbio.2015.1009.1039.
- C. Liu, Q. Guo, J. Li, S. Wang, Y. Wang, B. Li and H. Yang, PLoS One, 2014, 9, e108239 Search PubMed.
- J. Li, C. Liu, Q. Guo, H. Yang and B. Li, PLoS One, 2014, 9, e91799 Search PubMed.
- E. Cukierman, R. Pankov, D. R. Stevens and K. M. Yamada, Science, 2001, 294, 1708–1712 CrossRef CAS PubMed.
- G. J. Her, H. C. Wu, M. H. Chen, M. Y. Chen, S. C. Chang and T. W. Wang, Acta Biomater., 2013, 9, 5170–5180 CrossRef CAS PubMed.
- B. N. Mason, A. Starchenko, R. M. Williams, L. J. Bonassar and C. A. Reinhart-King, Acta Biomater., 2013, 9, 4635–4644 CrossRef CAS PubMed.
- L. S. Wang, J. Boulaire, P. P. Chan, J. E. Chung and M. Kurisawa, Biomaterials, 2010, 31, 8608–8616 CrossRef CAS PubMed.
- S. Murikipudi, H. Methe and E. R. Edelman, Biomaterials, 2013, 34, 677–684 CrossRef CAS PubMed.
- A. Banerjee, M. Arha, S. Choudhary, R. S. Ashton, S. R. Bhatia, D. V. Schaffer and R. S. Kane, Biomaterials, 2009, 30, 4695–4699 CrossRef CAS PubMed.
- R. S. Stowers, S. C. Allen and L. J. Suggs, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1953–1958 CrossRef CAS PubMed.
- B. P. Partlow, C. W. Hanna, J. Rnjak-Kovacina, J. E. Moreau, M. B. Applegate, K. A. Burke, B. Marelli, A. N. Mitropoulos, F. G. Omenetto and D. L. Kaplan, Adv. Funct. Mater., 2014, 24, 4615–4624 CrossRef CAS PubMed.
- A. P. Balgude, X. Yu, A. Szymanski and R. V. Bellamkonda, Biomaterials, 2001, 22, 1077–1084 CrossRef CAS PubMed.
- R. A. Marklein and J. A. Burdick, Soft Matter, 2010, 6, 136–143 RSC.
- K. Ziv, H. Nuhn, Y. Ben-Haim, L. S. Sasportas, P. J. Kempen, T. P. Niedringhaus, M. Hrynyk, R. Sinclair, A. E. Barron and S. S. Gambhir, Biomaterials, 2014, 35, 3736–3743 CrossRef CAS PubMed.
- K. E. Smith, S. L. Hyzy, M. Sunwoo, K. A. Gall, Z. Schwartz and B. D. Boyan, Biomaterials, 2010, 31, 6131–6141 CrossRef CAS PubMed.
- J. Cui, K. Kratz, B. Hiebl, F. Jung and A. Lendlein, Polym. Adv. Technol., 2011, 22, 126–132 CrossRef CAS.
- K. Ren, T. Crouzier, C. Roy and C. Picart, Adv. Funct. Mater., 2008, 18, 1378–1389 CrossRef CAS PubMed.
- F. Wang, Z. Li, J. L. Lannutti, W. R. Wagner and J. Guan, Acta Biomater., 2009, 5, 2901–2912 CrossRef CAS PubMed.
- T. H. Kim, D. B. An, S. H. Oh, M. K. Kang, H. H. Song and J. H. Lee, Biomaterials, 2015, 40, 51–60 CrossRef CAS PubMed.
- S. Mahadevaiah, K. G. Robinson, P. M. Kharkar, K. L. Kiick and R. E. Akins, Biomaterials, 2015, 62, 24–34 CrossRef CAS PubMed.
- S. Wang, D. H. R. Kempen, G. C. W. de Ruiter, L. Cai, R. J. Spinner, A. J. Windebank, M. J. Yaszemski and L. Lu, Adv. Funct. Mater., 2015, 25, 2715–2724 CrossRef CAS.
Footnote |
| † Soochow University (South Campus), 708 Renmin Rd, Rm 308 Bldg 1, Suzhou, Jiangsu 215007, China. E-mail: E-mail: binli@suda.edu.cn; +86-512-6778-1163; +86-512-6778-1163 |
| This journal is © The Royal Society of Chemistry 2016 |