 Open Access Article
Open Access ArticleBiomacromolecule-tagged nanoscale constructs for crossing the blood–brain barrier
Tyler L.
Odom
 ,
Hayden D.
LeBroc
,
Hayden D.
LeBroc
 and
Cassandra E.
Callmann
and
Cassandra E.
Callmann
 *
*
Department of Chemistry, The University of Texas at Austin, 105 E. 24th St, Austin, TX 78712, USA. E-mail: ccallmann@utexas.edu
First published on 20th January 2024
Abstract
Access to the brain is restricted by the low permeability of the blood–brain barrier (BBB), greatly hampering modern drug delivery efforts. A promising approach to overcome this boundary is to utilize biomacromolecules (peptides, nucleic acids, carbohydrates) as targeting ligands on nanoscale delivery vehicles to shuttle cargo across the BBB. In this mini-review, we highlight the most recent approaches for crossing the BBB using synthetic nanoscale constructs decorated with members of these general classes of biomacromolecules to safely and selectively deliver therapeutic materials to the brain.
Introduction
The blood–brain barrier (BBB) is a notoriously impermeable boundary that separates the brain from systemic circulation, making drug delivery to this organ a substantial challenge.1–3 The BBB is a complex network of capillaries and endothelial cells that are connected by a series of tight junctions.4,5 The primary role of the BBB is to maintain homeostasis of the central nervous system (CNS) and prevent entry of neurotoxic substances.6 While this is a critical physiological function, it is this same exclusivity that prevents therapeutics from efficiently entering the brain.7 Many approaches to circumvent this barrier rely on invasive and/or destructive exogenous techniques (e.g., intracranial injection, ultrasound, and osmotic shock).8–10 As an alternative to these aggressive techniques, substantial research has focused on less invasive, endogenous delivery mechanisms.11,12 A particularly promising strategy that has gained momentum in recent years is to leverage naturally-occurring active transport mechanisms on the BBB to shuttle therapeutic cargo.11,13,14 Targeting ligands derived from the three classes of natural biomacromolecules (peptides, nucleic acids, and carbohydrates) are particularly promising tools in this regard, as they are inherently biocompatible, degradable, and readily conjugated to synthetic materials (Fig. 1).15–17 In general, a ligand is first identified for a corresponding receptor that is specific to, or overexpressed on, the BBB. These ligands are then presented on the surface of nanoscale constructs, which are subsequently shuttled across the BBB via transcytosis mechanisms.18–20 This has been demonstrated with a variety of nanoscale systems, including exosomes, viral vectors, nanoparticulate systems, and drug-ligand conjugates.21–25 In this mini-review, we highlight the most recent advances in the design and application of synthetic nanoscale constructs that utilize biomacromolecule-based ligands to target and cross the BBB. We define these targeting ligands as being derived from one of the main classes of biomacromolecule (peptides, nucleic acids, or carbohydrates) and as having a molecular weight of less than 15 kDa. We also provide a brief summary and perspective on the future directions of the field.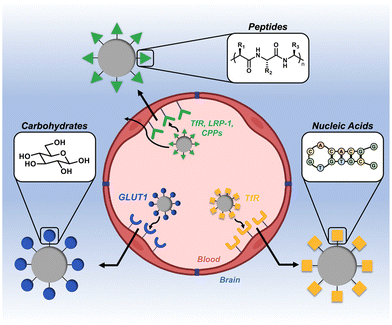 | ||
| Fig. 1 Overview of biomacromolecular targeting strategies for penetrating the blood–brain barrier (BBB). | ||
Peptides
Peptides are the most established biomacromolecule used to target the brain (Fig. 2a).7,26 Peptide-based brain delivery systems were first reported in the mid 1980s.27,28 Early work focused on the basic discovery of peptide sequences that could efficiently and selectively penetrate the BBB for use in simple constructs (e.g., peptide-drug conjugates).28,29 However, these systems suffer from rapid enzymatic degradation, short circulation lifetime, and off-target delivery.30,31 To address these issues, modern research has focused on conjugating targeting peptides to nanoscale materials, including liposomes, polymeric nanoparticles, and inorganic nanoparticles.32–34 These materials extend circulation lifetimes and can encapsulate therapeutic cargo. To date, a multitude of peptide-based BBB delivery systems have been developed using a wide range of platforms. The historically utilized peptide-drug conjugates will not be discussed herein (for an expansive list of peptide sequences used for brain delivery, there are several reviews we recommend30,31); instead, we focus on the newest nanoscale approaches.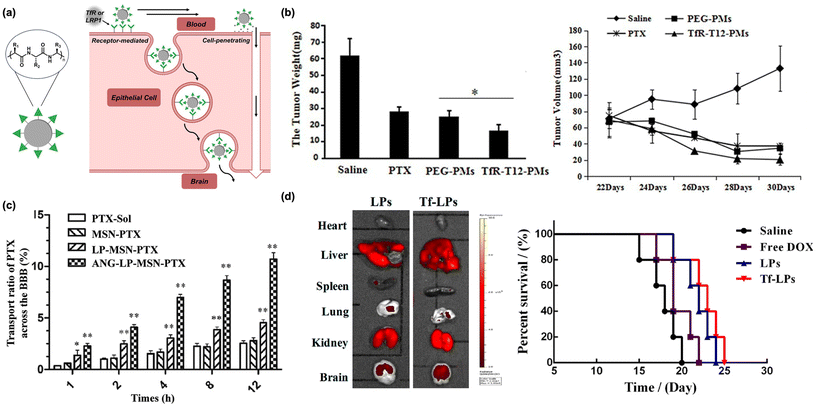 | ||
| Fig. 2 Peptides as biomacroolecular targeting ligands. (a) Illustration of peptide-based nanomaterials crossing the BBB through receptor- or absorptive-mediated transcytosis. (b) A decrease in glioblastoma tumor weight (left) and volume (right) is observed in animals administered transferrin-tagged micelles containing paclitaxel. Originally published by and used with permission from Dove Medical Press Ltd, International Journal of Nanomedicine 2020 15 6673–6687 (ref. 32). (c) Transport efficacy of silica-based nanoparticles across the BBB is improved with Angiopep-2. Reproduced with permission from ref. 46. (d) Biodistribution (left) and percent survival (right) of animals administered DOX-loaded nanoparticles tagged with TfR and R8. Reproduced with permission from ref. 50. | ||
The peptides most often utilized for brain delivery cross the BBB through receptor-mediated transcytosis.35 Most proteins are impermeable to the BBB, but there are several that take part in receptor-mediated transport across this barrier. The most notable of these is transferrin, which is overexpressed on the BBB and is responsible for delivering iron into the brain through receptor-mediated transcytosis with the transferrin receptor (TfR).36 This receptor is also overexpressed in metastatic brain cancers, including glioma,37 due to the high metabolic activity of these tissues, increasing the attractiveness of this target receptor. Both the native transferrin protein and TfR antibodies have been shown to be promising routes for selective brain delivery but are inaccessible through chemical synthesis,38 restricting their application scope. As such, researchers have focused on identifying short, synthetically accessible peptide fragments which have been historically challenging to systematically screen. However, advances in phage-displayed peptide libraries, wherein libraries of millions of unique peptides are generated via combinatorial expression on macrophage surfaces,39 have enabled high throughput screening of peptide targeting ligands. Indeed, this technique has allowed for the discovery of several synthetically accessible TfR-binding peptides.30 Conjugation of TfR-targeting peptides to polymeric micelles loaded with anti-cancer cargo, including paclitaxel and siRNA, has been shown to induce cancer cell death and reduce overall tumor size in animal models of glioblastoma (Fig. 2b).32,40 Beyond cancer therapy, TfR-targeting peptides on the surface of polymer and lipid-based nanocarriers have been developed to treat tuberculosis, microtubule accumulation in Alzheimer's disease models, and neuroinflammation in animal models.41–43
While TfR is the most well-represented receptor for brain targeting in modern literature, multiple other peptide classes have been utilized for brain delivery of nanoscale materials. Apolipoproteins are a class of proteins responsible for trafficking lipids and cholesterol derivatives into the brain and have been identified as promising BBB targets.44 Angiopep-2, a 19-residue peptide, is a synthetic peptide that is designed to interact with low-density lipoprotein receptor-related protein 1 (LRP-1) on the BBB.45 This sequence was found through derivatization of native proteins and currently represents the most successful bio-inspired brain delivery peptide.26 Nanoparticles decorated with Angiopep-2 are capable of crossing the BBB and delivering chemotherapeutics to glioblastoma tumors (Fig. 2c).46 As with the transferrin peptide, the utility of Angiopep-2 is broad in scope. Nanomaterials decorated with Angiopep-2 have also been utilized for treatment of ischemic stroke and epilepsy, among other ailments.45,47,48
As an alternative to peptides that permeate the BBB through receptor-mediated transcytosis, cell-penetrating peptides (CPPs) penetrate the BBB through adsorptive-mediated transcytosis.49 CPPs are typically highly positively charged and interact strongly with negatively charged cell surfaces, inducing transcytosis even at the highly restrictive BBB.49 While effective, CPPs are unable to selectively target the brain and suffer from nonspecific distribution when used in delivery systems, limiting their applicability.30 However, one of the major benefits to using a nanoscale platform is the ability to conjugate multiple functional moieties to the nanoparticle surface. Recently, it has been shown that doxorubicin-loaded liposomes tagged with both transferrin and the CPP octa-arginine (R8) improved brain delivery efficiency and exhibited a synergistic transport effect, efficiently treating glioblastoma in mice (Fig. 2d).50 Similar results were seen with the dual conjugation of Angiopep-2 and the TAT peptide (a CPP) to the surface of small extracellular vesicles. In mouse models of glioblastoma, a net improvement in transport was observed.51
Nucleic acids
While peptide-based nanoassemblies are a well-established method of penetrating the BBB, the utilization of polynucleotides (both DNA and RNA) as a targeting motif is a recently growing area of research (Fig. 3a).52,53 This interest is due primarily to advances in DNA and RNA aptamer libraries,54 an expansive screening technique for novel nucleotide sequences with high binding affinities for specific targets. These libraries enable the exploration of previously undiscovered chemical space and can facilitate the discovery of complex three-dimensional structures that selectively bind proteins of interest. This methodology is conceptually analogous to the aforementioned phage-displayed peptide libraries and has been proven to be a similarly powerful tool in permeating the highly selective BBB.52,55,56 Here, we summarize the research that utilizes DNA aptamers as targeting ligands on nanoscale delivery systems for brain delivery.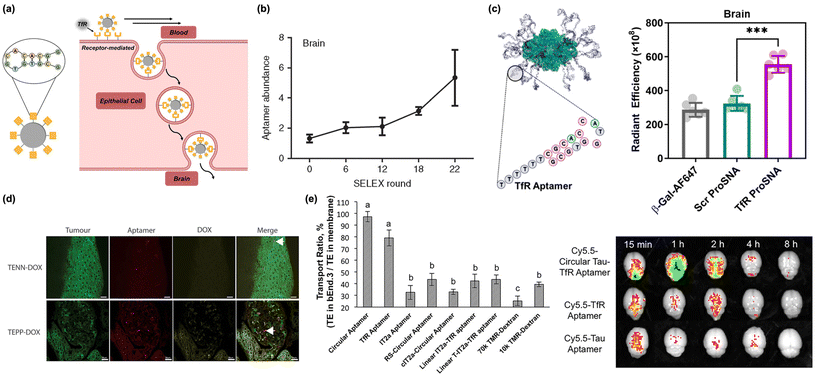 | ||
| Fig. 3 Nucleic acids (DNA/RNA) as biomacromolecular targeting ligands. (a) Illustration of peptide-based nanomaterials crossing the BBB through receptor-mediated transcytosis. (b) Assessment of aptamer enrichment in the brain following in vivo SELEX of RNA library to identify brain-penetrating sequences. Reproduced with permission from ref. 62. (c) Proteins conjugated with TfR aptamers (Tfr-ProSNA) show preferential uptake in the brain relative to scrambled controls. Reproduced with permission from ref. 17. (d) Nanoparticles conjugated with TfR and EpCAM-binding moieties colocalize in tumor tissue in the brain. Arrow indicates colocalization of aptamer with tumor cell. Reproduced with permission from ref. 55. (e) Conjugating the TfR aptamer to a Tau aptamer increased the transcytosis across in vitro endothelial monolayers (left) and increased brain accumulation in vivo (right). Reproduced with permission from ref. 56. | ||
Screening of DNA/RNA aptamer libraries uses Systematic Evolution of Ligands by Exponential Enrichment (SELEX), where DNA or RNA sequences with high binding affinities are isolated and amplified using polymerase chain reaction (PCR).57 This process is repeated over several iterations, serving to separate weakly binding sequences from ones that demonstrate a high binding affinity and specificity. SELEX allowed for the initial discovery of several aptamers that target TfR.36,58 Initial candidate sequences for TfR aptamers were identified in 2008 and were structurally improved and optimized, yielding several highly efficient TfR aptamers, including TfRA4 and TfRA15T.59–61 This approach was further generalized in 2013. Researchers performed in vivo SELEX, where an aptamer library was systemically injected in mice. All sequences capable of penetrating the BBB were isolated from brain tissue, then amplified using PCR and reinjected into mice (Fig. 3b).62 Through 22 rounds of selection, three top sequences emerged that were able to effectively bind endogenous cortical proteins and permeate the BBB.62 Recent progress in lab-on-a-chip technology has accelerated this identification process even further, with researchers using chip-based BBB models to quickly screen for useful aptamer sequences while avoiding the lengthy and laborious process of in vivo studies.63 This work, alongside others, has built a small library of accessible BBB penetrating aptamers, several of which have been utilized in nanoscale delivery systems.
An inherent problem with DNA and RNA aptamers is that they undergo rapid enzymatic degradation and have short circulation times due to rapid renal clearance. To address this issue, multiple copies of the aptamer can be conjugated to a nanoscale platform. These higher molecular weight materials serve to greatly extend circulation time, as well as mitigate the rate of degradation. In the case of nucleic acid-based nanomaterials, the high density of negative charges in the exterior corona creates a robust shell of water and positively-charged counterions, inhibiting enzymatic access.64
Conjugation of a transferrin aptamer to a liposomal carrier has shown significant permeability when transporting obidoxime cargo across the BBB to neutralize toxic organophosphorus compounds.65 However, this approach is not limited to synthetic nanoparticles. It was recently shown that the transport of a typically BBB-impermeable protein (β-galactosidase) could be achieved through the conjugation the protein's cysteine residues with a TfR DNA aptamer (Fig. 3c).17 This study supports the possibility of delivering entire therapeutic proteins to the central nervous system (CNS).
As mentioned above, nanoscale delivery systems are not necessarily limited to a single functional ligand. Many nanosystems can incorporate highly specific aptamers alongside other unique binding moieties in tandem. This idea has been applied to brain cancer, where the chemotherapeutic drug doxorubicin (DOX) was conjugated to a bifunctional aptamer containing both TfR and EpCAM binding moieties (Fig. 3d).55 Researchers showed that the multipurpose nanocomplexes can successfully cross the BBB in vivo and selectively target EpCAM-positive cancerous tissue. Thus, they were able to reinforce the success and therapeutic potential of multifunctional aptamer systems. This has also been recently demonstrated with a dimerized TfR-Tau aptamer as a bifunctional therapeutic material (Fig. 3e).56 The TfR aptamer facilitated transport across the BBB, and the Tau aptamer inhibited the Tau 441 protein which has been implicated in the accumulation of microtubules associated with neurodegenerative diseases, such as Alzheimer's.56
Carbohydrates
Similar to nucleic acids, carbohydrates have classically been underutilized as BBB targeting motifs relative to their peptide counterparts. This is because oligosaccharides are very challenging and laborious to synthesize, polysaccharide libraries have higher complexity than their peptide and nucleic acid counterparts, and, until recently, there has not been a clear BBB receptor to target.66,67 Together, this has historically meant that even if an efficient BBB receptor was discovered whose ligand is a complex polysaccharide, synthetic access may be impossible or only accessible by a few highly specialized research groups. While these challenges have hindered progress in the field, researchers have begun to use the ubiquitous metabolite glucose as a BBB transport ligand. This approach runs counterintuitively to the previously discussed peptide and oligonucleotide delivery systems that rely on high affinity interactions with proteins that are expressed almost exclusively on the BBB.61,62 In contrast, glucose transporters are present on virtually every cell in the body, and typically have a dissociation constant in the mM range.68,69 The core observation that spurred interest in glucose as a targeting ligand is that even though the brain is only 2% of total body mass, it is responsible for 20% of physiological glucose utilization.70 This lends a significant statistical edge to glucose-containing delivery platforms and allows for BBB permeation.Glucose enters the brain through glucose transporter 1 (GLUT1) on the BBB (Fig. 4a).69 Binding affinity to GLUT1 is increased by the presence of multiple copies of individual glucose molecules in close proximity, a well-established phenomenon termed the multivalent effect.71,72 By utilizing a nanoscale architecture, it is possible to recapitulate this effect, where glucose molecules are artificially clustered by supramolecular assembly, improving multivalency.67,72,73 Herein, we highlight the major recent advances in this area of BBB delivery.
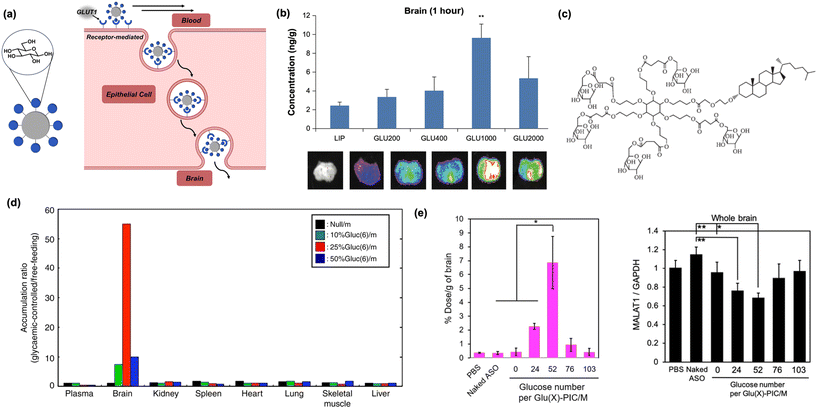 | ||
| Fig. 4 Carbohydrates as biomacromolecular targeting ligands. (a) Illustration of peptide-based nanomaterials crossing the BBB through receptor-mediated transcytosis. (b) Increasing the distance of glucose units from the nanoparticle core using PEG increases brain accumulation in vivo. Reproduced from ref. 74. (c) Increased glucose multivalency, as shown in the depicted dendrimer, increases brain uptake in mice models. Reproduced from ref. 72. (d) Glucose-tagged micelles administered to mice under glycaemic control selectively internalize into mice brain. Reproduced from ref. 67. (e) Brain internalization is optimal when glucose residues are dispersed on micelle surfaces (left). MALAT1 expression is decreased in the brain of upon delivery of complexed antisense RNA (right). Reproduced from ref. 75. | ||
In an early report, it was shown conjugating glucose to cholesterol via a polyethylene glycol (PEG) linker, then anchoring it into a lipid micelle, afforded a construct capable of penetrating the BBB.74 In addition, it was found longer chain lengths of PEG led to higher rates of nanoparticle internalization, suggesting that increased conformational flexibility of glucose and distance from the liposomal surface can improve binding and BBB permeation (Fig. 4b). In a subsequent study, the role of multivalency was more deeply investigated by modifying each cholesterol-PEG unit with one to five individual copies of glucose in a pendant fashion. The surface density of glucose, previously obtained through supramolecular clustering of individual chains, becomes supplemented by the covalent attachment of multiple glucose units to an adjacent center, serving to greatly increase overall density.72 As previously hypothesized, GLUT1 binding is positively correlated to increased multivalent presentation, with the liposomes containing ligands of five glucose molecules accumulating in the brain most efficiently (Fig. 4c).72 However, while this work provided a convenient route for BBB delivery using a carbohydrate ligand, significant off-target delivery was still observed.
To improve the selectivity of glucose targeting, it was recently reported that systemically injected zwitterionic copolymer micelles tagged with glucose into mice under glycemic control (e.g., a state of starvation) significantly increased brain targeting efficiency. The vulnerable metabolic state drastically increased the fraction of nanoparticles entering the brain compared to previous work, demonstrating for the first time that selectivity can be achieved while using a relatively promiscuous carbohydrate ligand such as a glucose (Fig. 4d).67 With this newfound methodology and flexible nanoparticle template identified, the potential of this system has been increasingly realized in recent years. A modified version of the original micelle model was used to perform gene knockdown in the brain.75 In this system, positively-charged polymers tagged with glucose were complexed with antisense oligonucleotides to form polyion complex micelles (PICs), which significantly reduced the expression of lung adenocarcinoma transcript 1, a brain-resident protein associated with cancer metastasis. Furthermore, to control micellar dissociation and nucleotide release, PICs were internally crosslinked with disulfide bonds which are degraded upon exposure to the reducing conditions of the brain (Fig. 4e).75 The same concept has also been utilized to deliver antibodies and antibody fragments to the brain to inhibit proteins involved in cancerous metastasis and Parkinson's disease, respectively, furthering the range of applications.76,77
Conclusions and outlook
Peptides continue to be the dominating biomacromolecular targeting ligand used in modern nanoscale BBB penetration and brain delivery. However, there has been a recent entry of two new players in this space: nucleic acids and carbohydrates as biomacromolecular targeting motifs (Table 1). From the basic discovery of BBB-relevant aptamers using SELEX to the advanced multifunctional systems discussed, the field of DNA/RNA-based systems as BBB delivery agents has gained momentum within the last decade.52,62 With the first examples of advanced applications only being published in the last few years, we anticipate that this field has potential for rapid expansion and will pave the way for entirely new therapeutic approaches, both as fundamental research as well as downstream clinical applications. Likewise, we have seen the emergence of a new subfield in the BBB transport space in recent years based on carbohydrates, with glucose-tagged materials being demonstrated efficient at brain permeation and capable of broad applications. However, this potential has not yet been fully realized, with only a few publications on the subject and only a small number of groups playing a role in research progression. As knowledge of carbohydrate-binding proteins and synthetic access to complex carbohydrate sequences increases,78 it is highly probable the field will experience continued growth and be expanded to include new targeting motifs available in the broad chemical space of sugars.| Entry | Advantages | Limitations | Ref. |
|---|---|---|---|
| Peptides | Synthesis well-established | Susceptible to enzymatic cleavage | 32, 40–48, 50, 51 |
| Large library screening available (phage display) | Receptor saturation possible | ||
| Moderate to high binding affinity to receptors | Potentially immunogenic | ||
| Cell penetrating variants can improves efficiency | |||
| Nucleic acids | Synthesis well-established | Susceptible to enzymatic cleavage | 17, 55, 56, 62, 65 |
| Large library screening available (SELEX) | High negative charge density | ||
| High binding affinity to receptors | |||
| Carbohydrates | High biocompatibility | Synthesis is arduous | 67, 72, 74–77 |
| Efficiency can be improved through glycemic control low susceptibility to enzymatic cleavage | Based on low-affinity interactions with receptors |
While the discovery and utilization of BBB penetrating biomacromolecular species has developed rapidly, there is still significant territory that is unexplored and many areas of ongoing research. The most cutting-edge research concerns the development of multifunctional biomacromolecular materials, e.g., utilizing several independent and orthogonal functionalities on the same nanoplatform.47,50,51,55,56,79 Likely there will continue to be advances in these multipurpose materials with increasingly improved systems serving to increase delivery selectivity and expand the therapeutic window.80
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge generous financial support from the Cancer Prevention Research Institute of Texas (Award #: RR210050) and the Welch Foundation (Award #: F-2093-20220331).References
- R. Pandit, L. Chen and J. Gotz, Adv. Drug Delivery Rev., 2020, 165–166, 1–14 CrossRef CAS PubMed.
- J. Xie, Z. Shen, Y. Anraku, K. Kataoka and X. Chen, Biomaterials, 2019, 224, 119491 CrossRef CAS PubMed.
- W. M. Pardridge, NeuroRx, 2005, 2, 3–14 CrossRef PubMed.
- N. J. Abbott, L. Ronnback and E. Hansson, Nat. Rev. Neurosci., 2006, 7, 41–53 CrossRef CAS PubMed.
- N. J. Abbott, A. A. Patabendige, D. E. Dolman, S. R. Yusof and D. J. Begley, Neurobiol. Dis., 2010, 37, 13–25 CrossRef CAS PubMed.
- J. Keaney and M. Campbell, FEBS J., 2015, 282, 4067–4079 CrossRef CAS PubMed.
- M. M. Patel and B. M. Patel, CNS Drugs, 2017, 31, 109–133 CrossRef CAS PubMed.
- S. Rathi, J. I. Griffith, W. Zhang, W. Zhang, J. H. Oh, S. Talele, J. N. Sarkaria and W. F. Elmquist, J. Intern. Med., 2022, 292, 3–30 CrossRef PubMed.
- K. H. Song, B. K. Harvey and M. A. Borden, Theranostics, 2018, 8, 4393–4408 CrossRef CAS PubMed.
- S. I. Rapoport, Cell. Mol. Neurobiol., 2000, 20, 217–230 CrossRef CAS PubMed.
- S. Reddy, K. Tatiparti, S. Sau and A. K. Iyer, Drug Discovery Today, 2021, 26, 1944–1952 CrossRef CAS PubMed.
- B. Xiong, Y. Wang, Y. Chen, S. Xing, Q. Liao, Y. Chen, Q. Li, W. Li and H. Sun, J. Med. Chem., 2021, 64, 13152–13173 CrossRef CAS PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- J. Niewoehner, B. Bohrmann, L. Collin, E. Urich, H. Sade, P. Maier, P. Rueger, J. O. Stracke, W. Lau, A. C. Tissot, H. Loetscher, A. Ghosh and P. O. Freskgard, Neuron, 2014, 81, 49–60 CrossRef CAS PubMed.
- T. Tian, H. X. Zhang, C. P. He, S. Fan, Y. L. Zhu, C. Qi, N. P. Huang, Z. D. Xiao, Z. H. Lu, B. A. Tannous and J. Gao, Biomaterials, 2018, 150, 137–149 CrossRef CAS PubMed.
- R. Ansari, S. M. Sadati, N. Mozafari, H. Ashrafi and A. Azadi, Eur. Polym. J., 2020, 128, 109607 CrossRef CAS.
- C. D. Kusmierz, C. E. Callmann, S. Kudruk, M. E. Distler and C. A. Mirkin, Bioconjugate Chem., 2022, 33, 1803–1810 CrossRef CAS PubMed.
- T. Tashima, Chem. Pharm. Bull., 2020, 68, 316–325 CrossRef PubMed.
- G. C. Terstappen, A. H. Meyer, R. D. Bell and W. Zhang, Nat. Rev. Drug Discovery, 2021, 20, 362–383 CrossRef CAS PubMed.
- A. R. Neves, J. F. Queiroz, S. A. C. Lima and S. Reis, Bioconjugate Chem., 2017, 28, 995–1004 CrossRef CAS PubMed.
- M. Qu, Q. Lin, L. Huang, Y. Fu, L. Wang, S. He, Y. Fu, S. Yang, Z. Zhang, L. Zhang and X. Sun, J. Controlled Release, 2018, 287, 156–166 CrossRef CAS PubMed.
- J. Korbelin, G. Dogbevia, S. Michelfelder, D. A. Ridder, A. Hunger, J. Wenzel, H. Seismann, M. Lampe, J. Bannach, M. Pasparakis, J. A. Kleinschmidt, M. Schwaninger and M. Trepel, EMBO Mol. Med., 2016, 8, 609–625 CrossRef PubMed.
- K. B. Johnsen, M. Bak, F. Melander, M. S. Thomsen, A. Burkhart, P. J. Kempen, T. L. Andresen and T. Moos, J. Controlled Release, 2019, 295, 237–249 CrossRef CAS PubMed.
- A. Vilella, D. Belletti, A. K. Sauer, S. Hagmeyer, T. Sarowar, M. Masoni, N. Stasiak, J. J. E. Mulvihill, B. Ruozi, F. Forni, M. A. Vandelli, G. Tosi, M. Zoli and A. M. Grabrucker, J. Trace Elem. Med. Biol., 2018, 49, 210–221 CrossRef CAS PubMed.
- M. I. Nounou, C. E. Adkins, E. Rubinchik, T. B. Terrell-Hall, M. Afroz, T. Vitalis, R. Gabathuler, M. M. Tian and P. R. Lockman, Pharm. Res., 2016, 33, 2930–2942 CrossRef CAS PubMed.
- M. Sanchez-Navarro and E. Giralt, Pharmaceutics, 2022, 14, 1874 CrossRef CAS PubMed.
- W. M. Pardridge, Annu. Rev. Physiol., 1983, 45, 73–82 CrossRef CAS PubMed.
- W. M. Pardridge, A. K. Kumagai and J. B. Eisenberg, Biochem. Biophys. Res. Commun., 1987, 146, 307–313 CrossRef CAS PubMed.
- W. M. Pardridge, Pharmacol. Toxicol., 1992, 71, 3–10 CrossRef CAS PubMed.
- X. Zhou, Q. R. Smith and X. Liu, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1695 Search PubMed.
- B. Jafari, M. M. Pourseif, J. Barar, M. A. Rafi and Y. Omidi, Expert Opin. Drug Delivery, 2019, 16, 583–605 CrossRef CAS PubMed.
- P. Sun, Y. Xiao, Q. Di, W. Ma, X. Ma, Q. Wang and W. Chen, Int. J. Nanomed., 2020, 15, 6673–6688 CrossRef CAS PubMed.
- Y. Zhang, H. Qu and X. Xue, Biomater. Sci., 2022, 10, 423–434 RSC.
- Y. Cheng, Q. Dai, R. A. Morshed, X. Fan, M. L. Wegscheid, D. A. Wainwright, Y. Han, L. Zhang, B. Auffinger, A. L. Tobias, E. Rincon, B. Thaci, A. U. Ahmed, P. C. Warnke, C. He and M. S. Lesniak, Small, 2014, 10, 5137–5150 CrossRef CAS PubMed.
- J. Song, C. Lu, J. Leszek and J. Zhang, Int. J. Mol. Sci., 2021, 22, 10118 CrossRef CAS PubMed.
- P. Aisen, Int. J. Biochem. Cell Biol., 2004, 36, 2137–2143 CrossRef CAS PubMed.
- B. Voth, D. T. Nagasawa, P. E. Pelargos, L. K. Chung, N. Ung, Q. Gopen, S. Tenn, D. T. Kamei and I. Yang, J. Clin. Neurosci., 2015, 22, 1071–1076 CrossRef CAS PubMed.
- K. Ulbrich, T. Hekmatara, E. Herbert and J. Kreuter, Eur. J. Pharm. Biopharm., 2009, 71, 251–256 CrossRef CAS PubMed.
- J. Pande, M. M. Szewczyk and A. K. Grover, Biotechnol. Adv., 2010, 28, 849–858 CrossRef CAS PubMed.
- Y. Wei, Y. Sun, J. Wei, X. Qiu, F. Meng, G. Storm and Z. Zhong, J. Controlled Release, 2021, 337, 521–529 CrossRef CAS PubMed.
- R. R. de Castro, F. A. do Carmo, C. Martins, A. Simon, V. P. de Sousa, C. R. Rodrigues, L. M. Cabral and B. Sarmento, Int. J. Pharm., 2021, 602, 120655 CrossRef CAS PubMed.
- L. Kong, X. T. Li, Y. N. Ni, H. H. Xiao, Y. J. Yao, Y. Y. Wang, R. J. Ju, H. Y. Li, J. J. Liu, M. Fu, Y. T. Wu, J. X. Yang and L. Cheng, Int. J. Nanomed., 2020, 15, 2841–2858 CrossRef CAS PubMed.
- V. Perumal, A. R. Ravula, A. Agas, M. Kannan, X. Liu, S. S. Ilangovan, S. Vijayaraghavalu, J. Haorah, Y. Zhang and N. Chandra, Micro, 2023, 3, 84–106 CrossRef.
- I. Ramasamy, Clin. Chem. Lab. Med., 2014, 52, 1695–1727 CAS.
- S. Habib and M. Singh, Polymers, 2022, 14, 712 CrossRef CAS PubMed.
- J. Zhu, Y. Zhang, X. Chen, Y. Zhang, K. Zhang, H. Zheng, Y. Wei, H. Zheng, J. Zhu, F. Wu, J. G. Piao, Z. Zhu and F. Li, Biochem. Biophys. Res. Commun., 2021, 534, 902–907 CrossRef CAS PubMed.
- Y. Dang, C. An, Y. Li, D. Han, X. Liu, F. Zhang, Y. Xu, H. Zhong, M. K. Khan, F. Zou and X. Sun, RSC Adv., 2019, 9, 1299–1318 RSC.
- X. Ying, Y. Wang, J. Liang, J. Yue, C. Xu, L. Lu, Z. Xu, J. Gao, Y. Du and Z. Chen, Angew. Chem., Int. Ed., 2014, 53, 12436–12440 CrossRef CAS PubMed.
- S. Stalmans, N. Bracke, E. Wynendaele, B. Gevaert, K. Peremans, C. Burvenich, I. Polis and B. De Spiegeleer, PLoS One, 2015, 10, e0139652 CrossRef PubMed.
- X. Wang, Y. Zhao, S. Dong, R. J. Lee, D. Yang, H. Zhang and L. Teng, Molecules, 2019, 24, 3540 CrossRef CAS PubMed.
- Z. Zhu, Y. Zhai, Y. Hao, Q. Wang, F. Han, W. Zheng, J. Hong, L. Cui, W. Jin, S. Ma, L. Yang and G. Cheng, J. Extracell. Vesicles, 2022, 11, e12255 CrossRef CAS PubMed.
- M. R. Dunn, R. M. Jimenez and J. C. Chaput, Nat. Rev. Chem., 2017, 1, 0076 CrossRef CAS.
- P. Rothlisberger and M. Hollenstein, Adv. Drug Delivery Rev., 2018, 134, 3–21 CrossRef CAS PubMed.
- T. K. Sharma, J. G. Bruno and A. Dhiman, Biotechnol. Adv., 2017, 35, 275–301 CrossRef CAS PubMed.
- J. Macdonald, D. Denoyer, J. Henri, A. Jamieson, I. J. G. Burvenich, N. Pouliot and S. Shigdar, Nucleic Acid Ther., 2020, 30, 117–128 CrossRef CAS PubMed.
- X. Li, Y. Yang, H. Zhao, T. Zhu, Z. Yang, H. Xu, Y. Fu, F. Lin, X. Pan, L. Li, C. Cui, M. Hong, L. Yang, K. K. Wang and W. Tan, J. Am. Chem. Soc., 2020, 142, 3862–3872 CrossRef CAS PubMed.
- M. Darmostuk, S. Rimpelova, H. Gbelcova and T. Ruml, Biotechnol. Adv., 2015, 33, 1141–1161 CrossRef CAS PubMed.
- H. Kawabata, Free Radicals Biol. Med., 2019, 133, 46–54 CrossRef CAS PubMed.
- C.-h. B. Chen, K. R. Dellamaggiore, C. P. Ouellette, C. D. Sedano, M. Lizadjohry, G. A. Chernis, M. Gonzales, F. E. Baltasar, A. L. Fan, R. Myerowitz and E. F. Neufeld, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15908–15913 CrossRef CAS PubMed.
- D. Porciani, G. Signore, L. Marchetti, P. Mereghetti, R. Nifosi and F. Beltram, Mol. Ther.–Nucleic Acids, 2014, 3, e144 CrossRef CAS PubMed.
- J. Macdonald, P. Houghton, D. Xiang, W. Duan and S. Shigdar, Nucleic Acid Ther., 2016, 26, 348–354 CrossRef CAS PubMed.
- C. Cheng, Y. H. Chen, K. A. Lennox, M. A. Behlke and B. L. Davidson, Mol. Ther.–Nucleic Acids, 2013, 2, e67 CrossRef PubMed.
- J. W. Choi, M. Seo, K. Kim, A. R. Kim, H. Lee, H. S. Kim, C. G. Park, S. W. Cho, J. H. Kang, J. Joo and T. E. Park, ACS Nano, 2023, 17, 8153–8166 CrossRef CAS PubMed.
- D. S. Seferos, A. E. Prigodich, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 308–311 CrossRef CAS PubMed.
- Y. Zhang, J. He, L. Shen, T. Wang, J. Yang, Y. Li, Y. Wang and D. Quan, J. Controlled Release, 2021, 329, 1117–1128 CrossRef CAS PubMed.
- C. Rademacher and J. C. Paulson, ACS Chem. Biol., 2012, 7, 829–834 CrossRef CAS PubMed.
- Y. Anraku, H. Kuwahara, Y. Fukusato, A. Mizoguchi, T. Ishii, K. Nitta, Y. Matsumoto, K. Toh, K. Miyata, S. Uchida, K. Nishina, K. Osada, K. Itaka, N. Nishiyama, H. Mizusawa, T. Yamasoba, T. Yokota and K. Kataoka, Nat. Commun., 2017, 8, 1001 CrossRef CAS PubMed.
- A. Lundqvist and P. Lundahl, J. Chromatogr., A, 1997, 776, 87–91 CrossRef CAS PubMed.
- B. Thorens and M. Mueckler, Am. J. Physiol. Endocrinol. Metab., 2010, 298, E141–E145 CrossRef CAS PubMed.
- P. Mergenthaler, U. Lindauer, G. A. Dienel and A. Meisel, Trends Neurosci., 2013, 36, 587–597 CrossRef CAS PubMed.
- J. L. Jiménez Blanco, C. Ortiz Mellet and J. M. García Fernandez, Chem. Soc. Rev., 2013, 42, 4518–4531 RSC.
- B. Qu, X. Li, M. Guan, X. Li, L. Hai and Y. Wu, Eur. J. Med. Chem., 2014, 72, 110–118 CrossRef CAS PubMed.
- M. H. Stenzel, Macromolecules, 2022, 55, 4867–4890 CrossRef CAS.
- F. Xie, N. Yao, Y. Qin, Q. Zhang, H. Chen, M. Yuan, J. Tang, X. Li, W. Fan, Q. Zhang, Y. Wu, L. Hai and Q. He, Int. J. Nanomed., 2012, 7, 163–175 CrossRef CAS PubMed.
- H. S. Min, H. J. Kim, M. Naito, S. Ogura, K. Toh, K. Hayashi, B. S. Kim, S. Fukushima, Y. Anraku, K. Miyata and K. Kataoka, Angew. Chem., Int. Ed., 2020, 59, 8173–8180 CrossRef CAS PubMed.
- T. Yang, Y. Mochida, X. Liu, H. Zhou, J. Xie, Y. Anraku, H. Kinoh, H. Cabral and K. Kataoka, Nat. Biomed. Eng., 2021, 5, 1274–1287 CrossRef CAS PubMed.
- A. Amano, N. Sanjo, W. Araki, Y. Anraku, M. Nakakido, E. Matsubara, T. Tomiyama, T. Nagata, K. Tsumoto, K. Kataoka and T. Yokota, J. Nanobiotechnol., 2023, 21, 36 CrossRef CAS PubMed.
- M. Guberman and P. H. Seeberger, J. Am. Chem. Soc., 2019, 141, 5581–5592 CrossRef CAS PubMed.
- L. Lu, H. Chen, L. Wang, L. Zhao, Y. Cheng, A. Wang, F. Wang and X. Zhang, Int. J. Nanomed., 2020, 15, 8875–8892 CrossRef CAS PubMed.
- P. Y. Muller and M. N. Milton, Nat. Rev. Drug Discovery, 2012, 11, 751–761 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |



