Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fabrication of polymeric biomaterials: a strategy for tissue engineering and medical devices
Ferdous
Khan
*a,
Masaru
Tanaka†
b and
Sheikh Rafi
Ahmad
c
aSenior Polymer Chemist, ECOSE-Biopolymer, Knauf Insulation Limited, P.O. Box 10, Stafford Road, St. Helens, WA10 3NS, UK. E-mail: ferdous.khan0@gmail.com; Ferdous.Khan@knaufinsulation.com; Tel: +44 (0)1744 766894
bBiomaterials Science Group, Department of Biochemical Engineering, Graduate School of Science and Engineering, Yamagata University, Jonan 4-3-16, Yonezawa, Japan. E-mail: tanaka@yz.yamagata-u.ac.jp; Tel: +81-238-26-1096
cCentre for Applied Laser Spectroscopy, CDS, DEAS, Cranfield University, Shrivenham, Swindon, Wiltshire SN6 8LA, UK. E-mail: sheikhrafiahmad@hotmail.com
First published on 21st August 2015
Abstract
Polymeric biomaterials have a significant impact in today's health care technology. Polymer hydrogels were the first experimentally designed biomaterials for human use. In this article the design, synthesis and properties of hydrogels, derived from synthetic and natural polymers, and their use as biomaterials in tissue engineering are reviewed. The stimuli-responsive hydrogels with controlled degradability and examples of suitable methods for designing such biomaterials, using multidisciplinary approaches from traditional polymer chemistry, materials engineering to molecular biology, have been discussed. Examples of the fabrication of polymer-based biomaterials, utilized for various cell type manipulations for tissue re-generation are also elaborated. Since a highly porous three-dimensional scaffold is crucially important in the cellular process, for tissue engineering, recent advances in the effective methods of scaffold fabrication are described. Additionally, the incorporation of factor molecules for the enhancement of tissue formation and their controlled release is also elucidated in this article. Finally, the future challenges in the efficient fabrication of effective polymeric biomaterials for tissue regeneration and medical device applications are discussed.
1. Introduction
The use of polymers as biomaterials has been the subject of intense investigation over the past fifty years.1,2 Different chemical structures and functional groups in such polymers govern their morphology and properties, and allow precise control of the creation of desired molecular architectures for a wide range of applications in the biomedical field. For example, biocompatible polymers have been used successfully as artificial organs and drug delivery systems.3,4 However, it is to be noted that the degree of success in such applications depends on the self-organization and biocompatibility of the formulated molecular architecture.The biomaterials which are derived from polymers generally fall into two categories: naturally occurring and human-made synthetic materials. Collagens, alginate and chitosan based materials are the best examples of biomaterials derived from natural resources. The polymers derived from synthetic origins are divided into two classes: non-biodegradable and biodegradable synthetic polymers. Recently, the biodegradable polymers have become highly important in the field of biomaterials and tissue engineering, due to the avoidable additional surgery to remove the implants or scaffolds. Thus, much attention needs to be undertaken on the synthesis of biodegradable polymers.
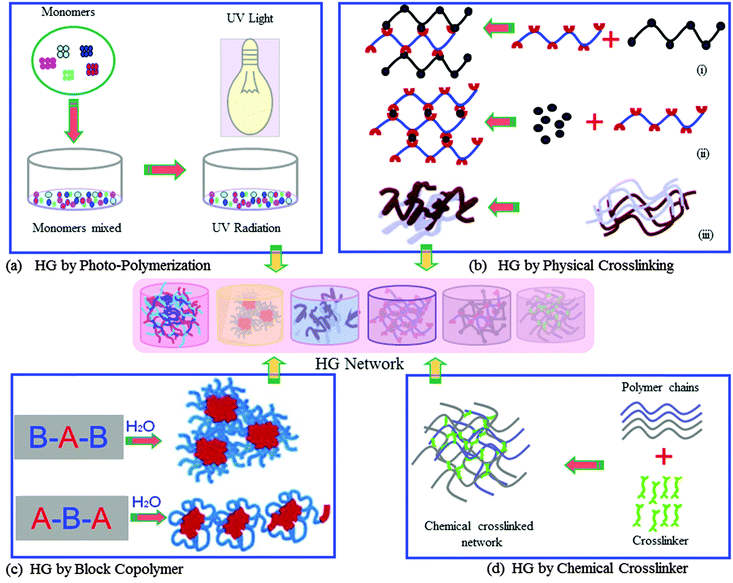
In medical applications there is an on-going research and development (R&D) effort for the improvement of methodologies and devices for more efficient and effective processing of biomaterials. The outcome of such R&D has recently been applied to successfully treat many diseases.5–7 Amongst the wide range of biomaterials which have been synthesised in recent time for potential use in medicine, majority of these do not have suitable properties to interact effectively with biological tissues or cells. However, it is deemed possible to improve their intrinsic proprieties using required and appropriate process engineering for optimum results. Crosslinking of biopolymers is one of the examples of process engineering, which has provided a means to improve the quality of biomaterials for wider medical applications. For example, the crosslinked form of soft polymers, classified as hydrogels,8 is a class of new generation of exciting biomaterials that has demonstrated the ability to form scaffolds for a variety of uses, such as tissue engineering, delivery of active molecules, and biosensors and actuators. Hydrogels are 3D structured polymeric materials, “swell gels”, which are formed via crosslinking reactions of polymers (Fig. 1).
The hydrogels can be synthesised with required properties depending on the chemical structure, composition and confirmation of starting materials, density of linking of polymer chains, hydrophobicity and hydrophilicity for a particular biomedical application.
The 3D structural-integrity and properties of hydrogels are mainly dependent on their method of preparation such as physical or chemical crosslinking reaction.3,4 Hydrogels from chemical crosslinking form permanent junction-type networks. The examples of this type of hydrogel include polymerisation of the acryloyl group, ionising radiation-induced crosslinking (photo-polymerisation, Fig. 1a), small molecule crosslinking with a polymer chain (glutaraldehyde, Fig. 1d) and polymer–polymer crosslinking by condensation reaction. The physical crosslinking of hydrogels allows forming transient junction-type networks, such as polymer chain entanglements or physical interactions (e.g. ionic interactions, as demonstrated in Fig. 1b), hydrogen bonds, or hydrophobic interactions. Indeed, there are varieties of different polymer structures, which can form physical and chemical hydrogel networks. These polymer structures include linear homopolymers, linear copolymers, and block, random or graft copolymers; polyion–multivalent ion, polyion–polyion or H-bonded complexes; hydrophilic networks stabilized by hydrophobic domains; interpenetrating polymer networks (IPNs) or physical blends; specific molecular recognition; and self-assembling of polymers or polypeptides.
Hydrogels can be synthesised both from natural and synthetic polymers. The examples of hydrogels from natural polymers are: collagen, gelatin, hyaluronic acid, chondroitin sulfate, chitin and chitosan, alginate, starch, cellulose, and their derivatives. Hydrogels from natural polymers have many advantages over the synthetically derived ones such as low toxicity and good biocompatibility because of their chemical structures and are very akin to the structure of glycosaminoglycan (GAG) molecules present in the native extracellular matrix (ECM). Hydrogels from synthetic polymers are prepared by chemical polymerisation methods. Various types of monomers, for examples, acrylates, methacrylates, acrylamides, esters, carboxylic acid and polyfunctional monomers, can be utilised for the preparation of synthetic hydrogels.9 The detailed description of the preparation of hydrogels is beyond the scope of this review. This topic has been covered in depth by several researchers.9–11
In this review, we describe the recent developments of polymeric biomaterials and 3D structure generation by utilizing a variety of advanced techniques and methods with the emphasis on various types of tissue engineering. Several strategies for the 3D scaffold fabrication, which include lithography and printing techniques, patterning by self-organisation of polymers, self-assembling of peptides, and cellular compatibility of polymer-based biomaterials and hydrogels, are presented. The advantages and drawbacks in the 3D scaffold fabrication methods are also discussed. Additionally, we describe the applications of polymeric biomaterials and scaffolds in tissue engineering, particularly to the cartilage, bone and neural tissue regeneration. Furthermore the approaches for the incorporation of bioactive factor molecules in biomaterials via physical encapsulation and chemical crosslinking, their functions and specific applications in tissue regeneration have been discussed.
2. Tissue engineering (TE)
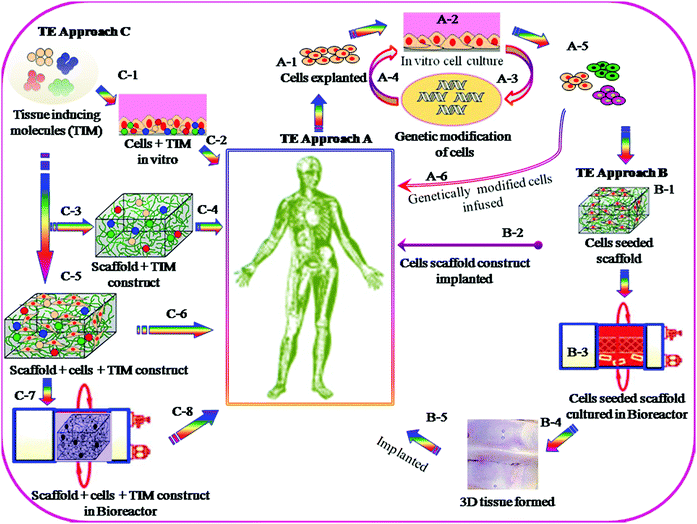
The objectives for the TE approach are to replace, repair or regenerate damaged tissues, or to create artificial tissues for transplantation, when normal physiologic reaction fails to take place and the surgical procedure becomes essential. A number of strategies for TE have been schematically presented in Fig. 2. Currently two different standards are used, e.g. autografts and allografts. Each of them, however, has several limitations, including donor-site morbidity in the case of using autografts and the associated potential risk of disease transmission in the case of using allografts. In recent time, considerable research effort has been made worldwide to overcome the inherent limitations of current standards and to improve the biomedical technology by employing 3D biomaterial scaffold-based TE strategies. In the scaffold-based TE approach, it is essential that the interactions of 3D-scaffold materials and cells take place by means of biocompatibility, cell adhesion, proliferation, growth, differentiation and matrix deposition. Scaffolds must be designed with an appropriate surface chemistry and morphology to promote cellular functions and with sufficient structural and physical properties such as mechanical strength, porosity and pore sizes. Such scaffolds can be fabricated from the original biodegradable and non-biodegradable polymers. In the case of a biodegradable 3D scaffold, it must be designed in such a way so that it maintains structural integrity, and functions and degrades in a controlled manner, until the new tissues are formed and the function continues.Biomaterial scaffolds have been synthesised from different types of organic and inorganic polymers and materials including polymers of natural and synthetic origin, ceramics, and their composites. Scaffold materials must be designed to mimic the 3D structure of the native tissue and have the ability to act as delivery agents for growth factors, drugs/antibiotics, and chemotherapeutic agents, depending on the nature of the tissue to be repaired. Biomaterial scaffolds can be pre-fabricated either solid structure or injectable forms that harden in situ (hydrogels) which essentially will depend on the nature of specific tissue engineering application.
3. 3D scaffold fabrication for TE
There are several strategies in TE currently under investigation; examples are schematically described in Fig. 2. Most of these utilize cells, which are seeded onto 3D scaffolds. Scaffolds are generally designed to be fabricated with a wide range of properties which include: appropriate surface chemistry, porosity with pore dimensions from macro- to submicron and interconnectivity networks, which allow cell–cell communication and migration, cell proliferation and differentiation, and finally to maintain the biocompatibility and structural integrity throughout the tissue regeneration process.The methods of fabrication of biocompatible 3D scaffolds with appropriate architectures are divided into two classes: (i) conventional and (ii) rapid prototyping. The former class of fabrication methods often does not provide sufficient physical and mechanical properties, and consequently such types of scaffolds undergo deformation because of cell motility. Whereas the rapid prototyping methods do not have such disadvantages and can provide all essential characteristics for specific TE application. 3D nano/micro pattern scaffolds fabricated by rapid prototyping showed a significant influence on cellular morphology, cell proliferation and differentiation and also on the functioning of various cell types.12–14 The scaffold fabrication by conventional methods includes phase separation,15 porogen leaching,16 gas foaming,17 fibre meshing18 and supercritical fluid processing.19 The second category is more advanced and examples of this prototyping technique include the selective laser sintering,20 3D printing21 and lithography.22 More recently, self-organized honeycomb porous structures using block-copolymers23 have been developed. The following section highlights on the recent development in scaffold fabrication by lithography and 3D printing, and also elaborates on self-organization methods, as well as self-assembly of peptides, specifically for the enhancement of cellular functioning in tissue engineering applications.
3.1. 3D scaffold fabrication by lithography and printing techniques
Polymer patterning of 3D surfaces in biomedical research to study cellular behaviour and TE24–26 has generated a great deal of interest of the academic and industrial researchers world-wide. Because of this, a great deal of advancement has taken place in this technology in recent time, in particular, polymeric biomaterials and crosslinked hydrogels have found a wide range of applications in micro-devices using various approaches. In the following section the recent development in hydrogel patterning using photolithography, dip-pen lithography, nanoimprinting, contact printing, solid-free form, robotic deposition and their application in TE have been described.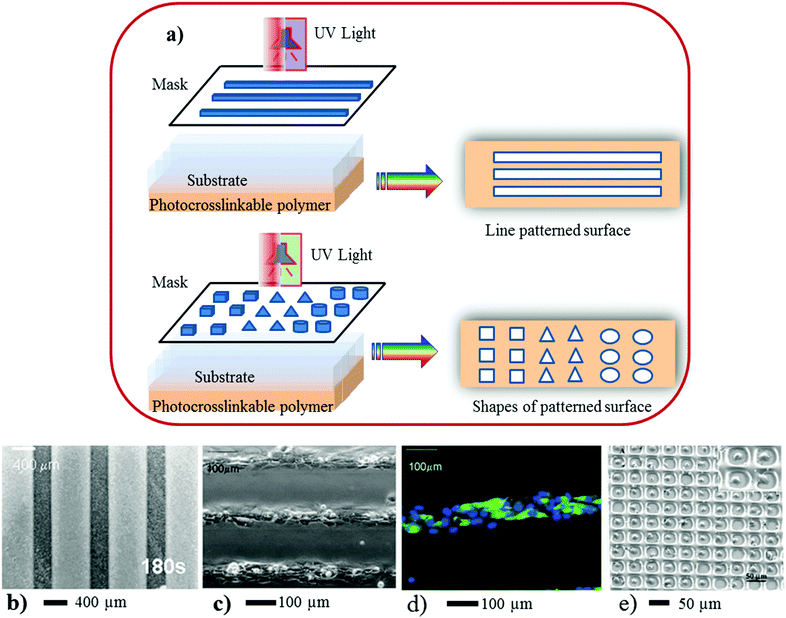 | ||
| Fig. 3 (a) Scheme represents 3D surface patterning of using photo-induced crosslinkable polymers. In the above illustration (a) a mask containing a variety of patterns was placed on top of the polymer coated coverslip. UV light was focused onto the mask for a certain period followed by repeated washes in PBS to remove the non-polymerized gel and expose the underlying glass substrate within the patterned regions. (b) A micrograph of a line pattern, (c and d) neonatal rat cardiomyocytes were seeded on chitosan patterned glass surfaces at 8 days of culture. The cells adhered to glass and formed confluent cell lanes that exhibited spontaneous contractions. (d) Patterned cardiomyocytes express cardiac troponin I (green) and exhibit a developed contractile apparatus. (e) Patterning of 3T3 fibroblasts and primary rat hepatocytes in 30 × 30 μm PEG wells, 10 × 10 array of single fibroblasts with 91% cell occupancy (×150). The inset shows a higher-magnification image of confined fibroblasts (×1200). (Reproduced with permission.35 Copyright 2003, ACS.) | ||
Photolithography is a high-throughput technique, and is suitable for a large-area of 3D surface pattern generation with good alignment (Fig. 3a–c) and topography. This technique can provide a broad range of features, varying from micrometres to sub-microns (e.g. 100 nanometres). However, for high-resolution 3D pattern surface generation special types of nonconventional masks, photoactive chemicals (e.g. monomers-, or oligomers or polymers),43 short wavelengths of radiation, advanced optical techniques and special set-ups for lithography are needed.44
3D pattern surfaces created by this technique are used as templates, and subsequently functionalised with other functional materials. Traditionally patterned polymer surfaces are used in the semiconductor industry. In recent years polymer patterned surfaces have found many applications such as LEDs,45 liquid-crystal displays,41 photonic crystals,46 sensors and actuators,47 and biomedical applications including microarrays of cells, proteins and peptides.35–39 Here, we focus on the use of this technology for cellular application as discussed below.
3D surface patterns that are created by the photolithographic process have the ability to manipulate cellular behaviour, and interactions of cells between themselves and with the polymer matrix.32–36 The patterns processed by photolithography provide confined geometry as well as lateral features for cellular adhesion. Due to the multiple features of the patterns suppresses the detrimental effects of cell arrays when cultured for a longer time, in contrast to those of other patterning techniques. This method has been employed to create 3D pattern surfaces using chitosan.37 Karp and co-workers37 have demonstrated the generation of 3D patterned surfaces of various shapes (e.g., lanes, squares, triangles and circles) by coating a thin layer of a photocrosslinkable chitosan on a glass slide. Subsequently cardiac fibroblasts were cultured on these patterned surfaces, which formed stable patterns for up to 18 days of culture period. Researchers have also demonstrated that when cardiomyocytes were cultured in lanes patterned with 68–99 μm width, they showed expression of cardiac troponin I and responsiveness towards electrical field stimulation. Osteoblasts (SaOS-2) were also cultured in squares, triangles, or circles (0.063–0.5 mm2), and the cells were localized in the patterned regions. SaOS-2 proliferated to confluence in 5 days, expressed alkaline phosphatase and produced a mineralized matrix.
Photolithography has also been utilised by a variety of other polymeric biomaterials such as polyethylene glycol (PEG),39 poly(N-isopropyl acrylamide) (PNIPAAm),36 and PEG-peptide Arg–Gly–Asp (RGD) hybrid hydrogels35 for cell patterning and their functional studies.
A high-density murine 3T3 fibroblast array was generated (Fig. 3), and cells were encapsulated in 3D confined hydrogel micro-wells.35 Encapsulation of hepatocytes within the PEG-diacrylate hydrogel via photo-induced patterning yielded about 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cell clusters per 100 mm2 gel as a living cell array with a precise control of cell positioning, in which the duration of cell viability was up to a few weeks.39 Albrecht and co-workers have investigated40 the multicellular organization in photo-induced patterned 3D hydrogel containing cells viable up to two weeks, which regulates the bovine articular chondrocyte.40
000 cell clusters per 100 mm2 gel as a living cell array with a precise control of cell positioning, in which the duration of cell viability was up to a few weeks.39 Albrecht and co-workers have investigated40 the multicellular organization in photo-induced patterned 3D hydrogel containing cells viable up to two weeks, which regulates the bovine articular chondrocyte.40
However, in photolithographic systems there are some challenges remain to be solved, such as: (i) economic viability of the processing method, (ii) lack of resolution, (iii) lack of original properties following the generation of patterns, and (iv) unsuitability of UV-sensitive biological materials for pattering.
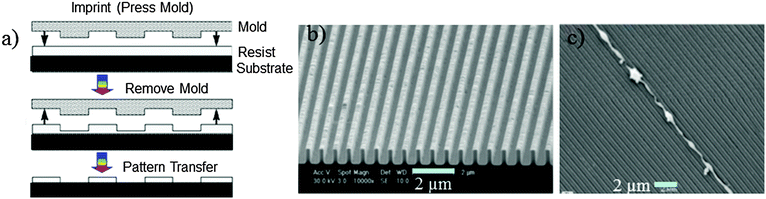 | ||
| Fig. 4 (a) Scheme of nanoimprinting. (b) NIL results using a 250 nm line width PDMS-b-PS grating. (c) 400 nm width and 800 nm pitch. SEM images show that the axons grow on the ridge edges, and not in the grooves. Reprinted with permission.54,58 Copyright 2007 and 2004, Wiley-VCH. | ||
In this method designing of thermoplastic materials to act as a suitable resist is a critical factor to obtain high-resolution and defect free 3D pattern surfaces. For example, some polymers such as poly(methyl methacrylate) (PMMA) and polystyrene (PS) are susceptible to lead fracture on the 3D pattern surface.53 Therefore, multifunctional copolymers, either block or graft-copolymers, are the preferred class of materials for defect free patterns.55 It has been demonstrated that using the polydimethylsiloxane (PDMS)-block-PS copolymer as a resist, a 250 nm line width grating pattern has been constructed (Fig. 4)54 with excellent mould releasing properties and without defects. Another feature of this technology is the formulation using UV-induced polymerisation; particularly those developed using acrylic and methacrylic monomers via free radical polymerization due to their high-reactivity. However, the environmental oxygen can lead to detrimental effects to the polymerisation reaction at the surface layer of the resist. To resolve this problem, either an inert atmosphere during processing or a UV-sensitive cationic crosslinking of cycloaliphatic epoxides has been developed.55
Other advanced chemical methods had been adopted for cell-based patterning,56 tissue engineering57,58 and the cellular response to the surface morphology and structures.59 For these, surface topography has been found to play an important role, as most attached cell types are reactive even on a few nm scale differences of the topographic structure.60 Development of groove pattern structures with varying width in the range between 100 nm and 400 nm with a constant depth of 97 nm depths has been reported and this template was utilised for nerve cell guiding.57 Researchers have demonstrated that cells do not follow the continuity of grooves and ridges, and the pattern surface influenced the shape of the cells by rearranging the cytoskeleton57 as well as induced gene regulation.59 Similarly, osteoblast cells cultured on groove surfaces with a depth of 150 nm, and found similar alignment behaviour of cells.58 The depth of the groove is a highly important parameter as this determines the wettability of the cells to be aligned.58
The challenges, such as controlling mould geometry, selection and formulation of thermoplastic resist materials, precise control of the process parameters and suitable photosensitive material selection, still remain reproducible for 3D pattern generation, which eventually will dictate future exploitation of this technique in the biomedical arena.
3.1.3.1. Microcontact printing (μCP) with UV-induced 3D patterning. Microcontact printing is a remarkable surface patterning technique with spatial resolution down to a nano-meter range, developed about a decade ago.61–66 This technique has drawn enormous attention from communities belonging to materials and chemical science, tissue engineering and biological sciences. In the past few years a significant improvement in the process, particularly, in the design technologies is commensurate with biomedical applications.61 Using this method, a high-quality 3D pattern has been achieved by selecting appropriate conditions with no contamination, without deformation of stamps and lateral diffusion of the ink; the more details on μCP patterning have been extensively reviewed in the open literature.22,61 In μCP a poly(dimethylsiloxane) (PDMS) stamp has relief features to transfer an inked material to a substrate, as demonstrated in Fig. 5a. Due to the elastomeric properties of PDMS, the stamp deforms macroscopically allowing increasing features over large areas (a few cm2). PDMS has low surface energy due to the flexibility of the siloxane chain and low intermolecular forces between the methyl groups,63 which facilitate peeling of the stamp from the substrate after printing. Researchers have reported patterns with features less than 50 nm using μCP with PDMS stamps.67 In high-resolution patterning, the deterioration of the surface features could be minimised by using functional polymers that interact with the surface. The examples of such polymers are: poly(acrylic acid), poly(ethyleneimine) and small heavy weight macromolecules (e.g. dendrimers). This technique has been employed enormously in various applications such as plastic electronics, optics, surface sciences and biological fields. For more details on the μCP technique the readers can refer to published reviews.38,61
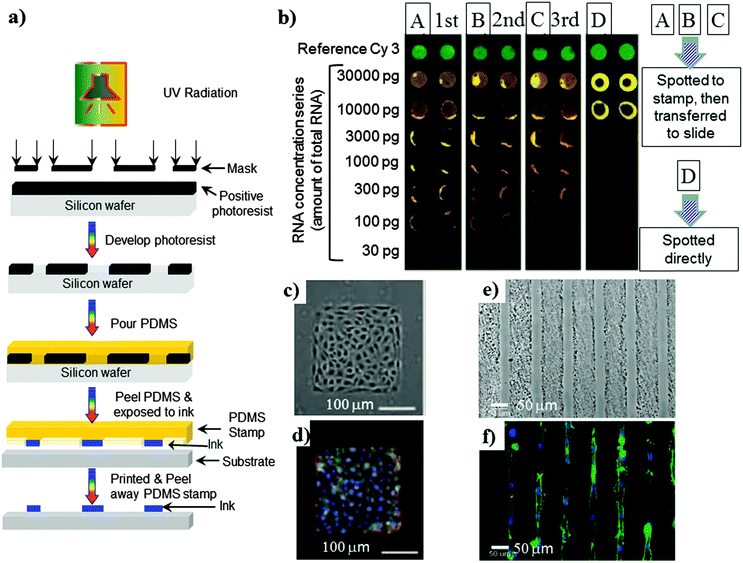 | ||
| Fig. 5 (a) Schematic representation of microcontact printing. (b) DNA arrays made by a μCP spotted PDMS stamp three in times in succession or by spotting directly. The arrays show hybridizations using different concentrations of the RNA starting material. (c and d) A monolayer of bovine pulmonary artery endothelial cells cultured on 250 mm squares of fibronectin. (e) Optical micrograph of the OEGMA/MA line pattern on chitosan film, (c) alignment of the cytoskeleton and nuclei in NIH3T3 fibroblasts cultured on 30 μm wide lines of PLGA substrates after 24 h. Actin microfilaments (green) were visualized by Alexa 488-labeled phalloidin. Cell nuclei were visualized by DAPI (blue). Reprinted with permission from (b)68 (Copyright 2004 ACS), (c and d)69 (Copyright 2005 National Academy of Sciences), (e)62 (Copyright 2003 ACS) and (f)73 (Copyright 2005 Elsevier Science). | ||
In biological fields, the μCP technique has been utilised for patterning DNA,68 the immobilization of proteins and peptides on substrates for cellular adhesion,62,69,70 or protein resistant polymers,64–66,71 as demonstrated in Fig. 5. Researchers have demonstrated cell patterning on silicon based substrates,72 PS,66 and on bio-resorbable polymers,59,62 which could potentially be applied in biomedical fields. Several researchers have demonstrated73–76 that this technique has the ability to manipulate polymeric biomaterials between microns to the nanometre scale to obtain various types of pattern shapes such as rectangular and lines (Fig. 5e), which has a significant positive influence in cellular functioning, regenerative medicine and the drug delivery system.
For example, Lin and co-workers created line patterns of proteins and cells using μCP on biodegradable polymers such as poly(lactic acid) (PLA) and poly(lactide-co-glycolide) (PLGA) substrates (Fig. 5f),73 which are routinely used as scaffolds in tissue engineering. Site-specific immobilisation of proteins and NIH3T3 fibroblasts was achieved by printing a protein resistance polymer such as poly(oligoethylene glycol methacrylate) in a particular area, thus creating a line pattern for cellular attachment. Cells remain confined within the line patterns on the PLGA and PLA films for up to 14 days, and aligned their actin cytoskeleton along the line patterns. This suggests that this method could have a significant influence in cell-based tissue engineering applications for controlling the spatial morphology and distribution of cells on synthetic biomaterials.
Although this method has few drawbacks, such as multilayer and multicomponent pattern process, which makes it less economically viable. Routinely generated micrometre size features using μCP is expected to have an important role in the polymer and biomaterial 3D pattern generation when combining with other techniques,77–79e.g. photolithography, dip-pen lithography or with self-assembly polymeric systems.
3.1.3.2. Contact printing without UV. Microstructure generation by contact printing is a very recent approach used for the deposition of organic solvents onto a solid polymer film surface.80 The schematic diagram of contact printing by solid pins is shown in Fig. 6a.80 In this process two layers of polymer coated glass slides in which the bottom layer is chitosan (CS) (thickness ∼ 1 μm) and the top layer is the polystyrene surface with a thickness either 1.2 μm or 2.4 μm measured using scanning electron microscope were used. The mechanism of microwell fabrication by this technique is totally different than the previously described other techniques (photo- and soft-lithography, μCP, etc.), as in this case the polymer is locally transferred from the centre to the edge region that allowed the formation of rims very akin to the explanation of micro-fluidic flow proposed for the formation of a “ring-shaped coffee stain” when dropped onto a solid surface.81,82 This technique has several advantages over other techniques, such as (i) ease of processing, (ii) no bulk flow of solvents required unlike lithography, (iii) the dimensions of microwells can easily be controlled by tuning physical and chemical parameters, and (iv) high density (several hundreds e.g. 600 per cm2) of microwell features can be generated in a single experiment. The well-defined and desired dimension of microwell fabrication will certainly depend on the selection of polymers and their solubility, solvents, sizes of the solid pins, the amount of solvent deposited onto the polymer surface, printing temperature and humidity. However, the fabrication of microwells in a nano-scale range is required to be investigated.
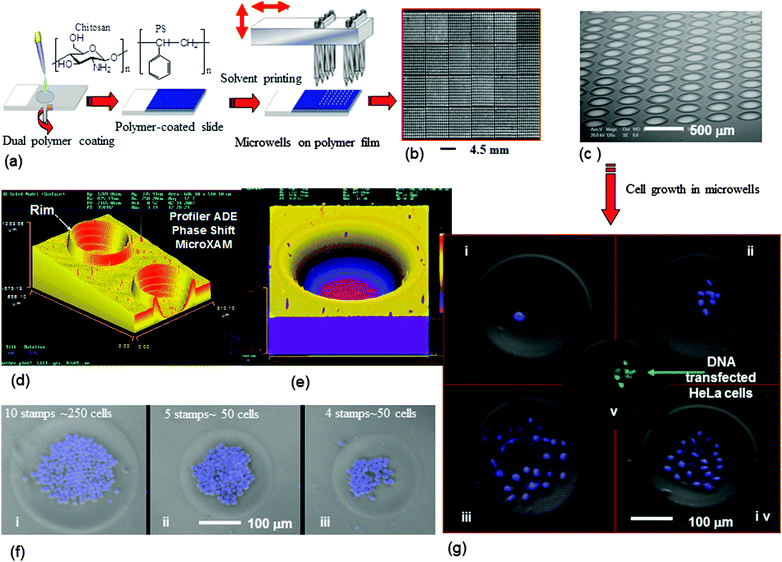 | ||
| Fig. 6 Microwell fabrication by contact printing and their cellular application: (a) general process of polymer microwell fabrication by contact printing. Images of the fabricated microwell arrays (b and c), (b) a low-resolution image obtained using a BioAnalyzer 4F/4S white light fluorescent-based scanner showing an array of 600 wells per cm2 (PS film thickness = 2.4 μm, printing pin diameter 150 μm) and (c) the SEM image of a microwell array at 70° angle with 490 wells per cm2 (PS film thickness = 1.2 μm, printing pin diameter 150 μm). 3D images of microwells fabricated on PS films (d) and (e). (d) Generated using 4 solvent stamps on a 1.2 μm PS film with solid pins of a diameter of 150 μm (K2783). (e) A single microwell fabricated on a PS film (2.4 μm thickness) by stamping acetophenone/ethyl acetate 8 times with a 150 μm diameter solid pin (K2783). (f) Microwells hosting a monolayer of K562 suspension cells. Composite digital image: phase contrast and DAPI-staining. (g) HeLa cells growth in microwells: culture period 24 h (i), 48 h (ii), 96 h (iii) and (iv), and DNA transfection to HeLa cells (v). Reprinted with permission.80 Copyright 2007 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
The microwells that have been generated allowed various cell type manipulation, encapsulation and growth,80 for example, cervical carcinoma (HeLa) and human leukemia (K562) cells, and DNA transfection to the cells has been demonstrated in Fig. 6.
This technique is also extensively utilised to fabricate polymer microarrays by dispensing pre-form polymer solution onto a solid surface, which has been described in detail in our recent published literature.83–85 Such polymer microarrays are immobilised with desired type of cell culture, allowing the identification of cell-compatible polymeric biomaterials for subsequent scaffold fabrication and implantation.86,87
This technique has been utilised in 3D patterning of photoresists, polyelectrolytes, conjugated polymers, biopolymers, photocurable oligomers and monomers, and polymer colloids.
This is a simple method of producing 3D micro-patterns with flexible size and shape. However, it is crucially important particularly for ink-jet printing of polymers to identify the well-defined rheological properties of the polymer solutions utilised for patterning, surface tension and the boiling temperature of the solvent.92 In this method the selection of the polymer and solvent can be crucial, due to their solubility interaction and viscoelastic properties of polymer solutions which influence the break-down of jets into droplets.92 Structural and molecular architecture of polymers, their chain length and polydispersity index, and concentration of the polymer in solution will eventually dictate the viscoelastic properties governing the printing pattern. This method has been investigated by research groups to obtain 3D patterning of polymer-arrays using both non-biodegradable90,93 and biodegradable (PLGA)88 polymers, for applications in sensors93 and cell patterning.88,94,95
Sanjana and co-workers95 have generated neuron-adhesive patterns using biodegradable polymers, namely the collagen and poly(D-lysine) (PDL) mixture via selectively ink-jet printing on the poly(ethylene glycol) (PEG) surface. In these patterns PEG acts as the cell-repulsive material, while the collagen/PDL mixture acts as the cell-adhesive material. The inkjet printing technique is used for the construction of synthetic biodegradable scaffolds via printing of crosslinkers onto liquid alginate/gelatin solutions, which formed 3D hydrogel scaffolds that have potential application in tissue engineering.96
To achieve a controllable pattern, the substrate on which the solvent and the polymer are ink-jet printed and the underlying layer should be prevented from dissolution and swelling.
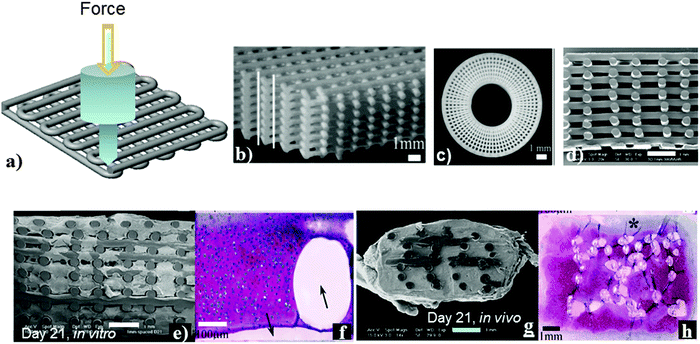 | ||
| Fig. 7 Robotic deposition (a) two layers scaffold produced by 3D plotting. Optical images illustrating (b) a 3-D periodic structure with a simple tetragonal symmetry reveal the high integrity interfaces formed between layers and (c) a 3D radial structure comprised of alternating layers deposited using radial and concentric fill patterns. (d) SEM section of 3D deposited scaffolds with homogeneous 1 mm fibre spacing showing typical fibre diameters and pore geometries 20×. SEM (e and g) and safranin-O stained (f and h) of 3D-deposited 300/55/45 scaffolds following (e and f) 21 days dynamic culture in vitro; (g and h) 21 days subcutaneous implantation in nude mice; (arrows indicate PEGT/PBT fibre, * indicates fibrous capsule). Scale bar = 1 mm (b–e, g and h), 100 μm (f). Reprinted with permission.103,104 Copyright 2002 and 2004, Elsevier Science. | ||
Many research groups have demonstrated that this technique is very useful for generating complex geometry using various bioactive polymers and copolymers derived from natural and synthetic origins.101–106 Some examples of such polymers are: poly-L-lactide (PLLA), poly(ε-caprolactone) (PCL),101 poly(lactide-co-glycolide) (PLGA),102 poly(ethylene glycol terephthalate-b-butylene terephthalate),103 agarose, gelatin,104 CS105 and polyelectrolytes.106 In the case of polyelectrolytes, the solution blend of cationic and anionic polymers was deposited from a nozzle and rapidly coagulated in an alcohol–water solution to obtain a 3D periodic structure. Optical microscopic images of 3D lattices and radial structures with a resolution of 1 μm show high integrity surfaces, as shown in Fig. 7b–d.107
This technique has several advantages for 3D heterogeneous structure generation with high efficiency, with features from a submicron to micron range in contrast to conventional lithographic methods. Furthermore, this method does not use UV radiation for curing and causes no damage to light-sensitive molecules. Therefore, light sensitive bioactive molecules can be used in the fabrication of devices. Several research groups have demonstrated in their published literature reports107–110 that this method can be used to fabricate suitable devices in photonics, microfluidics, bio-mineralization90–92 and the most promising is the fabrication of the scaffold-matrix for cellular attachment, proliferation and differentiation for tissue regeneration.110 Seol et al.26 have demonstrated that it is essential to have appropriate porosity and pore sizes with well-defined shape, mechanical integrity and biocompatibility over a time period for cells to function during tissue regeneration.26 Articular chondrocytes were cultured on PEG-based block copolymer scaffolds for skeletal tissue regeneration. In this system due to the suitable porosity and pore sizes cells were homogenously distributed throughout the scaffolds and supported the formation of the cartilage tissue.101 A separate investigation111 showed that human-bone-marrow-derived osteoprogenitor cells, cultured on scaffolds fabricated using PCL and PCL–hydroxyapatite biodegradable composites, developed osteogenic lineage.111 As an example, Woodfield et al.103 have shown that when cellular compatible scaffolds were used for tissue regeneration, due to the attachment, proliferation of expanded human chondrocytes throughout the scaffold and matrix deposition by the cells led to the filling of pores with high cell viability (Fig. 7).103
3.2. 3D-scaffold fabrication and patterning by self-organization
As discussed before, many techniques and methods have been developed for biomedical applications, particularly for the applications of cell-based tissue engineering and biomedical devices. Each of the above mentioned techniques require multiple steps, highly expensive and limited resources of starting materials needed for scaffold and device production. Therefore, there is a need for suitable alternative approaches for 3D structure generation. If we look at our biological nature, certainly one can see a number of examples of fabrication of self-organization of organic and inorganic components under ambient conditions. As an example, one can see that butterfly wings have established interference patterns showing the self-cleaning properties similar to leaves and photonic crystals.112 Another example is the Gecko feet which consists of about 5 × 105 setae and can generate a strong adhesive force which has drawn significant research interest.113,114 Inspired by the 3D pattern existing in biological structures, polymer and biomaterial scientists have developed 3D hierarchical and sophisticated architecture in the order of micron- to nano-structure from functional polymers and biomaterials112–125 alternative to the existing lithography techniques.There are many advantages of generating a suitable 3D structure by the self-organization method. Some examples are as follows: (i) a structure can be generated under physiological conditions, (ii) no toxic chemicals or initiators are needed and, (iii) no requirement of high temperature or UV radiation for curing. Therefore, this self-organization technique can be employed in a variety of biomedical applications. Many research groups have developed reproducible 3D structures of the self-organized honeycomb-pattern with highly regular porous networks using a number of different types of polymers under various conditions,122–132 and their porous network structures have been identified by scanning electron microscopy (SEM), as presented in Fig. 8a and b. This revealed a 3D pattern of a highly regular and uniform honeycomb hexagonal pore structure. This structure comprises top and bottom layers, which are laterally interconnected with nano-scale side pores. The tilted SEM image (Fig. 8b) clearly shows a side-view of two hexagonal lattices connected at the vertices of the hexagons by vertical columns. This double-layered structure reflects the 3D surface morphology of the template, which is self-organized and hexagonally packed. The mechanisms of these hexagonal structures have been described in several published literature reports.124–127 In brief description, a water-immiscible solvent was used to dissolve a polymer, followed by the casting of polymer solution onto a substrate surface and then by immediately evaporative cooling of humid air used. This allowed condensation of water droplets to be deposited onto the surface of casted polymer solution. These water droplets acted as a temporary template for pore generation. The condensed water droplets were unstable and it was essential to stabilise water droplets in order to achieve a highly regular honeycomb pattern surface. For achieving water droplet stability, the amphiphilic polymers were used, which act as surfactants and contribute to the stabilization of the water droplets at the interface of the polymer solution and water, resulting in a highly reproducible and uniform structures. A number of experimental parameters are required to optimise water droplet stabilisation, which includes selection of polymers, concentration of polymers in the water-immiscible solvent, suitability of the solvent and its rate of evaporation, casting volume polymer solution. These parameters ultimately govern the porous network structure, pore sizes and distribution. Researchers126 have demonstrated that uniform pore size can be achieved by altering the parameters of polymer solution casting. The amount of polymer solution used for casting was found to influence the pore size of the fabricated honeycomb films, because the size of condensed water droplets increased with the evaporation time.
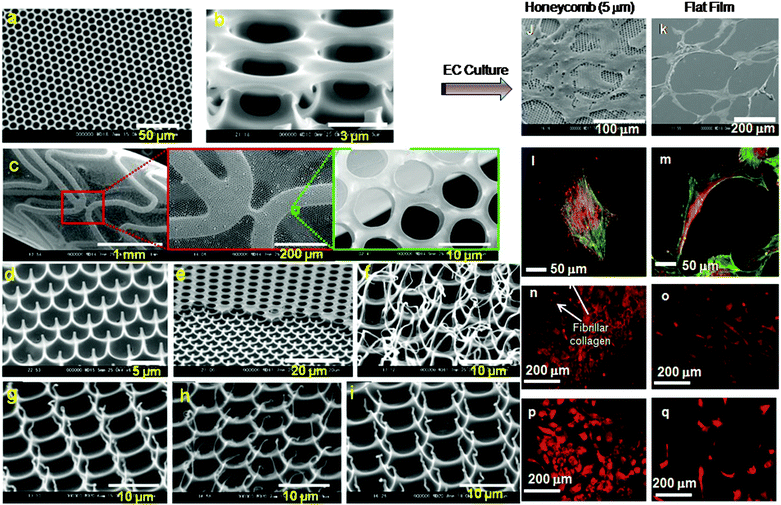 | ||
| Fig. 8 SEM images of the structure of the self-organized honeycomb pattern surfaces and cellular application: (a and b) regular and uniform hexagonal pores (a) top view and (b) tilted view, (c) bile duct stent covered with a vertically open-pored honeycomb film. SEM images of the surface topography of arrays of polymer pincushions arrays (d–i). (d) Tilt-angle (55°) scanning electron micrographs of the surface topography of arrays of PS pincushion arrays formed on the glass surface. (e) A polymer pincushion surface formed from PTFHMA. Only half of the honeycomb film has been formed into a pincushion pattern. Tilt-angle scanning electron micrographs of the polymer pincushions (f) PCL, (g) PLA, (h) PLGA and (i) PHB. SEM images of endothelial cells (ECs) cultured on the honeycomb (j) and flat films (k) for 5 days, (l); (m) cultured on the honeycomb film (pore size, about 5 μm) for 5 days. CLSM images of ECs cultured on honeycomb (l) and flat (m) films. The cytoskeletal protein actin filaments (green) and vinculin (red) are stained using immunofluorescence. Immunofluorescence CLSM images of ECs cultured on the honeycomb film type IV collagen expression (arrow indicates collagen generation) (n), and laminin (p), and on flat film type IV collagen (o) and laminin (q). (a–i) Reprinted with permission.141 Copyright 2009 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (j–q) Reprinted with permission.133 Copyright © 2007 American Scientific Publishers. | ||
For the cell-based biomedical application of the 3D porous network structure, it is critically important to investigate not only cellular attachment, viability and growth after culturing on the scaffold matrix, but also other events such as cell spreading, cell migration, and differentiated cell functions. Thus, the physico-chemical and biocompatible properties of 3D scaffold substrates play a significant role in determining the cellular response. It has been demonstrated in several published papers133–139 that the 3D honeycomb structure of scaffolds has a strong influence on cell proliferation, cytoskeleton, focal adhesion, and extracellular protein generation. As an example, hepatocytes formed spheroids, and synthesised albumin and urea when cultured on the 3D honeycomb scaffold. Researchers have also found that the pore sizes of the scaffold has a significant influence on gene regulation.134 Culturing of endothelial cells (ECs) on the honeycomb scaffolds with 5 μm pore size allowed to provide high levels of proliferation.133 Similarly neural stem cells (NSCs) were cultured on 3D honeycomb materials with a pore size of 3 μm, and were found to undergo accelerated proliferation while such a 3D structure did not support differentiation of NSCs into neurons.136 In addition, the pore size of the honeycomb pattern also affects the mesenteric-visceral adipocyte function and that a honeycomb film with a pore size of 20 μm had the highest cell functions.
It is interesting to note that studies on the growth of cancer cells on the 3D honeycomb surface were also conducted in recent time. It was found that the growth of such cancer cells was much lower as compared to that of a control 2D surface. Hence the surface topography of honeycomb scaffolds possibly has an anticancer effect while culturing cancer cells. Thus, the effects of honeycomb structures on cellular phenotypes depend on the cell lineage type, e.g. ECs, NSCs and other normal, cancer and stem cells, and culture conditions. These were achieved in a culturing media, which do not contain growth factors. The results of the investigations suggest that the honeycomb structure with different pore sizes could regulate the cell adhesion, morphologies, and functions while no growth factors were used. Recently, a vertically open-pored film support for the tubule given by a metallic tubular mesh has been commercialized as a bile duct stent (Fig. 8c). Co-culture of ECs and smooth muscle cells on the inner and outer side of the tubular honeycomb film are expected to find applications in novel cardiovascular stents and artificial vessels.
Using a simple method of polymer solution casting on a glass substrate and peeling off adhesive tape a completely different 3D architecture of polymer pincushion arrays can be achieved.140 Such 3D surfaces, having nano-and micro-structures, are suitable for specific cell-based tissue engineering and drug delivery. The mechanical properties and biodegradability of such scaffolds should resemble those of healthy tissues during tissue regeneration. Hexagonal arrays of biodegradable polymer pincushions was developed using biodegradable, and biocompatible polymers such as polycaprolactone (PCL), poly(lactic acid) (PLA), poly(D,L-lactide-co-glycolide) (PLGA), and poly(3-hydroxybutyrate) (PHB),141 which are U.S. Food and Drug Administration (FDA) approved materials. Thus the use of such materials in medical devices can avoid FDA hurdles in clinical application.
The pincushion structures are confirmed by microscopic analysis (Fig. 8d and e), which shows that each pore is surrounded by six pincushions with a diameter of approximately 0.1–0.5 μm. The tilted SEM analysis of pincushion structures (Fig. 8f–i) showed vertically and hairy aligned morphologies. The heights, widths, and distances of separation of the pincushions were dependent on the type of polymer used and the pore size of the original honeycomb film. For example, PCL pincushions (Fig. 8f) showed an elongated hair-like morphology as compared to the pincushions generated from other polymers. Both sharp and hairy pincushion structures could be controlled by peeling off at a certain temperature above or below glass transition temperature. Such structural and morphological differences could be attributed to the polymers viscoelastic and mechanical properties, and their interaction with glass surfaces.
The generation of 3D pincushions was performed under physiological conditions with simplicity, flexibility and cost effectiveness and different from other techniques. It has been demonstrated that the nano-structured surfaces are utilized for the long-term maintenance of the stem cell phenotype and multipotency,142 and such structures have a positive influence on cell- and material-based therapeutic applications.87
As discussed earlier that the honeycomb structure has a significant influence on cellular behaviour as compared to that of the flat surface, an example is presented in Fig. 8j–q for endothelial cell growth and differentiation. Immunohistochemical analysis (Fig. 8n–q) revealed a remarkably high extracellular matrix protein production when EC cultured on honeycomb film with 5 μm pore surface as compared to that of the flat surface.133
3.3. 3D scaffolds by self-assembly peptides
Peptides are naturally inspired materials, synthesised from the sequence of the amino acid monomers that carry a carboxyl and an amine functional group on the chain. The peptides are designed both from natural and synthetic amino acids; they link together to form short peptides and then long polypeptide chains in a controlled manner.143 The functional groups such as amines (NH) and carbonyls (CO) present in the peptide chain allow further chemical reactions to be performed with functional groups such as thiols and alcohols, and can be combined with a wide range of materials such as lipids, sugars, nucleic acids, metallic nanocrystals and many more.144 Moreover, the peptides have excellent properties such as biocompatibility, resistance to extreme conditions of high and low temperatures, detergents and denaturants.144 Thus the peptides are capable of a wide range of chemical interactions and molecular recognitions, forming various non-covalent interactions in water, including hydrogen bonding, ionic, π–π interactions, hydrophilic and hydrophobic. These interactions lead to the formation of supramolecular self-assemblies that can give rise to a variety of 3D nano-structures such as nano-fibers, nanotubes, and nanoparticles.145,146In the last two decades, significant advances have been made on the self-assembly peptides (SAPs), and continue to expand rapidly worldwide as a fundamental part of nano-structure generation.143,147 Now, these SAP systems are reaching a wide range of applications in biology, drug delivery, nanobiotechnology and nanoelectronics.
However, their use in technological applications is facing several challenges, which include (i) the precise positioning of peptide-based nanostructures, (ii) their controlled assembly and positioning, and (iii) their integration into microsystems. Until now, the positioning of the SAPs has been limited on flat surfaces and the fabrication of peptide arrays.
Dinca et al.144 demonstrated that SAPs, with unique physical and chemical stability, are capable of functioning as a template for the fabrication of low resistance, and conducting nanowires. In this research, they proposed a methodology for the precise, 3D patterning of amyloid fibrils with a combination of laser technology and biotin–avidin mediated assembly on a polymer surface. They also suggested that this method can be applied from molecular electronics to tissue engineering. In this section, we focused on the use of SAPs for cell-based tissue regeneration.
In TE, SAPs with low-molecular-weight peptides (oligopeptides) are capable of creating microenvironments suitable for cell culture,148,149 and tissue regeneration.150,151 Several researchers152,153 have reported that SAP nanofibre scaffolds promoted optic nerve regeneration. These SAP nanofibre scaffolds are formed spontaneously from individual peptides by interacting with physiological salts and, are entirely biocompatible.152 Such scaffolds composed of Arg–Ala–Asp–Ala (RADA) oligopeptides utilised in in vitro PC12 cell culture which promoted neurite outgrowth and synapse formation by hippocampal neurons.153
Kisiday et al.148 have investigated the SAP hydrogel constructed with positively charged lysines (K), negatively charged aspartic acids (D) and hydrophobic leucines (L) of twelve units, termed as KLD-12. This hydrogel is utilised for encapsulation of chondrocytes. Chondrocytes seeded within the SAP hydrogel retained their morphology and developed a cartilage-like ECM rich in proteoglycans and type II collagen, in 28 days of in vitro culture period. They have also demonstrated that the SAP hydrogel is a potential scaffold for the biosynthesis of the extracellular matrix (ECM) and glycosaminoglycan (GAG) accumulation within a 3D cell culture for cartilage tissue repair. The SAP-based hydrogels can also be used to incorporate bioactive molecules via chemical conjugation to different moieties to allow signaling to cell surface receptors and to enhance cellular adhesion and function.
3.4. Polymeric biomaterial mediated cell manipulation
Research on various types of cell encapsulation methods in a variety of polymeric biomaterials, particularly hydrogels derived from Matrigel,154 collagen,155 alginate156 and blends of CS and polyethylene imine (PEI)8 have been investigated. Polymeric scaffolds and biomaterials used in TE to mimic the natural extracellular protein matrix and to provide structural support and cellular functions required for new tissue generation.157Hydrogels are capable of assisting neural regeneration158 (Fig. 9a), allowing human neural stem cells (hNSCs) to differentiate between neurons and glial cells. The conditions of gel formation are needed to be optimised for Matrigel and PuraMatrix, and mechanical properties are also important for such a gel to support hNSCs following transplantation into the injured brain or spinal cord. Several other studies have demonstrated human embryonic stem cell (hESC) culturing in well-defined 3D settings by using a variety of scaffolds for cellular functioning, cell viability and lineage guidance. The hydrogels synthesised from naturally derived polysaccharides such as hyaluronic acid (HA) supported hESC growth in vitro (Fig. 9b),155 because they co-regulate gene expression, signalling, proliferation, motility, adhesion, metastasis, and morphogenesis of hESCs in vivo. In humans, the HA content is the greatest in undifferentiated cells and during early embryogenesis and then decreases at the onset of differentiation, where it plays a crucial role in the regulation of the angiogenic process. It has been demonstrated155 that when the hESC is encapsulated in 3D hydrogels, prepared from HA, hESCs maintained their undifferentiated state (Fig. 9b),155 and preserved their normal chromosome state in the cell nuclei. hESCs in hydrogels maintained their full differentiation capacity by embryoid body formation while these cells can be differentiated within the same hydrogel by incorporating soluble factor molecules. Thus HA hydrogels, with their developmentally relevant composition, tuneable porosity, pore sizes and mechanical strength, provide a unique microenvironment for the self-renewal and differentiation of hESCs. The 3D structural biomaterials, developed from synthetic materials, had been tested for self-renewal of hESCs for a limited period. This has shown that much research and development is needed to design a robust synthetic material system associated with relevant bioactive molecules to support long-term the hESCs.
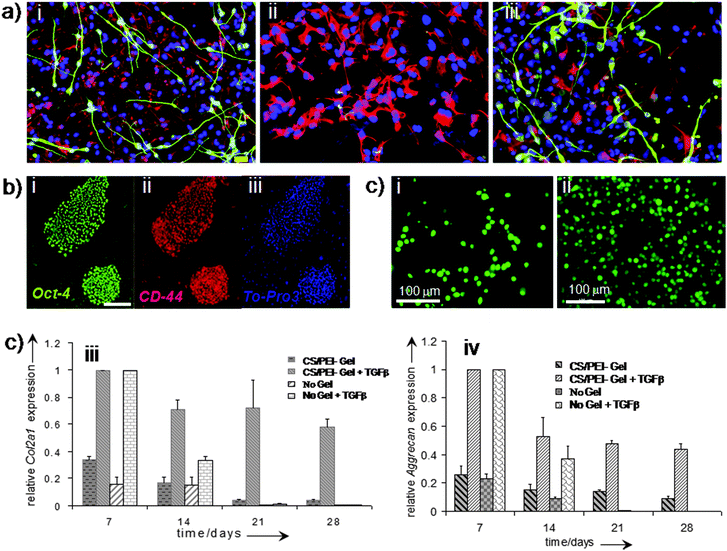 | ||
| Fig. 9 Polymer hydrogels for cellular function. Differentiation capacity of human neural stem cells beneath Matrigel and PuraMatrix. Immunofluorescence images of human neural stem cells primed for 5 days and then differentiated for 7 days in the absence of a hydrogel (control, a-i); in contact with 20% Matrigel (a-ii); and in contact with 0.25% PuraMatrix (a-iii). Overlay of DAPI (blue), GFAP (red) and Tuj1 (green) and overlay. Scale bar = 20 μm (a). Hyaluronic acid (HA) plays a role during hESC culture on MEFs (b). Staining of hESCs (H1 line) grown on MEFs for HA binding site (green), undifferentiated membrane marker TRA-1–81 (red), and nuclei (blue). Scale bar = 100 μm (b). (c) Human fetal skeletal cells, labeled with CellTracker Green, grown in the hydrogel scaffold (chitosan/PEI 40:60): (c-i) day 7 and (c-ii) day 21. Analysis of chondrogenic gene expression (Col2a1 and Aggrecan) by fetal skeletal cells cultured within chitosan/PEI hydrogels and in monolayers over a course of 28 days with and without TGF-β3. Relative gene expression levels were normalised to the expression of β-actin, which served as a house-keeping gene. The group with the highest expression was assigned a value of 1 and expression levels in the remaining groups were determined relative to that group. Fold relative expression levels were expressed as the mean ± SD for plotting as bar graphs, n = 4 for monolayer cultures and n = 3 for hydrogel cultures. (a) Reprinted with permission.158 Copyright © 2007 Elsevier B.V. (b) Reprinted with permission.155 Copyright © 2007 by The National Academy of Sciences of the USA. (c) Reprinted with permission,8 Copyright © 2009 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Significant research interest has been drawn both in academia and in biotechnology industries to replace fully or partially biologically derived native materials with synthetics. Materials of biological origins have several drawbacks, such as high cost, batch to batch variation and, sometimes, uncertainty of component identification. While synthetic materials are highly reproducible without variations between batches, and economically viable.
Fischbach et al.159 developed a synthetic 3D polymer scaffold to engineer 3D human tumour models using carcinoma cells. Human umbilical vein endothelial cells (HUVECs) were cultured on 2D and 3D surfaces produced from poly(lactide-co-glycolic acid) (PLGA) and Matrigel biodegradable polymers, and their analysis was performed by means of proliferation and differentiation. They exhibited angiogenic potential and cell proliferation of 3D surfaces to be remarkably higher than those from other cultured conditions.159
Recently we have demonstrated8 that water soluble polymer blending of chitosan and PEI can provide scaffold degradation behaviour after implantation. Polymer solutions can be mixed with cells before the gelling process can take place allowing cell migration and proliferation throughout the 3D hydrogel scaffold. CS and PEI have been found to support the growth of human fetal skeletal cells within the 3D gel with suitable mechanical properties. The porosity of gels facilitated cell proliferation and prevented dedifferentiation of the skeletal cells into fibroblasts by maintaining these cells in a chondrocyte-like spherical morphology (Fig. 9c).8
3.5. Cartilage
Cartilages are tough, flexible tissues, which act as shock absorbers. These cover the surface of joints found throughout the body and facilitate bones to slide over one another with reduced friction, and damage. There is no blood supply through cartilage unlike other tissues such as skin or muscle, which makes it difficult to regenerate damaged cartilage tissue. Articular cartilage lies between joints such as knee joints where the most common and serious damage occurs, resulting in pain, swelling, and some loss of mobility. Therefore, it is essential to develop a 3D scaffold matrix for repairing cartilage tissues for clinical applications.Culturing of mesenchymal stem cells (MSCs) on a variety of TE scaffolds facilitating chondrogenesis and formation of cartilage have been reported in several research papers.160–164 However, there appears to have some limitations in achieving identical properties of native cartilages. Additionally, generation of functional cartilage by MSCs has been found to be troublesome, as it depends on the viable cell source for extracellular cartilage matrix production, leading to high quality cartilage regeneration.164 It is noted that the uniform distribution of such a matrix, generated by MSCs is essential for the optimum mechanical strength of the tissue. Therefore, appropriate design of 3D structured biomaterials to support uniform distribution of formed tissue is essential for effective cartilage formation by MSCs.
Studies performed both in vitro and in vivo165 have shown that culturing of MSCs on a functionalised HA-based hydrogel by the crosslinking method maintains chondrocyte viability and chondrogenic differentiation. It was, however, reported that the ECM distribution was not homogeneous due to the unmet degradation rate of hydrogels as a function of ECM production. In an ideal scenario, the tissue engineering of 3D scaffold degradation should match with ECM production by the cells and their accumulation. The degradation rate of scaffolds affects the diffusion of nutrients and waste, cell–cell communication, cell–material interactions, and the distribution and retention of the ECM. Therefore, to control the rate of degradation of the 3D scaffold it is important to select an appropriate crosslinking procedure out of the following: UV-induced crosslinking, chemical crosslinking, and to select appropriate density of crosslinking, or of the functional group (if copolymerised) onto the backbone of HA. The MSCs, cultured within HA functionalized hydrogels, showed a rounded cell morphology.166 It is also reported that the tuning of physical and mechanical properties of scaffolds can control neocartilage formation. For tissue regeneration, the hydrogel scaffolds must control two important properties, i.e. mechanical stability and the degradation rate. These can be achieved via crosslinking of acrylate and aldehyde groups, which will lead to the repair of cartilage.167 Modified CS biodegradable hydrogels have been developed,168,169 and the biocompatibility was assessed by culturing chondrocytes on the hydrogel scaffold in which cells exhibited clustered growth and produced the extracellular matrix on CS gel under in vitro conditions. This CS gel–chondrocytes promoted cartilage regeneration defects in rabbits.168 However, development of hydrogels with high mechanical strength for cell encapsulation and 3D culture is a challenging task for cartilage tissue engineers. Therefore, double network and/or interpenetrating network structures of polymer hydrogels170–172 are now considered to be potential candidates for cartilage TE.
Polymer substances of natural origin, such as collagen, alginate, silk fibroin, agarose, etc., were also used to design and fabricate scaffolds in a wide variety of forms, including meshes, sponges, foams, hydrogels, glues, composite layers, biotextiles, nanofibers and microspheres.171–176 Various synthetic polymeric materials have been used to fabricate scaffolds for cartilage repair. These included PLA, PGA, and PLGA copolymers, and PEG or PPO polymers. These were found to form gels, ceramic composites and hydrogels containing PEG polymer-based derivatives at different temperatures.177 A list of polymer scaffolds fabricated by using a variety of techniques and used in pre-clinical animal and clinical human trials in cartilage tissue engineering178–186 is presented in Table 1.
| Materials types | Scaffold fabrication technique | Application | |
|---|---|---|---|
| Pre-clinical animal trial | Human clinical trial | ||
| Poly(L-lactide), poly(L-lactide-co-glycolide)177 | Leaching | Cartilage in mice | — |
| Poly(L-lactide-co-glycolide)/collagen178 | Freeze drying | Cartilage in mice | — |
| Poly(glycolic acid), PCL, poly(hydroxyl butyrate)179 | Solvent casting/leaching | Neocartilage in mice | — |
| Poly(ethylene glycol)–poly(butyl terephthalate) copolymer180 | Robotic deposition | Cartilage in mice | — |
| Hyaluronan-based181 | — | — | Chondrocyte transplantation for cartilage tissue |
| Collagen, HA, alginate, PLA182 | — | — | Cartilage tissue |
| Hyaluronan-based184,185 | — | — | Chondrocyte implantation; transplantation |
| Collagen244 | Freeze-drying | Tibia defects/bone in rats | — |
| Collagen/hyaluronate245 | Cross-linking | Cranial defects/bone in rats | — |
| PLGA/poly(vinyl alcohol)246 | Leaching | Cranial defects/bone in rabbits | — |
| Poly(propylene glycol-co-fumaric acid)247 | Gas foaming with effervescent reaction (in vivo) | Cortical defects bone in rats | — |
| PCL/PLLA179,248–250 | Solvent evaporation | Trabecular bone in mice and sheep | — |
| CS/poly(vinyl acetate)/PLLA blends87 | Freeze drying | Trabecular bone in mice | |
| Gelatin277 | Leaching | Artificial skin in mice | — |
| Poly(ethylene glycol), cystamine and PCL280 | Crosslinking | Connective tissue in rats | — |
| Silk281 | Freeze drying | Ligament in pig. | — |
3.6. Bone
The research and development in bone tissue engineering,187–197 using a combination of cells, factor molecules, and supportive 3D matrices, have gained momentum in recent years. Biomimetic and biodegradable polysaccharide scaffolds derived from chitosan,198–201 hyaluronic acid (HA),202–206 and alginate207–210 have been developed for bone tissue engineering application. However such materials, in their pure form, have mechanical weakness, instability and lack of remaining predefined shape and thus have limited applications211 in TE. Therefore, to improve their properties several research groups have developed copolymers,212,213 blends and composites214–218 of CS, HA and alginates for bone TE.Recently, several research groups have reported219–227 on a variety of biodegradable synthetic polymer scaffolds for bone TE, such scaffolds include PCL,219–221 poly(lactic acid),222,223 and their copolymers.224–229 It has been concluded that these polymer based scaffolds have some advantages over ceramic and glass based ones, primarily because the properties of the polymer based scaffolds can easily be processed and tailored to obtain suitable geometry for implantation. The major drawbacks with polymer scaffolds are low mechanical strength, shape retention failure, insufficient cell adhesion and growth, and hence, require surface modification with functional groups or incorporation of bioactive materials to form multicomponent biocompatible composite bone scaffolds230–243 to enhance osteogenicity244 for ultimate bone tissue engineering.
Recently very promising polymer based scaffolds have been developed and pre-clinical trials have been conducted (see Table 1).245–250 This showed that superior biocompatibility, biodegradability and high mechanical strength, and growth factors can be incorporated within the scaffold materials to enhance bone formation.86,87,249,250 Naturally derived polymers, particularly polysaccharides, have found a wide range of applications in biomedical technology as signalling molecules such as peptides, and proteins can easily be incorporated in these via chemical processing. Additionally, these are found to interact well with inorganic components and provide a very akin environment for cells to grow. Gels, crosslinked with inorganic components, themselves tend to be processed using simple chemical processes and can be introduced into the body through a minimally invasive surgery.189 In recent years, various designed material constructs have been developed in our group, using a blending approach of multi-component polymers for bone tissue engineering.86,87,249,250
3.7. Neural tissue engineering (NTE)
Physical injuries to the central nervous system (CNS), which can be caused by severe accidents and neurodegenerative diseases like Parkinson's and Alzheimer's diseases, disrupt brain architecture. As a result severe functional disorders may ensue due to the loss of neuronal cell bodies, axons, and associated glia supports. Regeneration of damaged neural tissue, because of their complex structure and functioning, is a highly challenging task in the global healthcare system in the field of tissue engineering applications.Currently the autologous nerve grafting approach has been used clinically to repair nerve defects. It is well known that such a clinical approach has two major disadvantages: (i) loss of function in the donor nerve graft sensory distribution and (ii) the geometrical mismatch between the damaged nerve and the nerve graft. Thus, there is a need for the neural TE strategy to be developed, focusing on 3D scaffold generation with favourable neural cell growth that facilitates regeneration. Several researchers have utilised scaffolds for enhancing regeneration within the CNS, and generated promising results.251 With the aim of nerve regeneration, several research groups have, independently, developed a variety of polymeric templates.251–259 For example, Tsai et al.251 have synthesised the poly(2-hydroxyethyl methacrylate) (PHEMA)–methyl methacrylate (MMA) hydrogel, which enables to incorporate growth factor molecules. This copolymer system combined with growth factors has allowed spinal cord injury repairing in animal models.
Recent studies suggested that stimuli responsive soft materials, especially electrically stimulated hydrogels, have played a significant role in the proliferation and differentiation of nerve cells.252–254 The neurite extension and outgrowth were substantially enhanced on electrically conducting polymer hydrogels in different culturing media. The effect was found to be more prominent in negatively charged polymeric materials than in positively charged or neutral252 ones. The neutral polymeric hydrogels (e.g. PEG and PHEMA), functionalized with ionic compounds to form ionic hydrogels, are able to bridge a spinal cord lesion when implanted inside a hemi-section cavity. HEMA-based hydrogels with charged functional groups, either cationic or anionic, have the ability to enhance axonal regeneration inside the implant, and surprisingly, no charge was observed when minimal axons infiltrate hydrogels.253 Researchers have also found that implanted hydrogels with positively charged groups increased axonal ingrowth into the central part of the implant. Astrocytes infiltrate only those hydrogel implants comprising negative charge or the neutral group, most of which are found only in the peripheral zones. Functional groups on the backbone of the HEMA hydrogel with different surface charges and density of charge influence the interaction between cells and materials and cellular functioning254 and consequently improve the quality of nerve regeneration. Therefore, conductive polymer-based material-aligned scaffolds260–272 and the incorporation of carbon-based nanomaterials270–277 into polymeric scaffolds have been investigated for neural tissue growth. Such acrylate-based hydrogel polymers are classified as non-biodegradable materials, due to the lack of desirable features to be used in TE as a scaffold.
Biodegradability of polymer scaffolds plays an important role in TE. They act as temporary scaffolds holding the growing tissue in place until the natural ECM has sufficiently developed. The scaffold breakdowns into nontoxic degradable products which are capable of being disposed of by the body leaving behind the newly formed tissue. There are a number of natural and synthetic biodegradable polymers such as collagen, HA, chitin and chitosan, PLLA and PLGA that are explored as scaffolds for NTE application.278,279
Biocompatible polymeric hydrogels and scaffolds have also been investigated for the regeneration of various other tissues, as shown in Table 1, such as artificial skin,280 connective tissue and ligaments.281,282
3.8. Growth factors (GFs) incorporated in the hydrogel and angiogenesis
A growth factor is defined as a naturally occurring protein or steroid hormone that binds to specific receptors on the surface of their target cells. GFs can play a role in a variety of physiological processes, such as new blood vessel generation, phenotypic activities of cells, tissue development and healing, wound healing and treatment of myocardial and hind limb ischemia.283–292 However, the stability of GFs is a critical factor in the above processes when administered in vivo. Therefore, a suitable delivery system to improve stability is needed in order to promote neo-vascularization at a local tissue site. The hydrogel polymer has been found to influence controlled release of such factor molecules.293,294For optimum performance of GFs, it is necessary to combine these with carrier molecules in order to release it in a controlled manner. Although some success of the current clinically available GF delivery devices have been reported in some TE fields, these are not even near enough to an ideal system, demanding further research on efficient and sustainable delivery devices. The clinical technologist and the researchers within the biotechnology industry are enthusiastically looking for systems for controlled and efficient delivery using lower doses of GFs and for the production of a more sustained release pattern to serve as a more effective 3D scaffold surface with a structural support in tissue engineering. An in-depth understanding of tissue-healing processes is, therefore, needed to allow us to design new suitable delivery systems for GFs. Additionally, the processes of normal tissue-healing needed to be biologically optimized so that there are sequential overlapping stages for the transition from immature to mature (definite) tissues. Logically, mimicking both the structures and the sequence of the tissue-healing process should be the best option for designing biomaterials for TE. This is mainly because of their ability to initiate the body's natural tissue-healing cascades at the site of injury. A number of GFs that have been studied in biomedical applications to enhance TE and angiogenesis in recent time are included in Table 2.
| Growth factors | Tissue engineering application | Function |
|---|---|---|
| Note: Ang (angiopoietin), BMP (bone morphogenetic protein), EGF (epidermal growth factor), EPO (erythropoietin), FGF (fibroblast growth factor), G-CSF (granulocyte-colony stimulating factor), GM-CSF (granulocyte-macrophage colony-stimulating factor), HGF (hepatocyte growth factor), IGF (insulin-like growth factor), GDF (growth and differentiation factor), NGF (nerve growth factor), PDGF (platelet-derived growth factor), TGF (transforming growth factor), TPO (thrombopoietin) and VEGF (vascular endothelial growth factor). | ||
| Ang-1283 | Blood vessel, heart, and muscle | Promote maturation and stability of blood vessel. |
| Ang-2283 | Blood vessel | Destabilize, regress and disassociate ECs from surrounding tissues. |
| BMP-2283,296,297,301 | Bone and cartilage | Promote differentiation and migration of osteoblasts. |
| BMP-7283,296,297 | Bone, cartilage and kidney | Enhance differentiation and migration of osteoblasts, as well as renal development. |
| BMP-9307 | Bone | Enhance osteogenic differentiation and bone formation. |
| EGF283 | Dermal tissue, epithelia tissue, and nerve healing | Maintaining epithelial cell growth, proliferation and differentiation. |
| FGF-2283 | Blood vessel, bone, skin, nerve, spine, and muscle | Induce angiogenesis; enhance the formation of blood vessels; migration, proliferation and survival of ECs; inhibition of differentiation of embryonic stem cells. |
| EPO283 | Nerve, spine, and wound healing | Promoting the survival of red blood cells and development of precursors to red blood cells. Protect the myocardium from ischemic injury. |
| G-CSF308 | Prevents common chemotherapy complications | Peripheral artery disease and critical limb ischemia. |
| GM-CSF308 | Epithelial tissues | Circulating leukocytes, act as a paracrine fashion to recruit circulating neutrophils, monocytes and lymphocytes to enhance their functions in host defence. |
| HGF283 | Bone, liver, and muscle | Promoting proliferation, migration and differentiation of MSCs. |
| IGF-1283 | Muscle, bone, cartilage, liver, lung, kidney, nerve, and skin | Cell proliferation and inhibition of cell apoptosis. |
| GDF-5309 | Central nervous system | Increasing the survival of neurons. |
| GDF-8 (myostatin)310 | Muscle | Control the growth and differentiation of muscle cells. |
| GDF-9311 | — | Regulating ovarian follicular growth. |
| GDF-10312 | Skeletal | Skeletal morphogenesis. |
| GDF-11313 | Muscle | Regenerative capacity of satellite cells. |
| NGF283 | nerve, spine, and brain | Survival and proliferation of neural cells. |
| PDGF-AB (or BB)283 | Blood vessel, muscle, bone, cartilage, and skin | Function for embryonic development, proliferation, migration, growth of ECs. |
| TGF-α283 | Brain and skin | Assisting proliferation of basal cells or neural cells. |
| TGF-β283 | Bone and cartilage | Enhancing proliferation and differentiation of bone-forming cells, antiproliferative factor for epithelial cells. |
| TPO314 | Liver | Hepatic progenitors during fetal liver development. |
| VEGF283,295 | Blood vessel | Migration, proliferation and survival of ECs. |
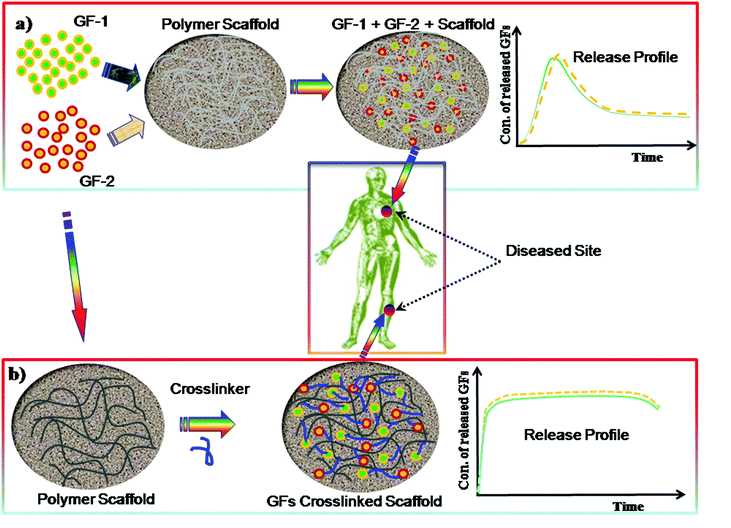
Polymeric hydrogels play a significant role as ECM scaffolds in serving as a matrix for bioactive molecule delivery to the cells as well as regulating cellular activity. The GF can be incorporated within the hydrogel matrix by crosslinking during the preparation of the gel and can control the sustained release, as demonstrated in Fig. 10. Polymer hydrogels are highly porous network structures through which cell migration, cell proliferation and cell–cell communication take place. Lowe and co-workers293 described that cells communicate with the ECM via signalling pathways through integrins which eventually can alter gene expression, resulting in cell migration, differentiation, proliferation or apoptosis. Several studies157,295 show that vascular endothelial growth factor (VEGF) is one of the most important growth factors for repairing many types of tissues. Other examples of GFs are bone morphogenic proteins, such as BMP-2 and BMP-7, which have shown bone formation in clinical use.296,297 However, such factors still remains unsafe298,299 and highly expensive.300
Protein based hydrogels have been used to deliver bone morphogenetic protein 2 (BMP-2) for skull defects.301 However the main disadvantages of the protein based hydrogels are the control of their degradation as most of them are derived from animal products, such as Matrigel. Therefore, current research has been devoted to the development of synthetic based polymer hydrogels. A class of synthetic based materials, namely injectable polymer hydrogels derived from the oligo(poly(ethylene glycol)fumarate) (OPF), have been developed by Park and co-workers,302 for the delivery of GFs to the cells. The hydrolysis of OPF hydrogels can degrade to the ester bonds in the fumarate group.
Recently, the thermo-reversible polymer hydrogels have attracted considerable attention both in academia and industries, particularly for TE scaffold technology and drug delivery.303 Cells cultured in thermo-reversible hydrogels demonstrated higher viability and enhanced cellular functions.297 The research has shown that the thermo-reversible polymers were useful as an injectable hydrogels. However, very limited studies in ‘in vivo’ tests for TE, involving growth factors incorporation have been conducted so far.304 Therefore, an injectable, in situ crosslinkable, biodegradable and thermo-reversible, hydrogel is needed for minimally invasive delivery of therapeutic molecules to the localised cells and tissues. The in situ crosslinking approach of injectable hydrogels, with or without cells into the infracted myocardium, shows improvement in neovascularization and heart function and enhanced angiogenic response.292 In this research, Researchers reported that injectable alginate-based hydrogels with and without RGD – modified alginate, increased the arteriole density as compared to that of controlled one with the RGD modified alginate having the greatest angiogenic response. In this case alginate biopolymers act as synthetic ECMs and RGD peptides as a cell–matrix mediator, which increase cell binding affinity and the effect on cell behaviour through integrin–ligand interactions. Several pre-clinical studies have demonstrated that angiogenic GFs can stimulate the development of collateral arteries in animal models of peripheral and myocardial ischemia.303
An in vivo study shows that the CS based hydrogels could be useful for gradual release of the fibroblast growth factor-2 (FGF-2) molecules as they biodegraded in vivo.304 Researchers also described that releasing FGF-2 molecules from the hydrogels caused induction of angiogenesis and collateral circulation occurred in healing impaired diabetic (db/db) mice and in the ischemic limbs of rats. However the sustained release and local delivery of GFs will certainly depend on the nature and characteristics of the hydrogel and the method of fabrication and process. The GFs can be crosslinked with the gel materials by either physical interactions or chemical reactions. Recently it has been demonstrated that photo-induced crosslinkable and biodegradable pluronic/heparin composite hydrogels were synthesised with a specific objective for local and sustained delivery of basic fibroblast growth factor (bFGF) to induce angiogenesis.305
However, there are still several concerns that GFs have adverse effects, especially for using high levels of doses. Exposure of the myocardium to high local levels of GFs can cause hemangioma-like tumors, vascular malformations, and neointimal development.306 To minimize such adverse effects, dose reduction of GFs and their controlled delivery would be important strategies in this field.
4. Conclusions and future prospects
Numerous efforts have been made globally in the last two decades to bring laboratory-based ideas into the clinical trial stage followed by clinical procedures in biomedical applications. Currently biomaterial technology within the overall healthcare system is receiving benefits as a result of the multidisciplinary field of research, albeit often with disappointing outputs. For achieving the ultimate goal, however, many challenges need to be addressed and overcome. For TE applications, the production of more complex scaffold materials with biomimetic properties and mechanical stability is necessary. Designing and fabrication of biomimetic synthetic scaffolds, aimed at producing biofunctional synthetic matrices to enhance the cellular function and leading to high quality tissue development, are also issues to be addressed. Combining multiple physical and chemical approaches by incorporating suitable functionalities into the molecular chain of polymeric materials is expected to provide complex scaffolds with architectural-hierarchy, which will enable them to mimic the cellular environment, exchange information with cells and enhance cell–cell communication.To date, the examples of technologically advanced biomaterials have been the multi-component polymer hydrogels derived from various functional monomers, polymers or oligomers, synthesized either by physical or chemical crosslinking. Such hydrogel systems are expected to find potential use in a variety of areas including the regeneration of tissues, and the delivery of bioactive molecules (e.g. growth factors, drugs). However, a number of hurdles, such as biocompatibility, mechanical strength, rate of degradation, etc., also need to be addressed and overcome for effective TE applications. For such applications, the materials with controllable mechanical properties, degradation profiles and 3D structure, which could easily be modified to suit a particular tissue purpose, need to be developed. Now we can generate 3D synthetic scaffolds with appropriate structural properties that actively support cells to form tissue in defect sites. For example, the polymer scaffolds have been found to generate new bone formation when implanted into a defect site in rat femora without the use of expensive growth factors.86,87,249,250 Naturally derived polymeric biomaterials are employed in a wide range of applications; some device materials are commercially available and such polymers most likely to remain a best candidate for further research and evaluation in biotechnology. In the future, the research is expected to continue on the processing of the 3D structure and product development using naturally derived polymers, perhaps by combining this with synthetic polymers, for the appropriate tissue type.
To generate the 3D structure of scaffolds, various production techniques and methods using different types of polymeric materials need to be employed. Each technique has a particular processing method and multiple steps and has advantages and disadvantages. Therefore, the choice of the technique for 3D fabrication will depend on the nature and type of material, its structure and properties, interaction with the substrate and finally, the intended applications. The robotic technique is considered to be the best choice in an application where a complex 3D architecture scaffold is needed as the generation of such a structure is not possible by other techniques. However, some of which are still in an early stage of development and require significant improvement in the bio-chemical technology as well as an in-depth understanding of the basic processes involved. The latter will provide tools to generate structures with best performance. The challenges are the improvement of 3D patterning efficiency with high resolution and low-cost pattering, with good levels of performance. To achieve cost-effective 3D patterning and to achieve patterns on multiple length scales, a combination of different techniques will be necessary. Recently, self-organization techniques have shown to be a very promising approach for the cost-effective fabrication of the 3D honeycomb structure with exiting in vitro and in vivo results, but more research is needed in this area. Self-assembly peptides nanostructures are very promising biomimetic materials and could be used in various fields of TE, but still in their early stage of development. Incorporation of bioactive molecules into the 3D polymer hydrogels/scaffolds and their release in a controlled manner towards targeted tissue will provide a powerful methodology to study and manipulate developmental and regenerative processes. This will depend on the biological demand for the target tissue.
Harnessing the potential of this technology for clinical use strongly depends upon more research studies and multidisciplinary approaches that combine engineering, biomaterials, medicine and the technical expertise of medical specialists. Working in close collaboration between polymer chemists, materials scientists, tissue engineers and reconstructive surgeons may eventually help to achieve clinical excellence and products for a range of degenerative diseases, for ultimate improvement in the quality of life.
Acknowledgements
We greatly acknowledge the financial support from BBSRC, Scottish Stem Cells network (SSCN), and Funding Program for Next Generation World-Leading Researchers (NEXT Program, Japan).Notes and references
- O. Wichterle and D. Lim, Nature, 1960, 185, 117 CrossRef PubMed.
- I. C. Kwon, Y. H. Bae and S. W. Kim, Nature, 1991, 354, 291 CrossRef CAS PubMed.
- A. S. Hoffman, Adv. Drug Delivery Rev., 2002, 43, 3 CrossRef.
- D. Shi, Introduction to Biomaterials, World Scientific, Tsinghua University Press, 2006 Search PubMed.
- W. G. Liu, M. Griffith and F. Li, J. Mater. Sci.: Mater. Med., 2008, 19, 3365 CrossRef CAS PubMed.
- F. Yang, Y. Wang, Z. Zhang, B. Hsu, E. W. Jabs and J. H. Elisseeff, Bone, 2008, 43, 55 CrossRef CAS PubMed.
- E. Eljarrat-Binstock, F. Orucov, J. Frucht-Pery, J. Pe'er and A. J. Domb, J. Ocul. Pharmacol. Ther., 2008, 24, 344 CrossRef CAS PubMed.
- F. Khan, R. S. Tare, R. O. C. Oreffo and M. Bradley, Angew. Chem., Int. Ed., 2009, 48, 978 CrossRef CAS PubMed.
- C. R. Nuttelman, M. A. Rice, A. E. Rydholm, C. N. Salinas, D. N. Shah and K. S. Anseth, Prog. Polym. Sci., 2008, 33, 167 CrossRef CAS PubMed.
- W. E. Hennink and C. F. van Nostrum, Adv. Drug Delivery Rev., 2002, 54, 13 CrossRef CAS.
- K. R. Kamath and K. Park, Adv. Drug Delivery Rev., 1993, 11, 59 CrossRef CAS.
- S. J. Hollister, Nat. Mater., 2005, 4, 518 CrossRef CAS PubMed.
- A. S. G. Curtis and C. D. W. Wilkinson, Biomaterials, 1997, 18, 1573 CrossRef CAS.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997 CrossRef CAS PubMed.
- A. Asefnejad, M. T. Khorasani, A. Behnamghader, B. Farsadzadeh and S. Bonakdar, Int. J. Nanomed., 2011, 6, 2375 CrossRef CAS PubMed.
- R. M. Allaf and I. V. Rivero, J. Mater. Sci.: Mater. Med., 2011, 22, 1843 CrossRef CAS PubMed.
- C. Z. Wang, M. L. Ho, W. C. Chen, C. C. Chiu, Y. L. Hung, C. K. Wang and S. C. Wu, Mater. Sci. Eng., C, 2011, 31, 1141 Search PubMed.
- A. Saraf, L. S. Baggett, R. M. Raphael, F. K. Kasper and A. G. Mikos, J. Controlled Release, 2010, 143, 95 CrossRef CAS PubMed.
- E. Reverchon, S. Cardea and C. Rapuano, J. Supercrit. Fluids, 2008, 45, 365 CrossRef CAS PubMed.
- B. Duan, M. Wang, W. Y. Zhou, W. L. Cheung, Z. Y. Li and W. W. Lu, Acta Biomater., 2010, 6, 4495 CrossRef CAS PubMed.
- F. Khan and S. R. Ahmad, in Biomaterials and Stem Cells in Regenerative Medicine, ed. M. Ramalingam, S. Ramakrishna and S. Best, CRC Press, Taylor & Francis, USA, 2012, ch. 5, pp. 101–121 Search PubMed.
- B. D. Gates, Q. Xu, M. Stewart, D. Ryan, C. G. Willson and G. M. Whitesides, Chem. Rev., 2005, 105, 1171 CrossRef CAS PubMed.
- G. Widawski, M. Rawiso and B. Francois, Nature, 1994, 369, 387 CrossRef CAS PubMed.
- M. Théry, V. Racine, A. Pépin, M. Piel, Y. Chen, J.-B. Sibarita and M. Bornens, Nat. Cell Biol., 2005, 7, 947 CrossRef PubMed.
- M. Théry, V. Racine, M. Piel, A. Pépin, A. Dimitrov, Y. Chen, S. J. B. Sibarita and M. Bornens, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19771 CrossRef PubMed.
- Y. J. Seol, T. Y. Kang and D. W. Cho, Soft Matter, 2012, 8, 1730 RSC.
- M. R. Beaulieu, N. R. Hendricks and J. J. Watkins, ACS Photonics, 2014, 1, 799 CrossRef CAS.
- S. Park, D. Kim, S. Y. Ko, J. O. Park, S. Akella, B. Xu, Y. Zhang and S. Fraden, Lab Chip, 2014, 14, 1551 RSC.
- G. Comina, A. Suska and D. Filippini, Lab Chip, 2014, 14, 424 RSC.
- R. Voelkel, U. Vogler, A. Bramati, M. Hennemeyer, R. Zoberbier, A. Voigt, G. Grutzner, N. Unal and U. Hofmann, Microsyst. Technol., 2014, 20, 1839 CrossRef CAS.
- M. Muller, J. Becher, M. Schnabelrauch and M. Zenobi-Wong, J. Visualized Exp., 2012, 67, e50632 Search PubMed.
- J. J. Balowski, Y. Wang and N. L. Allbritton, Adv. Mater., 2013, 25, 4107 CrossRef CAS PubMed.
- Y. Lu and S. Chen, Methods Mol. Biol., 2012, 868, 289 CAS.
- M. A. K. Liebschner and M. Wettergreen, Methods Mol. Biol., 2012, 868, 71 CAS.
- A. Revzin, R. G. Tompkins and M. Toner, Langmuir, 2003, 19, 9855 CrossRef CAS.
- M. Yamato, C. Konno, M. Utsumi, A. Kikuchi and T. Okano, Biomaterials, 2002, 23, 561 CrossRef CAS.
- J. M. Karp, Y. Yeo, W. Geng, C. Cannizarro, K. Yan, D. S. Kohane, G. Vunjak-Novakovic, R. S. Langer and M. Radisic, Biomaterials, 2006, 27, 4755 CrossRef CAS PubMed.
- E. Menard, M. A. Meitl, Y. Sun, J. U. Park, D. J. L. Shir, Y. S. Nam, S. Jeon and J. A. Rogers, Chem. Rev., 2007, 107, 1117 CrossRef CAS PubMed.
- D. R. Albrecht, V. L. Tsang, R. L. Sah and S. N. Bhatia, Lab Chip, 2005, 5, 111 RSC.
- D. R. Albrecht, G. H. Underhill, T. B. Wassermann, R. L. Sah and S. N. Bhatia, Nat. Methods, 2006, 3, 369 CrossRef CAS PubMed.
- E. Menard, M. A. Meitl, Y. Sun, J. U. Park, D. J. L. Shir, Y.-S. Nam, S. Jeon and J. A. Rogers, Chem. Rev., 2007, 107, 1117 CrossRef CAS PubMed.
- J. H. Moon, J. Ford and S. Yang, Polym. Adv. Technol., 2006, 17, 83 CrossRef CAS PubMed.
- T. M. Bloomstein, M. Rothschild, R. R. Kunz, D. E. Hardy, R. B. Goodman and S. T. Palmacci, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.–Process., Meas., Phenom., 1998, 16, 3154 CrossRef CAS.
- T. M. Bloomstein, M. F. Marchant, S. Deneault, D. E. Hardy and M. Rothschild, Opt. Express, 2006, 14, 6434 CrossRef CAS.
- C. D. Müller, A. Falcou, N. Reckefuss, M. Rojahn, V. Wiederhirn, P. Rudati, H. Frohne, O. Nuyken, H. Becker and K. Meerholz, Nature, 2003, 421, 829 CrossRef PubMed.
- M. Campbell, D. N. Sharp, M. T. Harrison, R. G. Denning and A. G. Turberfield, Nature, 2000, 404, 53 CrossRef CAS PubMed.
- J. Hoffmann, M. Plotner, D. Kuckling and W. J. Fischer, Sens. Actuators, A, 1999, 77, 139 CrossRef CAS.
- Z. Liu, D. G. Bucknall and M. G. Allen, J. Micromech. Microeng., 2011, 21, 065036 CrossRef.
- Z. Liu, D. G. Bucknall and M. G. Allen, Nanotechnology, 2011, 22, 225302 CrossRef PubMed.
- D. Xu, K. P. Chen, K. Ohlinger and Y. Lin, Nanotechnology, 2011, 22, 035303 CrossRef PubMed.
- Z. Zhendong, B. Benfeng, D. Huigao, Z. Haosu, Z. Mingqian, Y. Oubo, L. Qunqing, T. Qiaofeng, W. Jia and F. Shoushan, Small, 2014, 10, 1603 CrossRef PubMed.
- T. Chen, I. Amin and R. Jordan, Chem. Soc. Rev., 2012, 41, 3280 RSC.
- C. Chao and L. J. Guo, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.–Process., Meas., Phenom., 2002, 20, 832 Search PubMed.
- L. J. Guo, Adv. Mater., 2007, 19, 495 CrossRef CAS PubMed.
- X. Cheng, L. J. Guo and P. F. Fu, Adv. Mater., 2005, 17, 1419 CrossRef CAS PubMed.
- R. S. Kane, S. Takayama, E. Ostuni, D. E. Ingber and G. M. Whitesides, Biomaterials, 1999, 20, 2363 CrossRef CAS.
- F. Johansson, P. Carlberg, N. Danielsen, L. Montelius and M. Kanje, Biomaterials, 2006, 27, 1251 CrossRef CAS PubMed.
- S. Lenhert, L. Zhang, J. Mueller, H. P. Wiesmann, G. Erker, H. Fuchs and L. Chi, Adv. Mater., 2004, 16, 619 CrossRef CAS PubMed.
- M. J. Dalby, M. O. Riehle, S. J. Yarwood, C. D. W. Wilkinson and A. S. G. Curtis, Exp. Cell Res., 2003, 81, 274 Search PubMed.
- A. S. G. Curtis, Eur. Cells Mater., 2004, 8, 27 CAS.
- A. P. Quist, E. Pavlovic and S. Oscarsson, Anal. Bioanal. Chem., 2005, 381, 591 CrossRef CAS PubMed.
- G. Kumar, Y. C. Wang, C. Co and C. C. Ho, Langmuir, 2003, 19, 10550 CrossRef CAS.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103 CrossRef CAS PubMed.
- J. Hyun, H. Ma, Z. Zhang, T. Beebe and A. Chilkoti, Adv. Mater., 2003, 15, 576 CrossRef CAS PubMed.
- A. Khademhosseini, S. Jon, K. Y. Suh, T. N. T. Tran, G. Eng, J. Yeh, J. Seong and R. Langer, Adv. Mater., 2003, 15, 1995 CrossRef CAS PubMed.
- V. A. Liu, W. E. Jastromb and S. N. Bhatia, J. Biomed. Mater. Res., 2002, 60, 126 CrossRef CAS PubMed.
- H. W. Li, B. V. O. Muir, G. Fichet and W. T. S. Huck, Langmuir, 2003, 19, 1963 CrossRef CAS.
- S. A. Lange, V. Benes, D. P. Kern, J. K. H. Horber and A. Bernard, Anal. Chem., 2004, 76, 1641 CrossRef CAS PubMed.
- C. M. Nelson, R. P. Jean, J. L. Tan, W. F. Liu, N. J. Sniadecki, A. A. Spector and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11594 CrossRef CAS PubMed.
- J. M. Corey and E. L. Feldman, Exp. Neurol., 2003, 184(suppl 1), S89 CrossRef.
- M. Mrksich, L. E. Dike, J. Tien, D. E. Ingber and G. M. Whitesides, Exp. Cell Res., 1997, 235, 305 CrossRef CAS PubMed.
- M. Scholl, C. Sprossler, M. Denyer, M. Krause, K. Nakajima, A. Maelicke, W. Knoll and A. Offenhausser, J. Neurosci. Methods, 2000, 104, 65 CrossRef CAS.
- C. C. Lin, C. C. Co and C. C. Ho, Biomaterials, 2005, 26, 3655 CrossRef CAS PubMed.
- H. Bae, H. Chu, F. Edalat, J. M. Cha, S. Sant, A. Kashyap, A. F. Ahari, C. H. Kwon, J. W. Nichol and S. Manoucheri, J. Tissue Eng. Regener. Med., 2014, 8, 1 CrossRef CAS PubMed.
- H. Hwang, J. Park, C. Shin, Y. K. Do and Y. K. Cho, Biomed. Microdevices, 2013, 15, 627 CrossRef CAS PubMed.
- M. F. Hsieh, C. Sheu and H. Yang, Adv. Mater. Res., 2013, 647, 170 CrossRef.
- S. Danmark, M. Gladnikoff, T. Frisk, M. Zelenina, K. Mustafa, A. Russom and A. Finne-Wistrand, Biomed. Microdevices, 2012, 14, 885 CrossRef CAS PubMed.
- A. P. Zhang, X. Qu, P. Soman, K. C. Hribar, J. W. Lee, S. Chen and S. He, Adv. Mater., 2012, 24, 4266 CrossRef CAS PubMed.
- A. Koroleva, A. A. Gill, I. Ortega, J. W. Haycock, S. Schlie, S. D. Gittard, B. N. Chichkov and F. Claeyssens, Biofabrication, 2012, 4, 025005 CrossRef CAS PubMed.
- F. Khan, R. Zhang, A. Unciti-Broceta, J. J. Diaz-Mochon and M. Bradly, Adv. Mater., 2007, 19, 3524 CrossRef CAS PubMed.
- E. Bonaccurso, H. J. Butt, B. Hankeln, B. Niesenhaus and K. Graf, Appl. Phys. Lett., 2005, 89, 124101 CrossRef PubMed.
- R. D. Deegan, O. Bakaljin, T. F. Dupont, G. Huber, S. R. Nagel and T. A. Witten, Nature, 1997, 389, 827 CrossRef CAS.
- C. N. Medine, F. Khan, S. Pernagallo, R. Zhang, O. Tura, M. Bradley and D. C. Hay, in Biomaterials and Stem Cells in Regenerative Medicine, ed. M. Ramalingam, S. Ramakrishna and S. Best, CRC Press, Taylor & Francis, USA, 2012, ch. 1, pp. 1–30 Search PubMed.
- R. S. Tare, F. Khan, G. Tourniaire, S. M. Morgan, M. Bradley and R. O. C. Oreffo, Biomaterials, 2009, 30, 1045 CrossRef CAS PubMed.
- F. Khan, S. Valere, S. Fuhrmann, V. Arrighi and M. Bradley, J. Mater. Chem. B, 2013, 1, 2590 RSC.
- F. Khan, R. S. Tare, J. M. Kanczler, R. O. C. Oreffo and M. Bradley, Biomaterials, 2010, 31, 2216 CrossRef CAS PubMed.
- F. Khan, J. O. Smith, J. M. Kanzcler, R. S. Tare, R. O. C. Oreffo and M. Bradley, Adv. Funct. Mater., 2013, 23, 2850 CrossRef CAS PubMed.
- J. D. Kim, J. S. Choi, B. S. Kim, Y. C. Choi and Y. W. Cho, Polymer, 2010, 51, 2147 CrossRef CAS PubMed.
- B. J. de Gans, P. C. Duineveld and U. S. Schubert, Adv. Mater., 2004, 16, 203 CrossRef CAS PubMed.
- R. Zhang, A. Liberski, F. Khan, J. J. Diaz-Mochon and M. Bradley, Chem. Commun., 2008, 1317 RSC.
- T. Billiet, M. Vandenhaute, J. Schelfhout, S. V. Vlierberghe and P. Dubruel, Biomaterials, 2012, 33, 6020 CrossRef CAS PubMed.
- Y. Christanti and L. M. Walker, J. Non-Newtonian Fluid Mech., 2001, 100, 9 CrossRef CAS.
- J. C. Carter, R. M. Alvis, S. B. Brown, K. C. Langry, T. S. Wilson, M. T. McBride, M. L. Myrick, W. R. Cox, M. E. Grove and B. W. Colston, Biosens. Bioelectron., 2006, 21, 1359 CrossRef CAS PubMed.
- E. A. Roth, T. Xu, M. Das, C. Cregory, J. J. Hickman and T. Boland, Biomaterials, 2004, 25, 3707 CrossRef CAS PubMed.
- N. E. Sanjana and S. B. Fuller, J. Neurosci. Methods, 2004, 136, 151 CrossRef PubMed.
- T. Boland, X. Tao, B. J. Damon, B. Manley, P. Kesari, S. Jalota and S. Bhaduri, Mater. Sci. Eng., C, 2007, 27, 372 CrossRef CAS PubMed.
- S. Z. Guo, M. C. Heuzey and D. Therriault, Langmuir, 2014, 30, 1142 CrossRef CAS PubMed.
- C. Gao, M. N. Rahaman, Q. Gao, A. Teramoto and K. Abe, J. Biomed. Mater. Res., Part A, 2013, 101A, 2027 CrossRef CAS PubMed.
- H. Yun, S. Kim, Y. Hyun, S. Heo and J. Shin, Chem. Mater., 2007, 19, 6363 CrossRef CAS.
- M. Xu, G. M. Gratson, E. B. Duoss, R. F. Shepherd and J. A. Lewis, Soft Matter, 2006, 2, 205 RSC.
- G. Vozzi, A. Previti, D. De Rossi and A. Ahluwalia, Tissue Eng., 2002, 8, 1089 CrossRef CAS PubMed.
- G. Vozzi, C. Flaim, A. Ahluwalia and S. Bhatia, Biomaterials, 2003, 24, 2533 CrossRef CAS.
- T. B. F. Woodfield, J. Malda, J. deWijn, F. Peters, J. Riesle and C. A. van Blitterswijk, Biomaterials, 2004, 25, 4149 CrossRef CAS PubMed.
- R. Landers, U. Hubner, R. Schmelzeisen and R. Mulhaupt, Biomaterials, 2002, 23, 4437 CrossRef CAS.
- L. Geng, W. Feng, D. W. Hutmacher, Y. S. Wong, H. T. Loh and J. Y. H. Fuh, Rapid Prototyping J., 2005, 11, 90 CrossRef.
- G. M. Gratson, M. Xu and J. A. Lewis, Nature, 2004, 428, 386 CrossRef CAS PubMed.
- D. Therriault, S. R. White and J. A. Lewis, Nat. Mater., 2003, 2, 62 CrossRef PubMed.
- M. Xu, G. M. Gratson, E. B. Duoss, R. F. Shepherd and J. A. Lewis, Soft Matter, 2006, 2, 205 RSC.
- G. M. Gratson, F. Garcia-Santamaria, V. Lousse, M. Xu, S. Fan, J. A. Lewis and P. V. Braun, Adv. Mater., 2006, 18, 461 CrossRef CAS PubMed.
- D. W. Hutmacher, Biomaterials, 2000, 21, 2529 CrossRef CAS.
- M. Endres, D. W. Hutmacher, A. J. Salgado, C. Kaps, J. Ringe, R. L. Reis, M. Sittinger, A. Braddwood and J. T. Schantz, Tissue Eng., 2003, 9, 689 CrossRef CAS PubMed.
- A. Campo and E. Arzt, Chem. Rev., 2008, 108, 911 CrossRef PubMed.
- P. Vukusic and J. P. Sambles, Nature, 2003, 424, 852 CrossRef CAS PubMed.
- K. Autumn, Y. A. Liang, S. T. Hsieh, W. Sesch, W. P. Chan, T. W. Kenny, R. Fearing and R. J. Full, Nature, 2000, 405, 681 CrossRef CAS PubMed.
- R. M. Duffy and A. W. Feinberg, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 178 CrossRef CAS PubMed.
- M. R. Badrossamay, K. Balachandran, A. K. Capulli, H. M. Golecki, A. Agarwal, J. A. Goss, H. Kim, K. Shin and K. K. Parker, Biomaterials, 2014, 35, 3188 CrossRef CAS PubMed.
- M. Koepf, F. Cherioux, J. A. Wytko and J. Weiss, Coord. Chem. Rev., 2012, 256, 2872 CrossRef CAS PubMed.
- H. Aubin, J. W. Nichol, C. B. Hutson, H. Bae, A. L. Sieminski, D. M. Cropek, P. Akhyari and A. Khademhosseini, Biomaterials, 2010, 31, 6941 CrossRef CAS PubMed.
- C. Suwanchawalit, A. J. Patil, R. K. Kumar, S. Wongnawa and S. Mann, J. Mater. Chem., 2009, 19, 8478 RSC.
- Z. Nie and E. Kumacheva, Nat. Mater., 2008, 7, 74 CrossRef PubMed.
- A. Mahdavi, L. Ferreira, C. Sundback, J. W. Nicho, E. P. Chan and D. J. D. Carter, et al. , Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2307 CrossRef CAS PubMed.
- J. R. McMillan, M. Akiyama, M. Tanaka, S. Yamamoto, M. Goto, R. Abe, D. Sawamura, M. Shimomura and H. Shimizu, Tissue Eng., 2007, 13, 789 CrossRef CAS.
- M. Birch, M. Tanaka, G. Kirmizidis, S. Yamamoto and M. Shimomura, Tissue Eng., Part A, 2013, 19, 2087 CrossRef CAS PubMed.
- H. Choi, M. Tanaka, T. Hiragun, M. Hide and K. Sugimoto, Nanomedicine, 2014, 10, 313 CrossRef CAS PubMed.
- E. Kitakami, M. Aoki, C. Sato, H. Ishihata and M. Tanaka, BiMed. Res. Int., 2014, 2014, 102648 Search PubMed.
- M. Tanaka, M. Takebayashi, M. Miyama, J. Nishida and M. Shimomura, Biomed. Mater. Eng., 2004, 14, 439 Search PubMed.
- Y. Fukuhira, E. Kitazono, T. Hayashi, H. Kaneko, M. Tanaka, M. Shimomura and Y. Sumi, Biomaterials, 2006, 27, 1797 CrossRef CAS PubMed.
- N. Maruyama, T. Koito, T. Sawadaishi, O. Karthaus, K. Ijiro, N. Nishi and M. Shimomura, Thin Solid Films, 1998, 327–329, 854 CrossRef CAS.
- M. H. Stenzel-Rosenbaum, T. P. Davis, A. G. Fane and V. Chen, Angew. Chem., Int. Ed., 2001, 40, 1408 CrossRef.
- H. Yabu, M. Tanaka, K. Ijiro and M. Shimomura, Langmuir, 2003, 19, 694 CrossRef.
- M. Tanaka, K. Yoshizawa, A. Tsuruma, H. Sunami, S. Yamamoto and M. Shimomura, Colloids Surf., A, 2008, 313–314, 515 CrossRef PubMed.
- Y. Fukuhira, M. Ito, H. Kaneko, Y. Sumi, M. Tanaka, S. Yamamoto and M. Shimomura, J. Biomed. Mater. Res., Part B, 2008, 86B, 353 CrossRef CAS PubMed.
- M. Tanaka, A. Takayama, E. Ito, H. Sunami, S. Yamamoto and M. Shimomura, J. Nanosci. Nanotechnol., 2007, 7, 763 CrossRef CAS PubMed.
- M. Tanaka, K. Nishikawa, H. Okubo, H. Kamachi, T. Kawai, M. Matsushita, S. Todo and M. Shimomura, Colloids Surf., A, 2006, 284–285, 464 CrossRef PubMed.
- M. Shimomura, T. Nishikawa, A. Mochizuki and M. Tanaka, JP 2001/157574, 2001.
- A. Tsuruma, M. Tanaka, S. Yamamoto and M. Shimomura, Colloids Surf., A, 2008, 313–314, 536 CrossRef PubMed.
- S. Yamamoto, M. Tanaka, H. Sunami, S. Yamashita, Y. Morita and M. Shimomura, Langmuir, 2007, 23, 8114 CrossRef CAS PubMed.
- H. Ishihata, M. Tanaka, N. Iwama, M. Ara, M. Shimonishi, M. Nagamine, N. Murakami, S. Kanaya, E. Nemoto, H. Shimauchi and M. Shimomura, J. Biomech. Sci. Eng., 2010, 5, 252 CrossRef PubMed , Special Issue on Micro. Nanobiotech. for Cells.
- T. Sato, M. Tanaka, S. Yamamoto, E. Ito, K. Shimizu, Y. Igarashi, M. Shimomura and J. Inokuchi, J. Biomater. Sci., Polym. Ed., 2010, 21, 1947 CrossRef CAS PubMed.
- H. Yabu, M. Takebayashi, M. Tanaka and M. Shimomura, Langmuir, 2005, 21, 3235 CrossRef CAS PubMed.
- M. Tanaka, M. Takebayashi and M. Shimomura, Macromol. Symp., 2009, 279, 175 CrossRef CAS PubMed; M. Tanaka, Biochim. Biophys. Acta, 2011, 1810, 251 CrossRef PubMed.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. C. Oreffo and M. J. Dalby, Nat. Mater., 2011, 10, 637 CrossRef CAS PubMed.
- Z. Xiaojun and Z. Shuguang, Macromol. Biosci., 2007, 7, 13 CrossRef PubMed.
- V. Dinca, E. Kasotakis, J. Catherine, A. Mourka, A. Ranella, A. Ovsianikov, B. N. Chichkov, M. Farsari, A. Mitraki and C. Fotakis, Nano Lett., 2007, 8, 538 CrossRef PubMed.
- E. Gazit, Chem. Soc. Rev., 2007, 36, 1263 RSC.
- S. Scanlon and A. Aggeli, Nano Today, 2008, 3, 22 CrossRef CAS.
- S. Zhang, Mater. Today, 2003, 6, 20 CrossRef CAS.
- J. Kisiday, M. Jin, B. Kurz, H. Hung, C. Semino, S. Zhang and A. J. Grodzinsky, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9996 CrossRef CAS PubMed.
- F. Gelain, D. Bottai, A. Vescovi and S. Zhang, PLoS One, 2006, 1, e119 Search PubMed.
- S. Zhang, F. Gelain and X. Zhao, Semin. Cancer Biol., 2005, 15, 413 CrossRef PubMed.
- R. G. Ellis-Behnke, Y. X. Liang, S.-W. You, D. K. C. Tay, S. Zhang, K.-F. So and G. E. Schneider, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5054 CrossRef CAS PubMed.
- S. Zhang, T. Holmes, C. Lockshin and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3334 CrossRef CAS.
- T. C. Holmes, S. de Lacalle, X. Su, G. Liu, A. Rich and S. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6728 CrossRef CAS.
- J. M. Jukes, L. Moroni, C. A. van Blitterswijk and J. de Boer, Tissue Eng., Part A, 2008, 14, 135 CAS.
- S. Gerecht, J. A. Burdick, L. S. Ferreira, S. A. Townsend, R. Langer and V.-G. Novakovic, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11298 CrossRef CAS PubMed.
- J. Elisseeff, A. Ferran, S. Hwang, S. Varghese and Z. Zhang, Stem Cells Dev., 2006, 15, 295 CrossRef CAS PubMed.
- J. J. Rice, M. M. Martino, L. D. Laporte, F. Tortelli, P. S. Briqiez and J. A. Hubbell, Adv. Healthcare Mater., 2013, 2, 57 CrossRef CAS PubMed.
- J. R. Thonhoff, D. I. Lou, P. M. Jordan, X. Zhao and P. Wu, Brain Res., 2008, 1187, 42 CrossRef CAS PubMed.
- C. Fischbach, R. Chen, T. Matsumoto, T. Schmelzle, J. S. Brugge, P. J. Polverini and D. J. Mooney, Nat. Methods, 2007, 4, 855 CrossRef CAS PubMed.
- C. Chung and J. A. Burdick, Adv. Drug Delivery Rev., 2008, 60, 243 CrossRef CAS PubMed.
- A. Karkhaneh, Z. Naghizadeh, M. A. Shokrgozar, S. Bonakdar, A. Solouk and N. Haghighipour, Int. J. Artif. Organs, 2014, 37, 142 CrossRef PubMed.
- A. Karkhaneh, Z. Naghizadeh, M. A. Shokrgozar and S. Bonakdar, J. Appl. Polym. Sci., 2014, 131, 40635 CrossRef PubMed.
- P. M. Favi, R. S. Benson, N. R. Neilsen, R. L. Hammonds, C. C. Bates, C. P. Stephens and M. S. Dhar, Mater. Sci. Eng., C, 2013, 33, 1935 CrossRef CAS PubMed.
- R. L. Mauck, X. Yuan, R. S. Tuan and B. Chief, Osteoarthritis Cartilage, 2006, 14, 179 CrossRef CAS PubMed.
- C. Chung and J. A. Burdick, Tissue Eng., Part A, 2009, 15, 243 CrossRef CAS PubMed.
- C. Chung, M. Beecham, R. L. Mauck and J. A. Burdick, Biomaterials, 2009, 30, 4287 CrossRef CAS PubMed.
- Y. H. Jeon, J. H. Choi, J. K. Sung, T. K. Kim, B. C. Cho and H. Y. Chung, J. Craniofac. Surg., 2007, 18, 1249 CrossRef PubMed.
- Z. Chen, M. Zhao, K. Liu, Y. Wan, X. Li and G. Feng, J. Mater. Sci.: Mater. Med., 2014, 25, 1903 CrossRef CAS PubMed.
- S.-Y. Lee, A.-S. Wee, C.-K. Lim, A. A. Abbas, L. Selvaratnam, A. M. Merican, T. S. Ahmad and T. Kamarul, J. Mater. Sci.: Mater. Med., 2013, 24, 1561 CrossRef CAS PubMed.
- C. Fan, L. Liao, C. Zhang and L. Liu, J. Mater. Chem. B, 2013, 1, 4251 RSC.
- D. A. Rennerfeldt, A. N. Renth, Z. Talata, S. H. Gehrke and M. S. Detamore, Biomaterials, 2013, 34, 8241 CrossRef CAS PubMed.
- F. Yu, X. Cao, L. Zeng, Q. Zhang and X. Chen, Carbohydr. Polym., 2013, 97, 188 CrossRef CAS PubMed.
- L. R. Almeida, A. R Martins, E. M. Fernandes, M. B. Oliveira, V. M. Correlo, I. Pashkuleva, A. P. Marques, A. S. Ribeiro, N. F. Duraes and C. J. Silva, Acta Biomater., 2013, 9, 8167 CrossRef CAS PubMed.
- L. D. Solorio, E. L. Vieregge, C. D. Dhami and E. Alsberg, Tissue Eng., Part B, 2013, 19, 209 CrossRef CAS PubMed.
- W. Zhao, X. Jin, Y. Cong, Y. Liu and J. Fu, J. Chem. Technol. Biotechnol., 2013, 88, 327 CrossRef CAS PubMed.
- I. Martin, S. Miot, A. Barbero, M. Jakob and D. Wendt, J. Biomech., 2007, 40, 750 CrossRef PubMed.
- S. R. Frenkel and P. E. Di Cesare, Ann. Biomed. Eng., 2004, 32, 26 CrossRef.
- Y. Tanaka, H. Yamaoka, S. Nishizawa, S. Nagata, T. Ogasawara, Y. Asawa, Y. Fujihara, T. Takato and K. Hoshi, Biomaterials, 2010, 31, 4506 CrossRef CAS PubMed.
- W. Dai, N. Kawazoe, X. Lin, J. Dong and G. Chen, Biomaterials, 2010, 31, 2141 CrossRef CAS PubMed.
- S. J. Shieh, S. Terada and J. P. Vacanti, Biomaterials, 2004, 25, 1545 CrossRef CAS.
- T. B. F. Woodfield, J. Malda, J. de Wijn, F. Peters, J. Riesle and C. A. van Blitterswijk, Biomaterials, 2004, 25, 4149 CrossRef CAS PubMed.
- E. Kon, A. Di Martino and G. Filardo, et al. , Eur. J. Radiol., 2011, 79, 382 CrossRef CAS PubMed.
- C. He, W. Nie and W. Feng, J. Mater. Chem. B, 2014, 2, 7828 RSC; J. A. Goding, A. D. Gilmour, P. J. Martens, L. A. Poole-Warren and R. A. Green, J. Mater. Chem. B, 2015, 3, 5058 RSC.
- J. Iwasa, L. Engebretsen, Y. Shima and M. Ochi, Knee Surg. Sports Traumatol. Arthrosc., 2009, 17, 561 CrossRef PubMed.
- M. Marcacci, E. Kon, S. Zaffagnini, G. Filardo, M. Delcogliano, M. P. Neri, F. Iacono and A. P. Hollander, Knee Surg. Sports Traumatol. Arthrosc., 2007, 15, 610 CrossRef PubMed.
- M. Marcacci, S. Zaffagnini, E. Kon, A. Visani, F. Iacono and I. Loreti, Knee Surg. Sports Traumatol. Arthrosc., 2002, 10, 154 CrossRef CAS PubMed.
- L. E. Niklason, Nat. Biotechnol., 2000, 18, 929 CrossRef CAS PubMed.
- A. U. Daniels, K. P. Andriano, W. P. Smutz, M. K. O. Chang and J. Heller, J. Appl. Biomater., 1994, 5, 51 CrossRef CAS PubMed.
- A. J. Thornton, E. Alsberg, M. Albertelli and D. J. Mooney, Transplantation, 2004, 77, 1798 CrossRef CAS.
- Z. Li, H. R. Ramay, K. D. Hauch, D. Xiao and M. Zhang, Biomaterials, 2005, 26, 3919 CrossRef CAS PubMed.
- A. Tampieri, M. Sandri, E. Landi, G. Celotti, N. Roveri, M. Mattioli-Belmonte, L. Virgili, F. Gabbanelli and G. Biagini, Acta Biomater., 2005, 1, 343 CrossRef CAS PubMed.
- Y. Khan, M. J. Yaszemski, A. G. Mikos and C. T. Laurencin, J. Bone Joint Surg. Am., 2008, 90, 36 CrossRef PubMed.
- A. Hussein, A. Nadia and A. K. Numa, Res. J. Pharm., Biol. Chem. Sci., 2014, 5, 295 Search PubMed.
- N. Sultana, Appl. Mech. Mater., 2014, 554, 42 CrossRef.
- Y. Hu, X. Gu, Y. Yang, J. Huang, M. Hu, W. Chen, Z. Tong and C. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 17166 CAS.
- H. Y. Mi, S. M. Palumbo, X. Jing, L. S. Turng, W. J. Li and X. F. Peng, J. Biomed. Mater. Res., Part B, 2014, 102B, 1434 CrossRef CAS PubMed.
- J. Minton, C. Janney, R. Akbarzadeh, C. Focke, A. Subramanian, T. Smith, J. McKinney, J. Liu, J. Schmitz and P. F. James, J. Biomater. Sci., Polym. Ed., 2014, 25, 1856 CrossRef CAS PubMed.
- A. R. Costa-Pinto, R. L. Reis and N. M. Neves, Tissue Eng., Part B, 2011, 17, 331 CrossRef CAS PubMed.
- W. B. Tsai, Y. R. Chen, H. L. Liu and J. Y. Lai, Carbohydr. Polym., 2011, 85, 129 CrossRef CAS PubMed.
- A. M. Martins, C. M. Alves, K. F. Kurtis, A. G. Mikos and R. L. Reis, J. Mater. Chem., 2010, 20, 1638 RSC.
- A. R. Costa-Pinto, V. M. Correlo, P. C. Sol, M. Bhattacharya, P. Charbord, B. Delorme, R. L. Reis and N. M. Neves, Biomacromolecules, 2009, 10, 2067 CrossRef CAS PubMed.
- M. S. Bae, D. H. Yang, J. B. Lee, D. N. Heo, Y. D. Kwon, I. C. Youn, K. Choi, J. H. Hong, G. T. Kim and Y. S. Choi, Biomaterials, 2011, 32, 8161 CrossRef CAS PubMed.
- W. R. Walsh, R. M. Oliver, G. Gage, Y. Yu, D. Bell, J. Bellemore and H. D. Adkisson, Tissue Eng., Part A, 2011, 17, 213 CrossRef CAS PubMed.
- T. B. L. Nguyen, Y. K. Min and B. T. Lee, J. Mater. Sci., 2013, 48, 4233 CrossRef CAS.
- G. Hulsart-Billstroem, P. K. Yuen, R. Marsell, J. Hilborn, S. Larsson and D. Ossipov, Biomacromolecules, 2013, 14, 3055 CrossRef CAS PubMed.
- F. D. Ivan, A. Marian, C. E. Tanase, M. Butnaru and L. Verestiuc, Key Eng. Mater., 2014, 587, 191 CrossRef CAS.
- M. Neufurth, X. Wang, H. C. Schroeder, Q. Feng, B. Diehl-Seifert, T. Ziebart, R. Steffen, S. Wang and W. E. G. Mueller, Biomaterials, 2014, 35, 8810 CrossRef CAS PubMed.
- A. Moshaverinia, C. Chen, X. Xu, K. Akiyama, S. Ansari, H. H. Zadeh and S. Shi, Tissue Eng., Part A, 2014, 20, 611 CrossRef CAS PubMed.
- R. Nakaoka, Y. Hirano, D. J. Mooney, T. Tsuchiya and A. Matsuoka, J. Artif. Organs, 2013, 16, 284 CrossRef CAS PubMed.
- A. Moshaverinia, S. Ansari, C. Chen, X. Xu, K. Akiyama, M. L. Snead, H. H. Zadeh and S. Shi, Biomaterials, 2013, 34, 6572 CrossRef CAS PubMed.
- N. Shanmugasundaram, P. Ravichandran, P. Neelakanta Reddy, N. Ramamurty, S. Pal and K. P. Rao, Biomaterials, 2001, 22, 1943 CrossRef CAS.
- J. K. Park, J. H. Shim, K. S. Kang, J. Yeom, H. S. Jung, J. Y. Kim, K. H. Lee, T. H. Kim, S. Y. Kim and D. W. Cho, Adv. Funct. Mater., 2011, 21, 2906 CrossRef CAS PubMed.
- F. Khan and S. R. Ahmad, Macromol. Biosci., 2013, 13, 395 CrossRef CAS PubMed.
- N. Siddiqui and K. Pramanik, J. Appl. Polym. Sci., 2014, 131, 41025 CrossRef PubMed.
- S. Mathews, R. Bhonde, P. K. Gupta and S. Totey, J. Biomed. Mater. Res., Part B, 2014, 102, 1825 CrossRef PubMed.
- T. B. L. Nguyen, Y. K. Min and B. T. Lee, J. Mater. Sci., 2013, 48, 4233 CrossRef CAS.
- J. H. Park, E. J. Lee, J. C. Knowles and H. W. Kim, J. Biomater. Appl., 2014, 28, 1079 CrossRef PubMed.
- R. A. Perez, M. Kim, T. H. Kim, J. H. Kim, J. H. Lee, J. H. Park, J. C. Knowles and H. W. Kim, Tissue Eng., Part A, 2014, 20, 103 CrossRef CAS PubMed.
- N. Thadavirul, P. Pavasant and P. Supaphol, J. Biomed. Mater. Res., Part A, 2014, 102A, 3379 CrossRef CAS PubMed.
- J. Lim, M. S. K. Chong, J. K. Y. Chan and S. H. Teoh, Small, 2014, 10, 2495 CrossRef CAS PubMed.
- G. Z. Jin, T. H. Kim, J. H. Kim, J. E. Won, S. Y. Yoo, S. J. Choi, J. K. Hyun and H. W. Kim, J. Biomed. Mater. Res., Part A, 2013, 101A, 1283 CrossRef CAS PubMed.
- T. Jacobs, H. Declercq, N. Geyter, R. Cornelissen, P. Dubruel, C. Leys, A. Beaurain, E. Payen and R. Morent, J. Mater. Sci.: Mater. Med., 2013, 24, 469 CrossRef CAS PubMed.
- H. Mahjoubi, J. M. Kinsella, M. Murshed and M. Cerruti, ACS Appl. Mater. Interfaces, 2014, 6, 9975 CAS.
- A. Das, S. Tanner, D. A. Barker, D. Green and E. A. Botchwey, J. Biomed. Mater. Res., Part A, 2014, 102A, 1210 CrossRef CAS PubMed.
- M. Bao, X. Lou, Q. Zhou, W. Dong, H. Yuan and Y. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 2611 CAS.
- A. Sadiasa, T. H. Nguyen and B. T. Lee, J. Biomater. Sci., Polym. Ed., 2014, 25, 150 CrossRef CAS PubMed.
- E. Ko, K. Yang, J. Shin and S.-W. Cho, Biomacromolecules, 2013, 14, 3202 CrossRef CAS PubMed.
- N. Y. C. Yu, A. Schindeler, L. Peacock, K. Mikulec, J. Fitzpatrick, A. J. Ruys, J. J. Cooper-White and D. G. Little, Eur. Cells Mater., 2013, 25, 190 CAS.
- C. Zhou, Q. Shi, W. Guo, L. Terrell, A. T. Qureshi, D. J. Hayes and Q. Wu, ACS Appl. Mater. Interfaces, 2013, 5, 3847 CAS.
- B. Torabinejad, J. Mohammadi-Rovshandeh, S. M. Davachi and A. Zamanian, Mater. Sci. Eng., C, 2014, 42, 199 CrossRef CAS PubMed.
- T. Lou, X. Wang, G. Song, Z. Gu and Z. Yang, Int. J. Biol. Macromol., 2014, 69, 464 CrossRef CAS PubMed.
- J. M. Fernandez, M. S. Cortizo and A. M. Cortizo, J. Biomater. Tissue Eng., 2014, 4, 227 CrossRef CAS PubMed.
- X. Zhang, W. Chang, P. Lee, Y. Wang, M. Yang, J. Li, S. G. Kumbar and X. Yu, PLoS One, 2014, 9, e85871 Search PubMed.
- C. Zong, X. Qian, Z. Tang, Q. Hu, J. Chen, C. Gao, R. Tang, X. Tong and J. Wang, J. Biomed. Nanotechnol., 2014, 10, 1091 CrossRef CAS PubMed.
- W. Lu, K. Ji, J. Kirkham, Y. Yan, A. P. Boccaccini, M. Kellett, Y. Jin and X. B. Yang, Cell Tissue Res., 2014, 356, 97 CrossRef CAS PubMed.
- N. Sultana and T. H. Khan, J. Bionanosci., 2013, 7, 169 CrossRef CAS PubMed.
- K. R. Remya, J. Joseph, S. Mani, A. John, H. K. Varma and P. Ramesh, J. Biomed. Nanotechnol., 2013, 9, 1483 CrossRef CAS PubMed.
- W. Chang, X. Mu, X. Zhu, G. Ma, C. Li, F. Xu and J. Nie, Mater. Sci. Eng., C, 2013, 33, 4369 CrossRef CAS PubMed.
- A. B. Kutikov and J. Song, Acta Biomater., 2013, 9, 8354 CrossRef CAS PubMed.
- W. Y. Choi, H. E. Kim and Y. H. Koh, J. Porous Mater., 2013, 20, 701 CrossRef CAS.
- C. Gao, B. Yang, H. Hu, J. Liu, C. Shuai and S. Peng, Mater. Sci. Eng., C, 2013, 33, 3802 CrossRef CAS PubMed.
- M.-J. Chern, Y.-K. Shen and H.-H. Hung, Adv. Sci. Lett., 2013, 19, 2572 CrossRef CAS PubMed.
- F. Khan and S. R. Ahmad, in Biomimetics: Advancing Nanobiomaterials and Tissue Engineering, ed. M. Ramalingam, X. Wang, G. Chen, P. Ma and F. Z. Cui, Scrivener Publishing, Wiley, 2013, ch. 5, pp. 91–117 Search PubMed.
- R. A. Thibault, A. G. Mikos and F. K. Kasper, Adv. Healthcare Mater., 2013, 2, 13 CrossRef CAS PubMed.
- L. B. Rocha, G. Goissis and M. A. Rossi, Biomaterials, 2002, 23, 449 CrossRef CAS.
- L. S. Liu, A. Y. Thompson, M. A. Heidaran, J. W. Poser and R. C. Spiro, Biomaterials, 1999, 20, 1097 CrossRef CAS.
- S. H. Oh, S. G. Kang, E. S. Kim, S. H. Cho and J. H. Lee, Biomaterials, 2003, 24, 4011 CrossRef CAS.
- D. J. Trantolo, S. T. Sonis, B. M. Thompson, D. L. Wise, K. U. Lewandrowski and D. D. Hile, Int. J. Oral. Maxillofac. Implants, 2003, 18, 182 Search PubMed.
- M. Bradley, F. Khan, R. O. C. Oreffo and R. S. Tare, WO 2010/023463 A2, 2010.
- J. O. Smith, E. R. Tayton, F. Khan, A. Aarvold, R. B. Cook, A. Goodship, M. Bradley and R. O. C. Oreffo, J. Tissue Eng. Regener. Med., 2014 DOI:10.1002/term.2007.
- E. C. Tsai, P. D. Dalton, M. S. Shoichet and C. H. Tator, Biomaterials, 2006, 27, 519 CrossRef CAS PubMed.
- M. Dadsetan, A. M. Knight, L. Lu, A. J. Windebank and M. J. Yaszemski, Biomaterials, 2009, 30, 3874 CrossRef CAS PubMed.
- A. Hejcl, P. Lesny, M. Pradny, J. Sedy, J. Zamecnik, P. Jendelova, J. Michalek and E. Sykova, J. Mater. Sci.: Mater. Med., 2009, 20, 1571 CrossRef CAS PubMed.
- S. R. Hynes, M. F. Rauch, J. P. Bertram and E. B. Lavik, J. Biomed. Mater. Res., 2009, 89A, 499 CrossRef CAS PubMed.
- C. Y. Yang, B. Song, Y. Ao, A. P. Nowak, R. B. Abelowitz, R. A. Korsak, L. A. Havton, T. J. Deming and M. V. Sofroniew, Biomaterials, 2009, 30, 851 Search PubMed.
- D. R. Nisbet, D. Moses, T. R. Gengenbach, J. S. Forsythe, D. I. Finkelstein and M. K. Horne, J. Biomed. Mater. Res., 2009, 89A, 24 CrossRef CAS PubMed.
- P. Krsko, T. E. McCann, T. T. Thach, T. L. Laabs, H. M. Geller and M. R. Libera, Biomaterials, 2009, 30, 721 CrossRef CAS PubMed.
- D. R. Nisbet, K. E. Crompton, M. K. Horne, D. I. Finkelstein and J. S. Forsythe, J. Biomed. Mater. Res., Part B, 2008, 87B, 251 CrossRef CAS PubMed.
- M. C. Dodla and R. V. Bellamkonda, Biomaterials, 2008, 29, 33 CrossRef CAS PubMed.
- R. Balint, N. J. Cassidy and S. H. Cartmell, Acta Biomater., 2014, 10, 2341 CrossRef CAS PubMed.
- H. Xu, J. M. Holzwarth, Y. Yan, P. Xu, H. Zheng, Y. Yin, S. Li and P. X. Ma, Biomaterials, 2014, 35, 225 CrossRef CAS PubMed.
- J. E. Collazos-Castro, G. R. Hernandez-Labrado, J. L. Polo and C. Garcia-Rama, Biomaterials, 2013, 34, 3603 CrossRef CAS PubMed.
- Z. Zhou, P. Yu, H. M. Geller and C. K. Ober, Biomacromolecules, 2013, 14, 529 CrossRef CAS PubMed.
- P. Kumar, Y. E. Choonara, L. C. du Toit, G. Modi, D. Naidoo and V. Pillay, Int. J. Mol. Sci., 2012, 13, 13966 CrossRef CAS PubMed.
- S. Guan, X. L. Zhang, X. M. Lin, T. Q. Liu, X. H. Ma and Z. F. Cui, J. Biomater. Sci., Polym. Ed., 2013, 24, 999 CrossRef CAS PubMed.
- J. F. Cherry, A. L. Carlson, F. L. Benarba, S. D. Sommerfeld, D. Verma, G. Loers, J. Kohn, M. Schachner and P. V. Moghe, Biointerphases, 2012, 7, 22 CrossRef CAS PubMed.
- S. N. Alhosseini, F. Moztarzadeh, M. Mozafari, S. Asgari, M. Dodel, A. Samadikuchaksaraei, S. Kargozar and N. Jalali, Int. J. Nanomed., 2012, 7, 25 CAS.
- M. P. Prabhakaran, L. Ghasemi-Mobarakeh, G. Jin and S. Ramakrishna, J. Biosci. Bioeng., 2011, 112, 501 CrossRef CAS PubMed.
- Y.-S. Lee, G. Collins and A. T. Livingston, Acta Biomater., 2011, 7, 3877 CrossRef CAS PubMed.
- L. Ghasemi-Mobarakeh, M. P. Prabhakaran, M. Morshed, M. H. Nasr-Esfahani, H. Baharvand, S. Kiani, S. S. Al-Deyab and S. Ramakrishna, J. Tissue Eng. Regener. Med., 2011, 5(4), e17 CrossRef CAS PubMed.
- V. Lundin, A. Herland, M. Berggren, E. W. H. Jager and A. I. Teixeira, PLoS One, 2011, 6(4), e18624 CAS.
- Q. Tu, L. Li, Y. Zhang, J. Wang, R. Liu, M. Li, W. Liu, X. Wang, L. Ren and J. Wang, Biomaterials, 2011, 32, 3253 CrossRef CAS PubMed.
- S. H. Ku, M. Lee and C. B. Park, Adv. Healthcare Mater., 2013, 2, 244 CrossRef CAS PubMed.
- X. Luo, C. L. Weaver, S. Tan and X. T. Cui, J. Mater. Chem. B, 2013, 1, 1340 RSC.
- K. Zhou, G. A. Thouas, C. C. Bernard, D. R. Nisbet, D. I. Finkelstein, D. Li and J. S. Forsythe, ACS Appl. Mater. Interfaces, 2012, 4, 4524 CAS.
- G. Z. Jin, M. Kim, U. S. Shin and H. W. Kim, Neurosci. Lett., 2011, 501, 10 CrossRef CAS PubMed.
- G. Z. Jin, M. Kim, U. S. Shin and H. W. Kim, Cell Biol. Int., 2011, 35, 741 CrossRef CAS PubMed.
- J. Ai, A. Kiasat-Dolatabadi, S. Ebrahimi-Barough, A. Ai, N. Lotfibakhshaiesh, A. Norouzi-Javidan, H. Saberi, B. Arjmand and H. R. Aghayan, Arch. Neurosci., 2014, 1, 15 CrossRef.
- T. Freier, R. Montenegro, H. Shan Koh and M. S. Shoichet, Biomaterials, 2005, 26, 4624 CrossRef CAS PubMed.
- S. B. Lee, Y. H. Kim, M. S. Chong, S. H. Hong and Y. M. Lee, Biomaterials, 2005, 26, 1961 CrossRef CAS PubMed.
- Z. A. A. Hamid, A. Blencowe and B. Ozcelik, et al. , Biomaterials, 2010, 31, 6454 CrossRef CAS PubMed.
- H. Fan, H. Liu, L. T. Siew and J. C. H. Goh, Biomaterials, 2009, 30, 4967 CrossRef CAS PubMed.
- K. Lee, E. A. Silva and D. J. Mooney, J. R. Soc., Interface, 2011, 8, 153 CrossRef CAS PubMed.
- K. Hori, C. Sotozono, J. Hamuro, K. Yamasaki, Y. Kimura, M. Ozeki, Y. Tabata and S. Kinoshita, J. Controlled Release, 2007, 118, 169 CrossRef CAS PubMed.
- T. Wang, X. Jiang, T. Lin, S. Ren, X. Li, X. Zhang and Q. Tang, Biomaterials, 2009, 30, 4161 CrossRef CAS PubMed.
- D. M. Yoon, E. C. Hawkins, S. Francke-Carroll and J. P. Fisher, Biomaterials, 2007, 28, 299 CrossRef CAS PubMed.
- L. R. Rodino-Klapac, A. M. Haidet, J. Kota, C. Handy, B. K. Kaspar and J. R. Mendell, Muscle Nerve, 2009, 39, 283 CrossRef CAS PubMed.
- S. J. Jhaveri, M. R. Hynd, N. Dowell-Mesfin, J. N. Turner, W. Shain and C. K. Ober, Biomacromolecules, 2009, 10, 174 CrossRef CAS PubMed.
- L. W. Hosack, M. A. Firpo, J. A. Scott, G. D. Prestwich and R. A. Peattie, Biomaterials, 2008, 29, 2336 CrossRef CAS PubMed.
- B. Sullenbarger, J. H. Bahng, R. Gruner, N. Kotov and L. C. Lasky, Exp. Hematol., 2009, 37, 101 CrossRef CAS PubMed.
- X. Guo, H. Park, G. Liu, W. Liu, Y. Cao, Y. Tabata, F. K. Kasper and A. G. Mikos, Biomaterials, 2009, 30, 2741 CrossRef CAS PubMed.
- J. Yu, Y. Gu, K. T. Du, S. Mihardja, R. E. Sievers and R. J. Lee, Biomaterials, 2009, 30, 751 CrossRef CAS PubMed.
- S. B. Lowe, V. T. G. Tan, A. H. Soeriyadi, T. P. Davies and J. J. Gooding, Bioconjugate Chem., 2014, 25(9), 1581 CrossRef CAS PubMed.
- N. Yonet-Tanyeri, M. H. Rich, M. Lee, M.-H. Lai, J. H. Jeong, R. J. DeVolder and H. Kong, Biomaterials, 2013, 34, 8416 CrossRef CAS PubMed.
- S. P. Herbert and D. Y. Stainier, Nat. Rev. Mol. Cell Biol., 2011, 12, 551 CrossRef CAS PubMed.
- O. P. Gautschi, S. P. Frey and R. Zellweger, ANZ J. Surg., 2007, 77, 626 CrossRef PubMed.
- M. K. Nagai and J. M. Embil, Expert Opin. Biol. Ther., 2002, 2, 211 CrossRef CAS PubMed.
- I. J. Frieden, Pediatr. Dermatol., 2008, 25, 590 CrossRef PubMed.
- E. J. Carragee, E. L. Hurwitz and B. K. Weiner, Spine J., 2011, 11, 471 CrossRef PubMed.
- K. R. Garrison, S. Donell, J. Ryder, I. Shemilt, M. Mugford, I. Harvey and F. Song, Health Technol. Assess., 2007, 11, 1 CAS.
- D. H. R. Kempen, M. C. Kruyt, L. Lu, C. E. Wilson, A. V. Florschutz, L. B. Creemers, M. J. Yaszemski and W. J. A. Dhert, Tissue Eng., Part A, 2009, 15, 587 CrossRef CAS PubMed.
- H. Park, J. S. Temenoff, Y. Tabata, A. I. Caplan, R. M. Raphael, J. A. Jansen and A. G. Mikos, J. Biomed. Mater. Res., 2009, 88A, 889 CrossRef CAS PubMed.
- R. Baffour, J. L. Garb, J. Kaufman, J. Berman, S. W. Rhee, M. A. Norris and P. Friedmann, J. Surg. Res., 2000, 93, 219 CrossRef CAS PubMed.
- S. Nakamura, M. Ishihara, K. Obara, K. Masuoka, T. Ishizuka, Y. Kanatani, B. Takase, T. Matsui, H. Hattori, T. Sato, Y. Kariya and T. Maehara, J. Biomed. Mater. Res., 2006, 78A, 364 CrossRef CAS PubMed.
- J. J. Yoon, H. J. Chung and T. G. Park, J. Biomed. Mater. Res., 2007, 83A, 597 CrossRef CAS PubMed.
- Y. Liu, L. Sun, Y. Huan, H. Zhao and J. Deng, J. Surg. Res., 2006, 136, 85 CrossRef CAS PubMed.
- L. Chen, W. Jiang, J. Huang, B. C. He, G. W. Zuo and W. Zhang, et al. , J. Bone Miner. Res., 2010, 25, 2447 CrossRef CAS PubMed.
- B. Wolach, L. J. van der Laan, N. A. Maianski, T. A. Tool, R. van Bruggen, D. Roos and T. W. Kuijpers, Exp. Hematol., 2007, 35(4), 541 CrossRef CAS PubMed.
- Q. Zeng, X. Li, G. Beck, G. Balian and F. H. Shen, Bone, 2007, 40, 374 CrossRef CAS PubMed.
- A. C. McPherron, A. M. Lawler and S. J. Lee, Nature, 1997, 387, 83 CrossRef CAS PubMed.
- J. R. Silva, R. van den Hurk, H. T. van Tol, B. A. Roelen and J. R. Figueiredo, Mol. Reprod. Dev., 2005, 70, 11 CrossRef CAS PubMed.
- N. S. Cunningham, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, A. H. Reddi and S. J. Lee, Growth Factors, 1995, 12, 99 CrossRef CAS.
- R. E. Andersen and D. A. Lim, Cell Res., 2014, 24, 1381 CrossRef CAS PubMed.
- E. Schmelzer, A. Deiwick, H. Bruns, H. C. Fiegel and A. Bader, Eur. J. Gastroenterol. Hepatol., 2008, 20, 209 CrossRef CAS PubMed.
Footnote |
| † Present address: Soft Materials Chemistry, Institute for Materials Chemistry and Engineering, Kyushu University, 744 Motooka Nishi-ku, 819-0395, Japan. E-mail: masaru_tanaka@ms.ifoc.kyushu-u.ac.jp; Web: http://www.soft-material.jp/ |
| This journal is © The Royal Society of Chemistry 2015 |