Novel biosensing methodologies for ultrasensitive detection of viruses
Ming Soon Cheng and Chee-Seng Toh*
Division of Chemistry and Biological Chemistry, School of Physical and Mathematical Sciences, Nanyang Technological University, 21 Nanyang Link, Singapore 637371. E-mail: cstoh@ntu.edu.sg; Fax: +65-6791 1961; Tel: +65-6592 2553
First published on 23rd August 2013
Abstract
Various infectious diseases caused by the spread of viruses create adverse implications on global biosecurity. Increasing demands for virus surveillance and effective control of the spread of diseases reveal the need for rapid and sensitive virus diagnostic devices. Due to the remarkable sensitivity and specificity of biosensors, they appear as a potential and promising tool for accurate and quantitative detection of viruses. Furthermore, recent advancements in transduction systems, nanotechnology and genetic engineering offer various strategies to improve the detection performance of biosensors. This review presents an overview of the current states of novel biosensing methodologies for the ultrasensitive detection of viruses with highly promising applications for future disease diagnosis. Additionally, a brief summary of the recent state-of-the-art virus diagnostic molecular technologies is included.
 Ming Soon Cheng | Ming Soon Cheng received his BSc (Hons) in Chemistry from National University of Singapore in the year 2010. He is currently pursuing his PhD under the supervision of Prof. Chee-Seng Toh at the Nanyang Technological University, Singapore. His research interests focus on analytical chemistry, electrochemistry and biosensors. |
 Chee-Seng Toh | Chee-Seng Toh obtained his PhD from University of Southampton, United Kingdom. He later continued his postdoctoral work at the California Institute of Technology, United States of America. At present, he is an assistant professor at the Nanyang Technological University. His research interests include environmental sustainability, biosensors and nanotechnology. |
1. Introduction
In recent decades, there have been increasing cases of emerging and re-emerging diseases brought about by infectious viruses including dengue virus (DENV), influenza virus, hepatitis virus, human immunodeficiency virus (HIV), Japanese encephalitis virus (JEV) and severe acute respiratory syndrome coronavirus. These viruses are capable of fast spreading and thus create ongoing threats to global public health. Viruses utilise different mechanisms to invade hosts, by relying on the machinery and metabolism of host cells for self-replication. The ability of viruses to mutate rapidly along with a complex interplay between different aspects such as global movement of human and animals, demographic shift, climatic and ecological changes contribute to the emergence of numerous infectious diseases.1 Therefore, facing the challenges and threatening consequences caused by the spread of infectious diseases, high throughput virus surveillance and diagnosis to achieve effective disease control have become the major concerns of the public and organisations.2At present, biosensing techniques have been developed into sensitive, specific and rapid diagnostic tools for different pathogens. Furthermore, attributed to inexpensive instrumentation, minimal or no sample pre-treatment, easy operation and rapid analysis capability, biosensors are envisaged as a competent technique to modernise virus diagnostic methods.3 This review will look at the current state and future perspective of biosensors based on electrochemical and optical transducers in virus diagnosis. Furthermore, different approaches to improve the analytical performance of biosensors such as surface functionalisation, nanotechnology, strategies for the labelling and immobilisation of biorecognition elements will be discussed in detail (in this review, the term “biorecognition elements” is used interchangeably with “affinity reagents”). Besides, a concise summary of the novel molecular technologies with regard to virus detection is also included in this review.
2. State-of-the-art molecular technologies
Conventional virus diagnostic methods include virus isolation, immunofluorescence microscopy, enzyme immunoassays and conventional qualitative polymerase chain reaction (PCR) techniques are becoming obsolete for routine clinical practices. Enzyme immunoassays such as enzyme-linked immunosorbent assay (ELISA),4 enzyme-linked immunospotting (ELISPOT)5 and others have been widely employed for high throughput analysis of multiple samples. However, the laboratory usage of these methods are limited by time-consuming analysis (3–5 h), false negative results attributed to low virus titre during the early stages of infection and the need to obtain multiple serum samples from patients for serological diagnosis during different phases of viral infection.Recent laboratory virus diagnostic tests rely exclusively on real-time PCR (RT-PCR) and multiplex PCR techniques. RT-PCR assays usually involve RNA extraction, reverse transcription and polymerase chain reaction. The advantages of RT-PCR assays include high sensitivity and reduced chances of generating false results. Nevertheless, the diagnostic application of RT-PCR is limited due to the fact that only a single gene or pathogen can be detected in a single test. As a result, multiplex PCR assays emerged as a more promising technique, which is capable of detecting multiple agents in a single specimen and simultaneous amplification of nucleic acids. These assays incorporate multiple primers with each primer set designed in such a way that an amplicon with specific size is generated for a particular pathogen. Ultimately, multiplex PCR has been integrated with RT-PCR for the simultaneous detection of multiple specific sized amplicons.6,7
In addition, a new generation of isothermal nucleic acid amplification methods such as loop-mediated isothermal amplification, nucleic acid sequence-based amplification, helicase-dependent amplification and insulated isothermal PCR methods are gaining popularity as alternative molecular tests. These methods involve simple procedures, easy handling and rapid reaction. Additionally, the need for temperature cycling and expensive instrumentation can be avoided.8,9 Furthermore, different multiplex technologies have emerged, some of which permit rapid detection of multiple pathogens in a single test. For example, RespPlex technology engages PCR for the purification and amplification of nucleic acids followed by detection using a liquid-phase bead-based array technique. Other related integrated molecular systems available nowadays include Infiniti system, Jaguar system, film array system, scalable target analysis routine technology and PLEX-ID technology (see ref. 10 for the detailed operation of each of these technologies).10
3. Affinity reagents and viral targets
Based on the different types of affinity reagents and viral targets, virus biosensors using affinity interactions can be classified into four main categories namely immuno- (antibody-), DNA-, antigen- and cell-based biosensors (see Fig. 1). These affinity reagents and viral targets are categorised in Fig. 2. Immunosensors are a subclass of biosensors, which relies on the specific interactions between antigens and antibodies to produce measurable responses. Immunosensing assays are broadly used in clinical applications since 1970s to identify antigenic components and diagnose pathogenic diseases. Antibodies, also known as immunoglobulins, are produced in humans and animals by injecting small sub-lethal doses of antigens into the hosts' bodies to trigger their immune responses that generate specific antibodies against the invading antigens.11 Moreover, successful isolation of specific antibodies for a wide spectrum of analytes has quickened the growth of immunosensors over recent decades.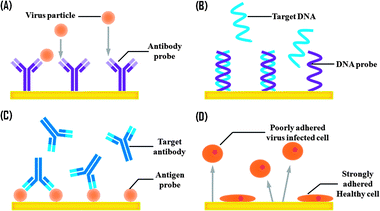 | ||
| Fig. 1 Schematic of virus biosensors based on the different types of affinity reagents for the relevant targets. (A) Immuno- (or antibody-), (B) DNA-, (C) antigen- and (D) cell-based biosensors. | ||
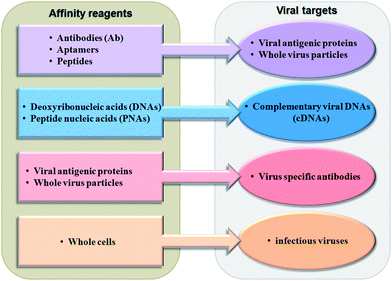 | ||
| Fig. 2 List of affinity reagents with their corresponding viral targets. | ||
Aptamers are single-stranded oligonucleic acid (ssDNA and ssRNA) or peptide molecules (∼40–60 bases in length) that recognise target viral antigens with high selectivity and affinity. Aptamers are usually selected from a pool of completely random oligonucleic acid using the Cell-SELEX (systematic evolution of ligands by exponential enrichment) technique.12 DNA aptamers recognise virus targets by adopting a three-dimensionally preferred orientation that differentiates targets based on subtle structural differences.13 Whereas peptide aptamers imitate the function of antibodies, and are made up of engineered protein with specific target binding sites located at peptide surfaces or loops.14
DNA-based biosensors operate based on the principle of nucleic acid hybridisation. These are constructed by immobilising short single stranded DNAs (ssDNAs, ∼20–40 base pairs) on the sensor surface with retained stability, reactivity and accessibility toward target DNA strands. Recent efforts in DNA-based biosensing applications emphasise the use of peptide nucleic acids (PNAs). PNAs, first reported by Nielsen et al. in 1991, are a structural mimic of DNA in which the sugar phosphate backbone is replaced by the pseudopeptide backbone comprising N-(2-aminoethyl) glycine units. PNAs exhibit excellent stability, and are capable of rapid, specific and strong hybridisation, making these a promising tool in DNA detection.15,16
Biosensors based on antigenic probes such as whole virus particles and surface antigens (capsids, envelopes, nucleocapsid proteins) have been reported lately.17,18 The diagnostic applications of these biosensors commonly involve the detection of virus-specific antibodies derived from human sera of patients. However, similar to the conventional serological tests and enzyme-linked immunoassays, the utility and reliability of these biosensors are restricted by the amount of antibodies produced during different phases of infection.
Apart from that, cell-based biosensing assays have been utilised as common alternatives to conventional animal testing in investigating infectious diseases. Sensors with pre- or post-infected cells adhered onto the sensor surface are employed to provide a detailed profile of cell adhesion in response to viral infection. Infections induce degenerative morphological changes, detachment, membrane degradation and eventual cell death in cell cultures, which are collectively known as cytopathic effects (CPEs).19 These effects are reflected quantitatively in the detected signals of these cell-attached sensors such as changes in conductance, resistance (electrochemical)20 and optical signals, thus allowing viral infection to be diagnosed, monitored and studied in a real-time fashion. The assay is highly advantageous in being capable of identifying infectious virus particles.
A comprehensive list of virus biosensors reported within the recent three years and the analytical performance in terms of limits of detection (LOD) and linear working range are summarised in Table 1.
| Viral target | Affinity reagent | Approach | Transducer (s) | LOD | Linear range | Reference |
|---|---|---|---|---|---|---|
| a Abbreviations—Ab: antibody, AEP: acetone-extracted propolis, AgNP: silver nanoparticle, AuNP: gold nanoparticle, C protein: capsid protein, cDNA: complementary DNA, CNT: carbon nanotube, CTAB: cetyltrimethylammonium bromide, DENV: dengue virus, E protein: envelope protein, EBV: Epstein–Barr virus, EIS: electrochemical impedance spectroscopy, FRET: fluorescence resonance energy transfer, GA: glutaraldehyde, GBP: gold-binding polypeptide, GCE: glassy carbon electrode, GE: gold electrode, GlyP: glycoprotein, HA protein: hemagglutinin protein, HAU: hemagglutination unit, HRP: horseradish peroxidase, HV: hepatitis virus, HBV: hepatitis B virus, HCV, hepatitis C virus, HIV, human immunodeficiency virus, HPV: human papillomavirus, IAV: influenza A virus, IDA: interdigited electrode array, IgG: immunoglobulin G, JEV: Japanese encephalitis virus, LAMP, loop-mediated isothermal amplification, LMP-1: latent membrane protein 1, mAb: monoclonal antibody, MWCNT: multi-walled carbon nanotube, N protein: nucleocapsid protein, NP: nanoparticle, NS1: non-structural protein 1, pAb: polyclonal antibody, NuP: nucleoprotein, PDI: N,N-bis-(1-aminopropyl-3-propylimidazol salt)-3,4,9,10-perylene tetracarboxylic acid diimide, PICA: poly(indole-6-carboxylic acid), pfu: plaque forming unit, PNA: peptide nucleic acid, QD: quantum dot, RT: reverse transcriptase, SAM: self-assembled monolayer, RABV: rabies virus, S-OIV: swine-origin influenza virus, SARS: severe acute respiratory syndrome coronavirus, SPE: screen-printed electrode, SPR: surface plasmon resonance, ssDNA: single stranded DNA, SWCNT: single-walled carbon nanotube, TADC: 3-thiophene-acetamide-diaza-18-crown-6, VACV: vaccinia virus, VSV: vesicular somatitis virus. | ||||||
| AIV, surface antigen | pAb/IgG | GBP/multi-spot Au capped NP array chip | SPR | 1 pg mL−1 | 1 pg mL−1 to 1 μg mL−1 | Park et al.21 |
| AIV, antigen | mAb | QD based fluorescence quenching immunoassay | Fluorescence | 0.09 ng mL−1 | 0.27–12 ng mL−1 | Li et al.22 |
| AIV, cDNA | DNA | Aptamer/hybrid nanomaterial modified GE | Voltammetry | 0.43 pM | 5 pM to 1 nM | Liu et al.13 |
| AIV, cDNA | DNA | SAM thiolated ssDNA probe modified GE | Voltammetry | 24 pM | 10–60 pM | Malecka et al.23 |
| AIV, HA protein | DNA aptamer | Portable DNA aptamer SPR biosensor | SPR | 0.128 HAU | 0.128–1.28 HAU | Bai et al.24 |
| AIV, cDNA | DNA | Sandwich hybridisation assay-QD induced FRET | FRET | 0.27 nM | 0.5–50 nM | Chou et al.25 |
| DENV, NS1 | mAb, IgG | Fuel cell Prussian blue nanotube membrane | Amperometry | 0.04 pfu mL−1 | 3–45 pfu mL−1 | Toh et al.26 |
| DENV, NS1 | mAb, IgG | Nanoporus alumina membrane electrode | Voltammetry | 1 pfu mL−1 | 1–103 pfu mL−1 | Toh et al.27 |
| DENV, NS1 | mAb | Protein A coated gold CD trode | Voltammetry | 0.33 ng mL−1 | 1–100 ng mL−1 | Cavalcanti et al.28 |
| DENV, NS1 | mAb, IgG | Nanoporus alumina membrane electrode | EIS | 1 pfu mL−1 | 1–900 pfu mL−1 | Toh et al.29 |
| DENV, cDNA | DNA | Nanoporus alumina membrane electrode | Voltammetry | 9.55 pM | 1 pM–1 μM | Toh et al.30 |
| DENV, cDNA | DNA | Pencil graphite electrode | Voltammetry | 0.92 nM | 1–40 nM | Souza et al.31 |
| EBV, LMP-1 | Ab | QD/silica NP label-sandwiched immunoassay | Voltammetry | 1 pg mL−1 | 1 pg mL−1 to 10 ng mL−1 | Chen et al.32 |
| HV, antigens | mAb | Protein A/nanogold modified GE | Voltammetry | ≤1.0 ng mL−1 | ∼1–350 ng mL−1 | Tang et al.33 |
| HBV, IgG | E protein | AuNP based magnetosandwich immunoassay | Chronoamperometry | 3 mIU mL−1 | 5–69.2 mIU mL−1 | Escosura-Muniz et al.17 |
| HBVs pAb | Surface antigen | GBP/surface antigen–gold surface | SPR | 1 pg mL−1 | 1 pg mL−1 to 10 μg mL−1 | Zheng et al.18 |
| HBV, surface antigen | Single chain Ab | GBP/single chain Ab fusion protein—GE | EIS | 0.14 ng mL−1 | 1 ng mL−1 to 50 μg mL−1 | Heo et al.34 |
| HBV, surface antigen | Single chain Ab | GBP/single chain Ab fusion protein–gold surface | SPR | 0.1 ng mL−1 | 0.1 ng mL−1 to 10 μg mL−1 | Zheng et al.18 |
| HBV, surface antigen | mAb | Monoclonal Ab modified gold nanorod | SPR | 0.01 IU mL−1 | 0.01–1 IU mL−1 | Wang et al.35 |
| HBV, cDNA | DNA | Multiprobe modified DNA–AuNP aggregate | Voltammetry | 5 aM | 10 aM to 100 fM | Li et al.36 |
| HBV, cDNA | DNA | PICA–MWCNT composite modified GCE | Voltammetry | 2 fM | 10 fM to 1 pM | Nie et al.37 |
| HBV, cDNA | DNA | SWCNT–AuNP array | EIS | 1 aM | 1 aM to 1 μM | Wang et al.38 |
| HBV, cDNA | DNA | Polycarbonate based SPR LAMP sensing cartridge | SPR | 2 fg mL−1 | 2 fg mL−1 to 10 μg mL−1 | Chuang et al.39 |
| HBV, cDNA | DNA | Fluorescein/ssDNA-CTAB/gold nanorod | FRET | 15 pM | 45 pM to 6 nM | Lu et al.40 |
| HCV, core antigen | Ab | Nanocomposite modified GCE | Voltammetry | 0.17 ng mL−1 | 2–512 ng mL−1 | Ma et al.41 |
| HCV, cDNA | DNA | AuNP coupled hairpin DNA probe | Voltammetry | ∼1 pM | 0.01–2 nM | Cai et al.42 |
| HCV, cDNA | PNA | SAM thiolated PNA probe modified GE | Voltammetry | 0.57 pM | 1–50 nM | Hejazi et al.15 |
| HCV, cDNA | PNA | SAM thiolated PNA probe modified GE | Voltammetry | 1.8 pM | 10–100 pM | Pournaghi-Azar et al.16 |
| HCV, cDNA | DNA | BamHI–thionine/HRP/GHS modified GCE | Voltammetry | 1 pM | 0.01 nM to 8 μM | Tang et al.43 |
| HCV, protease | Peptide | Ferrocene conjugated peptide modified GE | EIS | 5 pM | 10–100 pM | Sowole et al.44 |
| HCV, cDNA | DNA | PCR + thionine/AuNP/DNA probe modified GCE | Voltammetry | 0. 31 zM | ∼1 zM to 0.1 nM | Cai et al.45 |
| HCV, RNA | PNA | TADC SAM PNA probe modified GE | EIS | 23 pM | <10–500 nM | Park et al.46 |
| HIV, C protein | pAb | AuNP/MWCNT/AEP film modified GE | Voltammetry | 6.4 pg mL−1 | 0.01–60 ng mL−1 | Kheiri et al.47 |
| HIV, C protein | mAb | AuNP electroplated GCE | Voltammetry | 8 pg mL−1 | 0.01–100 ng mL−1 | Zheng et al.48 |
| HIV, C protein | mAb | Magnetism controlled CNT modified SPE | Voltammetry | 0.32 μg mL−1 | 0.6–160 μg mL−1 | Gan et al.49 |
| HIV, cDNA | DNA | AgNP/CNT modified gold microelectrode | Voltammetry | 0.5 pM | 1–100 pM | Wang et al.50 |
| HIV, cDNA | DNA | Long range SAM DNA nanostructures | Voltammetry | 2 aM | 2 aM–10 pM | Chen et al.51 |
| HIV, cDNA | DNA | Paramagnetic microparticles/CNT modified SPE | Voltammetry | 20 ng mL−1 | 40–1250 ng mL−1 | Adam et al.52 |
| HIV, cDNA | DNA | Chitosan/Fe3O4 NP modified SPE | Voltammetry, EIS | 50 pM | 50–300 pM | Lam et al.53 |
| HIV, cDNA | DNA | PDI/graphene modified GCE | EIS | 0.55 pM | 1 pM to 1 μM | Hu et al.54 |
| HIV, cDNA | DNA | Molecular beacon–nucleic acid dye SYBR Green I | Fluorescence | 30 pM | 80 pM to 8 nM | Xiang et al.55 |
| HIV, RT | Peptide | Ferrocene-labelled lipoic acid–AuNP/SPCE | Voltammetry, EIS | 0.8 pg mL−1 | 1–500 pg mL−1 | Labib et al.56 |
| HPV, C protein | Peptide aptamer | Polyaniline/MWCNT modified platinum IDA | Voltammetry | 0.49 nM | 10–50 nM | Tran et al.14 |
| HPV, cDNA | DNA | Thiolated probe/bipodal thiol-GE array | Amperometry | 0.17 nM | 0.1–12 nM | Civit et al.57 |
| HPV, cDNA | DNA | SWCNT–AuNP array | EIS | 1 aM | 1 aM to 1 pM | Wang et al.38 |
| IAV, whole virus | pAb | Neutravidin/biotinylated Ab/thiol modified GE | EIS | 8 ng mL−1 | 0–64 ng mL−1 | Hassen et al.58 |
| IAV, cDNA | DNA | Avidin/biotinylated DNA probe/modified GCE | Voltammetry | 85.1 fM | 0.1 nM to 0.1 pM | Chung et al.59 |
| IAV, cDNA | DNA | Paramagnetic microparticles/CNT based SPE | Voltammetry | 12 ng mL−1 | 40–1250 ng mL−1 | Adam et al.52 |
| IAV, cDNA | DNA | QD/ssDNA-oxidised CNT | FRET | 9.39 nM | 0.01–20 μM | Tian et al.60 |
| JEV, E protein | mAb/IgG | MWCNT/magnetic beads/HRP and magnetic GE | Voltammetry | 2 × 103 pfu mL−1 | 2 × 103 to 5 × 105 pfu mL−1 | Li et al.61 |
| JEV, antigen | Serum Ab | Protein A/GA-silanised interdigited electrode | EIS | 0.75 μg mL−1 | 1–10 μg mL−1 | Huy et al.62 |
| RABV, NuP/GlyP | mAb/IgG | Monoclonal Ab mixture linked flow cell on chip | SPR | 70 pg mL−1 | ∼1.96–125 mg mL−1 | Xu et al.63 |
| SARS, N protein | RNA aptamer | QD conjugated RNA aptamer on glass chip | Fluorescence | 0.1 pg mL−1 | ∼0.1–5 pg mL−1 | Roh et al.64 |
| S-OIV, NuP | mAb | Dual channel-paired surface plasmon waves | SPR | 30 pfu mL−1 | 18–1.8 × 106 pfu mL−1 | Su et al.65 |
| S-OIV, HA protein | pAb | Localised surface plasmon/fluorescence/fibre optic | Fibre optic | 13.9 pg mL−1 | 5–50 ng mL−1 | Chang et al.66 |
| VACV, whole virus | DNA aptamer | SAM thiolated primer/aptamer-AuNP/SPE | Voltammetry | 2 × 103 pfu mL−1 | 5 × 103 to 3 × 104 pfu mL−1 | Berezovski et al.67 |
| VSV, whole virus | DNA aptamer | Aptamer mediated immunoshielding of virus | EIS | 104 pfu mL−1 | 5 × 104 to 5 × 106 pfu mL−1 | Berezovski et al.68 |
4. Electrochemical biosensors
An electrochemical biosensor incorporates the sensitivity of electroanalytical techniques and the specificity of biorecognition elements to detect chemical and biological analytes. Since the first biosensor was invented in 1962 by Clark and Lyons for the detection of blood glucose using a glucose oxidase coated electrode,69 there has been increasing interest in utilising biosensors for environmental monitoring, clinical, agricultural and industrial applications. Biosensors with electrochemical transducers possess advantages such as high sensitivity, cost effectiveness, simple instrumentation and the possibility of miniaturisation with micro-litre sample volume. In principle, the biorecognition processes in electrochemical biosensors generally produce quantifiable current at varying potential (voltammetry), changes in conductivity of solutions (conductometry), measurable potential without drawing appreciable current (potentiometry) or opposition of a circuit to the current flow (impedance). Virus detection methodologies based on voltammetric and impedimetric techniques will be mainly focused in this review because these are sensitive to the affinity interactions between probes and targets.4.1. Voltammetric biosensors
Voltammetry comprises a group of electroanalytical techniques that depend on the measurement of current as a function of applied potential. Voltammetric techniques consist of linear sweep voltammetry, cyclic voltammetry, differential pulse voltammetry, square wave voltammetry, hydrodynamic voltammetry and stripping voltammetry.70 In addition, voltammetry has proven very useful as an initial screening tool for acquiring information about a novel or complicated system.71 Voltammetric biosensors often utilise screen-printed electrodes (SPEs) because of the disposability, low cost, and minimal sample requirement. Also, problems commonly encountered in electrochemical measurements such as electrode fouling and memory effects in analysing multiple biological samples are avoided.Nanoscale materials exhibit tunable chemical and spectral properties by varying sizes and shapes.72 Furthermore, because these nanomaterials are also electro-active, functionalisation of these materials with biorecognition elements is frequently employed for electronic signal amplification. Nanoparticles such as gold (AuNP), silver (AgNP) and iron oxide (Fe3O4NP) offer appealing features such as excellent biocompatibility, conductivity, and adsorption capability.36,48 By using a nanocomposite material comprising Fe3O4 nanoparticles and chitosan as the stabilising agent, Lam et al. have reported a square wave voltammetric biosensor for the detection of HIV DNA with a dynamic range of 50–300 pM.53 In another work, Chen et al. have described a voltammetric sandwich immunoassay using cadmium telluride quantum dot (QD) capped silica (SiO2) nanoparticle labels to improve the detection sensitivity. A low LOD of 1 pg mL−1 Epstein–Barr virus (EBV) antigen is achieved by the authors.32
Electrode surface modification is regularly performed to improve the conductivity, reversibility and sensitivity of virus biosensing assays. Lately, nanocomposite films constructed from AuNPs and carbon nanotubes (CNTs) offer a promising platform for the sensitive detection of viral antigens and DNAs. A CNT exhibits excellent electrical conductivity, mechanical strength, thermal and chemical stability.73 A AuNP–CNT film provides an appropriate surface for the immobilisation of biorecognition elements and eliminates non-specific adsorption of interfering biocomponents. Furthermore, the synergistic interactions of CNT and AuNP significantly increase the electron transfer efficiency at the electrode–solution interface.47 Another nanocomposite film made up of zirconia (ZrO2) nanoparticles, AuNPs and chitosan has been reported by Ma et al. to detect hepatitis C virus (HCV). ZrO2 nanoparticles display high thermal stability, chemical inertness and high binding affinity toward proteins.41
Our research group has fabricated platinum electrodes modified with ∼0.3–0.5 μm thick nanoporous alumina layers, which provide a suitable surface for the direct immobilisation of affinity reagents. Based on the described platform, ultrasensitive detection of DENV antigen and DNA with low LODs of 1 pfu mL−1 and 9.55 pM respectively have been reported (see Fig. 3 for the scheme of operation).27,30 Our research group also recently reported a novel fuel cell DENV biosensor using a DENV specific antibody coated Prussian blue nanotube (PB-nt) membrane electrode. The biosensor operates without the need for voltage power input but depends on the presence of hydrogen peroxide for powering the PB-nt biosensor. By employing amperometry, an electroanalytical technique which measures current at a fixed potential,70 we demonstrated an excellent LOD of 0.04 pfu mL−1 and a dynamic range of 3–45 pfu mL−1.26
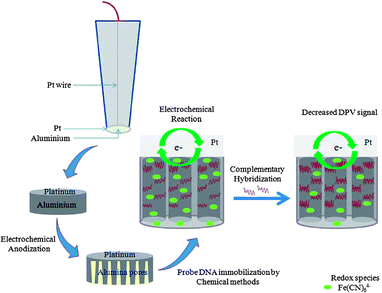 | ||
| Fig. 3 Scheme of construction and operation of a nanoporous alumina membrane based DNA biosensor. Reproduced from ref. 30. | ||
Magnetic beads (MBs) are recognised as a versatile tagging tool for different affinity reagents.74 Target analytes are preconcentrated onto the surface of these affinity reagent modified MBs and subsequently separated from the biological or chemical media by applying a magnetic field. In this way, signal interferences arising from matrix effects can be notably reduced. By immobilising hepatitis B virus (HBV) onto MBs, the immunoglobulin G (IgG) antibody specific to HBV can be detected down to 3 mIU mL−1 based on a magnetic sandwich chronoamperometric immunoassay.17 Chronoamperometry measures current as a function of time while potential is stepped. The advantage of this technique is that time resolution is only limited by the data collection frequency of the electrochemical instrument. However, the reliability and accuracy of the quantitative detection system are restricted by the low chemical selectivity.70
Alkaline phosphatase (ALP)75 and horseradish peroxidase (HRP)43 are among the most widely used enzymes to amplify signals in electrochemical assays. Compared to ALP, HRP exhibits greater stability and higher turnover rate. Moreover, HRP uses a wide spectrum of organic compounds as electron donors or acceptors. In a recent work, Tang et al. reported a novel HCV DNA detection system based on dual enzymatic activity: site specific cleavage by BamHI endonuclease and signal amplification by HRP encapsulated in a nanogold hollow sphere. Authors demonstrated that the high enzymatic activity and electron transfer efficiency are preserved through encapsulation.43
4.2. Impedimetric biosensors
Electrochemical impedance spectroscopy (EIS) has recently gained popularity for the diagnostic applications. It measures the resistance and reactance in the biosensor by employing very small sinusoidal potential excitation signals, which cause only minimal perturbation to an electrochemical system. EIS measurements are conducted over a range of alternating current (AC) frequencies to generate impedance spectra, from which information about the electron transfer kinetics and diffusional characteristics can be extracted.76 In spite of complicated data analysis and expensive instruments, EIS has demonstrated the potential capabilities, which include non-invasive measurements, useful on high resistive materials, availability of time-dependent and quantitative data.To address the problems of random orientation and weak adsorption of affinity reagents on the surface of an electrode, deposition of a thiol SAM onto a gold electrode is often performed. Thiol groups (–SH) are chemisorbed onto the gold surface through strong Au–S bonds.77 These groups allow covalent immobilisation of affinity reagents onto the surface of the electrode, forming an oriented, highly ordered and densely packed monolayer.58 Another electrode surface modification approach using silanisation similarly produces a well-organised SAM of silane. However, the target electrode surface has to be polar, for instance, possessing hydroxyl and oxide groups.78
Apart from that, intermediate linker molecules are also broadly utilised to capture affinity reagents in right orientation on the electrode surface. Among them, avidin, streptavidin and neutravidin, which are derived from both avians and amphibians, are the most popularly used linkers due to the considerable affinity toward biotin. Avidin–biotin interaction is known as the strongest non-covalent interaction (dissociation constant = 10−15 M).79 Moreover, an avidin–biotin complex is unaffected by the extreme of temperature, pH, denaturating agents and organic solvents. Neutravidin–biotin based detection of influenza A virus (IAV) antigens has been reported by Hassen et al. lately with a reported LOD of 8 ng mL−1. Authors have demonstrated insignificant interferences related to non-target proteins based on the described immunosensor.58 Besides, immunosensing techniques commonly employ protein A molecules as the intermediate linkers between antibody and SAM modified electrode. Protein A possesses great loading capability, biocompatibility, and binding affinity toward antibody molecules. A recent work has been performed using a protein A/glutaraldehyde coated silanised immunosensor in detecting JEV antigens, with an acceptable LOD of 750 ng mL−1.62
To date, the electrochemical cell–substrate impedance sensing (ECIS) technique has shown great potential in automated, real-time investigation of cellular behaviour, and response toward drugs and viruses. The operation involves initial culturing of cells on the electrode surface. Virus induced CPEs on cell's growth and adhesion are then monitored in a real-time fashion by applying small amplitude AC electric field. Cell membranes are essentially non-conducting and therefore changes in impedance signal responses can be correlated with the alterations in size or coverage of cells on the electrode surface.80 A microfluidic system based on conductive polymer microelectrode array has been reported by Killerich-Pederson et al. for the monitoring of cytomegaloviral infection of human foreskin fibroblast cells. Infection induced signal changes are detectable within 3 hours post-infection, a significant advantage in terms of reduced time frame of virus diagnostics compared to conventional inspection methods.20
Extensive research on aptamer-based electrochemical virus biosensors has been conducted by Berezovski and co-workers. The fabricated biosensing platforms generally comprise a thiolated DNA primer which hybridised to the complementary end of a virus specific aptamer. This hybrid is subsequently self-assembled onto the surface of the electrode (see Fig. 4). Electrode surface modification and binding of virus are characterised using both EIS and voltammetric techniques. Authors have demonstrated the feasibility of these electrochemical aptasensors for the application in quantification of virus, viability assessment and virus immunoshielding.67,81
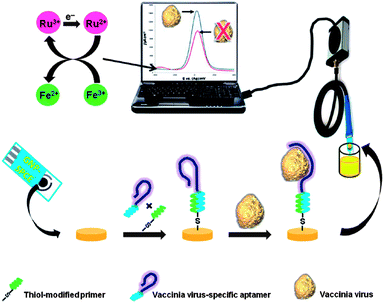 | ||
| Fig. 4 Schematic diagram of the electrochemical detection protocol adopted in this study. A thiolated DNA primer is hybridised with a complementary end of a vaccinia virus (VACV)-specific aptamer and the hybrid was self-assembled onto a gold nanoparticle-modified screen-printed carbon electrode (GNP-SPCE). Binding of the virus to the immobilised aptamer causes an increase in the redox current, measured via square wave voltammetry. Reproduced from ref. 67. | ||
5. Optical biosensors
Similar to other sensing technologies, biosensors with an optical type transducer rely on the specific interaction between viral target and affinity reagents. Because of the advantages of high sensitivity, rapid response and capability of real-time monitoring, optical biosensing techniques have been widely utilised to investigate DNA–DNA hybridisations, and antigen–antibody and protein–protein interactions.18,40,66 The operation of optical biosensors usually involves monitoring of the changes in luminescence intensity and reflective index and the difference between angles of incident and reflected lights.82 The detection performance and current states of optical virus biosensing methodologies including surface plasmon resonance (SPR)-, fibre optic- and fluorescence-based, will be discussed in this review. Without the need for direct electrical connection, these techniques provide a promising platform for the sensitive and rapid virus detection.5.1. Surface plasmon resonance (SPR)-based biosensors
Surface plasmons (SPs) refer to the light stimulated longitudinal electron density waves that exist at the metal–dielectric interface. Whereas SPR represents a collective electron oscillation which takes place upon frequency matching between incident light and oscillating electrons. Surface plasmon waves (SPWs) are able to penetrate into the dielectric medium ∼200 nm from the interface and highly sensitive to the changes in effective refractive index of the dielectric medium.83 In principle, total internal reflection of incident light in a glass prism will eventually establish a SP that propagates along the ambient-metal interface.The SPR techniques have been in use for over 3 decades since Nylander and Liedberg demonstrated the use of SPR in biosensing applications and gas detection in the year 1982.84 However, the limits on the assay sensitivity of conventional SPR methods render them incapable of detecting ultra trace amounts of analytes that generate negligible changes in the effective refractive index.85 Nanotechnology has provided a brand new range of useful approaches to improve the performance of SPR biosensing assays. For instance, SPR sensitivity can be significantly improved by using metal nanoparticles, which enhance plasmonic field and increase index contrast.86 In the context of nano-sized structures, the term “localised SPR (LSPR)” is technically employed. According to the Mie theory, movement of electrons in the internal framework of nano-sized metal particles is restricted, and this collective charge density oscillation is denoted as LSPR.87 Recent work conducted by Wang et al. in detecting HBV using the LSPR biosensor with monoclonal antibody coated gold nanorods (AuNRs) reported a LOD value of 0.01 IU mL−1. In comparison to AuNPs, AuNRs display higher light absorption and cross-sectional scattering capability, making it a superior tool in amplifying signal responses.35 In another novel work, Yu et al. reported a label free LSPR detection system based on nanoislands to detect adenovirus. LOD and sensitivity are significantly enhanced by the nanoisland platform, which localises and excites a SP, and controls related evanescent near fields. However, the synthesis of nanoislands produces random and uncontrollable patterns of which only some can generate improved results.88
A genetic fusion protein with metal binding property provides an alternative approach for the direct protein–gold surface interaction apart from the classic gold–thiol methodology. Gold-binding polypeptides (GBPs) are composed of repeats of 14–30 amino acid sequences in which the polar groups of peptides i.e. serine and threonine are believed to be responsible for the interaction with gold.89 Recent application of GBPs in virus diagnosis is demonstrated by Zheng et al. This label free LSPR method has been successfully applied to detect both HBV surface antigen and antigen specific antibody at LODs of 100 and 1 pg mL−1 respectively.18
Recently, the label free SPR-based Biacore technology made simultaneous profiling of up to 400 protein interactions and high throughput screening of over thousand analytes possible by using a single commercially available assay setup. In addition, Commercial Biacore capture kits equipped with sensor chips of different surfaces, either pre-immobilised with recognition elements or modified by different immobilisation methods, are available. Applications of Biacore sensor chips for the detection of different biomolecules including viruses have been reviewed by Gopinath.90
5.2. Optical fibre-based biosensors
An optical fibre is generally a transparent and flexible fibre. It consists of a cylindrical core and a cladding, both made of silica or plastic. A single optical fibre or bundles function as the dielectric waveguide where light is kept in the core by total internal reflection. Propagating light passing through an optical fibre is typically made up of a guide field in the core and an exponentially decaying evanescent field in the cladding.91Recent efforts focus on the SPR optical fibre-based transduction system that monitors the excitation of SPWs in a novel way. Lately, Francois et al. developed a SPR optical fibre-based biosensor for the label free, rapid and sensitive detection of IAV. In this study, an optical fibre is coated with a silver film at different sections, which function as sensing regions. Detection of IAV is conducted by collecting re-emitted lights (produced by the plasmonic surface scattering of SPWs) using an optical fibre which was placed within antibodies immobilised sensing regions. The combination of collection and spectral properties of evanescent fields in sensing regions noticeably improves the signal-to-noise ratio.92 Moreover, detection of S-OIV serotype H1N1 by a localised surface plasmon coupled fluorescence optical fibre-based biosensor (LSPCF-FOB) has been reported by Chang and co-workers. In this work, a low LOD of 13.9 pg mL−1 and a dynamic range from 5 to 50 ng mL−1 of S-OIV hemagglutinin protein are achieved using a sandwich immunoassay with a secondary antibody labelled LSPCF probe. These achievements can be attributed to the strong background signals, which are induced by the coaxial propagation of laser beams.66
5.3. Fluorescence-based biosensors
In general, these biosensors measure luminescent signals generated from fluorescence probes conjugated to affinity reagents or viral targets. A wide variety of specific fluorescence probes i.e. quantum dots,22,64 fluorescent nanoparticles93 and organic fluorescent dyes55 contributes to the recent development in fluorescence-based analytical techniques. Application of QDs as the fluorescence signal amplifier in detecting AIV and SARS-CoV has been reported.22,64 QDs possess excellent optical features such as bright, tunable, broad absorption and narrow emission spectra.94 Labelling of a DNA sample with Alexa Fluor® 555 dye doped SiO2 nanoparticles has been performed by Ricco et al. to enhance the signal intensity in detecting human papillomavirus (HPV). These luminescent nanoparticles display appealing features such as excellent stability in water, cost effectiveness, ease of fabrication and biocompatibility. Moreover, these nanoparticles are advantageous over QDs in terms of sensitivity improvement.93 In another work, He et al. made use of a dual colour fluorescence (SYBR Green I and 5-carboxyl-X-rhodamine dyes) system for the detection of HIV DNA. Molecular beacons which exhibit great affinity toward ssDNA hybridisation function as the DNA detection probe in this study. Fluorescence quantitative detection is performed by using synchronous scanning fluorescence spectrometry. Based on the described platform, sensitivity is significantly improved and a low LOD of 30 pM HIV DNA is attained.55Current biosensors based on fluorescence resonance energy transfer (FRET) have shown remarkable sensitivity for the quantitative detection of viruses. Principally, this physical phenomenon involves a non-contact, radiationless and distance-dependent energy transfer between two fluorophores, which takes place only when the donor fluorophore and the acceptor fluorophore have both spatial and spectral proximity. Thus, FRET introduces secondary specificity for the in situ monitoring of biomolecular interactions. FRET-based detections are usually conducted either by donor-quenching or emission-shift strategy. Donor-quenching detection methodologies i.e. FRET from fluorescein to AuNRs40 and QDs to oxidised CNTs60 have been reported for the detection of HBV antigens (LOD = 15 pM) and IAV DNA (LOD = 9.39 nM) respectively. In both cases, an ssDNA probe modified donor fluorophore is quenched by an electron acceptor which has its absorption peak overlapped with the emission peak of a donor fluorophore. On the other hand, a novel QD induced FRET-based nucleic acid sandwich hybridisation assay is reported by Chou et al. The detection assay exhibits two-dimensional signals, that is an increase in the target DNA concentration give rises to reduced capturing probe modified QD (donor) emission and enhanced reporter probe modified Alexa Fluor 660 dye (acceptor) emission (the scheme of operation is demonstrated in Fig. 5).25 Based on the measurement of emission shift, authors revealed a low LOD value of 0.27 nM AIV DNA.25
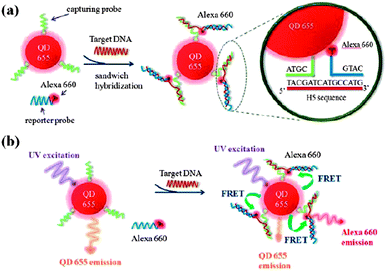 | ||
| Fig. 5 Schematic illustration of the sandwich hybridisation assay with a QD-induced FRET reporter system for hemagglutinin H5 target DNA detection: (a) sandwich hybridisation with a label-free H5 sequence (target) by the capturing probes conjugated on QD655 (FRET donor) and the reporter probes labelled with an Alexa Fluor 660 (FRET acceptor); (b) FRET emission shift before and after the sandwich hybridisation. Reproduced from ref. 25. | ||
6. Conclusion
It is clear that biosensing methodologies have achieved huge improvements for the virus detection in terms of LOD, linear detection range, sensitivity, specificity and response time. In spite of these achievements, biosensing techniques still remain nascent in diagnosis of infectious diseases. Increasing demands for effective virus surveillance and high throughput screening test signify the need to develop biosensors into point-of-care diagnostic devices which are rapid, cost effective and sensitive in analysing multiple samples. Among all the available detection techniques, biosensors are possibly the best candidates for development into hand-held pen-size testing tools. In urban and suburban areas, problems exist where infectious diseases spread well beyond initial affected regions and the control of diseases at this stage has often become a huge logistic problem for the veterinary services to deal with. Hence, rapid pen-size field test may become essential for the implementation of initial disease control. This would eliminate the delay in diagnosis caused by sending routine samples to specialised laboratories and time taken to deliver the test results. In addition, further research to discover and develop new materials i.e. nano-sized materials, composites and polymers with considerable biocompatibility may enhance the specificity and stability of affinity reagents. Apart from that, more studies have to be conducted to overcome the common problems encountered by most of the biosensors such as adsorption of non-specific biomolecules and interfering substances present in the complex biological samples (human sera and blood) in order to meet the demands of clinical chemistry. Recent technological advancements would certainly accelerate the development of biosensors into an essential, well-established diagnostic tool to accomplish the need of medical laboratories and hospitals for the routine examination of viral infections.Acknowledgements
The authors thank Singapore Immunology Network-Agency for Science, Technology and Research (SigN-A*STAR) for a research grant and M. S. Cheng acknowledges NTU for a PhD research scholarship.Notes and references
- A. Fenton and A. B. Pedersen, Emerging Infect. Dis., 2005, 11, 1815–1821 CrossRef PubMed.
- E. Spackman, Avian Pathol., 2012, 41, 251–258 CrossRef CAS PubMed.
- S. B. Shinde, C. B. Fernandes and V. B. Patravale, J. Controlled Release, 2012, 159, 164–180 CrossRef CAS PubMed.
- M. A. Hoque, L. F. Skerratt, S. Garland, G. W. Burgess and P. Selleck, Indian J. Virol., 2012, 23, 261–269 CrossRef PubMed.
- E. M. Linares, C. S. Pannuti, L. T. Kubota and S. Thalhammer, Biosens. Bioelectron., 2013, 41, 180–185 CrossRef CAS PubMed.
- J. J. Maurer, in Annual Review of Food Science and Technology, ed. M. P. Doyle and T. R. Klaenhammer, Annual Reviews, Palo Alto, 2011, vol. 2, pp. 259–279 Search PubMed.
- S. Olofsson, R. Brittain-Long, L. M. Andersson, J. Westin and M. Lindh, Expert Rev. Anticancer Ther., 2011, 9, 615–626 CrossRef CAS PubMed.
- F. Sidoti, M. Bergallo, C. Costa and R. Cavallo, Mol. Biotechnol., 2013, 53, 352–362 CrossRef CAS PubMed.
- Y. L. Tsai, H. T. T. Wang, H. F. G. Chang, C. F. Tsai, C. K. Lin, P. H. Teng, C. Su, C. C. Jeng and P. Y. Lee, PLoS One, 2012, 7 DOI:10.1371/journal.pone.0045278.
- A. M. Caliendo, Clin. Infect. Dis., 2011, 52, S326–S330 CrossRef PubMed.
- K. D. Elgert, Immunology: understanding the immune system, 2nd edn, John Wiley & Sons, Hoboken, New Jersey, 2009 Search PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- X. G. Liu, Z. Q. Cheng, H. Fan, S. Y. Ai and R. X. Han, Electrochim. Acta, 2011, 56, 6266–6270 CrossRef CAS PubMed.
- L. D. Tran, D. T. Nguyen, B. H. Nguyen, Q. P. Do and H. L. Nguyen, Talanta, 2011, 85, 1560–1565 CrossRef CAS PubMed.
- M. S. Hejazi, M. H. Pournaghi-Azar and F. Ahour, Anal. Biochem., 2010, 399, 118–124 CrossRef CAS PubMed.
- M. H. Pournaghi-Azar, F. Ahour and M. S. Hejazi, Anal. Bioanal. Chem., 2010, 397, 3581–3587 CrossRef CAS PubMed.
- A. de la Escosura-Muniz, M. Maltez-da Costa, C. Sanchez-Espinel, B. Diaz-Freitas, J. Fernandez-Suarez, A. Gonzalez-Fernandez and A. Merkoci, Biosens. Bioelectron., 2010, 26, 1710–1714 CrossRef CAS PubMed.
- S. Zheng, D. K. Kim, T. J. Park, S. J. Lee and S. Y. Lee, Talanta, 2010, 82, 803–809 CrossRef CAS PubMed.
- C. E. Campbell, M. M. Laane, E. Haugarvoll and I. Giaever, Biosens. Bioelectron., 2007, 23, 536–542 CrossRef CAS PubMed.
- K. Kiilerich-Pedersen, C. R. Poulsen, T. Jain and N. Rozlosnik, Biosens. Bioelectron., 2011, 28, 386–392 CrossRef CAS PubMed.
- T. J. Park, S. J. Lee, D. K. Kim, N. S. Heo, J. Y. Park and S. Y. Lee, Talanta, 2012, 89, 246–252 CrossRef CAS PubMed.
- X. P. Li, D. L. Lu, Z. H. Sheng, K. Chen, X. B. Guo, M. L. Jin and H. Y. Han, Talanta, 2012, 100, 1–6 CrossRef CAS PubMed.
- K. Malecka, I. Grabowska, J. Radecki, A. Stachyra, A. Gora-Sochacka, A. Sirko and H. Radecka, Electroanalysis, 2012, 24, 439–446 CrossRef CAS.
- H. Bai, R. H. Wang, B. Hargis, H. G. Lu and Y. B. Li, Sensors, 2012, 12, 12506–12518 CrossRef CAS PubMed.
- C. C. Chou and Y. H. Huang, Sensors, 2012, 12, 16660–16672 CrossRef CAS PubMed.
- Y. Y. Wei, L. P. Wong and C. S. Toh, Anal. Chem., 2013, 85, 1350–1357 CrossRef CAS PubMed.
- M. S. Cheng, J. S. Ho, C. H. Tan, J. P. S. Wong, L. C. Ng and C. S. Toh, Anal. Chim. Acta, 2012, 725, 74–80 CrossRef CAS PubMed.
- I. T. Cavalcanti, M. I. F. Guedes, M. Sotomayor, H. Yamanaka and R. F. Dutra, Biochem. Eng. J., 2012, 67, 225–230 CrossRef CAS PubMed.
- T. T. N. Binh, A. E. K. Peh, C. Y. L. Chee, K. Fink, V. T. K. Chow, M. M. L. Ng and C. S. Toh, Bioelectrochemistry, 2012, 88, 15–21 CrossRef PubMed.
- V. Rai, H. C. Hapuarachchi, L. C. Ng, S. H. Soh, Y. S. Leo and C. S. Toh, PLoS One, 2012, 7 DOI:10.1371/journal.pone.0042346.
- E. Souza, G. Nascimento, N. Santana, D. Ferreira, M. Lima, E. Natividade, D. Martins and J. Lima, Sensors, 2011, 11, 5616–5629 CrossRef CAS PubMed.
- L. Y. Chen, Z. J. Qi, R. J. Chen, Y. Li and S. Q. Liu, Clin. Chim. Acta, 2010, 411, 1969–1975 CrossRef CAS PubMed.
- D. P. Tang, J. Tang, B. L. Su, J. J. Ren and G. N. Chen, Biosens. Bioelectron., 2010, 25, 1658–1662 CrossRef CAS PubMed.
- N. S. Heo, S. Zheng, M. Yang, S. J. Lee, S. Y. Lee, H. J. Kim, J. Y. Park, C. S. Lee and T. J. Park, Sensors, 2012, 12, 10097–10108 CrossRef CAS PubMed.
- X. H. Wang, Y. A. Li, H. F. Wang, Q. X. Fu, J. C. Peng, Y. L. Wang, J. A. Du, Y. Zhou and L. S. Zhan, Biosens. Bioelectron., 2010, 26, 404–410 CrossRef PubMed.
- H. Li, Z. Y. Sun, W. Y. Zhong, N. Hao, D. K. Xu and H. Y. Chen, Anal. Chem., 2010, 82, 5477–5483 CrossRef CAS PubMed.
- G. M. Nie, Z. M. Bai, J. Chen and W. Y. Yu, ACS Macro Lett., 2012, 1, 1304–1307 CrossRef CAS.
- S. Wang, L. Li, H. L. Jin, T. Yang, W. W. Bao, S. M. Huang and J. C. Wang, Biosens. Bioelectron., 2013, 41, 205–210 CrossRef CAS PubMed.
- T. L. Chuang, S. C. Wei, S. Y. Lee and C. W. Lin, Biosens. Bioelectron., 2012, 32, 89–95 CrossRef CAS PubMed.
- X. C. Lu, X. Dong, K. Y. Zhang, X. W. Han, X. Fang and Y. Z. Zhang, Analyst, 2013, 138, 642–650 RSC.
- C. X. Ma, G. M. Xie, W. Zhang, M. Liang, B. Liu and H. Xiang, Microchim. Acta, 2012, 178, 331–340 CrossRef CAS.
- W. Li, P. Wu, H. Zhang and C. X. Cai, Chem. Commun., 2012, 48, 7877–7879 RSC.
- D. P. Tang, J. Tang, B. L. Su, Q. F. Li and G. N. Chen, Chem. Commun., 2011, 47, 9477–9479 RSC.
- M. A. Sowole and H. B. Kraatz, Analyst, 2012, 137, 1120–1124 RSC.
- S. N. Liu, P. Wu, W. Li, H. Zhang and C. X. Cai, Anal. Chem., 2011, 83, 4752–4758 CrossRef CAS PubMed.
- J. Y. Park, Y. S. Lee, B. Y. Chang, B. H. Kim, S. Jeon and S. M. Park, Anal. Chem., 2010, 82, 8342–8348 CrossRef CAS PubMed.
- F. Kheiri, R. E. Sabzi, E. Jannatdoust, E. Shojaeefar and H. Sedghi, Biosens. Bioelectron., 2011, 26, 4457–4463 CrossRef CAS PubMed.
- L. Zheng, L. Y. Jia, B. Li, B. Situ, Q. L. Liu, Q. Wang and N. Gan, Molecules, 2012, 17, 5988–6000 CrossRef CAS PubMed.
- N. Gan, N. X. Luo, T. H. Li, L. Zheng and M. J. Ni, Chin. J. Anal. Chem., 2010, 38, 1556–1562 Search PubMed.
- R. B. Wang, C. H. Xue, M. Gao, H. L. Qi and C. X. Zhang, Microchim. Acta, 2011, 172, 291–297 CrossRef CAS.
- X. Chen, C. Y. Hong, Y. H. Lin, J. H. Chen, G. N. Chen and H. H. Yang, Anal. Chem., 2012, 84, 8277–8283 CrossRef CAS PubMed.
- V. Adam, D. Huska, J. Hubalek and R. Kizek, Microfluid. Nanofluid., 2010, 8, 329–339 CrossRef CAS.
- D. T. Lam, H. N. Binh, V. H. Nguyen, V. T. Hoang, L. N. Huy and X. N. Phuc, Mater. Sci. Eng., C, 2011, 31, 477–485 CrossRef PubMed.
- Y. W. Hu, K. K. Wang, Q. X. Zhang, F. H. Li, T. S. Wu and L. Niu, Biomaterials, 2012, 33, 1097–1106 CrossRef CAS PubMed.
- D. S. Xiang, G. H. Zhou, M. Luo, X. H. Ji and Z. K. He, Analyst, 2012, 137, 3787–3793 RSC.
- M. Labib, S. Martic, P. C. Shipman and H. B. Kraatz, Talanta, 2011, 85, 770–778 CrossRef CAS PubMed.
- L. Civit, A. Fragoso, S. Holters, M. Durst and C. K. O'Sullivan, Anal. Chim. Acta, 2012, 715, 93–98 CrossRef CAS PubMed.
- W. M. Hassen, V. Duplan, E. Frost and J. J. Dubowski, Electrochim. Acta, 2011, 56, 8325–8328 CrossRef CAS PubMed.
- D. J. Chung, K. C. Kim and S. H. Choi, Appl. Surf. Sci., 2011, 257, 9390–9396 CrossRef CAS PubMed.
- J. P. Tian, H. M. Zhao, M. Liu, Y. Q. Chen and X. Quan, Anal. Chim. Acta, 2012, 723, 83–87 CrossRef CAS PubMed.
- F. Li, L. Mei, Y. M. Li, K. H. Zhao, H. C. Chen, P. Wu, Y. G. Hu and S. B. Cao, Biosens. Bioelectron., 2011, 26, 4253–4256 CrossRef CAS PubMed.
- T. Q. Huy, T. H. H. Nguyen, N. T. Thuy, P. Van Chung, P. T. Nga and M. A. Tuan, Talanta, 2011, 86, 271–277 CrossRef CAS PubMed.
- J. Xu, J. Y. Wan, S. T. Yang, S. F. Zhang, N. Xu, N. Li, J. P. Li, H. Y. Wang, X. Bai and W. S. Liu, Acta Vet. Brno, 2012, 81, 107–111 CrossRef CAS.
- C. Roh and S. K. Jo, J. Chem. Technol. Biotechnol., 2011, 86, 1475–1479 CrossRef CAS.
- L. C. Su, C. M. Chang, Y. L. Tseng, Y. F. Chang, Y. C. Li, Y. S. Chang and C. E. Chou, Anal. Chem., 2012, 84, 3914–3920 CrossRef CAS PubMed.
- Y. F. Chang, S. F. Wang, J. C. Huang, L. C. Su, L. Yao, Y. C. Li, S. C. Wu, Y. M. A. Chen, J. P. Hsieh and C. Chou, Biosens. Bioelectron., 2010, 26, 1068–1073 CrossRef CAS PubMed.
- M. Labib, A. S. Zamay and M. V. Berezovski, Analyst, 2013, 138, 1865–1875 RSC.
- D. Muharemagic, M. Labib, S. M. Ghobadloo, A. S. Zamay, J. C. Bell and M. V. Berezovski, J. Am. Chem. Soc., 2012, 134, 17168–17177 CrossRef CAS PubMed.
- L. C. Clark and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 CrossRef CAS.
- D. A. Skoog, D. M. West, F. J. Holler and S. R. Crouch, Fundamentals of analytical chemistry, 8th edn, Thomson Brooks/Cole, Belmont, California, 2004 Search PubMed.
- A. J. Bard and L. R. Faulkner, Electrochemical methods: fundamentals and applications, 2nd edn, John Wiley and Sons, New York, 2001 Search PubMed.
- K. C. Halfpenny and D. W. Wright, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 277–290 CrossRef CAS PubMed.
- C. B. Jacobs, M. J. Peairs and B. J. Venton, Anal. Chim. Acta, 2010, 662, 105–127 CrossRef CAS PubMed.
- E. Palecek and M. Fojta, Talanta, 2007, 74, 276–290 CrossRef CAS PubMed.
- M. Bandilla, A. Zimdars, S. Neugebauer, M. Motz, W. Schuhmann and G. Hartwich, Anal. Bioanal. Chem., 2010, 398, 2617–2623 CrossRef CAS PubMed.
- D. A. Skoog, F. J. Holler and S. R. Crouch, Principles of instrumental analysis, 6th edn, Thomson Brooks/Cole, Belmont, California, 2007 Search PubMed.
- F. Davis and S. P. J. Higson, Biosens. Bioelectron., 2005, 21, 1–20 CrossRef CAS PubMed.
- D. Mandler and S. Kraus-Ophir, J. Solid State Electrochem., 2011, 15, 1535–1558 CrossRef CAS PubMed.
- N. M. Green, Biochem. J., 1963, 89, 599–609 CAS.
- K. Cheung, S. Gawad and P. Renaud, Cytometry, Part A, 2005, 65, 124–132 CrossRef PubMed.
- M. Labib, A. S. Zamay, D. Muharemagic, A. Chechik, J. C. Bell and M. V. Berezovski, Anal. Chem., 2012, 84, 1677–1686 CrossRef CAS PubMed.
- G. G. Daaboul, C. A. Lopez, A. Yurt, B. B. Goldberg, J. H. Connor and M. S. Unlu, IEEE J. Sel. Top. Quantum Electron., 2012, 18, 1422–1433 CrossRef CAS.
- F. Yu, D. F. Yao and W. Knoll, Anal. Chem., 2003, 75, 2610–2617 CrossRef CAS.
- J. Homola, S. S. Yee and G. Gauglitz, Sens. Actuators, B, 1999, 54, 3–15 CrossRef CAS.
- L. He, M. D. Musick, S. R. Nicewarner, F. G. Salinas, S. J. Benkovic, M. J. Natan and C. D. Keating, J. Am. Chem. Soc., 2000, 122, 9071–9077 CrossRef CAS.
- S. Moon, Y. Kim, Y. Oh, H. Lee, H. C. Kim, K. Lee and D. Kim, Biosens. Bioelectron., 2012, 32, 141–147 CrossRef CAS PubMed.
- W. L. Barnes, A. Dereux and T. W. Ebbesen, Nature, 2003, 424, 824–830 CrossRef CAS PubMed.
- H. Yu, K. Kim, K. Ma, W. Lee, J. W. Choi, C. O. Yun and D. Kim, Biosens. Bioelectron., 2013, 41, 249–255 CrossRef CAS PubMed.
- T. J. Park, M. S. Hyun, H. J. Lee, S. Y. Lee and S. Ko, Talanta, 2009, 79, 295–301 CrossRef CAS PubMed.
- S. C. B. Gopinath, Sens. Actuators, B, 2010, 150, 722–733 CrossRef CAS PubMed.
- H. Mukundan, A. S. Anderson, W. K. Grace, K. M. Grace, N. Hartman, J. S. Martinez and B. I. Swanson, Sensors, 2009, 9, 5783–5809 CrossRef CAS PubMed.
- A. Francois, J. Boehm, S. Y. Oh, T. Kok and T. M. Monro, Biosens. Bioelectron., 2011, 26, 3154–3159 CrossRef CAS PubMed.
- R. Ricco, A. Meneghello and F. Enrichi, Biosens. Bioelectron., 2011, 26, 2761–2765 CrossRef CAS PubMed.
- A. M. Smith and S. M. Nie, Nat. Biotechnol., 2009, 27, 732–733 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2013 |
