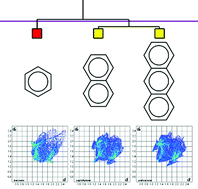
This paper describes a proof-of-concept example for a method that allows the calculation of a similarity index between whole molecular crystal structures; this has been termed ‘structural genetic fingerprinting’. It is based on the use of fingerprint plots derived from Hirshfeld surfaces coupled with cluster analysis and associated multivariate statistics. Using this formalism, it is possible to show quantitatively (using correlation coefficients) that, for example naphthalene is more similar to anthracene than to benzene, and moreover that benzodicoronene is more similar to anthrabenzonaphthopentacene than naphthalene is to anthracene. Whereas the correlation coefficients themselves obtained say nothing about the ways in which the patterns of intermolecular interactions are similar or different for two different structures, the fingerprint plots do contain such information. In principle this method for quantifying structural similarities of whole molecular crystal structures should be both robust and generally applicable. This method is potentially applicable to datasets consisting of many hundreds or even thousands of structures.