Recent progress of nanostructure-based enrichment of circulating tumor cells and downstream analysis
Lihua
Guo†
a,
Chang
Liu†
a,
Manlin
Qi
b,
Liang
Cheng
*b,
Lin
Wang
 b,
Chunxia
Li
b,
Chunxia
Li
 *c and
Biao
Dong
*c and
Biao
Dong
 *a
*a
aState Key Laboratory on Integrated Optoelectronics, College of Electronic Science and Engineering, Jilin University, Changchun, 130012, P. R. China. E-mail: dongb@jlu.edu.cn
bDepartment of Oral Implantology, Jilin Provincial Key Laboratory of Tooth Development and Bone Remodeling, School of Stomatology, Jilin University, Changchun, 130021, P. R. China. E-mail: chengliang@jlu.edu.cn
cInstitute of Molecular Sciences and Engineering, Shandong University, Qingdao, 266237, P. R. China. E-mail: cxli@sdu.edu.cn
First published on 30th January 2023
Abstract
The isolation and detection of circulating tumor cells (CTCs) play an important role in early cancer diagnosis and prognosis, providing easy access to identify metastatic cells before clinically detectable metastases. In the past 20 years, according to the heterogeneous expression of CTCs on the surface and their special physical properties (size, morphology, electricity, etc.), a series of in vitro enrichment methods of CTCs have been developed based on microfluidic chip technology, nanomaterials and various nanostructures. In recent years, the in vivo detection of CTCs has attracted considerable attention. Photoacoustic flow cytometry and fluorescence flow cytometry were used to detect CTCs in a noninvasive manner. In addition, flexible magnetic wire and indwelling intravascular non-circulating CTCs isolation system were developed for in vivo CTCs study. In the aspect of downstream analysis, gene analysis and drug sensitivity tests of enriched CTCs were developed based on various existing molecular analysis techniques. All of these studies constitute a complete study of CTCs. Although the existing reviews mainly focus on one aspect of capturing CTCs study, a review that includes the in vivo and in vitro capture and downstream analysis study of CTCs is highly needed. This review focuses on not only the classic work and latest research progress in in vitro capture but also includes the in vivo capture and downstream analysis, discussing the advantages and significance of the different research methods and providing new ideas for solving the heterogeneity and rarity of CTCs.
1 Introduction
Cancer-related diseases are currently a major global health problem with approximately eight million deaths each year.1 It is estimated that 28.4 million new cancer cases will occur in 2040, an increase of 47 percent over the corresponding 19.3 million cases in 2020.2 Pathological analysis based on tumor tissue is the “gold standard” for cancer diagnosis. It relies on invasive methods, including surgical resection and needle aspiration. These tissues contain key information, including histopathology and molecular atlas, which helps to classify, diagnose and predict the progression of cancer. However, invasive sampling methods have certain limitations. First, invasive sampling is expensive and the procedure is complicated. Second, invasive sampling methods may put patients at risk of injury. For example, advanced prostate cancer metastases are often found in hardened bones and require a larger drill to sample. In addition, tumors are recognized as having spatio-temporal heterogeneity, that is, the extent to which the biopsy sample can represent the tumor itself biologically and molecularly, depending on the sampling location and will change with the passage of treatment time. If the occurrence and metastasis of tumors can be detected earlier through non-invasive methods, the suffering of patients can be greatly reduced, tumors may be treated at an early stage, and the treatment and prognosis of cancer can be improved.Circulating tumor cells (CTCs) are cancer cells that shed from a primary tumor and spread to other organs through the bloodstream.3 CTCs may play a key role in cancer metastasis and is responsible for more than 90% of cancer-related deaths.4,5 CTCs are considered to contain the same genetic information with the CTCs of the primary tumor and metastatic sites. Therefore, a better understanding of CTCs is beneficial for exploring the mechanisms of primary tumor and metastasis. Isolation of these CTCs from blood, commonly referred to as a “liquid biopsy,” provides a minimally invasive method to obtain cancer cells for enumeration or molecular and cellular analysis.6,7 The number of CTCs has been shown to be predictive of cancer prognosis and may also assist with characterizing treatment efficacy.8 Indeed, an increase in CTCs may indicate that chemotherapy is no longer effective or that the cancer has changed, making tumor cells more resistant to current treatments.9 In particular, the number of CTCs can be used as an independent predictor and prognostic marker for various cancers, such as breast, prostate, liver, colorectal, and ovarian cancers.10–13 Molecular profiling of CTCs, such as monitoring of heterogeneous phenotypes, resistance from gene mutations, and functional protein activity, may provide insights into the mechanisms of cancer progression, and guide diagnosis and precise personalized therapy.14–19 Despite the enormous clinical value mentioned above, CTC analysis has not yet entered routine clinical practice. This is mainly due to the lack of practical techniques to isolate these rare, highly sensitive and selective tumor cells. Due to the difficulty of obtaining CTCs, understanding the mechanism of tumor metastasis through CTC analysis remains a great challenge.
CTCs carry a large amount of information about tumor occurrence, development, metastasis and drug resistance, and can provide real-time information about patient staging (metastatic and non-metastatic) and tumor molecular characteristics. The detection and diagnosis of CTCs provide extensive diagnosis and support for the early diagnosis of cancer metastasis, and might provide the entire development process of CTCs. From the earliest in vitro detection of blood collection based on the physical properties of CTCs, a lot of novel platforms and technologies have been developed for CTCs detection in the last two decades (Fig. 1). The method of in vitro testing refers to two processes, including extraction of blood from patients, as well as CTCs isolation from the whole blood. At present, the in vitro separation of CTCs is mostly based on microfluidic technology, which is captured according to the physical characteristics (size, density, dielectric properties, etc.) and biological characteristics of CTCs. In addition, there is a nanoscale interaction between nanostructures and CTCs, which is conducive to cell capture and culture. In recent years, there have been many enrichment methods of CTCs based on nanomaterials or nano-surfaces, such as using antibody-modified magnetic beads and cleverly designed antibodies to modify the microstructure. Compared with conventional invasive biopsy, in vitro test samples are easy to sample, and have the advantages of high specificity and good patient compliance. In vivo detection refers to implanting a designed capture platform into the body or injecting molecules for imaging into a patient's vein, and directly detecting CTCs in the blood in the patient's normal physiological environment. One example is inserting the retention needle of the antibody-modified outer wall into the venous blood vessel, which can stay in the blood for a long time. CTCs can be efficiently captured when a large amount of blood is flowing through it. There are also studies using photoacoustic imaging technology to directly observe CTCs in venous blood vessels. In some technologies, the cells captured and fixed in the vein can be killed in situ in the body by photothermal effect, which has important clinical significance for adjuvant cancer treatment and provides new ideas for the prevention of tumor metastasis. The advantage of in vivo testing is that the target blood volume is large (whole body blood ∼5 L), CTCs can be detected under normal physiological environment, and possible contamination during the sampling processing is avoided. At the same time, it can detect CTCs from various parts of the body, so that subsequent molecular level analysis can provide more comprehensive information. Because CTCs can provide real-time, comprehensive and accurate tumor biological information, it also has a lot of room for development in clinical application. At present, the downstream analytical applications of CTCs include prognosis assessment, adjuvant targeted therapy, and drug sensitivity testing.
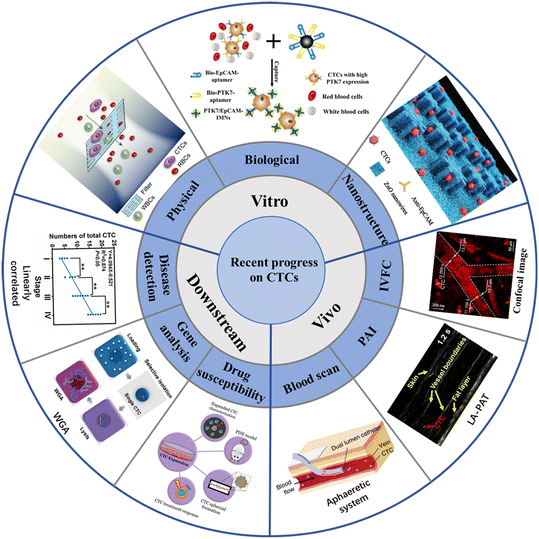 | ||
| Fig. 1 Schematic illustration of the recent progress of CTCs on enrichment in vitro, enrichment in vivo, and downstream analysis20–28 (reproduced with permission from ref. 20, Copyright 2022, Royal Society of Chemistry; reproduced with permission from ref. 21, Copyright 2022, American Chemical Society; reproduced with permission from ref. 22, Copyright 2020, Royal Society of Chemistry; reproduced with permission from ref. 23, Copyright 2018, John Wiley and Sons; reproduced with permission from ref. 24, Copyright 2018, SPIE; reproduced with permission from ref. 25, Copyright 2019, Springer Nature; reproduced with permission from ref. 26, Copyright 2020, MDPI; reproduced with permission from ref. 27, Copyright 2022, American Chemical Society; reproduced with permission from ref. 28, Copyright 2022, American Chemical Society). | ||
In this review, we introduce various recently developed CTC enrichment technologies that are categorized based on physical and biological characteristics, summarize various emerging detection technologies based on nanostructures, and provide detailed information about the latest CTC-based technologies and clinical applications. This article may be a useful guide for cancer research scientists and oncologists to understand various CTCs detection methods and downstream analysis techniques. A better understanding of CTC-based technologies and anticipated future developments promises to improve the treatment and diagnosis of cancer patients.
2 Technologies for CTC enrichment in vitro
2.1 Physical properties for isolation
After tumor cells enter the circulatory system, metabolic abnormalities and metabolic disorders, cell composition, gene expression and modification, as well as the protein synthesis and polymerization of certain polar particulate matter change. This causes the physical properties of CTCs entering the circulatory system to be different from other cells in the blood vessels, which makes it possible to separate CTCs from whole blood based on physical properties.Membrane microfilters and microfluidic devices are the most commonly used size-based CTCs separation technologies. According to the difference in size and deformability of tumor cells and blood cells, Hosokawa et al. designed a microfluidic device equipped with size-selective microcavity array for the efficient and rapid detection of tumor cells in blood.32 The device allows for detection of breast, stomach and colon tumor cell lines, including EpCAM-negative tumor cells, which cannot be separated by conventional immunomagnetic separation, but have more than 80% capture efficiency in microcavity arrays. Zheng et al. made a size-based filtration device for CTCs detection by using parylene-C, which can complete cell analysis in 10 minutes, and the recovery rate of CTCs is 90%.33 Based on the difference of size and deformation, Xu et al. separated CTCs from blood by microfilter, and used a Surface-Enhanced Raman Scattering (SERS) biological probe on the microfilter to realize the in situ isolating and direct detection of CTCs in peripheral blood.34 As shown in Fig. 2A, Li et al. designed an ultra-thin Si3N4 filter membrane with slit pores, which is prepared by standard microfabrication process and easy for large-scale production, and successfully achieved efficient separation of CTCs from whole blood.20 The pore diameter of the filter membrane is 6 nm, the thickness is 200 nm, and the porosity is as high as 34%. In the capture experiments of A549 cells and Hct-116 cells, the Si3N4 filter membrane showed high capture rate (96%), high leukocyte removal rate (99.99%) and high cell viability (90%). In the detection of clinical samples, the microfluidic chip can isolate CTCs from blood samples of cancer patients (5 patients with non-small cell lung cancer and 5 patients with colorectal cancer) and analyze the mutation of the EGFR gene, showing a good potential for clinical application.
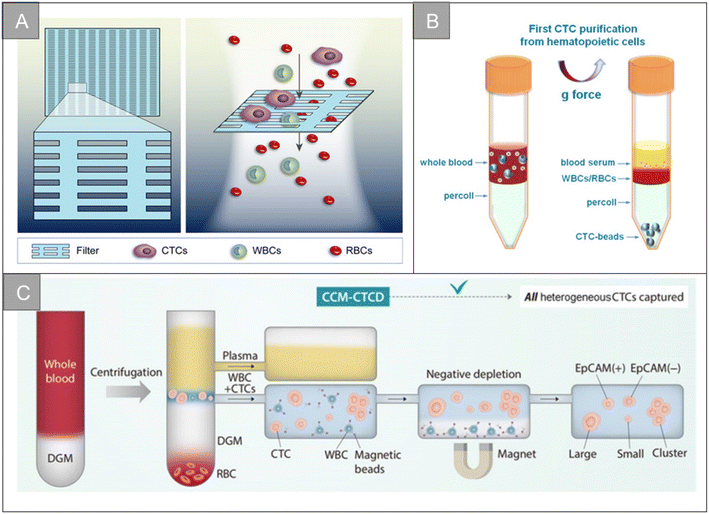 | ||
| Fig. 2 Physical properties for isolation | (A) schematic diagram of the filtering mechanism of slit-shaped pores for efficient enrichment of CTCs20 (reproduced with permission from ref. 20, Copyright 2022, Royal Society of Chemistry). (B) Schematic diagram of the centrifugation of live CTCs from hematopoietic cells by selective sedimentation with an improved centrifugation medium (Percoll) and CTC-beads38 (reproduced with permission from ref. 38, Copyright 2018, Ivyspring International Publisher (https://ivyspring.com/)). (C) Schematic diagram of the CCM-CTCD extraction strategy resulting in the capture of all polyphyletic CTCs39 (reproduced with permission from ref. 39, Copyright 2022, Ivyspring International Publisher (https://ivyspring.com/)). | ||
Circulating tumor microemboli (or CTC cluster) accounts for about 2–5% of all CTCs, and is a cluster formed by multiple CTCs or CTC and other blood components. Compared with a single CTC, the CTC cluster is more hidden, can survive longer in peripheral blood, and has stronger invasive ability. A. Fatih Sarioglu et al. designed a size-based microfluidic chip for capturing CTC clusters in unprocessed blood.35 The basic construction module of the chip is formed by three triangular columns, two of which form a narrowing channel that gathers cells into the opening, and the edge of the third column is positioned to bifurcate the laminar flow. With the blood flow, a single blood cell and a single CTCs flow into one of the two streamline lines, and the CTC clusters stay at the front of the bifurcation column under dynamic force balance. Mert Boya et al. designed the Cluster-Wells chip by combining the speed and practicability of membrane filtration with the sensitive and deterministic screening provided by the microfluidic chip.36 Cluster-Wells chips capture CTC clusters in micropores, and individual cells in the blood will pass through the micropores. Once the CTC cluster enters the hole, it is confined to the grid by a sloping sidewall, and the 2 μm wide grid line ensures that the cell cluster enters different openings; the cell flow rate of 65 μm s−1 (about 10 times lower than the physiological free flow velocity in human capillaries) can prevent the cell cluster from dissociating. The number of micropores of the Cluster-Wells chip is more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The micropores are uniformly distributed on the membrane with a diameter of 47 mm, which realizes the mild treatment of CTC clusters, while providing a processing rate of 25 mL h−1, which can meet the clinical requirements and allow the recovery of viable clusters from the device.
000. The micropores are uniformly distributed on the membrane with a diameter of 47 mm, which realizes the mild treatment of CTC clusters, while providing a processing rate of 25 mL h−1, which can meet the clinical requirements and allow the recovery of viable clusters from the device.
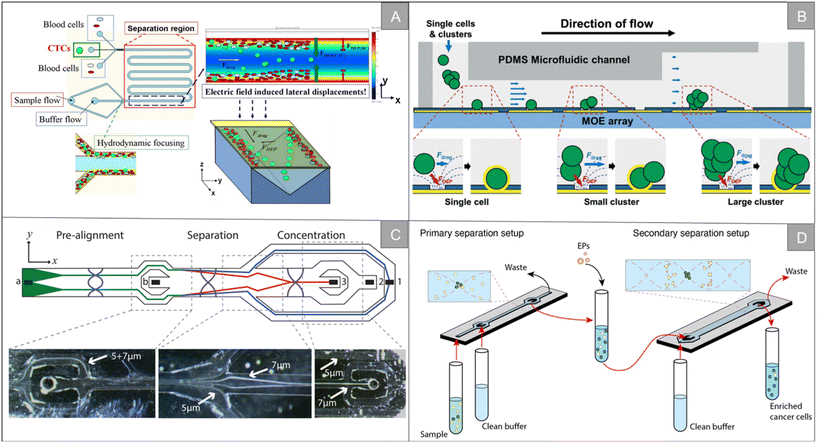 | ||
| Fig. 3 Physical properties for isolation | (A) schematic diagram of the LDEP CTC isolation system40 (reproduced with permission from ref. 40, Copyright 2015, Royal Society of Chemistry). (B) Schematic diagram of a microfluidic system for the simultaneous trapping of single and clustered cells by using a MOE device43 (reproduced with permission from ref. 43, Copyright 2022, Royal Society of Chemistry). (C) Schematic illustration of the chip from the top showing particle trajectories and inset photographs, showing the separation of 5 and 7 μm polystyrene particles45 (reproduced with permission from ref. 45, Copyright 2015, American Chemical Society). (D) Schematic illustration of the workflow and separation principle in A2 (ref. 47) (reproduced with permission from ref. 47, Copyright 2021, published by American Chemical Society). | ||
Inertial focusing utilizes fluid inertial effects in shaped microchannels to align particles and cells at high flow rates. When randomly dispersed particles flow through a channel with a particle Reynolds number of 1 or greater, they experience two counteracting inertial lift forces. The combination of these forces causes particles to migrate to two to four dynamic equilibrium positions located between the channel centerline and the wall.48 After focusing, by subsequently dividing the channel into multiple outlets, the focused cell scans are collected in a smaller volume and focused significantly in a size-dependent manner.49 Majid Ebrahimi Warkiani et al. used soft lithography technology to manufacture a spiral microfluidic chip for high-throughput separation and recovery of CTCs in blood (Fig. 4A).50 The chip makes use of the inertia and Dean resistance in the spiral microfluidic device to focus the larger CTCs and the smaller blood cells to different positions, and achieves a peak cell recovery rate of more than 85% of multiple cancer cell lines. Tian et al. developed an interfacial viscoelastic microfluidic system by using the competition between the inertial lift and interfacial elastic lift.51 This system can directly separate tumor cells from whole blood by size selection, with a separation efficiency of 95.1% and a recovery rate of 77.5%. The survival rate of tumor cells after separation is about 100%. R. Guglielmi et al. designed a spiral microfluidic chip, which separated WBC by inertial force and centrifugal force in a curved channel, thus realizing label-free enrichment of CTCs.52 Fang et al. designed a small centrifuge based on inertial microfluidics.53 The centrifuge is composed of four parallel inertial spiral channels and a two-stage serpentine channel connected in series, which realizes the efficient washing and concentration of biological samples (Fig. 4B). It has the advantages of simple manufacturing, convenient operation and small floor space. Experiments show that 10–20 μm particles can be eluted from the original sample, and the concentration is increased. The recovery rate is more than 93%. In addition, the centrifuge has been successfully applied to various biochemical experiments, including extracting target cells from original samples, purifying target biological particles from micro/nano-particle background, changing the culture medium of cultured MCF-7 breast cancer cells, extracting A549 lung cancer cells from calcein-AM staining solution, purifying WBC from cracked whole blood, and extracting target cells from a background of unbound magnetic microbeads.
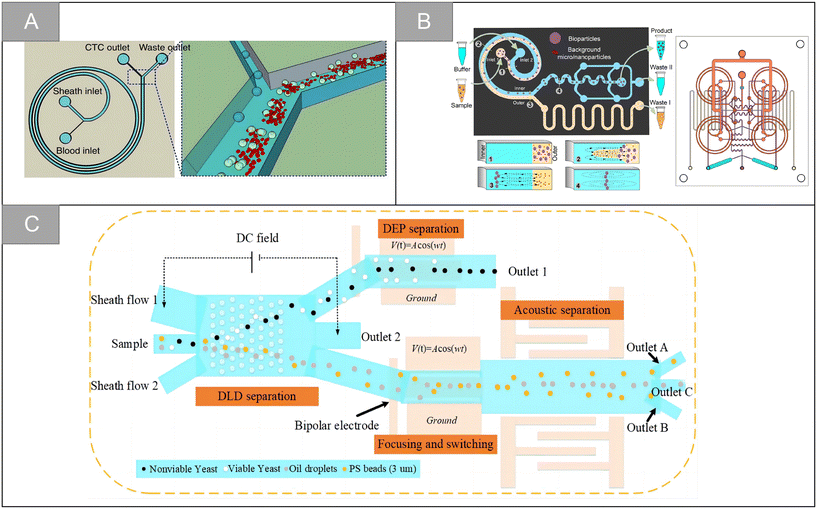 | ||
| Fig. 4 Physical properties for isolation | (A) schematic diagram of CTCs isolation using spiral microfluidics50 (reproduced with permission from ref. 50, Copyright 2015, Springer Nature). (B) Schematic diagram illustrating the working principle of miniaturized centrifuge and detailed structure of the integrated multilayer miniaturized centrifuge53 (reproduced with permission from ref. 53, Copyright 2022, Royal Society of Chemistry). (C) Schematic illustration of the integrated microfluidic system designed to achieve multitarget separation55 (reproduced with permission from ref. 55, Copyright 2021, American Chemical Society). | ||
Although great success has been achieved, the microfluidic sorter using only a single technology still has the limitation of low capture purity. By integrating two or more different separation technologies, hybrid separation technology inherits the advantages of different technologies, which is beneficial to improve the separation performance and solve the limitations of a single technology. In recent years, it has attracted increasingly more attention. Xiang et al. combined inertial microfluidic with deterministic lateral displacement (DLD) to design a two-stage microfluidic sorting device.54 A high-throughput spiral microfluidic channel is used as the first-stage sorter to remove most of the background cells to reduce the blocking risk of the next stage. The DLD sorting channel composed of triangular microcolumns is used in the second stage of sorting, so as to further remove the residual background cells and obtain high-purity target cells. The device improves the throughput and accuracy of cell sorting, the clearance rate of blood cells is 99.95%, and the average recovery rate of the tumor cells is 91.34%. Wu et al. designed a microfluidic chip with integrated sound field and electric field for multiple separation of mixed biological samples.55 As shown in Fig. 4C, the chip consists of four modules: DLD array, bipolar electrode (BPE) focusing module, surface acoustic wave (SAW) sorting module and DEP sorting module. It has about 90% separation efficiency for selecting various types of cells and particles.
2.2 Biological properties for isolation
The separation technology based on biological characteristics is widely used among the existing CTC enrichment technologies. These technologies rely on the unique antigen expression of CTCs, related to their tissue origin and different from other normal components circulating in the blood, to identify CTCs precisely. EpCAM is the most commonly used target, expressed in epithelial cancers, and does not appear in normal cells of the circulatory system. However, CTC has a high degree of heterogeneity and epithelial–mesenchymal transition (EMT) process. Only using EpCAM as a target cannot enrich multiple subtypes of CTC. Therefore, new targets such as N-cadherin, CD44, HER2, and MUC1 are developed to enrich CTC. In addition to traditional antibodies, many CTC enrichment schemes based on aptamers, peptides, and chemical molecules have emerged in recent years. This section introduces CTC enrichment technology based on different targeting molecules.The CTCs separation method based on immunomagnetic separation has made many improvements and breakthroughs in capture efficiency and purity, but the traditional preconcentration and detection method has the limitations of long detection time and tedious process. In recent years, the direct detection strategy of CTCs has received widespread attention. Wu et al. developed a cell-friendly one-step detection strategy based on immunomagnetic nanospheres (IMNs) and immunofluorescent nanospheres (IFNs).61 The detection of CTCs in whole blood of 1 mL was achieved only after 20 minutes of incubation, with a capture efficiency of 98.1 ± 0.8%, a capture purity of 59.5%, and a good reproducibility. On the basis of this research, based on the capture performance of multi-functional nanospheres, a microfluidic platform based on spectrally combined encoding (SCE) was proposed to realize the multiple phenotypic analysis of a single CTC (Fig. 5A).62 Different cellular biomarkers uniquely labeled by multi-functional nanospheres bar codes have the same magnetic tags and different optical features, which can separate heterogeneous CTCs in situ with an efficiency of more than 91.6%, and quantitatively analyze the type and expression level of biomarkers by composite spectral characteristics. Guo et al. designed a supersensitive platform for the direct detection of CTCs using lanthanide fluorescence nanoprobes, which effectively avoided the interference of autofluorescence and the detection limit was as low as 1 cell per well in 96-well plates. The detection rate of blood samples from cancer patients (n = 15) was 93.9%.63 Our research group combines the inverted microfluidic chip with immunomagnetic beads, and uses anti-EpCAM-Fe3O4@Ce6@silaneNPs to separate CTCs in the microcavity.64 The capture surface of the microfluidic chip has a three-dimensional porous inverse opal structure and modifies the antibody. This increases the contact area with cells, while enhancing the ability of specific capture, and the capture efficiency is improved to 95%. In addition, the double-layer IOPC structure made of YVO4:Yb3+ and Er3+ materials achieved 32-fold luminescence enhancement. The combination of the upconversion luminescence and immunomagnetic beads luminescence realized the real-time imaging of CTCs and real-time counting based on the fluorescence ratio. Liu et al. designed a dual-mode (electrochemical/visual) microfluidic device based on covalent organic framework (COF) to achieve the rapid and sensitive detection of PCC-CTC, as shown in Fig. 5B.65 The device captures and recognizes the target CTCs by binding to the specific immunogenicity of the norepinephrine transporter and somatostatin receptor overexpressed on the surface of PCC cells. The detection time is 5 min, with a low detection limit of 1 cell per mL and a wide linear range of 2–105 cell per mL.
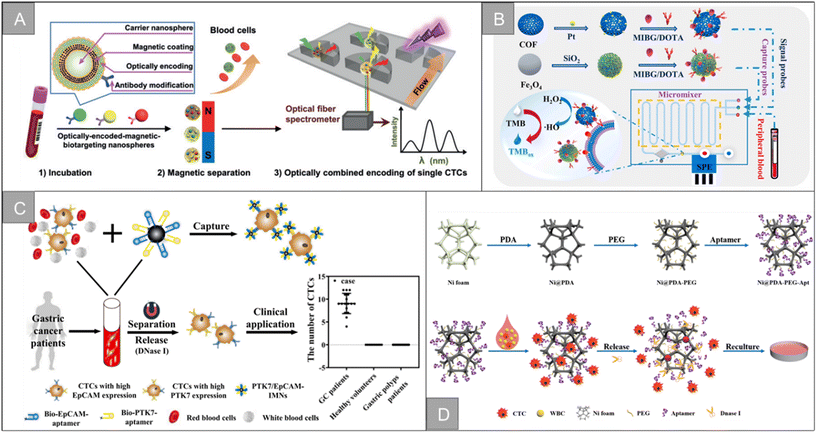 | ||
| Fig. 5 Biological properties for isolation | (A) schematic diagram of a microfluidic chip with magnet62 (reproduced with permission from ref. 62, Copyright 2020, John Wiley and Sons). (B) Schematic of the CTC capture, isolation, and quantification in the microfluidic chip65 (reproduced with permission from ref. 65, Copyright 2022, Elsevier). (C) Schematic illustrations of the preparation of dual-aptamer-modified IMNs and its clinical application21 (reproduced with permission from ref. 21, Copyright 2022, American Chemical Society). (D) Synthesis procedure of the Ni@PDA-PEG-Apt wafer, and capture and release of tumor cells with Ni@PDA-PEG-Apt wafer75 (reproduced with permission from ref. 75, Copyright 2022, American Chemical Society). | ||
The nanoparticles camouflaged by the biomimetic cell membrane have ideal characteristics, which can resist homologous adsorption, improve the biocompatibility and enhance the targeting ability of materials, and have been widely used in various biomedical detection strategies. Rao et al. fused platelet (PLT) and leukocyte (WBC) membranes to form hybrid membranes, which were coated on magnetic beads, and then modified their surface with specific antibodies.66 The PLT–WBC hybrid membrane is coated with immunomagnetic beads (HM-IMBs). HM-IMBs inherit the cancer cell binding ability enhanced by PLTs, reduce the adsorption of homologous WBC, and are further used for the efficient and highly specific separation of CTCs. Through the use of labeled blood samples, it was found that compared with commercial IMBs, the cell separation efficiency of HM-IMBs increased from 66.68% to 91.77%, and the cell purity increased from 66.53% to 96.98%. Wu et al. used white cell membranes to functionalize magnetic nanoparticles and modify antibodies on the surface of the membrane for the highly sensitive detection of CTCs.67 The experimental results showed that the modification of the white cell membrane effectively reduces the interference of homologous leukocytes, the capture efficiency of CTCs is increased to 96.82%, the capture purity is 90.68%, and the successful monitoring of clinical blood samples is realized.
Fang et al. designed a CTCs detection strategy combining upconversion nanoparticles and magnetic nanoparticles. In this study, aptamer-modified upconversion nanoparticles were used as a probe to identify tumor cells for the first time, and then enriched cells by attaching magnetic nanoparticles and stopping in a magnetic field to enhance the sensitivity of CTCs detection.68 Li et al. developed a DNA template magnetic nanoparticles-quantum dots-aptamer copolymer (MQAPs) for the rapid magnetic separation of blood CTCs.69 MQAPs can be detected in 20 min, the capture efficiency and purity are more than 80%, and can be used for single cell analysis. Wang et al. designed a fluorescence-open aptamer sensor FSC-D-P0 for the sensitive detection of CTCs and tumor imaging. FSC-D-P0 is composed of magnetic nanoparticles coated with two ssDNA and fluorescent P0 aptamers, which has stable targeting and can be used for selective capture, MR/FL imaging and release of CTCs.70 Li et al. designed a new type of anti-non-specific adsorption immunomagnetic platform Fe3O4@SiO2@PTMAO@aptamer, which can efficiently capture different phenotypes of CTCs in 10 min, and briefly analyzed the relationship between different phenotypes of CTCs and the progression, diagnosis, surgery and chemotherapy of colorectal cancer (CRC) patients.28 In addition, the team designed double aptamer (EpCAM and PTK7)-modified immunomagnetic Fe3O4 particles for the effective capture and downstream analysis of heterogeneous CTCs in patients with gastric cancer (Fig. 5C).21 After 20 minutes of incubation, more than 95% of the MGC-803 and BGC-823 cells could be captured internally, and the prepared IMNs were successfully applied to CTC capture in peripheral blood samples of 1.0 mL patients with gastric cancer. Ding et al. combined aptamer-modified near-infrared fluorescent Ag2S nanoparticles with immunomagnetic beads to design near-infrared fluorescence probes for efficient CTC capture, which greatly improved the imaging sensitivity.71 On this basis, the research team designed the magnetic nanoparticles HM-Fe3O4@SiO2 coated with the tumor cell membrane and leukocyte membrane by making use of the homologous repulsion characteristics of the white cell membrane and the homologous targeting ability of the tumor cell membrane.72 It was then connected with multivalent aptamer-functionalized Ag2S nanoparticles through the action of SA-biotin, which greatly improved the binding ability and anti-interference ability of the materials. The capture efficiency and purity of the CTCs are as high as 97.63% and 96.96%, respectively.
In addition to the traditional application of magnetic separation, a variety of cell detection strategies based on aptamers have emerged in recent years. Chen et al. designed a sensitive, simple and low-cost non-enzyme amplification CTCs detection strategy to achieve the uniform visual and fluorescence detection of A549 lung cancer cells in clinical blood samples.73 Based on the selective recognition of Ag+ and C–Ag+–C by CdTe QDs, the detection limit of 3 cell per mL for A549 cells was detected by using mucin 1 as the CTCs marker and the aptamer as the recognition probe, and A549 cells with a mass concentration of 100 cell per mL could be identified by naked eye. Peng et al. designed electrochemical biosensors modified by double aptamers for the detection of specific CTCs in complex whole blood matrix.74 Two aptamer hairpin probes can bind to two adjacent proteins (MUC1 and EpCAM) on the cell membrane, respectively, thus amplifying the electrochemical signal by rolling ring amplification reaction to achieve the sensitive quantification of specific CTCs, and the detection limit is as low as 3 cell per mL. Li et al. developed a low-cost and easy-to-manufacture aptamer-functionalized wafer Ni@PDA-PEG-Apt with three-dimensional (3D) interconnected porous structure (Fig. 5D).75 It realized the rapid capture and release of CTCs in 1 hour. The 3D interconnected porous structure of the Ni@PDA-PEG-Apt wafer provides enough channels for cells to flow through and improves the contact frequency between aptamers and cells, with a high capture efficiency of 78.25% and a release efficiency of 61.85%. Wang et al. designed a CTC capture strategy based on a double aptamer DNA network that realized the efficient CTC capture, 3D encapsulation and CRISPR/Cas responsive release, which is helpful for downstream analysis of living cells (Fig. 6A).76 Compared with the traditional cell enrichment strategy, the DNA network has higher capture efficiency and cell-friendly controlled release. The capture efficiency of the CEM cells and MCF-7 cells is 75% and 85%, respectively, and the post-60 min release efficiency is 80%. After 48 hours of culture, 91.8% of the cells have cell viability.
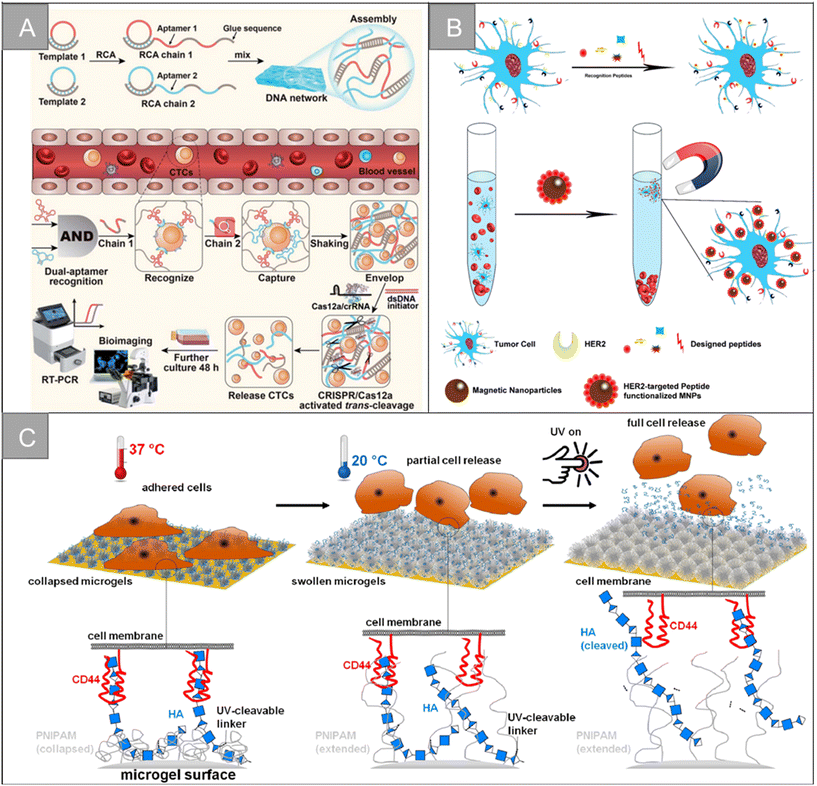 | ||
| Fig. 6 Biological properties for isolation | (A) schematic of a CRISPR/Cas12a-responsive DNA network for CTC capture and release76 (reproduced with permission from ref. 76, Copyright 2022, Royal Society of Chemistry). (B) Schematic illustration for HER2-targeting peptide screening and CTC isolation by Pep@MNPs79 (reproduced with permission from ref. 79, Copyright 2017, American Chemical Society). (C) Illustration of the surfaces coated with dual-stimuli responsive HA-functionalized microgels and the expected cell release by temperature and light stimulus93 (reproduced with permission from ref. 93, Copyright 2021, American Chemical Society). | ||
In addition, Han et al. designed an electrochemical biosensor for blood detection based on multifunctional peptides and the conducting polymer poly(3,4-ethylenedioxythiophene) (PEDOT).81 Multifunctional peptides are assembled to the surface of the gold electrode by Au–S bond, which has the functions of anchoring, doping, antifouling and recognition. Electrodeposition of PEDOT promotes the electron transfer at the sensor interface and improves the signal-to-noise ratio of detection, thus improving the sensitivity of the biosensor. The sensor has a wide linear range of more than 4 orders, and the detection limit is 17 cell per mL. Li et al. simulated the surface modification of folic acid (FA) and arginine–glycine–aspartic acid (RGD) peptides on the erythrocyte membrane.82 The capture surface consists of a polydopamine layer, phosphorylcholine zwitterionic polymer and polyethylene glycol (PEG), which can repel 99.999% blood cell adhesion, effectively improve the enrichment ability of HeLa cells (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times), and obtain high capture efficiency (91%) and high capture purity (89%) in labeled whole blood samples. Jiyoon Bu et al. developed a capture surface based on PD-L1-binding peptides to separate CTCs expressing PD-L1 from exosomes in the blood. This study examined the effects of the polyethylene glycol spacer, secondary peptide structure and peptide sequence modification on the capture efficiency.83 The optimized pPD-1 configuration was 1.5 and 1.2 times more efficient than their total antibody counterpart (aPD-L1) in capturing tumor cells and exocrine expressing PD-L1, respectively. It showed that the common analysis of two biomarkers can further improve the accuracy of the clinical application of the system.
000 times), and obtain high capture efficiency (91%) and high capture purity (89%) in labeled whole blood samples. Jiyoon Bu et al. developed a capture surface based on PD-L1-binding peptides to separate CTCs expressing PD-L1 from exosomes in the blood. This study examined the effects of the polyethylene glycol spacer, secondary peptide structure and peptide sequence modification on the capture efficiency.83 The optimized pPD-1 configuration was 1.5 and 1.2 times more efficient than their total antibody counterpart (aPD-L1) in capturing tumor cells and exocrine expressing PD-L1, respectively. It showed that the common analysis of two biomarkers can further improve the accuracy of the clinical application of the system.
Hyaluronic acid (HA) is a very rich polysaccharide in the extracellular matrix of all vertebrates, which can specifically target cancer cells that overexpress CD44 receptors. The CD44 receptor is overexpressed in breast cancer, prostate cancer, ovarian cancer, head and neck cancer, lung cancer and other cancer types, and tumor cells with a high expression of CD44 are more aggressive. Based on the specific recognition of HA and CD44 receptors and the high metastatic expression of CD44 cells, HA has great application potential in the fields of early tumor detection and drug therapy. Sun et al. designed silica magnetic beads with hierarchical structure to achieve efficient unlabeled cell capture.89 Graded magnetic beads improve hydrophilicity, biocompatibility and stability through HA functionalization, with a high cell capture efficiency of 87.9–98.7%. Zhang et al. designed the HA-functionalized immunomagnetic nanomaterial Fe3O4@SiO2-SS-HA, which realized the efficient capture and release of CTCs, benefiting from the high affinity between HA and CD44 overexpressed cancer cells.90 The material can specifically capture CD44-overexpressed MCF-7 cells with an efficiency of 92%, and 81.4% of the captured cells were released after DTT treatment. Li et al. grafted dopamine onto the hyaluronic acid chain and developed a microsphere HA–DA for capturing CTCs overexpressed by CD44.91 It has good selectivity and capture efficiency, and the detection limit is 10 cell per mL. Yin et al. designed the HA-functionalized silica coated magnetic beads HA-SiO2@MBs, and combined it with a microfluidic chip to achieve the efficient capture and separation of CD44-overexpression cells.92 The magnetic beads are specific to HeLa cells-overexpressing CD44 receptors, and the capture efficiency is as high as 92.0%. Melanie Schmidt et al. designed HA-functionalized double-response poly (N-isopropylacrylamide) (PNIPAM) microgel coating for CTC capture and release, as shown in Fig. 6C.93 The HA-modified PNIPAM microgel membrane can specifically capture the breast cancer cell line MDA-MB-231 expressing CD44, and the captured cells can be released by light stimulation and changing temperature.
Tannic acid (TA) is a widespread phenolic compound, which is composed of a large number of phenolic hydroxyl groups, catechol and galloyl groups. There is a strong hydrogen bond and hydrophobic interaction between the TA-containing galloyl group and glycolysis of tumor cells, which has excellent tumor-targeting ability. Moreover, TA shows good anti-leukocyte adhesion effect, which has great application potential in CTCs broad-spectrum capture. Yang et al. developed a simple and effective chemical strategy for capturing CTCs using TA functional membranes.94 Because of the priority of the TA membrane over PBMCs to cancer cells, the membrane can effectively capture many types of cancer cells without the help of capture antibodies, the capture efficiency is greater than 76% and the capture cell viability is up to 90%. Ding et al. designed a simple broad-spectrum separation strategy for the effective separation of heterogeneous CTCs from blood samples using magnetic nanoparticles (MNPs)-functionalized by TA.95 In the capture of artificial samples of seven kinds of cancer cells (HeLa, PC-3, T24, MAD-MB231, MCF-7, HT1080, A549), the capture efficiency of MNPs-TA is 62.3–93.7%. In addition, MNPs-TA was successfully used to test clinical blood samples from different cancer patients (n = 21). Zhou et al. designed a novel multi-functional platform based on double-response fluorescent magnetic Fe3O4/RhmB@ZIF-8-pTA nanoparticles (FR@Z-pTANPs) for the efficient separation and release of CTCs.96 The platform has excellent resistance to the non-specific adhesion of leukocytes and high sensitivity to cancer cells in the patient's blood. It can capture more than 88% of EpCAM positive cells (MCF-7, HepG2) and EpCAM negative cells (MDA-MB-231, HeLa), and effectively releases captured cells with high efficiency (>80%) and survival rate (>90%) under cell-friendly pH/ATP stimulation. It provides a good solution to solve the challenge of heterogeneous CTCs in clinical application.
3 Nanostructure-based enrichment of circulating tumor cells
With the development of nanotechnology, nanostructured substrates have shown great potential in separating and detecting rare cells. Because the nanomaterials have similar sizes to the microvilli and pseudopodia on the cell surface, they can enhance cell adhesion according to the nano-topological interaction. In addition, nanostructured substrates have a larger specific surface area and provide more binding sites, which contribute to cell attachment. In recent years, a variety of nanostructures have been used for CTC enrichment based on the physical and biological properties of CTC. Besides 0-dimensional nanoparticles, 1-dimensional nanostructures (such as nanowires, nanorods, nanofibers), 2-dimensional nanostructures (such as nanosheets), various biomimetic nanostructure substrates and microfluidic chips for channel design based on hydrodynamics have emerged, which significantly promote the development of CTC enrichment technology.3.1 Nanowire, nanopillar and nanorod
Silicon nanowires are generally obtained by chemical etching. The enhanced local morphology interaction between silicon nanowires and cell surface components (such as microvilli and filamentous pseudopodia) can effectively improve the efficiency of cell capture, and better CTCs separation can be obtained by cooperating with flow channel structure or immunomagnetic beads. Zhang et al. prepared layered assembled ITO nanowire arrays with horizontal and vertical nanowire branches, as shown in Fig. 7A, and the synergism between the terrain effect and specific molecular recognition significantly improved the detection time (35 min) and capture efficiency (89%).97 Our team combines silicon nanowires with multi-functional magnetic nanocomposite Fe3O4@C6/Ce6@silane to design an inverted microfluidic chip to realize real-time monitoring of CTCs and in situ photodynamic therapy (Fig. 7B).98 Under the action of composite markers and magnetic field, the capture purity of CTCs is increased to 90%, and the capture efficiency is 90.3%. Li et al. prepared a simple and highly selective bioseparation platform (A-BGC-SW).99 For the recognition and release of CTCs in complex biological samples, A-BGC-SW used silicon nanowires (SW) as a substrate, amyloid bovine serum albumin (BGC) as coating and aptamer modification as the recognition group. Compared with natural bovine serum albumin (BSA), BGC-SW has a stronger surface interaction. The contamination of HAS, PRP and WBC is reduced by 88.5%, 88.0% and 83.7%, respectively. About 6 cells can be captured from a fresh blood sample containing only 10 CTCs per milliliter. Song et al. fabricated silver nanorod arrays by oblique deposition and modified double-tetrahedral DNA (DTDN) probes on their surfaces for the efficient capture, highly sensitive detection and lossless release of CTCs.100 Under the best conditions, the capture efficiency of the biological interface is 90.2%. A total of 93.4% of the cells are released by Zn2+-assisted DNAzyme cutting, and the survival rate of the released CTCs is about 98.0%.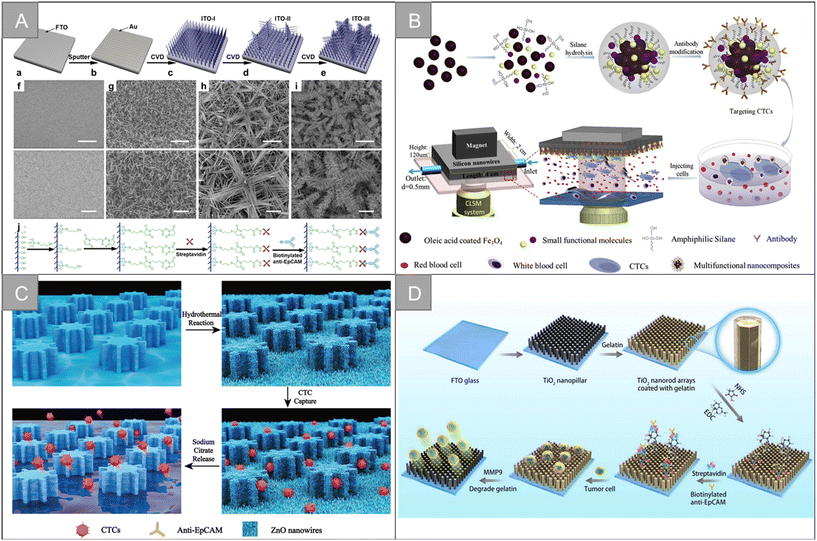 | ||
| Fig. 7 Nanowire, nanopillar and nanorod | (A) schematic of the fabrication and functionalization of hierarchical ITO nanowire arrays on FTO97 (reproduced with permission from ref. 97, Copyright 2016, American Chemical Society). (B) Schematic of the CTC capture platform, involving the synthesis of the anti-EpCAM-Fe3O4@C6/Ce6@silane nanocomposites and microfluidics chip under the confocal laser scanning system98 (reproduced with permission from ref. 98, Copyright 2017, Elsevier). (C) Diagram showing a ZnO-coated G-PDMS pillar microchip for the capture and release of cancer tumor cells22 (reproduced with permission from ref. 22, Copyright 2020, Royal Society of Chemistry). (D) Schematic of the modification of the TiO2 nanopillar arrays (TNA) coated with gelatin film and conjugated antibody for the capture and release of circulating tumor cells104 (reproduced with permission from ref. 104, Copyright 2019, IOP Publishing). | ||
Zinc oxide (ZnO) is a biocompatible metal oxide semiconductor material with excellent electrical, optical, mechanical and chemical properties, showing satisfactory performance in cell capture and release. Based on the characteristics, a variety of ZnO nanostructures, including nanowires and nanorods, have been widely used. Cui et al. grew ZnO nanowires vertically on the surface of a polydimethylsiloxane (PDMS) column substrate with gear structure to fabricate micro-nano biochips for the specific capture and lossless release of CTCs (Fig. 7C).22 ZnO nanowires provide more binding sites and rough surfaces for capture. The capture rate of CTCs is more than 90%, the release efficiency is 93.95%, and the cell viability is 96%. Vahideh Shirmohammadli et al. designed a patterned highly vertical ZnO nanorod substrate to achieve the efficient capture of two cell lines MCF-7 and MDA-MB-231, which adhere to different positions of the substrate, and the maximum capture efficiency is 98.4% and 86%, respectively.101 Su et al. designed a 3D graphene macrocellular foam substrate with ZnO nanorod array.102 Through the regular changes of electrical resistance and electrochemical impedance spectra of the foam substrate in the process of CTC adhesion, the specificity and non-invasive detection of CTCs were realized. Xu et al. constructed a sensing platform for CTC capture and detection based on CDS/ZnO nanorod arrays coated with polyaminophenylboric acid (APBA).103 The sensing platform takes the terminal sialic acid (SA) molecule in CTCs as the recognition site, and realizes cell capture and release based on the regeneration of the ester bond between phenylboric acid and SA. It has good sensitivity and specificity in the concentration range of 50–1 × 106 cells per mL.
Titanium dioxide (TiO2) has good stability, biocompatibility, and light transmittance. It is easy to modify the targeted molecules, so it is widely used in a variety of cell capture platforms based on nanostructures. Li et al. fabricated TiO2 nanocolumn arrays coated with gelatin film to achieve the effective capture and lossless release of CTCs, as shown in Fig. 7D.104 In this work, the interaction between the cell membrane and the nanostructure substrate improves the capture efficiency of CTCs up to 94.48%. The gelatin layer can be digested rapidly by matrix metalloproteinase-9 to achieve the lossless release of CTCs, with a release efficiency of nearly 100% and cell viability of 100%. Fan et al. coated a layer of gelatin film doped with gold nanorods on the TiO2 nanorod substrate to achieve the efficient capture of CTCs and photocontrolled release in response to near-infrared light.105 By adjusting the size and position of near-infrared light, selected cells can be locally released, while gelatin has good biocompatibility and low near-infrared phototoxicity so that the cells released by this method have high vitality.
3.2 Nanofiber
The nanofiber structure is a unique nano-structure imitating natural extracellular matrix, which has the characteristics of high specific surface area, high porosity, high mechanical and structural stability. It provides a large number of active sites and enough contact area for cell capture and chemical reaction, and is an ideal platform for capturing circulating tumor cells. Chitosan is the most commonly used natural polymer material in tissue engineering because of its low cost, non-toxicity and good biocompatibility. Sun et al. prepared a functional biological interface based on chitosan nanofibers by electrospinning, which was used for the specific capture and lossless release of CTCs in blood.106 The TiO2 nanofiber is a widely used inorganic nanofiber material with the advantages of easy deposition, accurate control of size and bulk density. Chen et al. developed a platform for the sensitive capture and efficient sorting of CTCs based on the asparagine–glycine–arginine (NGR) peptide and TiO2 nanofiber substrate.107 As shown in Fig. 8A, the cell trapping of TiO2 nanofibers densely arranged on the glass sheet provides a three-dimensional interface, which is conducive to enhancing the nano-topological interaction between the cell surface structure and the TiO2 substrate. Subsequently, dopamine and bovine serum albumin (BSA) were introduced into the interface of TiO2 nanofibers as adhesive and antifouling molecules for substrate surface modification. The formed TiO2-BSA-NGR substrate has high capture sensitivity and efficiency, and the detection limit is 10 cells per mL, which provides a basic technical route for CTC capture.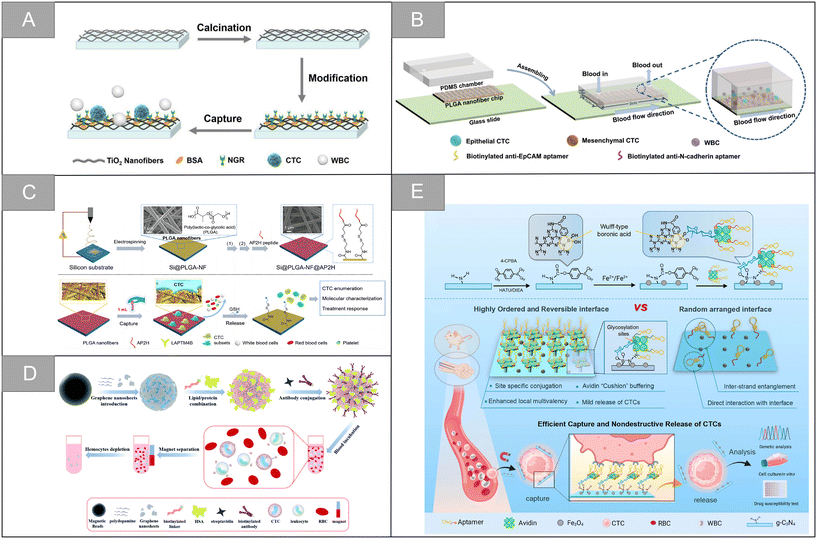 | ||
| Fig. 8 Nanofiber and nanosheets | (A) schematic illustration of the CTC capture using the NGR peptide modified TiO2 nanofiber substrate107 (reproduced with permission from ref. 107, Copyright 2020, Springer Nature). (B) Schematic illustration of the dual aptamer-modified PLGA nanofiber-based microfluidic device for the different phenotypic CTC capture strategies108 (reproduced with permission from ref. 108, Copyright 2021, Royal Society of Chemistry). (C) Schematic illustration synthesis and working principle of the Si@PLGA-NF@AP2H substrate111 (reproduced with permission from ref. 111, Copyright 2021, American Chemical Society). (D) Schematic illustration of the fabrication process of artificial cell membrane-camouflaged immunomagnetic nanoparticles and general application in circulating tumor cell isolation116 (reproduced with permission from ref. 116, Copyright 2022, Royal Society of Chemistry). (E) Scheme for the preparation of the CTC capture biointerface (Apt-Avi-BCN) and advantages of the highly ordered and reversible biointerface Apt-Avi-BCN with respect to randomly functionalized biointerface and its working principle for CTC capture and release119 (reproduced with permission from ref. 119, Copyright 2022, American Chemical Society). | ||
Synthetic polymers have high flexibility in synthesis, processing and modification. Polyvinyl alcohol (PVA), polycaprolactone (PCL), polylactic acid (PLA), poly(lactic-co-glycolic acid) (PLGA) and other synthetic polymers are often used to design nanofiber substrates with different structural properties. In addition, compared with natural polymers, the mechanical properties of synthetic polymers can be effectively adjusted, and the economic benefits are better. Wu et al. assembled the aptamer targeting epithelial cell adhesion molecule and N-cadherin onto the PLGA nanofiber chip surface, and modified bovine serum albumin (BSA) on the chip surface to prepare an electrospun PLGA nanofiber chip for CTC capture of ovarian cancer (Fig. 8B).108 The chip can improve the capture sensitivity and ensure the capture purity. The capture efficiencies of ovarian cancer A2780 cells and OVCAR-3 cells are 91% and 89%, and the release efficiency is 95% and 88%, respectively. Zhao et al. developed a simple method for CTC capture based on an electrospun PLA nanofiber substrate.109 In this study, HA was modified on the surface of randomly or orderly electrospun polylactic acid fibers, which improved the hydrophilicity and blood compatibility of the PLA nanofibers. Experiments showed that ordered nanofibers had a higher capture efficiency of cancer cells with high expression of CD44 receptor, reaching 95.3%. Zhu et al. developed an aptamer-modified PEG–PLGA-nanofiber microfluidic chip.110 The chip can be used for dynamic monitoring of CTCs mutation and rapid identification of rare subtypes, with excellent cell capture efficiency and high cell survival rate. The chip also has high application potential in dynamic CTCs counting, early tumor screening and histological change monitoring. Zhong et al. introduced lysosomal protein transmembrane 4 β (LAPTM4B)-targeting peptide as a new molecular target for the specific recognition of CTCs, and developed an amphiphilic and nanostructured platform for monitoring CTCs and capturing different CTC subtypes during treatment.111 As shown in Fig. 8C, the platform uses an electrospun PLGA fiber scaffold as the base, utilizes the characteristic of high stable expression of the LAPTM4B protein during the epithelial-mesenchymal transition of CTCs, and uses the AP2H peptide as the recognition molecule to specifically capture tumor cells, which overcomes the limitation that some CTC subtypes are insensitive to traditional epithelial markers, and significantly enhances the capture affinity and selectivity. Wang et al. developed a negative microfluidic detection platform targeting WBC to solve the problem that cells must be labeled or attached to the substrate in the positive sorting method, which is difficult to release and culture.112 In this research scheme, electrospun PLGA nanofibers were modified by streptavidin and embedded in a PDMS microfluidic chip. Through the interaction between biotin and streptavidin, the platform can sort the leucocytes labeled by biotinylated anti-CD45 antibody, and enrich CTCs and A549 cells from the blood of patients with non-small cell lung cancer. The leukocyte capture efficiency and cell recovery rate are ninety-seven percent and 97.5% respectively, which is an effective CTCs sorting and identification platform.
3.3 Nanosheets
Two-dimensional nanosheets are usually single-layer or laminated structures, which have the advantages of large surface area, flexible design, controllable morphology and excellent conductivity. Their surfaces can be modified with other materials, and their end groups are easy to functionalize, resulting in hybrid structures with enhanced electrical, catalytic or optical properties and detection capabilities, which are widely used in biosensing.113Graphene oxide has unique optical properties, controllable size and easy surface modification with polyethylene glycol-based chemicals. It is an attractive nanomaterial in drug delivery and biosensing. In 2013, Hyeun Joong Yoon et al. designed functionalized graphene oxide nanosheets, which realized the effective separation of CTCs from blood samples of patients with pancreatic cancer, breast cancer and lung cancer, and the capture efficiency was 73 ± 32.4% at low concentration of target cells (3–5 cells per milliliter of blood).114 Dou et al. synthesized a kind of aptamer functionalized and gold nano-array modified magnetic graphene nano-sheet, and realized multiple electrochemical detection of CTCs in blood.115 The potential and current intensity of the probe respectively reflect the type and quantity of CTCs. In the detection of Ramos and CCRF-CEM cells, the detection limit is 4 and 3 cells per milliliter. As shown in Fig. 8D, Zhou et al. combined graphene nanosheets (GNS) with magnetic nanoparticles, and constructed antibody-modified artificial cell membranes on the periphery, which achieved the efficient capture of CTCs in simulated and clinical blood samples, with an average capture rate of 87.0%.116
Besides graphene oxide, other types of materials are widely used in the capture of circulating tumor cells based on nanosheets. You et al. designed and prepared a Ti3C2TxMXene nanosheets-near-infrared responsive gelatin hydrogel membrane for the specific capture and release of CTCs.117 The surface of anti-EpCAM modified Ti3C2Tx@gelatin film can recognize highly specific EpCAM positive cells, and it has two release modes: temperature response release and near infrared response fixed-point release. The average effective release rate is 89%, and the cell vitality is 87%. Shen et al. developed an ultra-sensitive and specific sandwich cell sensor for detecting CTCs based on the aptamer-modified magnetic beads Apt@MBs and FA-functionalized carbon dot/cobalt oxyhydroxide nanosheet system FA@CDs/CoOOH.118 Wang et al. developed a new cell capture interface based on boric acid-functionalized g-C3N4 nanosheets (Fig. 8E).119 Compared with the free aptamer, the affinity of the engineered cell capture interface to the target CTCs is 100 times higher, and the captured CTCs can be released competitively through high-activity acidic fructose, which is beneficial to downstream cell culture and genome analysis, and further used for drug sensitivity test.
3.4 Bionic nanostructure
In addition to the imitation of mutual recognition between cells, there are many unique biological systems in nature. Compared with artificial systems, these systems have found their own ways to achieve the efficient recognition of specific targets. In recent years, a variety of biomimetic nanostructures have been constructed for CTC recognition. For example, pollen uses spiny structures to recognize and adhere to stigmas, and Wang et al. developed a biologically inspired pollen-like graded surface by assembling replicating pollen grains.120 After being modified with cancer cell-specific trapping agents, the pollen-like surface can capture target cancer cells with high efficiency and specificity, with an efficiency of 72.0% ± 1.5%. In addition, the pollen-like surface not only ensures the high viability of the captured cells, but also performs well in the cell mixing system and low cell density. Octopuses use long tentacles containing many suckers to hunt. Chen et al. mimicked the characteristics of the octopus and developed a device called “nano octopus” to isolate cancer cells in whole blood.121 The device consists of magnetic particles (MP) imitating the head of an octopus and a long single-stranded DNA sequence anchored to the surface of the MP as antennae. Their ultra-high sensitivity and specificity are attributed to the multivalent binding of tentacle DNA to cellular receptors without steric hindrance. It has the characteristics of simple manufacturing, rapid detection, non-invasive capture and release, allowing a wide range of downstream cell culture and molecular analysis. Tabular corals have evolved long and flexible tentacles, allowing repeated “prickle cells” to maximize contact with flowing targets in a multi-valent way. Inspired by the efficient predation mode of plate coral, Jia et al. designed a multivalent nano-plate aptamer functionalized biomimetic magnetic graphene oxide platform (MNPA-TCMMGO), which combines the high binding affinity of polyvalent nucleic acid aptamers with the self-targeting and lateral fluidity of a tumor cell membrane to improve the recognition and capture ability of cancer cells in biological samples (Fig. 9A).122 Inspired by the efficient predation mechanism of the sea urchin multitube foot and endoskeleton, Zhang et al. combined dual-multivalent-aptamers (DMAs) Sgc8 and SYL3C into AuNPs to form sea urchin-like nanoprobe sea urchin-DMA-AuNPs, and designed a super-efficient bionic single CTC recognition platform, as shown in Fig. 9B.123 After co-incubation of CTCs and sea urchin-DMA-AuNPs, the separation is carried out by a microfluidic device integrated with a spiral separation unit and hydraulic filtration purification unit. After separation, by measuring the amount of 197Au isotope in sea urchin-DMA-AuNPs, the biomarker protein in a single CTC can be analyzed without background by inductively coupled plasma mass spectrometry. The microchip is helpful to identify a single CTC, and the separation rate is 93.6% at a flow rate of 60 μL min−1. The measurement efficiency of a single CTC is 73.8 ± 5.0%.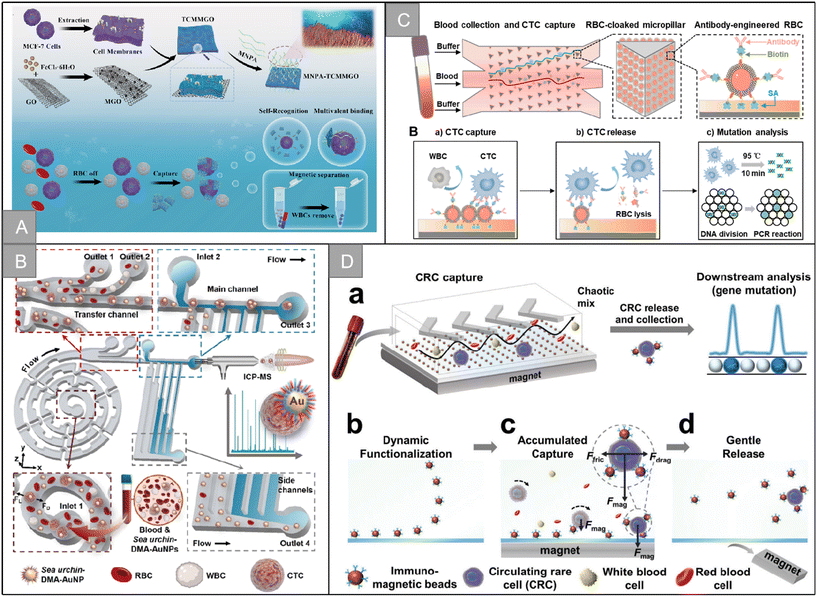 | ||
| Fig. 9 Bionic nanostructure and micro-nano channel structure | (A) schematic illustration of the preparation of the MNPA-TCMMGO nanoplatform and MNPA-TCMMGO for the efficient isolation and detection of rare tumor cells122 (reproduced with permission from ref. 122, Copyright 2023, Elsevier). (B) Schematic illustration of the dual-multivalent-aptamer-conjugated nanoprobes123 (reproduced with permission from ref. 123, Copyright 2021, American Chemical Society). (C) Schematic illustration of the RBC-chip127 (reproduced with permission from ref. 127, Copyright 2022, Elsevier). (D) Schematic illustration of the DynarFace-chip131 (reproduced with permission from ref. 131, Copyright 2021, John Wiley and Sons (https://www.wiley.com/)). | ||
Red rose petals are chosen as templates because of their microstructure size (the typical diameter of hemispherical micropetals is 20–30 μm) similar to CTCs, that promotes cell adhesion. In addition, many nanoscale folds are superimposed on these microstructures, which is beneficial for the cell pseudopod to grasp on the surface. Dou et al. prepared a hierarchical three-dimensional PDMS substrate composed of microcavities or microtops with nanostructured epidermal folds using rose petals as templates, and modified EpCAM antibodies on its surface for the effective capture and release of CTCs.124 Compared with planar PDMS, there is a morphological interaction between the hierarchical structure and cells, which provides more sites for specific cell adhesion. The graded capture substrate shows 6 times higher cell trapping ability than a flat PDMS surface at the concentration of 100 cell per mL. The effective release of captured cells can be achieved by adding a GSH reducer. Wang et al. designed a biologically stimulated three-dimensional epithelial cell adhesion molecule (EpCAM) aptamer-modified rose petal-derived ZnO microchip EpCAM-RPD-ZnO-chip, which is used to achieve the efficient capture and release of CTCs.125 The 3D rough spherical concave surface on the EpCAM-RPD-ZnO chip ensures the maximum contact with CTCs, and the capture efficiency is more than 90.5%. In addition, the simple release of ZnO was achieved by dissolving CTCs under moderately acidic conditions, with a release rate of 84.4% and a survival rate of more than 96.8%.
3.5 Micro-nano channel structure
Microfluidic technology, known as “lab on a chip”, is a technology that accurately controls and manipulates micro-scale fluids, which is mainly characterized by the manipulation of fluids in the micro-and nano-scale space. It has the ability to miniaturize the functions of sample preparation, reaction, separation and detection in the process of biological, chemical and medical analysis to a few square centimeters of chip. In 2007, Nagrath et al. reported a breakthrough development of applying microfluidic chip based on a microcolumn array to CTC capture. Since then, many microchip technologies for CTC capture and detection have been proposed.7In recent years, many works based on microfluidic channel design have been published, which has greatly promoted the development of in vitro detection of CTCs. Yang et al. designed a size-controlled microfluidic capture chip SDI-chip composed of triangular microcolumn arrays modified with EpCAM antibodies, which can make CTC fully contact with hydrodynamic optimized microcolumns.126 The capture efficiency in blood samples is more than 92%, and the purity is 82%. In the subsequent work, the team combined the erythrocyte interface with deterministic lateral displacement (DLD) to establish an antibody engineering erythrocyte affinity interface (RBC-chip) on a microfluidic chip for the efficient separation and release of CTCs (Fig. 9C).127 The erythrocyte layer reduces the collision between the cell and the microcolumn and avoids non-specific adsorption, and the capture efficiency of CTC is 96.5%. This work selectively cleaves RBC by simply changing the salt concentration, destroys the affinity interface, releases CTCs efficiently and gently, while avoiding nucleic acid contamination, and 96.1% CTCs maintain cell vitality. Mehdi Rahmanian et al. fabricated microfluidic chips with different geometric shapes (rhombus, rectangle, circle and triangle) microcolumns, and carried out numerical simulation and evaluation of the CTC capture efficiency and purity.128 The experimental results show that the optimized diamond microcolumn has high capture rate (>85%), high purity (>90%) and high activity (97%). In addition, the diamond microcolumn chip was successfully applied to the CTC detection of 12 patients with breast cancer to verify the effectiveness of the chip.
Brenda J. Green et al. designed a dual-mode microfluidic device PillarX consisting of a series of cylindrical devices and X-devices, which can analyze a single CTC and cluster in whole blood according to the size, deformability and epithelial marker expression of a single CTC and cluster.129 Larger, cohesive and less easily deformed clusters and larger individual cells are captured in the column device and classified according to the size of the column gap. Smaller, deformable clusters and individual cells are then captured in X-ray devices, and separated using functionalized magnetic nanoparticles according to the expression of epithelial markers. Aynur Abdulla et al. designed an antibody-functionalized microfluidic (AFM) chip for the rapid and accurate identification of CTCs in the whole blood of breast cancer patients.130 The AFM chip consists of an entrance, an exit, three buffers and four parallel capture areas, including equilateral triangular columns and periodic curvilinear obstacles. The chip can effectively capture EpCAM-expressing cancer cell lines (MCF-7, PC3 and A549). At the flow rate of 0.6 mL h−1, the capture efficiency reached 99.5%, 98.5% and 96.72% for MCF-7, PC3 and A549, respectively. The performance of the chip was evaluated with blood added by MCF-7, and the capture rate of the chip was 93%. Chen et al. designed a dynamic and reversible immunoaffinity interface (assembled by magnetic beads on the chip substrate by magnetic field) in the fishbone chip for efficient capture and release of CTCs (Fig. 9D).131 The chip has the advantages of convenient operation and reversible assembly, the capture efficiency is more than 98%, and the released cell vitality is more than 98%. Elyahb A. Kwizera et al. designed a kind of electric microfluidic chip, which realized the efficient capture and release of heterogeneous (EpCAM+ and CD44+) CTCs.132 The VIZA chip has two entrances and five continuous bifurcated structures. The main channel consists of parallel patterned microcolumns. The surface of the microcolumns was first coated with 4 nm thick coupled titanium films and 10 nm Au films (except for the microcolumns in the four side columns), and then decorated on the gold surface to capture antibodies. In addition, the chip controls the movement of cells between the capture area and the non-capture area in the main channel by controlling the duration of the AC electric field, with a maximum capture efficiency of 97.0 ± 0.8%. By injecting glutathione, the chip can achieve the efficient release of captured cells, with a release efficiency of 96.1 ± 1.2%.
4 Technologies for CTC enrichment in vivo
In vivo detection technology is an analysis method based on developing tumor cells that flow rapidly in blood vessels under physiological conditions. Because the tumor cells circulating in the body can be captured, even if the capture efficiency of the CTCs in vivo detection technology is as low as 0.02%, it is possible to detect one CTC in the body. In addition, it has many advantages, including the following: (1) it can dynamically monitor the changes in the number of CTCs in the patient's body in a formal physiological state; (2) it effectively avoids the influence of sample collection, pretreatment, transportation, storage, and other processes on the analysis results; (3) it is possible to capture CTCs from different parts of the body and different body fluids, which will promote a more comprehensive and accurate revealing of the tumor evolution process and mechanism, combining the subsequent molecular-level analysis. In recent years, in vivo detection technology has played a vital role in tumor staging, early detection, and predicting treatments' effects in metastatic cancer.4.1 Fluorescence in vivo flow cytometry
The fluorescence-based in vivo flow cytometry (IVFC) is an emerging tool to monitor circulating cells in vivo. As a noninvasive and real-time diagnostic technology, the fluorescence-based IVFC allows for the long-term monitoring of circulating cells without changing their native biological environment.133 Blood flow in the body is considered the fluid system of flow cytometry. When fluorescently labeled CTCs flow through the laser spot, the excited fluorescence is collected by a photomultiplier tube, producing a signal distinct from the blood background, enabling continuous, real-time, and long-term quantitative or qualitative analysis of CTCs. In 2004, the concept of in vivo flow cytometry (IVFC) was proposed for CTC detection.134,135 Blood flow is considered to be the fluid system of the cytometer, and the laser is non-invasively focused into the blood vessel to excite fluorescently labeled CTCs. The fluorescence of CTCs passing through the laser focus can then be excited, collected, and detected in vivo. In 2007, the observation of fluorescently labeled CTCs in mouse blood vessels using high-power femtosecond laser line scanning on a two-photon microscope was reported.136 In 2015, a specially designed fast-imaging confocal microscope was found to be able to capture images of injected cells, albeit at an insufficient imaging rate to capture all flowing cells.137 In 2018, Hu et al. reported a simple optical method to directly monitor CTCs in vivo by ordinary confocal microscopy without additional modification or design (Fig. 10A).23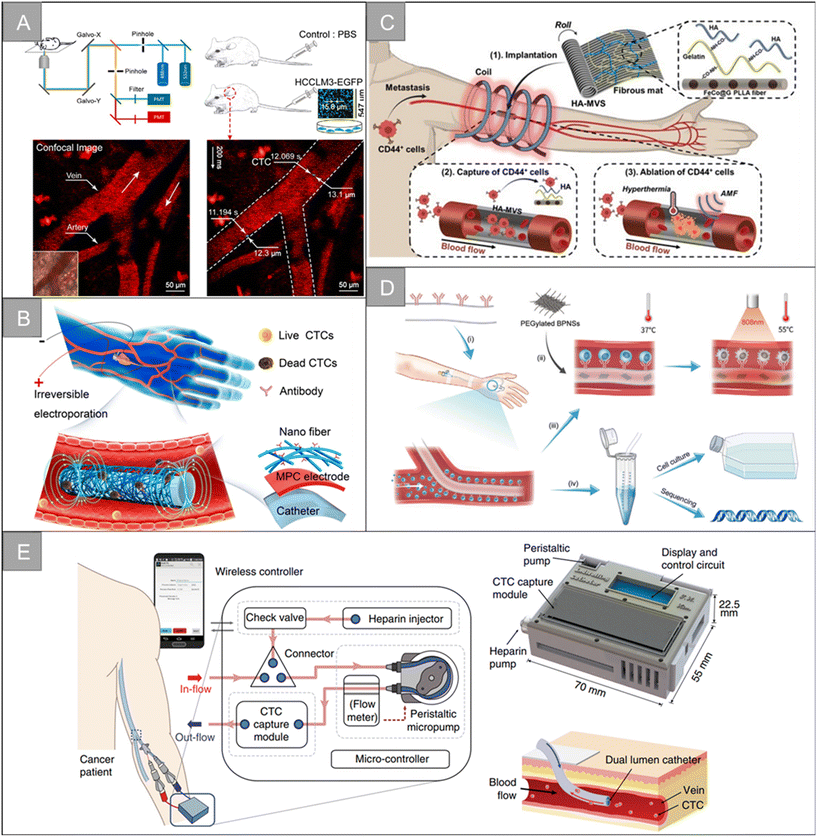 | ||
| Fig. 10 Technologies for CTC enrichment in vivo | (A) schematic diagram of the setup of CTC detection by the confocal microscope system23 (reproduced with permission from ref. 23, Copyright 2018, John Wiley and Sons). (B) Schematic diagram of the in vivo enrichment and elimination of CTCs using the flexible electronic catheter151 (reproduced with permission from ref. 151, Copyright 2022, American Chemical Society). (C) Schematic diagram of the CTC removal by vascular-like ITD152 (reproduced with permission from ref. 152, Copyright 2022, John Wiley and Sons). (D) Schematic diagram of the BPNSs-catheter based therapy155 (reproduced with permission from ref. 155, Copyright 2020, John Wiley and Sons (https://www.wiley.com/)). (E) Schematic diagram the in vivo aphaeretic CTC isolation system25 (reproduced with permission from ref. 25, Copyright 2019, Springer Nature). | ||
If the diameter of the vein (−80 μm) is too large compared to the CTC size (diameter: 10–20 μm), the CTCs may flow through the optical part of the scan line in the vessel. Therefore, such confocal line scans may miss those out-of-focus signals, resulting in reduced sensitivity of CTCs in whole vessels. However, confocal settings help suppress stray light from out-of-focus planes. The system then acquired each CTC signal with a higher signal-to-noise ratio. Fast line scanning with confocal microscopy enables continuous monitoring of CTCs in live mouse blood vessels for over 12 hours. In 2020, Patil et al. developed a new instrument called “in vivo diffusion flow cytometry” (DiFC) to count rare fluorescently labeled CTCs.138 EC-17, a FITC-folate conjugate that has been used in clinical trials for fluorescence-guided surgery, showed high affinity for FR+ L1210A and KB cells in vitro. In whole blood, 85.4% of L1210A and 80.9% of KB cells were labeled with EC-17 over a nonspecific background, and EC-17-labeled CTCs were easily detected in the circulation of DiFC mice. In 2019, Ding et al. established subcutaneous tumor and orthotopic tumor models, and fluorescence in vivo flow cytometry was used to monitor the changes in the number of circulating tumor cells in the blood vessels of mice before and after tumor resection.139 In the same year, Zhou et al. conducted label-free monitoring of melanoma cells in vivo based on the comparison of light absorption between melanin and other substances in blood in the near-infrared window.140 A subcutaneous model of melanoma was constructed in the ear of mice to explore differences in the number of circulating melanoma cells (melanoma is the deadliest type of skin cancer) detectable in blood vessels distal to and proximal to the tumor. Non-invasive CTC detection in small animal models is possible using multiphoton fluorescence imaging. However, most of these techniques require labeling of CTCs by transfection, and their clinical translation is hindered by the low transfection efficiency in vivo and the potential toxicity of labeled CTCs. In addition, the penetration depth of this optical method is relatively shallow and can only act on superficial vessels, limiting the amount of blood to be examined within a reasonable operative time, thereby reducing detection sensitivity. Due to strong light scattering in biological tissues, some non-invasive optical CTCs imaging methods rely on imaging specialized anatomical sites, such as the mouse ear.
4.2 Photoacoustic imaging
Photoacoustic imaging (PAI) combines thermally induced acoustic waves generated by optical probing and ultrasonic (US) detection of a subject by irradiation with short laser pulses.141,142 PAI allows the transfer of light energy absorbed by tissue, resulting in thermoelastic expansion. This expansion then generates ultrasound waves, which are detected by the transducer and produce a contrasting image of optical absorption within the tissue. Photoacoustic flow cytometry can achieve the signal recognition and counting of different cells. It can identify the target signal in the background based on the specific absorption of the target signal to a specific wavelength. Different types and shapes of cells or nanoparticles have specific differences in the absorption of different wavelengths of laser light. Based on this difference, a single or multi-wavelength laser can be selected for excitation to determine the location and quantity to be detected.143 Compared with fluorescence imaging, PAI has a centimeter-level penetration depth, can image surrounding blood vessels, and provides higher detection sensitivity by checking a relatively large blood volume at the same time. In 2020, Hai et al. used a linear array-based photoacoustic tomography system (LA-PAT) to detect melanoma CTCs, quantify their contrast-to-noise ratio (CNR), and measure their flow rates in most superficial veins in the human body.24 This approach relied on the production of melanin within melanoma cells, rather than the expression of receptors on the cell surface. The development of contrast agents for molecular photoacoustic imaging may extend the capabilities of LA-PAT to image cancer cells that do not express melanin.144However, PAI is facing some challenges in the clinical promotion. First, for cells that do not contain specific absorption substances, they need to be labeled with nanoparticles. The biological safety of these nanoparticles needs further study. Second, absorbers, including tissue and blood, act as a strong background for the PAI signal, severely reducing the specificity of detecting target molecules or cells, making it difficult to detect deeper blood vessels, and few CTCs in the effective vasculature in the early stages of cancer development. A new method called magnetodynamic photoacoustic (mmPA) imaging was developed to increase the specific contrast in PAI by suppressing background signals from intrinsic absorbers.145–148 mmPA senses objects labeled with composite particles combined with paramagnetic nanoparticles and optically absorbing components (e.g., gold nanorods), enabling magnetic manipulation to simultaneously target objects PA detection. By biocoupling the contrast agent to the target tissue or cells and magnetically manipulating it with an external magnetic field, the target region moves coherently with the magnetic field. This motion modulates the PAI signal spatially or temporally, allowing subsequent motion filtering of a series of recorded PA signals to suppress the magnetically insensitive background signal.145,146,148
4.3 Long term large blood volume scan
The low throughput of in vitro detection and the demand for a large number of CTCs in downstream analysis have promoted the development of CTC detection in vivo. In 2012, CellCollector, the world's first CTC in vivo capture product, was certified by CE and is currently the only in vivo CTC capture technology on sale that has been certified by CFDA. CellCollector technology inserts the wire coated with EpCAM into the elbow vein of the patient to collect CTC, which is taken out after 30 min, and the blood volume is up to 1500 mL, which is much higher than in vitro detection. In 2018, Vermesh et al. developed a flexible intravascular magnetic line MagWIRE for higher yields to retrieve rare biomarkers from the subject's in vivo blood.149 MagWIRE is an independent magnetic line, which has the characteristics of small diameter, good flexibility and good biocompatibility. It can achieve a high local field gradient in its whole length to effectively capture magnetic particles (MP) coated with injected antibodies in the target labeled in the blood. In the in vivo capture experiment of pig ear vein, although the highest capture efficiency of MagWIRE is 8%, the number of cells captured is equivalent to 80 tubes of blood extracted in vitro, which is 5000 times higher than that of passive immune capture in vivo. Cheng et al. designed an injectable and retractable flexible three-dimensional stent CTC-Net for intravascular collection of CTCs.150 CTC-Net has an interconnected spatial distribution network, which can hold a large number of fixed antibodies and increase the frequency of contact with cells. CTC-Net can be easily compressed and injected into blood vessels, and fully expanded to form a 3D “fishing net” structure for capturing CTCs. Dozens of CTCs have been successfully isolated and detected from tumor-bearing rats. In addition, the CTC-Net can be recompressed and retracted from the blood vessels to facilitate imaging and downstream analysis of the captured CTCs. As shown in Fig. 10B, Wang et al. combined electrospun nanofibers with liquid metal–polymer conductor (MPC) electrodes to develop a flexible electronic catheter that can efficiently capture and kill CTCs in vivo.151 The catheter can repeatedly screen the blood of the whole body and has good flexibility, mechanical strength, biocompatibility and electrical conductivity. It enhanced specific recognition and cell adhesion by surface-modified epithelial cell adhesion molecules of nanofibers. The number of leukocytes captured per square millimeter catheter is less than 10, and the capture efficiency is much higher than that of previous studies. In addition, the CTCs concentrated on the surface of the catheter was finally ablated by irreversible electroporation (IRE), which effectively reduced the content of CTCs in vivo and reduced the damage of normal tissue, providing a new method for the capture and elimination of tumor metastasis and prognostic markers such as CTCs and exocrine. As shown in Fig. 10C, Yin et al. proposed for the first time a vascular integrated trapped device (ITD), which is an implantable HA modified magnetic vascular stent (HA-MVS) for the permanent capture and magnetothermal ablation of CD44 positive (CD44+) CTCs under an alternating magnetic field (AMF).152 The HA-MVS can capture CTCs repeatedly in a dynamic environment (flow rate = 5.0 mL min−1). In the setting of extracorporeal closed-loop circulation, the capture efficiency reached 21.6 ± 1.5% after 120 cycles, and 13.26 ± 4.22% in rats after 3 hours of in vivo circulation. Moreover, the heat generated by HA-MVS under AMF can destroy the cell membrane and induce apoptosis, resulting in 91.3% of CTC death.Intravenous indwelling needle, which is widely used clinically, is a good framework for the construction of CTCs. Zhang et al. proposed the in vivo capture of CTC based on surface-modified intravenous indwelling needle transfusion.153 When the needle is used for intravenous infusion, CTCs is captured at the same time, which will not cause further physical harm and reduce the invasion of in vivo detection. Li et al. designed a medical needle coated with ZnO nano-flowers for the capture and killing of CTCs.154In vivo and in vitro experiments showed that about 90% of the captured CTC was killed and fell off the surface of the needle after death, enabling the ZNFs needle to continuously remove CTCs.
The catheter is usually made of polyurethane, which can be covalently modified with tumor-specific antibodies to capture CTC in the blood. In addition, due to its long indwelling time in vessel, a large amount of blood continuously flows through the surface of the retention needle with the aid of circulation. This greatly increases the contact rate between the cells in the blood vessel and the marker, which can effectively increase the capture efficiency. Wang et al. developed a minimally invasive therapeutic intravenous catheter (Fig. 10D) for in vivo enrichment and photothermal killing of CTCs.155 The surface of the catheter was modified with anti-EpCAM antibody and the interior was filled with black phosphorus nanosheets (BPNS). CTCs in peripheral blood are continuously captured by the catheter with the help of circulation. Captured CTCs are used for downstream analysis or eliminated in vivo by the near-infrared (NIR) photothermal effect of BPNS. A capture efficiency of 2.1% was obtained during a 5-minute treatment, and 100% of the captured CTCs were killed by NIR light irradiation in an in vitro closed-loop circulatory system and in an in vivo rabbit model. Kim et al. described an indwelling intravascular non-circulating CTC isolation system (Fig. 10E) to continuously collect CTCs directly from peripheral veins.25 A patient may wear the device for hours, and a relatively large amount of blood can be examined through the device. The system consisted of four main parts: a microcontroller, a peristaltic pump, a heparin injector, and a CTC capture module containing a microfluidic CTC capture chip. The system was designed to accommodate any type of CTCs isolator, as long as it is configured to fit the system manifold. The system allowed for periodic CTCs assessment by exchanging CTCs collection chips, while continuously monitoring blood flow. It could accommodate any type of CTCs separation device, as long as it was compatible with the system's manifold. The design of the interchangeable CTC chip solved the device blocking problem expected from saturation of the CTC capture module in the CTC capture module. This means the system can be left in place for longer periods of time, potentially allowing large blood samples to be interrogated with little harm. This allows for adequate patient mobility during surgery, and minimizes patient inconvenience. It is worth noting that the capture module currently used in this system is designed for CTC-immobilized EpCAM, which can be obtained by combining with other antibodies or aptamers or equipped with physical structures for higher capture efficiency.
In total, there has been little work on the in vivo enrichment of CTC based on nanostructures. Due to the complexity of the in vivo environment and the characteristics of CTC itself, the published research work, such as intravascular magnetic lines, temporary retention system, in vivo enrichment and killing of CTC, have further enriched the enrichment means of CTC, and explored the feasibility of CTC intervention in vivo, which have received great attention. However, the remarkable results have not been achieved. The low efficiency and the biosafety of in vivo capture still restrict the development of in vivo enrichment technology. Judging from the existing research, it is a possible development direction to improve the capture efficiency and detection efficiency based on in vivo intervention. In addition, the problems derived from in vivo capture technology, such as feasibility and safety, need further exploration by researchers.
5 Downstream analysis
With the progress of technology, the study of CTC has changed from simple counting to the era of molecular typing and genotyping. Because CTC is very rare and mixed in many peripheral blood cells, separating CTC from white blood cells, red blood cells, and other blood components is an indispensable step in CTC research and downstream analysis. Capture efficiency and capture purity are the two most basic indicators to evaluate the CTC enrichment technology platform. The enrichment platform with high capture efficiency can avoid the missed detection of CTC in samples to the greatest extent. That is, it has excellent peripheral blood CTC detection ability and detection rate. The enrichment platform with high capture purity maximally avoids the interference of other components (mainly white blood cells) in peripheral blood. It ensures the purity of genetic material extracted in downstream analysis. In addition, whether CTC can maintain high cell viability after release, the detection rate of authentic samples, and the number of captured CTC all affect the clinical application of CTC.With the development of new capture technology, the capture performance of CTC was dramatically improved. Recent CTCs enrichment strategies based on nanostructures and fundamental indicators are shown in Table 1. CTCs obtained by various enrichment methods are usually further processed for downstream analysis. The most commonly used methods for specific identification and downstream analysis of CTCs include tumor-specific antigen detection based on immunocytochemistry/immunofluorescence staining and various molecular analysis methods based on nucleic acid sequence identification (fluorescence in situ hybridization FISH, array comparative genomic hybridization aCGH, next-generation sequencing NGS, polymerase chain reaction PCR). Compared with ctDNA, capturing CTCs is relatively tricky. However, it has a complete cell morphology, which can be used for morphological and functional observation, and include complete genetic information such as proteome, transcriptome, and genome. In addition to CTCs counting, the analysis of CTCs helps identify specific molecular targets related to treatment, expanding people's understanding of basic molecular pathways related to invasion, migration, and immune surveillance, identifying more invasive tumor cells and promoting the development of personalized therapy.
| Category | Methods | Cell lines | Cell line isolation result | Clinical blood sample or animal model | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Yield | Purity | Viability | Ratio | CTC count | ||||
| Physical properties for isolation | ||||||||
| Size and morphology | Ultrathin Si3N4 filtering membrane with slit-shaped pores | A549, HCT-116 | C: 96% | 99.99% | 90% | 100% (n = 10) | 6–20 cells | 20 |
| Cluster-Wells microfluidic chip | HeyA8, LNCaP, MDA-MB-231, MCF-7 | C: 93.2%, R: 96% | 99% | 95.8% | 75% (n = 10) and 88.9 (n = 10) | 0.063 to 0.867 clusters per mL, 0.167 to 59.2 clusters per mL | 36 | |
| Density | Fully automated, negative depletion-based continuous centrifugal microfluidics | A549, PC9, H1975, MDA-MB-231, SK-BR-3, T24 | C: 92%, R: >90% | 99.9% | — | 100% (n = 30) | 12–120 cells | 39 |
| Electrical properties | Microwells fabricated on interdigitated electrodes | DU-145 | C: 90% | — | — | — | — | 43 |
| Acoustic properties | Initial acoustofluidic preseparation and negative selection acoustophoresis | DU145 | C: 94%, R: 42% | 99.5% | 96.8% | — | — | 47 |
| Fluid mechanics | A miniaturized centrifuge with four paralleled inertial spiral channels with a two-stage serpentine channel | MCF-7, A549 | C: 86.5% | 93% | — | — | — | 53 |
| Biological properties for isolation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antibody | Luminescent lanthanide nanoprobes | MCF-7 | C: 93.9% | — | — | 93.9% (n = 15) | 10–190 cells | 63 |
| Nanoprobes with gradient magnetic response | BT474, LNCaP, MDA-MB-231 | C: 90.2 ± 2% | 92.1 ± 2.4% | — | — | — | 60 | |
| Aptamer | Multivalent aptamer functionalized Ag2S nanodots and hybrid cell membrane-coated magnetic nanoparticles | MCF-7 | C: 97.63% | 96.96% | — | 100% (n = 8) | 6–10 cells per mL | 72 |
| Dual-aptamer-modified immunomagnetic particles | MGC-803, BGC-823 | C: 95% | — | — | 100% (n = 17) | 4–12 cells per mL | 21 | |
| Peptide | EpCAM recognition peptide functionalized iron oxide magnetic nanoparticles | MCF-7, SK-BR-3, PC3, Hep G2 | C: 90% | 93% | — | — | — | 78 |
| Folic acid | FA-functionalized bionic layer of RBC | HeLa, HCT116 | C: >80%, R: >70% | 90% | >90% | 100% (n = 5) | 3–21 cells | 86 |
| Hyaluronic acid | HA-functionalized redox responsive immunomagnetic nanocarrier | MCF-7 | C: 92%, R: 81.4% | — | — | — | — | 90 |
| Tannic acid | Dual-responsive fluorescent-magnetic nanoparticles | MCF-7, HepG2, MDA-MB-231, HeLa | C: >88%, R: >80% | 86.9% | >90% | 100% (n = 15) | 3–30 cells per mL | 96 |
| Nanostructure | ||||||||
|---|---|---|---|---|---|---|---|---|
| Nanorod | Double-tetrahedral DNA probe functionalized Ag nanorod biointerface | SGC-7901 | C: 90.2%, R: 93.4% | — | 98% | — | — | 100 |
| Nanofiber | Dual aptamer-modified PLGA nanofiber-based microfluidic devices | A2780, OVCAR-3 | C: 91%, R: 95% | — | >90% | 100% (n = 7) | 1–13 cells per mL | 108 |
| Nanosheets | Boronic acid-functionalized g-C3N4 nanosheets | HepG2, SK-HEP-1 | C: 97.64%, R: 99.17% | 99% | 98% | 100% (n = 20) | 5–43 cells per mL | 119 |
| Bionic nanostructure | Bioinspired 3D aptamer modified rose petal derived ZnO microchip | MCF-7, PC9, A549 | C: 90.5%, R: 84.4% | 94% | 96.8% | 100% (n = 21) | 3–36 cells per mL | 125 |
| Micro-nano channel structure | Antibody-engineered red blood cell affinity interface on microfluidic chip and DLD-patterned interaction model | SW480 | C: 96.5%, R: 96.9% | — | 96.10% | 100% (n = 20) | 2–58 cells per mL | 127 |
| A robust microfluidic chip containing virtually implemented zones surface-coated with gold | MCF-7, MDA-MB-231, MDA-MB-468 | C: 97%, R: 96.1% | — | >90% | 100% (n = 60) | 4–187 cells per mL | 132 | |
| In vivo enrichment of CTC | ||||||||
|---|---|---|---|---|---|---|---|---|
| C, capture efficiency; R, release efficiency; D, death rate. | ||||||||
| Venous catheter | Indwelling intravascular aphaeretic system | MCF-7 | C: 1–2% (2 h) | — | >90% | 762 cells in canine blood | 25 | |
| Black phosphorus and antibody functionalized intravenous catheter | HepG2 | C: 2.1% (5 min), D: 100% | — | — | 2100 cells in rabbit blood | 155 | ||
| HA-modified magnetic vascular scaffold | MDA-MB-231 | C: 13.26 ± 4.22% (3 h), D: 91.3% | — | — | 132![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 cells in rat blood 600 cells in rat blood |
152 | ||
5.1 Disease detection and prognosis evaluation
The prognostic value of CTCs has been confirmed in a variety of cancers. In addition, the value of CTCs in early detection of solid tumors, efficacy and disease recurrence monitoring has been gradually recognized. In recent years, based on a variety of emerging technologies, there has been a lot of research work on the use of CTCs in disease detection and prognosis evaluation. Wan et al. made a maze microfluidic device to effectively separate CTC from peripheral blood of patients with HCC, and studied the correlation between the number of CTCs and clinical HCC stage.156 In this work, blood samples from 42 HCC patients were selected and compared with 5 non-HCC healthy subjects. CTCs were identified in 88.1% of HCC patients at different tumor stages. The statistical results showed that a higher positive rate of CTCs was found with the later HCC stage. In addition, 55% of the patients had circulating tumor microthrombus (CTM), which was also related to the late HCC stage. Based on the immunomagnetic platform Fe3O4@SiO2@PTMAO@aptamer, Li et al. detected the number of epithelial CTC (E-CTC), stromal CTC (M-CTC) and mixed (epithelial and stromal) CTC (EM-CTC) in blood samples of patients with colorectal cancer, and analyzed the correlation between different phenotypes of CTCs and clinical information of CRC patients.28 The changes of the proportion of heterogeneous CTC in 40 patients with different stages of colorectal cancer before and after operation showed that the number and proportion of E-CTC or M-CTC was significantly lower than that of EM-CTC and M-CTC. This was positively correlated with the metastasis of colorectal cancer and the effect of operation and chemotherapy in patients with CRC. The proportion of M-CTC decreased in all stages after operation, but only in advanced patients. Based on the multi-functional nano-delivery system, Ren et al. realized genome editing and in situ evaluation of therapeutic response to in vitro CTCs at the single-cell level, providing accurate information for personalized cancer treatment.157 In this study, C-X-C motif chemokine receptor 4 (CXCR4), a typical target protein in cancer therapy, was selected as the representative target for genome editing in vitro. Single cell analysis showed that CXCR4 in CTCs of cancer patients was effectively downregulated, anti-cancer biomarkers such as p53 and p21 were upregulated, and the edited cancer cells showed inhibition of proliferation and inhibition of migration and invasion. Using the same targeted delivery vector to deliver molecular beacons to the liver, the therapeutic efficiency of cancer cell lines and genome editing in CTCs can be easily detected.5.2 Gene analysis and auxiliary targeted treatment
Molecular targeted therapy based on tissue biopsy is a typical case of cancer precision medicine. Although tissue biopsy can accurately obtain tumor genetic information, tissue biopsy is an expensive, invasive and painful process. Only some tumor sites can be sampled regularly. Obtaining tumor cells from blood sampling or liquid biopsy is a promising alternative to tumor biopsy. In recent years, CTC gene mutation profile analysis, as an auxiliary diagnostic tool, has been used to characterize tumor molecular heterogeneity and tumor genotype monitoring to guide treatment because of its low invasion, carrying primary and metastatic tumor characteristics and complete genetic information.Cheng et al. proposed a large-scale parallel analysis of hydrodynamics single-cell RNA sequencing (scRNA-seq) bar code technology Hydro-Seq for CTC capture and gene expression profile analysis.158 Hydro-Seq successfully sequenced the single cell RNA of 666 CTCs in 21 breast cancer patients. These CTCs were detected with ER\PR\AR and HER2 mutations, identifying the drug targets of hormone and targeted therapy for breast cancer, which can assist in the confirmation of targeted drug therapy. In addition, this study tracked individual cells expressing cancer stem cell (CSCs) and epithelial/mesenchymal cell state transition markers, which is beneficial to the study of tumor metastasis, treatment choice, and cell heterogeneity analysis. Kenji Kuroda et al. enriched CTCs by density gradient centrifugation and studied the blood samples of 100 patients with gastric cancer before gastrectomy.159 Immunohistochemical evaluation showed that the number of FGFR2-positive CTCs in peripheral blood was significantly correlated with the expression of FGFR2 in primary gastric cancer. The relapse-free survival rate of FGFR2-positive CTCs patients (≥5 cells/10 mL blood) was significantly lower than that of non-FGFR2-positive CTCs patients. Ruan et al. reported a single CTC mass spectrometry analysis platform DMF-scMS (Fig. 11A) based on digital microfluidic technology, which is used for single CTC isolation and multiple mutation analysis.27 The KRAS mutation spectrum of heterogeneous CTCs at the single cell level in 5 patients with colorectal cancer was detected. At present, DMF-scMS is only suitable for the detection of known mutations in a single CTC. This is because it requires specific primers for single base extension of the mutation site and the known mUniz signal as a reference, but it still has great clinical potential in guiding accurate molecular targeted therapy for known mutation sites.
 | ||
| Fig. 11 Downstream analysis | (A) schematic illustration of DMF-scMS for the efficient and multiplex gene mutation profiling of single CTC27 (reproduced with permission from ref. 27, Copyright 2022, American Chemical Society). (B) Schematic illustration of Multiomic characterization and drug testing162 (reproduced with permission from ref. 162, Copyright 2022, Elsevier). (C) Schematic illustration of workflow for biomarker independent isolation, expansion, and analysis of CTCs26 (reproduced with permission from ref. 26, Copyright 2020, MDPI). (D) Schematic representation of the synthesis of CMS164 (reproduced with permission from ref. 164, Copyright 2022, Royal Society of Chemistry). | ||
5.3 Ex vivo culture and testing of drug susceptibility
In vitro cultures from patients' tumor tissues can be used as an important alternative platform for evaluating different therapies, thus enabling clinicians to reliably predict individual patient responses. Compared with tumor tissue section culture, CTCs have a longer survival time and can be extracted from minimally invasive fluid biopsies in patients' peripheral blood, allowing regular samples to support real-time monitoring of clinical treatment efficacy and drug resistance development. In addition, CTC originated from the primary tumor, and the CTC line retained similar behavior and heterogeneous characteristics to cancer cells in the original tumor, with high uptake rate, rapid growth rate, similar tumor microenvironment and relatively low cost. The culture of CTCs in vitro not only provides a sufficient number of cells for other characterization, but also has great application potential in new drug screening and development, disease feature identification and biochemical mechanism research.As shown in Fig. 11C, Lianette Rivera-Báez et al. used a size-based inertial microfluidic separation device to isolate CTC from the blood of 10 patients with advanced pancreatic cancer, and amplified it in vitro and characterized its function.26 When tested with gemcitabine (gemcitabine) and 5FU (5-fluorouracil), CTC lines showed differences in drug sensitivity. Hu et al. used density gradient centrifugation and magnetic separation based on the CD45 antibody to separate HCC-CTC from patients' blood samples, and then used 3D culture to amplify the spheroids, and analyzed the relationship between clinicopathological variables of HCC patients and CTC spheres.160 Lin et al. use the developed eSelect system to extract CTCs from blood samples of patients with head and neck cancer, then amplified them to form organ-like organs in vitro, and analyzed the drug sensitivity of cisplatin treatment, so as to predict the clinical drug response of patients with head and neck cancer.161 Statistical analysis confirmed that the drug sensitivity of CTC-like organs in vitro was related to clinical response. The treatment accuracy of multivariate logical regression model for predicting chemotherapy response was 93.75%. The area under the curve (AUCs) of the prediction model was 0.8841 in the whole data set and 0.9167 in the cisplatin specific data set. As shown in Fig. 11B, Chen et al. separated CTCs based on a microfluidic chip modified by a biomimetic lipid membrane, established a CTC line in vitro, and carried out a multi-omics analysis and drug sensitivity test.162 Phenotype, genome, epigenome and transcriptome analysis and drug test results of metastatic breast cancer patients show that the drug sensitivity of CTCs reflects the drug sensitivity of metastatic tumors, while the reaction of cells sampled from primary tumors is similar to that of primary tumors, indicating that CTCs and metastatic tumors are more similar. Ajay Balakrishnan et al. developed a short-term CTCs culture method in which cancer cell clusters were attached and proliferated by Matrigel coated in 96-well plates, and drug sensitivity tests were carried out in individual patients.163 In the proof-of-concept experiment, CTCs were cultured from the blood of patients with baseline lung cancer and treated with the same drug regimen as patients with lung cancer. The survival rate of CTCs in the culture was 30.7%. The decrease in cell vitality was associated with the reduction of pleural effusion around the lungs of patients in actual treatment, showing the potential of this method in drug testing and personalized treatment. Smriti Arora et al. reported a cotton microfluidic substrate (CMS) that can effectively isolate CTCs and promote its expansion in vitro (Fig. 11D).164 CMS with nanostructured surface can effectively isolate CTCs and act as a matrix to promote the formation of 3D tumor spheres in capturing cells, and its size increases by 5 times from the 3rd day to the 10th day of culture. In addition, when treated with the anticancer drug cisplatin, the tumor size was reduced by almost 1 to 2 within 24 hours. On the second and third day after cisplatin treatment, the tumor shrank and stopped, followed by the gradual growth of tumor globules, indicating that the tumor acquired drug resistance and demonstrated the potential of CMS in evaluating the efficacy of chemotherapy in drug discovery and development.
6 Conclusion and outlook
The application of CTC detection technology in clinical oncology has shown great promise in addressing numerous unmet needs. As current knowledge and understanding of cancer continue to evolve, cancer biologists and medical scientists hope to characterize the dynamic biology of the disease. This paper reviews various methodological methods of new early diagnosis techniques for cancer patients and various methods of separating and detecting CTC from whole blood. As mentioned above, these CTC strategies have significant advantages in non-invasive or minimally invasive diagnosis by liquid biopsy, improving the comfort of cancer patients by avoiding unnecessary and invasive tumor sampling.Many kinds of CTC detection technologies have emerged, and dozens of different commercial CTC detection systems have been formed, such as CellSearch, CytoploRare, CTC-Biopay, CellScan, and CellCollector. However, it is a pity that these methods are inadequate to some extent. Label-free enrichment based on physical characteristics can enrich many subtypes of CTC, but still have some problems. These include the easy blockage of the filter membrane or micropore, loss of small CTC, the need for pretreatment steps such as centrifugation and dilution, and lack of sufficient specificity of isolated cells. Enrichment based on biological characteristics has strong specificity, but it also has the limitation of being unable to capture heterogeneous CTCs. Due to the heterogeneity of tumors and the EMT process of tumors, the expression patterns of CTCs are different. However, at present, immune recognition is mainly limited to epithelial CTCs and the capture base on the expression of EpCAM. Although double antibodies and other recognition methods targeting multiple targets appear, other CTCs phenotypes will still be missed. The capture interface based on various nanostructures effectively enhances cells' capture and release ability, but the inherent limitations of immune recognition still limit it. In addition, CTC enrichment in vitro cannot avoid the problems of low flux and spatiotemporal heterogeneity of CTCs, and can only provide a few CTCs for downstream analysis. In vivo capture of CTC solves the problem of low flux to some extent, and a large number of CTCs can be obtained. However, heterogeneity still limits identification, and the detectable flux of CTC capture in vivo is still low for the human body.
The future direction of CTC enrichment is to solve these problems. As the ultimate goal of CTC enrichment is to maximize clinical application, obtaining reliable clinical data for future use and general acceptance by doctors, patients, and the government responsible for medical care is necessary to achieve standardization and repeatability of CTCs technology. Aiming to improve capture purity, capture capacity, flux, and the requirement of heterogeneous CTCs detection, module integration, and multi-mode detection is a possible direction based on existing enrichment technology of nanostructures. Module integration can combine the advantages of different enrichment methods to improve the enrichment effect. Multi-mode detection improves data reliability by mutual verification of different detection methods. In vivo CTC enrichment and collection/killing is another possible direction. In vivo enrichment has high throughput characteristics, and more CTC can be obtained. Negative enrichment is also a possible direction. This method can sort WBC and detect a variety of CTC subtypes. In addition, broad-spectrum enrichment of CTCs based on the metabolic characteristics of cancer cells (such as PH and lactic acid) and co-expressed biomolecules (such as FA and TA recognition) is a possible direction. However, more research is still needed in terms of capture purity and capture performance. Besides the development of CTC enrichment technology, the downstream analysis of CTCs is also essential to research content. The improvement of molecular analysis technology and the further study of the clinical value of CTCs will significantly promote the development of the CTC field, which requires the joint efforts of researchers in various related fields.
The concept of circulating tumor cells has been put forward for more than one hundred years. However, the capture and analysis of circulating tumor cells in the blood and their clinical application have been paid increasingly more attention in the past ten years. Previous studies have shown that CTC with tumor-related information can make up for traditional biopsy puncture difficulties and cannot monitor early cancer detection, judging patients' prognoses, predicting drug efficacy, and exploring the mechanism of drug resistance. However, to make full use of CTC in the clinic, it is still necessary to systematically understand the physiological mechanisms of CTC, improve the performance of the CTC enrichment platform and continue to study the clinical value of CTC. Today, with the emergence of ctDNA, exosomes, and a variety of new fluid biopsy markers, the value and future of CTC research is a question that all relevant researchers must face. The answer to this question can only rely on more in-depth research, which needs researchers in various related fields to continue to study the physiological mechanism of CTC, improve the CTC enrichment platform's performance and expand CTC's clinical application scenarios.
Author contributions
Lihua Guo: investigation, writing – revised draft. Chang Liu: investigation, writing – original draft. Manlin Qi: data curation, validation, formal analysis. Liang Cheng: conceptualization, project administration, writing – review & editing. Lin Wang: conceptualization, supervision, writing – review & editing, funding acquisition. Chunxia Li: conceptualization, supervision, writing – review & editing. Biao Dong: supervision, writing – review & editing.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 52250077, 52272080, 82073475), National Science Foundation of Jilin Province (20210401059YY, 20220402005GH, 20200801017GH), Major Basic Research Projects of Shandong Natural Science Foundation (ZR2020ZD36), Funding of Jilin Province Development and Reform Commission (2021C035-1), and the Natural Science Foundation of Jilin Province (20210401059YY, 20220402005GH).References
- T. Gansler, P. A. Ganz, M. Grant, F. L. Greene, P. Johnstone, M. Mahoney, L. A. Newman, W. K. Oh, C. R. Thomas Jr, M. J. Thun, A. J. Vickers, R. C. Wender and O. W. Brawley, Ca-Cancer J. Clin., 2010, 60, 345–350 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- S. de Wit, G. van Dalum and L. W. M. M. Terstappen, Scientifica, 2014, 2014, 819362 CrossRef PubMed.
- S. Paget, Lancet, 1889, 133, 571–573 CrossRef.
- K. Pantel and C. Alix-Panabières, Trends Mol. Med., 2010, 16, 398–406 CrossRef PubMed.
- C. Alix-Panabières and K. Pantel, Clin. Chem., 2013, 59, 110–118 CrossRef PubMed.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy and A. Muzikansky, Nature, 2007, 450, 1235–1239 CrossRef CAS PubMed.
- J. S. De Bono, H. I. Scher, R. B. Montgomery, C. Parker, M. C. Miller, H. Tissing, G. V. Doyle, L. W. Terstappen, K. J. Pienta and D. Raghavan, Clin. Cancer Res., 2008, 14, 6302–6309 CrossRef CAS PubMed.
- E. Sollier, D. E. Go, J. Che, D. R. Gossett, S. O'Byrne, W. M. Weaver, N. Kummer, M. Rettig, J. Goldman and N. Nickols, Lab Chip, 2014, 14, 63–77 RSC.
- L. G. Martelotto, C. K. Ng, S. Piscuoglio, B. Weigelt and J. S. Reis-Filho, Breast Cancer Res., 2014, 16, 1–11 CrossRef PubMed.
- A. A. Powell, A. H. Talasaz, H. Zhang, M. A. Coram, A. Reddy, G. Deng, M. L. Telli, R. H. Advani, R. W. Carlson and J. A. Mollick, PLoS One, 2012, 7, e33788 CrossRef CAS PubMed.
- M. Labelle and R. O. Hynes, Cancer Discovery, 2012, 2, 1091–1099 CrossRef CAS PubMed.
- C. V. Pecot, F. Z. Bischoff, J. A. Mayer, K. L. Wong, T. Pham, J. Bottsford-Miller, R. L. Stone, Y. G. Lin, P. Jaladurgam and J. W. Roh, Cancer Discovery, 2011, 1, 580–586 CrossRef CAS PubMed.
- P. Paterlini-Brechot and N. L. Benali, Cancer Lett., 2007, 253, 180–204 CrossRef CAS PubMed.
- J. Chen, J. Li and Y. Sun, Lab Chip, 2012, 12, 1753–1767 RSC.
- Y.-F. Sun, X.-R. Yang, J. Zhou, S.-J. Qiu, J. Fan and Y. Xu, J. Cancer Res. Clin. Oncol., 2011, 137, 1151–1173 CrossRef PubMed.
- M. C. Miller, G. V. Doyle and L. W. Terstappen, J. Oncol., 2010, 2010, 617421 Search PubMed.
- S. Riethdorf, H. Fritsche, V. Müller, T. Rau, C. Schindlbeck, B. Rack, W. Janni, C. Coith, K. Beck and F. Jänicke, Clin. Cancer Res., 2007, 13, 920–928 CrossRef CAS PubMed.
- A. H. Talasaz, A. A. Powell, D. E. Huber, J. G. Berbee, K.-H. Roh, W. Yu, W. Xiao, M. M. Davis, R. F. Pease and M. N. Mindrinos, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3970–3975 CrossRef CAS PubMed.
- A. Li, X. He, J. Wu, J. Zhang, G. Xu, B. Xu, G. Zhao and Z. Shen, Lab Chip, 2022, 22, 3676–3686 RSC.
- C. Li, S. Yang, R. Li, S. Gong, M. Huang, Y. Sun, G. Xiong, D. Wu, M. Ji and Y. Chen, ACS Appl. Mater. Interfaces, 2022, 14, 7646–7658 CrossRef CAS PubMed.
- H. Cui, Q. Liu, R. Li, X. Wei, Y. Sun, Z. Wang, L. Zhang, X.-Z. Zhao, B. Hua and S.-S. Guo, Nanoscale, 2020, 12, 1455–1463 RSC.
- Y. Hu, W. Tang, P. Cheng, Q. Zhou, X. Tian, X. Wei and H. He, Cytometry, Part A, 2019, 95, 657–663 CrossRef PubMed.
- P. Hai, Y. Qu, Y. Li, L. Zhu, L. Shmuylovich, L. A. Cornelius and L. V. Wang, J. Biomed. Opt., 2020, 25, 036002 Search PubMed.
- T. H. Kim, Y. Wang, C. R. Oliver, D. H. Thamm, L. Cooling, C. Paoletti, K. J. Smith, S. Nagrath and D. F. Hayes, Nat. Commun., 2019, 10, 1–8 Search PubMed.
- L. Rivera-Báez, I. Lohse, E. Lin, S. Raghavan, S. Owen, R. Harouaka, K. Herman, G. Mehta, T. S. Lawrence and M. A. Morgan, Cancers, 2020, 12, 1011 CrossRef PubMed.
- Q. Ruan, J. Yang, F. Zou, X. Chen, Q. Zhang, K. Zhao, X. Lin, X. Zeng, X. Yu and L. Wu, Anal. Chem., 2021, 94, 1108–1117 CrossRef PubMed.
- C. Li, R. Li, X. Wu, Y. Zuo, G. Xiong, M. Huang, Y. Sun, R. Liao, Y. Xiao and L. Hu, Anal. Chem., 2022, 94, 15240–15249 CrossRef CAS PubMed.
- H. M. Shapiro, E. R. Schildkraut, R. Curbelo, C. W. Laird, B. Turner and T. Hirschfeld, J. Histochem. Cytochem., 1976, 24, 396–401 CrossRef CAS PubMed.
- P. Bankó, S. Y. Lee, V. Nagygyörgy, M. Zrínyi, C. H. Chae, D. H. Cho and A. Telekes, J. Hematol. Oncol., 2019, 12, 1–20 CrossRef PubMed.
- X. Xu, Z. Jiang, J. Wang, Y. Ren and A. Wu, Electrophoresis, 2020, 41, 933–951 CrossRef CAS PubMed.
- M. Hosokawa, T. Hayata, Y. Fukuda, A. Arakaki, T. Yoshino, T. Tanaka and T. Matsunaga, Anal. Chem., 2010, 82, 6629–6635 CrossRef CAS PubMed.
- S. Zheng, H. Lin, J.-Q. Liu, M. Balic, R. Datar, R. J. Cote and Y.-C. Tai, J. Chromatogr. A, 2007, 1162, 154–161 CrossRef CAS PubMed.
- X. Xu, J. Lin, Y. Guo, X. Wu, Y. Xu, D. Zhang, X. Zhang, X. Yujiao, J. Wang and C. Yao, Biosens. Bioelectron., 2022, 210, 114305 CrossRef CAS PubMed.
- A. F. Sarioglu, N. Aceto, N. Kojic, M. C. Donaldson, M. Zeinali, B. Hamza, A. Engstrom, H. Zhu, T. K. Sundaresan and D. T. Miyamoto, Nat. Methods, 2015, 12, 685–691 CrossRef CAS PubMed.
- M. Boya, T. Ozkaya-Ahmadov, B. E. Swain, C.-H. Chu, N. Asmare, O. Civelekoglu, R. Liu, D. Lee, S. Tobia and S. Biliya, Nat. Commun., 2022, 13, 1–13 Search PubMed.
- A. Lee, J. Park, M. Lim, V. Sunkara, S. Y. Kim, G. H. Kim, M.-H. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 11349–11356 CrossRef CAS PubMed.
- Q. Huang, F.-B. Wang, C.-H. Yuan, Z. He, L. Rao, B. Cai, B. Chen, S. Jiang, Z. Li and J. Chen, Theranostics, 2018, 8, 1624 CrossRef CAS PubMed.
- H. J. Woo, S.-H. Kim, H. J. Kang, S.-H. Lee, S. J. Lee, J. M. Kim, O. Gurel, S. Y. Kim, H. R. Roh and J. Lee, Theranostics, 2022, 12, 3676 CrossRef CAS PubMed.
- I.-F. Cheng, W.-L. Huang, T.-Y. Chen, C.-W. Liu, Y.-D. Lin and W.-C. Su, Lab Chip, 2015, 15, 2950–2959 RSC.
- T. Luo, L. Fan, Y. Zeng, Y. Liu, S. Chen, Q. Tan, R. H. Lam and D. Sun, Lab Chip, 2018, 18, 1521–1532 RSC.
- P. Modarres and M. Tabrizian, Sens. Actuators, B, 2019, 286, 493–500 CrossRef CAS.
- J. Park, C. Park, Y. Sugitani, T. Fujii and S. H. Kim, Lab Chip, 2022, 22, 3000–3007 RSC.
- P. Augustsson, C. Magnusson, M. Nordin, H. Lilja and T. Laurell, Anal. Chem., 2012, 84, 7954–7962 CrossRef CAS PubMed.
- M. Antfolk, C. Magnusson, P. Augustsson, H. Lilja and T. Laurell, Anal. Chem., 2015, 87, 9322–9328 CrossRef CAS PubMed.
- S. Karthick, P. N. Pradeep, P. Kanchana and A. K. Sen, Lab Chip, 2018, 18, 3802–3813 RSC.
- E. Undvall Anand, C. Magnusson, A. Lenshof, Y. Ceder, H. Lilja and T. Laurell, Anal. Chem., 2021, 93, 17076–17085 CrossRef CAS PubMed.
- S. C. Hur, H. T. K. Tse and D. Di Carlo, Lab Chip, 2010, 10, 274–280 RSC.
- A. J. Mach and D. Di Carlo, Biotechnol. Bioeng., 2010, 107, 302–311 CrossRef CAS PubMed.
- M. E. Warkiani, B. L. Khoo, L. Wu, A. K. P. Tay, A. A. S. Bhagat, J. Han and C. T. Lim, Nat. Protoc., 2016, 11, 134–148 CrossRef CAS PubMed.
- F. Tian, L. Cai, J. Chang, S. Li, C. Liu, T. Li and J. Sun, Lab Chip, 2018, 18, 3436–3445 RSC.
- R. Guglielmi, Z. Lai, K. Raba, G. van Dalum, J. Wu, B. Behrens, A. A. S. Bhagat, W. T. Knoefel, R. P. L. Neves and N. H. Stoecklein, Sci. Rep., 2020, 10, 1–9 CrossRef PubMed.
- Y. Fang, S. Zhu, W. Cheng, Z. Ni and N. Xiang, Lab Chip, 2022, 22, 3545–3554 RSC.
- N. Xiang, J. Wang, Q. Li, Y. Han, D. Huang and Z. Ni, Anal. Chem., 2019, 91, 10328–10334 CrossRef CAS PubMed.
- Y. Wu, R. Chattaraj, Y. Ren, H. Jiang and D. Lee, Anal. Chem., 2021, 93, 7635–7646 CrossRef CAS PubMed.
- C.-Y. Wen, L.-L. Wu, Z.-L. Zhang, Y.-L. Liu, S.-Z. Wei, J. Hu, M. Tang, E.-Z. Sun, Y.-P. Gong and J. Yu, ACS Nano, 2014, 8, 941–949 CrossRef CAS PubMed.
- Z. Wang, N. Sun, H. Liu, C. Chen, P. Ding, X. Yue, H. Zou, C. Xing and R. Pei, ACS Appl. Mater. Interfaces, 2019, 11, 39586–39593 CrossRef CAS PubMed.
- M. Hu, C. Li, Z. Wang, P. Ding, R. Pei, Q. Wang, H. Xu and C. Xing, Front. Bioeng. Biotechnol., 2022, 90 Search PubMed.
- W. Xia, H. Li, Y. Li, M. Li, J. Fan, W. Sun, N. Li, R. Li, K. Shao and X. Peng, Nano Lett., 2020, 21, 634–641 CrossRef PubMed.
- Z. Liao, L. Han, Q. Li, L. Li, Y. Liu, Y. Song, W. Tan and E. Song, Adv. Funct. Mater., 2021, 31, 2009937 CrossRef CAS.
- L.-L. Wu, C.-Y. Wen, J. Hu, M. Tang, C.-B. Qi, N. Li, C. Liu, L. Chen, D.-W. Pang and Z.-L. Zhang, Biosens. Bioelectron., 2017, 94, 219–226 CrossRef CAS PubMed.
- L.-L. Wu, Z.-L. Zhang, M. Tang, D.-L. Zhu, X.-J. Dong, J. Hu, C.-B. Qi, H.-W. Tang and D.-W. Pang, Angew. Chem., Int. Ed., 2020, 59, 11240–11244 CrossRef CAS PubMed.
- H. Guo, X. Song, W. Lei, C. He, W. You, Q. Lin, S. Zhou, X. Chen and Z. Chen, Angew. Chem., Int. Ed., 2019, 58, 12195–12199 CrossRef CAS PubMed.
- J. Lyu, H. Xu, B. Dong, C. Li, S. Yu, S. Hu, B. Zhou, L. Wang and H. Song, Chem. Eng. J., 2022, 448, 137761 CrossRef CAS.
- X. Liu, F. Wang, Y. Meng, L. Zhao, W. Shi, X. Wang, Z. He, J. Chao and C. Li, Biosens. Bioelectron., 2022, 207, 114208 CrossRef CAS PubMed.
- L. Rao, Q.-F. Meng, Q. Huang, Z. Wang, G.-T. Yu, A. Li, W. Ma, N. Zhang, S.-S. Guo and X.-Z. Zhao, Adv. Funct. Mater., 2018, 28, 1803531 CrossRef.
- X. Wu, Z. Lin, C. Zhao, L. Liu, K. Zhang, J. Lai, Q.-F. Meng, G. Yao, Q. Huang and X.-Z. Zhao, Biosens. Bioelectron., 2022, 114425 CrossRef CAS PubMed.
- S. Fang, C. Wang, J. Xiang, L. Cheng, X. Song, L. Xu, R. Peng and Z. Liu, Nano Res., 2014, 7, 1327–1336 CrossRef CAS.
- Z. Li, G. Wang, Y. Shen, N. Guo and N. Ma, Adv. Funct. Mater., 2018, 28, 1707152 CrossRef.
- Y. Wang, T. Huo, Y. Du, M. Qian, C. Lin, H. Nie, W. Li, T. Hao, X. Zhang and N. Lin, Biosens. Bioelectron., 2022, 215, 114530 CrossRef CAS PubMed.
- C. Ding, C. Zhang, X. Yin, X. Cao, M. Cai and Y. Xian, Anal. Chem., 2018, 90, 6702–6709 CrossRef CAS PubMed.
- C. Ding, C. Zhang, S. Cheng and Y. Xian, Adv. Funct. Mater., 2020, 30, 1909781 CrossRef CAS.
- P. Chen, Y. Wang, Y. He, K. Huang, X. Wang, R. Zhou, T. Liu, R. Qu, J. Zhou and W. Peng, ACS Nano, 2021, 15, 11634–11643 CrossRef CAS PubMed.
- Y. Peng, B. Lu, Y. Deng, N. Yang and G. Li, Biosens. Bioelectron., 2022, 201, 113973 CrossRef CAS PubMed.
- M. Li, J. Liu, X. Wang, J. Wang, L.-H. Huang, M. Gao and X. Zhang, Anal. Chem., 2022, 94, 15076–15084 CrossRef CAS PubMed.
- D.-X. Wang, J. Wang, Y.-X. Wang, J.-Y. Ma, B. Liu, A.-N. Tang and D.-M. Kong, Chem. Sci., 2022, 13, 10395–10405 RSC.
- N.-N. Lu, M. Xie, J. Wang, S.-W. Lv, J.-S. Yi, W.-G. Dong and W.-H. Huang, ACS Appl. Mater. Interfaces, 2015, 7, 8817–8826 CrossRef CAS PubMed.
- L. Bai, Y. Du, J. Peng, Y. Liu, Y. Wang, Y. Yang and C. Wang, J. Mater. Chem. B, 2014, 2, 4080–4088 RSC.
- J. Peng, Q. Zhao, W. Zheng, W. Li, P. Li, L. Zhu, X. Liu, B. Shao, H. Li and C. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 18423–18428 CrossRef CAS PubMed.
- F. Jia, Y. Wang, Z. Fang, J. Dong, F. Shi, W. Zhang, Z. Wang and Z. Hu, Anal. Chem., 2021, 93, 5670–5675 CrossRef CAS PubMed.
- R. Han, Y. Li, M. Chen, W. Li, C. Ding and X. Luo, Anal. Chem., 2022, 94, 2204–2211 CrossRef CAS PubMed.
- T. Li, N. Li, Y. Ma, Y.-J. Bai, C.-M. Xing and Y.-K. Gong, J. Mater. Chem. B, 2019, 7, 6087–6098 RSC.
- J. Bu, W. Jeong, R. Jafari, L. J. Kubiatowicz, A. Nair, M. J. Poellmann, R. S. Hong, E. W. Liu, R. H. Owen and P. A. Rawding, Biosens. Bioelectron., 2022, 114445 CrossRef CAS PubMed.
- L. Du, W. Chen, J. Wang, W. Cai, S. Kong and C. Wu, Sens. Actuators, B, 2019, 301, 127073 CrossRef CAS.
- Y. Hu, D. Chen, J. V. Napoleon, M. Srinivasarao, S. Singhal, C. A. Savran and P. S. Low, Sci. Rep., 2022, 12, 1–10 CrossRef PubMed.
- T. Zhang, W. Peng, W. Jiang, K. Gao and W. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 24543–24552 CrossRef CAS PubMed.
- P. Chen, Y. He, T. Liu, F. Li, K. Huang, D. Tang, P. Jiang, S. Wang, J. Zhou and J. Huang, Biosens. Bioelectron., 2022, 202, 114009 CrossRef CAS PubMed.
- Y. Xu, J. Lin, X. Wu, X. Xu, D. Zhang, Y. Xie, T. Pan, Y. He, A. Wu and G. Shao, J. Mater. Chem. B, 2022, 10, 3808–3816 RSC.
- S. Sun, R. Wang, Y. Huang, J. Xu, K. Yao, W. Liu, Y. Cao and K. Qian, Small, 2019, 15, 1902441 CrossRef PubMed.
- Y. Zhang, W. Wang, H. Guo, M. Liu, H. Zhu and H. Sun, Nanotechnology, 2021, 32, 475102 CrossRef CAS PubMed.
- X. Li, T. Cui, W. Zhang, Z. Zhai, F. Wu, Y. Zhang, M. Yang, W. Zhong and W. Yue, Colloids Surf., B, 2020, 196, 111281 CrossRef CAS PubMed.
- D. Yin, A. Shi, B. Zhou, M. Wang, G. Xu, M. Shen, X. Zhu and X. Shi, Langmuir, 2022, 38, 11080–11086 CrossRef CAS PubMed.
- M. Schmidt, A. Franken, D. Wilms, T. Fehm, H. J. Neubauer and S. Schmidt, ACS Appl. Bio Mater., 2021, 4, 6371–6380 CrossRef CAS PubMed.
- L. Yang, H. Sun, W. Jiang, T. Xu, B. Song, R. Peng, L. Han and L. Jia, Chem. Mater., 2018, 30, 4372–4382 CrossRef CAS.
- P. Ding, Z. Wang, Z. Wu, M. Hu, W. Zhu, N. Sun and R. Pei, ACS Appl. Mater. Interfaces, 2021, 13, 3694–3700 CrossRef CAS PubMed.
- Y. Zhou, X. Wang, Z. Luo, X. Liu, J. Hou and S. Zhou, Small Sci., 2022, 2, 2200061 CrossRef CAS.
- F. Zhang, Y. Jiang, X. Liu, J. Meng, P. Zhang, H. Liu, G. Yang, G. Li, L. Jiang and L.-J. Wan, Nano Lett., 2016, 16, 766–772 CrossRef CAS PubMed.
- H. Xu, B. Dong, S. Xu, S. Xu, X. Sun, J. Sun, Y. Yang, L. Xu, X. Bai and S. Zhang, Biomaterials, 2017, 138, 69–79 CrossRef CAS PubMed.
- S. Li, K. Wang, S. Hao, F. Dang, Z.-Q. Zhang and J. Zhang, Anal. Chem., 2022, 94, 6754–6759 CrossRef CAS PubMed.
- J. Li, Y. Yuan, H. Gan, C. Dong, B. Cao, J. Ni, X. Li, W. Gu, C. Song and L. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 32869–32879 CrossRef CAS PubMed.
- V. Shirmohammadli, N. Manavizadeh and Z. G. Bafghi, IEEE Sens. J., 2019, 20, 591–598 Search PubMed.
- H. Su, S. Yin, J. Yang, Y. Wu, C. Shi, H. Sun and G. Wang, Electrochim. Acta, 2021, 393, 139093 CrossRef CAS.
- J. Xu, C. Zhao, K. Niu, Z. Gao and Y.-Y. Song, Anal. Chim. Acta, 2021, 1142, 1–9 CrossRef CAS PubMed.
- W. Li, R. Li, B. Huang, Z. Wang, Y. Sun, X. Wei, C. Heng, W. Liu, M. Yu and S.-S. Guo, Nanotechnology, 2019, 30, 335101 CrossRef CAS PubMed.
- J. Fan, Z. Dang, T. Lu, J. Li, T. Chen, Y. Yang and X. Li, J. Mater. Sci., 2021, 56, 16634–16647 CrossRef CAS.
- N. Sun, M. Liu, J. Wang, Z. Wang, X. Li, B. Jiang and R. Pei, Small, 2016, 12, 5090–5097 CrossRef CAS PubMed.
- C. Chen, Z. Wu, P. Ding, N. Sun, H. Liu, Y. Chen, Z. Wang and R. Pei, Adv. Fiber Mater., 2020, 2, 186–193 CrossRef CAS.
- Z. Wu, Y. Pan, Z. Wang, P. Ding, T. Gao, Q. Li, M. Hu, W. Zhu and R. Pei, J. Mater. Chem. B, 2021, 9, 2212–2220 RSC.
- Y. Zhao, J. Xiong, X. Shi and F. Ko, Colloids Surf., A, 2019, 583, 123978 CrossRef CAS.
- Y. Zhu, C. Zou, J. Zhang, W. Jiang, F. Guan, K. Tang, S. Li, G. Li, J. Wang and Z. Ke, ACS Appl. Mater. Interfaces, 2020, 12, 5671–5679 CrossRef CAS PubMed.
- H. Zhong, C. Yuan, J. He, Y. Yu, Y. Jin, Y. Huang and R. Zhao, Anal. Chem., 2021, 93, 9778–9787 CrossRef CAS PubMed.
- M. Wang, Y. Tan, D. Li, G. Xu, D. Yin, Y. Xiao, T. Xu, X. Chen, X. Zhu and X. Shi, Adv. Fiber Mater., 2021, 3, 192–202 CrossRef CAS.
- R. Khan, A. Radoi, S. Rashid, A. Hayat, A. Vasilescu and S. Andreescu, Sensors, 2021, 21, 3369 CrossRef CAS PubMed.
- H. J. Yoon, T. H. Kim, Z. Zhang, E. Azizi, T. M. Pham, C. Paoletti, J. Lin, N. Ramnath, M. S. Wicha and D. F. Hayes, Nat. Nanotechnol., 2013, 8, 735–741 CrossRef CAS PubMed.
- B. Dou, L. Xu, B. Jiang, R. Yuan and Y. Xiang, Anal. Chem., 2019, 91, 10792–10799 CrossRef CAS PubMed.
- X. Zhou, Y. Zhang, K. Kang, N. Zhu, J. Cheng, Q. Yi and Y. Wu, J. Mater. Chem. B, 2022, 10, 3119–3125 RSC.
- Q. You, J. Peng, Z. Chang, M. Ge, Q. Mei and W.-F. Dong, Talanta, 2021, 235, 122770 CrossRef CAS PubMed.
- C. Shen, L. Zhong, L. Xiong, C. Liu, L. Yu, X. Chu, X. Luo, M. Zhao and B. Liu, Sens. Actuators, B, 2021, 331, 129399 CrossRef CAS.
- S. Wang, J. Cui, Q. Fan, J. Gan, C. Liu, Y. Wang, T. Yang, J. Wang and C. Yang, Anal. Chem., 2022, 94, 9450–9458 CrossRef CAS PubMed.
- W. Wang, G. Yang, H. Cui, J. Meng, S. Wang and L. Jiang, Adv. Healthcare Mater., 2017, 6, 1700003 CrossRef PubMed.
- Y. Chen, D. Tyagi, M. Lyu, A. J. Carrier, C. Nganou, B. Youden, W. Wang, S. Cui, M. Servos and K. Oakes, Anal. Chem., 2019, 91, 4017–4022 CrossRef CAS PubMed.
- L. Jia, X. Zhen, L. Chen, Q. Feng, W. Yuan, Y. Bu, S. Wang and X. Xie, J. Colloid Interface Sci., 2023, 631, 55–65 CrossRef CAS PubMed.
- X. Zhang, X. Wei, X. Men, C.-X. Wu, J.-J. Bai, W.-T. Li, T. Yang, M.-L. Chen and J.-H. Wang, ACS Appl. Mater. Interfaces, 2021, 13, 43668–43675 CrossRef CAS PubMed.
- X. Dou, P. Li, S. Jiang, H. Bayat and H. Schönherr, ACS Appl. Mater. Interfaces, 2017, 9, 8508–8518 CrossRef CAS PubMed.
- L. Wang, S. Huang, Q.-Y. Li, L.-Y. Ma, C. Zhang, F. Liu, M. Jiang, X. Yu and L. Xu, Chem. Eng. J., 2022, 435, 134762 CrossRef CAS.
- M. G. Ahmed, M. F. Abate, Y. Song, Z. Zhu, F. Yan, Y. Xu, X. Wang, Q. Li and C. Yang, Angew. Chem., Int. Ed., 2017, 56, 10681–10685 CrossRef CAS PubMed.
- H. Shen, R. Su, J. Peng, L. Zhu, K. Deng, Q. Niu, Y. Song, L. Yang, L. Wu and Z. Zhu, Bioact. Mater., 2022, 11, 32–40 CrossRef CAS PubMed.
- M. Rahmanian, O. S. Hematabad, E. Askari, F. Shokati, A. Bakhshi, S. Moghadam, A. Olfatbakhsh, E. A. S. Hashemi, M. K. Ahmadi and S. M. Naghib, J. Adv. Res., 2022, 08, 005 Search PubMed.
- B. J. Green, M. Marazzini, B. Hershey, A. Fardin, Q. Li, Z. Wang, G. Giangreco, F. Pisati, S. Marchesi and A. Disanza, Small, 2022, 18, 2106097 CrossRef CAS PubMed.
- A. Abdulla, Z. Zhang, K. Z. Ahmad, A. R. Warden, H. Li and X. Ding, Biosens. Bioelectron., 2022, 201, 113965 CrossRef CAS PubMed.
- X. Chen, H. Ding, D. Zhang, K. Zhao, J. Gao, B. Lin, C. Huang, Y. Song, G. Zhao and Y. Ma, Adv. Sci., 2021, 8, 2102070 CrossRef CAS PubMed.
- E. A. Kwizera, W. Ou, S. Lee, S. Stewart, J. G. Shamul, J. Xu, N. Tait, K. H. Tkaczuk and X. He, ACS Nano, 2022, 16, 11374–11391 CrossRef CAS PubMed.
- K. Pang, B. Gu, F. Liu, M. Dong, L. Zhu and X. Wei, J. Innovative Opt. Health Sci., 2019, 12, 1930008 CrossRef CAS.
- J. Novak, I. Georgakoudi, X. Wei, A. Prossin and C. P. Lin, Opt. Lett., 2004, 29, 77–79 CrossRef CAS PubMed.
- H. Lee, C. Alt, C. M. Pitsillides, M. Puoris'haag and C. P. Lin, Opt. Express, 2006, 14, 7789–7800 CrossRef PubMed.
- W. He, H. Wang, L. C. Hartmann, J.-X. Cheng and P. S. Low, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11760–11765 CrossRef CAS PubMed.
- H. Seo, Y. Hwang, K. Choe and P. Kim, Biomed. Opt. Express, 2015, 6, 2158–2167 CrossRef PubMed.
- R. A. Patil, M. Srinivasarao, M. M. Amiji, P. S. Low and M. Niedre, Mol. Imaging Biol., 2020, 22, 1280–1289 CrossRef CAS PubMed.
- N. Ding, Monitoring circulating tumor cells in prostate cancer model after tumor resection by in vivo flow cytometry, MPhil, Shanghai Jiao Tong University, 2019 Search PubMed.
- Y. Zhou, Developing in vivo photoacoustic flow cytometry technique for detection of circulating melanoma cells, MPhil, Shanghai Jiao Tong University, 2019 Search PubMed.
- C. Li and L. V. Wang, Phys. Med. Biol., 2009, 54, R59 CrossRef CAS PubMed.
- A. Karabutov, N. B. Podymova and V. S. Letokhov, Appl. Phys. B: Lasers Opt., 1996, 63, 545–563 CrossRef CAS.
- P. Yang, D. Wei, K. Pang, Q.-Y. Wang, Q.-Y. Zhou and X.-B. Wei, Laser Optoelectron. Prog., 2017, 54, 090001 CrossRef.
- J. Weber, P. C. Beard and S. E. Bohndiek, Nat. Methods, 2016, 13, 639–650 CrossRef CAS PubMed.
- C. Jia, S.-W. Huang, Y. Jin, C. H. Seo, L. Huang, J. F. Eary, X. Gao and M. O'Donnell, in Photons Plus Ultrasound: Imaging and Sensing 2010, SPIE, 2010, vol. 7564, pp. 275–280 Search PubMed.
- Y. Jin, C. Jia, S.-W. Huang, M. O'donnell and X. Gao, Nat. Commun., 2010, 1, 1–8 Search PubMed.
- M. Mehrmohammadi, J. Oh, S. Mallidi and S. Y. Emelianov, Mol. Imaging, 2011, 10, 7290.2010.00037 CrossRef.
- X. Hu, C.-W. Wei, J. Xia, I. Pelivanov, M. O'Donnell and X. Gao, Small, 2013, 9, 2046 CrossRef CAS PubMed.
- O. Vermesh, A. Aalipour, T. J. Ge, Y. Saenz, Y. Guo, I. S. Alam, S. Park, C. N. Adelson, Y. Mitsutake and J. Vilches-Moure, Nat. Biomed. Eng., 2018, 2, 696–705 CrossRef CAS PubMed.
- S.-B. Cheng, M. Wang, C. Zhang, M.-M. Chen, Y.-K. Wang, S. Tian, N. Zhan, W.-G. Dong, M. Xie and W.-H. Huang, Anal. Chem., 2020, 92, 5447–5455 CrossRef CAS PubMed.
- D. Wang, R. Dong, X. Wang and X. Jiang, ACS Nano, 2022, 16, 5274–5283 CrossRef CAS PubMed.
- Z. Yin, R. Shi, X. Xia, L. Li, Y. Yang, S. Li, J. Xu, Y. Xu, X. Cai and S. Wang, Adv. Mater., 2022, 2207870 CrossRef CAS PubMed.
- H. Zhang, Z. Jia, C. Wu, L. Zang, G. Yang, Z. Chen and B. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 20477–20484 CrossRef CAS PubMed.
- J. Li, X. Li, Y. Zhang, K. Jin, Y. Yuan, R. Ming, Y. Yang and T. Chen, Nano Res., 2022, 1–9 Search PubMed.
- D. Wang, C. Ge, W. Liang, Q. Yang, Q. Liu, W. Ma, L. Shi, H. Wu, Y. Zhang and Z. Wu, Adv. Sci., 2020, 7, 2000940 CrossRef CAS PubMed.
- S. Wan, T. H. Kim, K. J. Smith, R. Delaney, G.-S. Park, H. Guo, E. Lin, T. Plegue, N. Kuo and J. Steffes, Sci. Rep., 2019, 9, 1–11 CrossRef PubMed.
- X.-H. Ren, X.-Y. He, C. Xu, D. Han and S.-X. Cheng, Adv. Sci., 2022, 2105806 CrossRef CAS PubMed.
- Y.-H. Cheng, Y.-C. Chen, E. Lin, R. Brien, S. Jung, Y.-T. Chen, W. Lee, Z. Hao, S. Sahoo and H. M. Kang, Nat. Commun., 2019, 10, 1–11 CrossRef CAS.
- K. Kuroda, M. Yashiro, Y. Miki, T. Sera, Y. Yamamoto, A. Sugimoto, S. Nishimura, S. Kushiyama, S. Togano and T. Okuno, Cancer Sci., 2020, 111, 4500–4509 CrossRef CAS PubMed.
- C.-L. Hu, Y.-J. Zhang, X.-F. Zhang, X. Fei, H. Zhang, C.-G. Li and B. Sun, OncoTargets Ther., 2021, 14, 2673 CrossRef PubMed.
- K.-C. Lin, L.-L. Ting, C.-L. Chang, L.-S. Lu, H.-L. Lee, F.-C. Hsu, J.-F. Chiou, P.-Y. Wang, T. Burnouf, D. C.-Y. Ho, K.-C. Yang, C.-Y. Chen, C.-H. Chen, C.-Z. Wu and Y.-J. Chen, Cancers, 2021, 13, 6076 CrossRef CAS PubMed.
- J.-Y. Chen, H.-H. Chou, S. C. Lim, Y.-J. Huang, K.-C. Lai, C.-L. Guo, C.-Y. Tung, C.-T. Su, J. Wang and E. Liu, iScience, 2022, 25, 105081 CrossRef CAS PubMed.
- A. Balakrishnan, A. G. F. Thottian, K. Govind Babu and P. Kumar, Med. Oncol., 2023, 40, 1–6 CAS.
- S. Arora, A. D'Souza, G. Aland, N. Kale, B. Jadhav, T. Kad, P. Chaturvedi, B. Singh and J. Khandare, Lab Chip, 2022, 22, 1519–1530 RSC.
Footnote |
| † Theses authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |




