 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
ATR-FTIR spectroscopy coupled with multivariate analysis techniques for the identification of DENV-3 in different concentrations in blood and serum: a new approach
Marfran C. D. Santosa,
Yasmin M. Nascimentobc,
Josélio M. G. Araújo bc and
Kássio M. G. Lima
bc and
Kássio M. G. Lima *a
*a
aBiological Chemistry and Chemometrics, Institute of Chemistry, Federal University of Rio Grande do Norte, Natal 59072-970, Brazil. E-mail: kassiolima@gmail.com; Tel: +55 84 3342 2323
bLaboratory of Molecular Biology for Infectious Diseases and Cancer, Department of Microbiology and Parasitology, Federal University of Rio Grande do Norte, Natal 59072-970, Brazil
cLaboratory of Virology, Institute of Tropical Medicine, Federal University of Rio Grande do Norte, Natal 59072-970, Brazil
First published on 12th May 2017
Abstract
In most cases of virus infections the viral load is directly related to the intensity of the disease. Nowadays, the main routine diagnoses for dengue fever are only qualitative, they only inform us if the patient has dengue fever or not. However, it is important to be aware of the patient's viral load so that proper care can be taken. In this study we used attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) coupled with multivariate analysis techniques to identify and discriminate dengue serotype 3 (DENV-3) diluted in different concentrations in serum and blood samples with the purpose of developing a simple, fast and non-destructive methodology for a quantitative analysis of the dengue virus. Techniques such as principal component analysis-linear discriminant analysis (PCA-LDA), successive projection algorithm – linear discriminant analysis (SPA-LDA) and genetic algorithm – linear discriminant analysis (GA-LDA) were applied in this classification problem. Forty samples (40 for serum and 40 for blood) were infected with DENV-3 at different concentrations (ten samples for each concentration) and analyzed by IR spectroscopy. The results showed that the models were successful in classifying the virus, the best results being for blood samples. The results of the multivariate classification were tested based on sensitivity, specificity, positive and negative predictive values, Youden's index and positive and negative likelihood ratios, suggesting that ATR-FTIR spectroscopy coupled with multivariate analysis algorithms is an effective tool in quantifying the dengue virus in providing rapid results, in addition to being non-destructive to the sample.
Introduction
Dengue fever is the most transmitted viral disease in the world1 and dengue viruses (DENV) 1 to 4 are members of the genus Flavivirus of the family Flaviviridae. Infection by a serotype leaves the individual immune to subsequent infections by the same serotype throughout their life and immune to another serotype for only a few months.2,3 Dengue is an infection caused by a virus transmitted by arthropods. It is estimated that currently more than 50 million infections occur annually, of which 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lead to hospitalizations for dengue hemorrhagic fever (DHF), with a mortality rate of more than 5% in some regions.4
000 lead to hospitalizations for dengue hemorrhagic fever (DHF), with a mortality rate of more than 5% in some regions.4
Clinical complications caused by dengue virus infection may range from mild asymptomatic infection to dengue fever (DF) or more severe manifestations such as dengue hemorrhagic fever and dengue shock syndrome (DSS).5–7 In most cases, patients do not feel the symptoms or feel an undifferentiated fever with or without redness showing on the body. In the case of DF symptoms, they are usually a high fever, headache, muscle pain (myalgia), joint pain (arthralgia), pain behind the eyes and redness on the body. Patients with DHF and DSS present high fever, bleeding, thrombocytopenia and hemoconcentration as the main symptoms, and may develop other complications such as pleural effusion and gastrointestinal or gingival bleeding.7,8
Viral isolation, RNA detection, antigen detection and serological methods for detecting IgM and IgG are among the most widely used methods in laboratory diagnosis and in virological studies,9,10 but both of these methods have some limitations such as sample handling, requiring samples in the acute phase, and are time-consuming to achieve results, among others. Analyzing diagnostic methods, the most widely used methods in hospitals and diagnostic clinics are serological methods. These methods are qualitative, that is, they only inform if the patient is or is not infected. However, knowledge of the patient's viral load is of great importance, since in most cases the viral load is directly related to the intensity and stage of the disease. The more viruses circulating, the more severe the disease is normally, and the greater the care taken. Finally, identification of the viral load, for some viruses (HIV mainly), can be used to evaluate if the treatment is being effective (if the viral load decreases, it is assumed that the treatment is working). In the case of dengue, rapid determination of viral load can be used as a parameter to decide whether or not treatment is needed. With this in mind, a quantitative test that is inexpensive, quick and requires no sample handling is necessary so that clinical treatment can be started as quickly as possible.
Spectroscopy studies the behavior of samples against their interaction with radiation. Spectroscopic techniques are known to provide fast and reliable results and its use in biological studies has been defined by biospectroscopy.11 Several studies have been carried out using spectroscopy for virology purposes, such as detection and quantification of the poliovirus using FTIR spectroscopy,12 detection of hepatitis C virus infection using NMR spectroscopy,13 and diagnosis of HIV-1 infection using near-infrared spectroscopy,14 among others, showing promising results regarding the technique's ability to identify the presence of the virus.
The mid-infrared region comprises the 400 to 4000 cm−1 of the electromagnetic spectrum. This radiation is absorbed by molecules present in biological samples. The 900 to 1800 cm−1 range is known as the biomolecule fingerprint region, because spectral bands present referral tolipids (∼1750 cm−1), carbohydrates (∼1155 cm−1), proteins (amide I, ∼1650 cm−1, amide II, ∼1550 cm−1, amide III, ∼1260 cm−1) and DNA/RNA (∼1225 cm−1, 1080 cm−1). 15,16 In general, attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR) can be used for collecting spectra in this range.15
When interrogating different biological samples with FTIR, the generated spectra have a lot of information which can make interpreting difficult; therefore, it is convenient to use algorithms that aid and facilitate spectral interpretation. In this study we used principal component analysis (PCA – it reduces data dimensionality, making use of the components that explain data variability),16 successive projection algorithm and genetic algorithm (SPA and GA – they reduce the size of the data, selecting the variables that discriminate classes)17,18 and linear discriminant analysis (LDA – it provides a maximum separation of classes through the ratio of the variance between classes and within the classes) as chemometric algorithms of multivariate classification for discriminating classes via spectral information.
Classification by different concentrations of the dengue virus through ATR-FTIR spectra using the PCA-LDA, SPA-LDA and GA-LDA algorithms has never been studied. This study aims to quantitatively discriminate DENV-3 samples in serum and blood. The results were encouraging and show the potential of the technique to identify and quantify DENV-3, and it may be used in the future as ancillary tools for clinical diagnostics.
Material and methods
Subculture of cells
VERO E6 cells (African green monkey kidney cells) biological sample has been used by biomedical laboratories worldwide. It is very common for collaborating research groups to share this cell line. In this study, the VERO E6 cells was kindly provided by the Biotechnology Laboratory of Natural Polymer-Biopol, Department of Biochemistry, Federal University of Rio Grande do Norte (UFRN). VERO E6 cells (African green monkey kidney cells) cultivated at 37 °C in 5% CO2 were used in the study. Leibovitz's 15 (L-15) medium of the culture bottle (25 cm3) was discarded and subsequently 5 mL of PBS was poured in to remove cell debris (2 times, then discarding PBS). 1000 μL of trypsin was added and then incubated for 5 minutes at 37 °C. Then it was observed in a microscope to check if total monolayer displacement occurred. Upon observing total displacement, 10 mL of 10% L-15 medium was added to the bottle along with homogenates and 5 mL was transferred to a sterile bottle.Replication of dengue virus
Aliquots from cultivated DENV-3 were kindly provided by the staff from the Laboratory of Molecular Biology for Infectious Diseases and Cancer (LADIC/UFRN). Isolated DENV-3 was inoculated into VERO E6 cells with the purpose of promoting viral replication. Next, 100 μL viral sample was used in a 25 cm3 bottle of cells. It was incubated for 1 hour at 37 °C, being homogenized every 15 minutes. Then medium was added with 2% fetal bovine serum (FBS) in order to provide nutrients for the cell. Finally, it was incubated again at 37 °C where it remained for 7 days, and was observed daily. Confirmation of viral infection was performed by nested Reverse Transcriptase PCR assay (RT-PCR). After the viral infection was confirmed, the supernatant was transferred to a falcon and centrifuged for 5 minutes at 1000 turns per minute. Lastly, 20% fetal bovine serum was added to the virus isolate and stored at −70 °C. The titer of DENV-3 was determined by plaque assay at 1 × 104 PFU mL−1.RNA extraction and nested Reverse Transcriptase PCR assay (RT-PCR)
Viral RNA for the nested RT-PCR assay was extracted by using QIAamp® Viral RNA Mini kit, Qiagen (Catalog number 52906), in accordance with the manufacturer. The nested RT-PCR protocol for DENV detection and typing was performed as previously described by Lanciotti et al. (1992).Sample preparation
For IR measurement, healthy human serum and blood samples were infected with DENV-3 in conical-bottomed microtubes at four dilutions (1 × 103, 1 × 102, 1 and 0.1 PFU mL−1 for serum, and 1 × 103, 1 × 102, 10 and 0.1 PFU mL−1 for blood). All experiments were performed in compliance with the guideline “Biomedical research ethics review method involving people” (Brazil), and approved by the medical ethics committee at Federal University of Rio Grande do Norte/Brazil (protocol number # 51057015.5.0000.5537), informed consents were obtained from human participants of this study. The data set consisted of 40 samples (40 for serum and 40 for blood) divided into ten for each dilution. Another 10 non-infected serum and blood samples were used as a control.ATR-FTIR spectroscopy
Spectra acquisition was performed within 1 hour of sample contamination. ATR-FTIR spectra were performed using a BRUKER model FTIR VERTEX 70 equipped with ATR accessory. The spectra were obtained in the range of 400 to 4000 cm−1, with a resolution of 4 cm−1, with 16 scans and a measurement time of 13 seconds per spectrum. The ambient temperature was 22 °C. Air was used as background in obtaining the spectra of samples contaminated with DENV-3. Soon after each acquisition of the background, 2 μL of each sample was added to the ATR crystal, making sure that no air bubbles were trapped. At each acquisition, a small piece of aluminum foil was placed on the sample, following the studies of Cui et al. (2016) where it has been observed that this is an appropriate substrate for ATR-FTIR analyzes of various biological specimens because it does not interfere with the spectra, does not sacrifice important biochemical fingerprint information of the sample and is inexpensive.19 After the acquisitions, the crystal was then cleaned with alcohol (70% v/v) and dried using paper towel. In order to make sure the ATR crystal was clean, a spectrum was collected using the latest background collected for reference. BRUKER OPUS software was used to convert the spectra to absorbance.Chemometric methods and software
Principal Component Analysis (PCA) is a multivariate statistical method that seeks to determine a smaller set of variables from the linear combination of the original variables which summarize the original system, thus reducing the dimensionality and complexity of the data. A reduction of variables is performed in which the original spectral matrix X is decomposed as the product of the matrices scores and loadings,20 as shown in eqn (1):| X = TPT + E | (1) |
Successive Projection Algorithm (SPA) is a variable selection method (in this case, the variables are the wavenumbers) that uses simple operations in a vector space to minimize variable collinearity. SPA is a direct selection method, meaning it starts with 1 variable and then incorporates another variable in each interaction until it reaches a number of more discriminant N variables.20
Genetic algorithm (GA) was another technique used for variable selection. This algorithm makes use of techniques based on biological genetics and evolution. A population is created with n subsets, where each subset is composed by a random combination of variables (wavenumbers). Each subset is formed by m (the total number of variables that can be chosen), 1's (variables selected by the model) and 0's (unselected variables). Therefore, in genetic terms, each variable represents a gene, and a set of variables represents a chromosome. For example, for a selection problem with 10 variables, a chromosome could be 1001010110, where variables 1, 4, 6, 8 and 9 would be the variables selected for the model, and variables 2, 3, 5, 7 and 10 would be the variables to be optimized.21 In this study for the GA routine, the number of individuals (population) for each generation was 24, with the number of generations equal to 12. The genetic operator mutation and crossover were held constant at 10 and 60%, respectively. GA was repeated three times and the best result was used. In this study, in order to select the best optimal number of variables for SPA and GA we used a cost function calculated in the validation set,22 as shown in eqn (2):
 | (2) |
 | (3) |
Linear Discriminant Analysis (LDA) was another technique employed. It is a supervised technique that is based on the discriminant process developed by Fisher in 1936. LDA maximizes the ratio between the variance between classes and intraclass variation in any particular data set, thus ensuring maximum separability. LDA is efficient when combined with dimensionality reduction techniques (such as PCA, SPA and GA).
The results of the multivariate classification for PCA-LDA, SPA-LDA and GA-LDA were tested based on:sensitivity (confidence in obtaining a positive result for a truly positive sample):
 | (4) |
 | (5) |
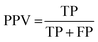 | (6) |
 | (7) |
| YOU = sens − (1 − spec) | (8) |
 | (9) |
 | (10) |
MATLAB R2012b software (Math-works, Natick, USA) was used for data import, pre-treatment, and construction of the chemometric classification models. The raw spectra were pre-processed with cuts between 900 and 1800 cm−1 (235 wavenumbers at 4 cm−1 spectral resolution), baseline-corrected and Savitzky–Golay smoothing (window 15 points). The samples were divided into three sets for the PCA-LDA, SPA-LDA and GA-LDA models (60% for training, 20% for validation and 20% for prediction) using the classic Kennard–Stone uniform sampling algorithm.23 The KS algorithm was applied separately for each class to maximize the Euclidean minimum distances between selected and unselected samples.
Results and discussion
In Fig. 1a and b we can see the raw spectra of infected serum (1 × 103, 1 × 102, 1 and 0.1 PFU mL−1) and infected blood samples (1 × 103, 1 × 102, 10 and 0.1 PFU mL−1), respectively. These spectra were subjected to pre-processing (cut between 900 and 1800 cm−1, baseline correction and smoothing) and the results can be seen in Fig. 2a and b. As can be seen, the spectra are very similar, being impossible the visual differentiation between them.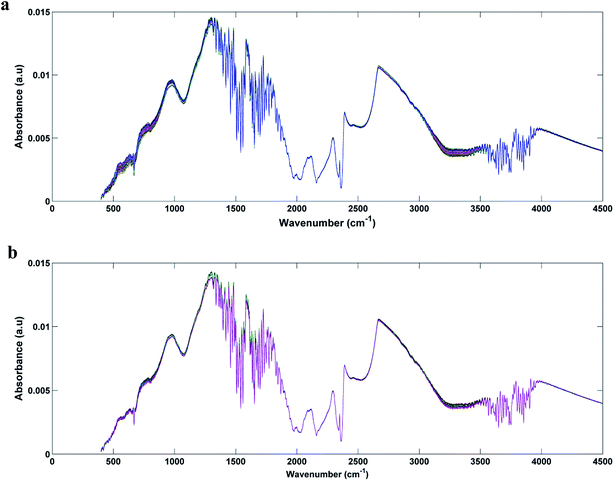 | ||
| Fig. 1 Raw spectra for each original class: (a) DENV-3 in serum (1 × 103, 1 × 102, 1 and 0, 1 PFU mL−1); (b) DENV-3 in blood (1 × 103, 1 × 102, 10 and 0, 1 PFU mL−1). | ||
Due to the great similarity between the spectra of the different classes, it is necessary to use algorithms (PCA-LDA, SPA-LDA and GA-LDA, in this case) that mathematically find spectral information capable of discriminating one class from another. The models were applied to classify the serum and blood samples by the concentration of DENV-3 present, and finally their performances were compared. The results are discussed below.
PCA-LDA
Fig. 3a and b show the plot of the Fisher scores in a two-dimensional space resulting from principal component analysis-linear discriminant analysis (PCA-LDA) for the infected serum and blood samples, respectively. Each point represents a spectrum of a sample. As can be seen, Fisher scores do not show good segregation between classes. For this study the PCA-LD was performed using 4 principal components (4 PC's, 94% of variance explained for serum samples and 95% for blood samples). | ||
Fig. 3 DF1 × DF2 discriminant function values calculated with 4 PC's for: (a) DENV-3 in serum ( 1 × 103, 1 × 103,  1 × 102, 1 × 102,  1 and 1 and  0,1 PFU mL−1); (b) DENV-3 in blood ( 0,1 PFU mL−1); (b) DENV-3 in blood ( 1 × 103, 1 × 103,  1 × 102, 1 × 102,  10 and 10 and  0.1 PFU mL−1). 0.1 PFU mL−1). | ||
SPA-LDA and GA-LDA
Then, SPA-LDA and GA-LDA were applied to select an optimal number of variables through the minimum cost of the G function. Fisher scores were obtained for both models, with a better segregation of the classes in relation to the PCA-LDA models.In the case of infected serum samples the SPA-LDA selected 20 wave numbers, while for the case of infected blood samples the SPA-LDA selected 19 wave numbers (Table 1). The selected wave numbers are like biological markers, selected by the models as the variables that most discriminate one class from another. Fig. 4a and b show the graphs of the selected variables by SPA-LDA for serum and blood DENV-3 samples, respectively. Using these selected variables, the Fisher scores were calculated (shown in Fig. 5). As can be seen, Fisher scores for SPA-LDA show a better segregation of classes than those calculated for PCA-LDA, being visually better separated for dengue-3 samples in blood than in serum.
| Chemometric analysis | Wavenumber selected (cm−1) |
|---|---|
| SPA-LDA for DENV-3 in serum | 908, 943, 989, 1041, 1080, 1124, 1144, 1194, 1302, 1315, 1360, 1441, 1477, 1500, 1541, 1578, 1632, 1695, 1724, 1801 |
| SPA-LDA for DENV-3 in blood | 918, 989, 1049, 1076, 1105, 1151, 1227, 1304, 1317, 1356, 1414, 1489, 1524, 1564, 1618, 1653, 1682, 1718, 1801 |
 | ||
| Fig. 4 Graph of the variables selected by SPA-LDA, marked in the average spectrum of: (a) DENV-3 in serum and (b) DENV-3 in blood. | ||
In analyzing the wavenumbers selected by SPA-LDA for the contaminated serum samples, we observed that the main biological changes of interest that discriminate the different concentrations are related to amide II of proteins (≈1500 cm−1) and RNA (≈1080 cm−1) vibrations. When examining the wavenumbers selected by SPA-LDA for infected blood samples, the main changes are related to carbohydrate (≈1151 cm−1), protein structures (amide I, ≈1653 cm−1, amide III, ≈1317 cm−1), and RNA (≈1227 cm−1, ≈1076 cm−1) vibrations.
In the case of the GA-LDA, 17 wavenumbers were selected for the infected serum samples and 11 wavenumbers for the infected blood samples (Table 2). The graph of the selected variables can be seen in Fig. 6. As can be seen in Fig. 7, Fisher scores showed good visual segregation between classes, mainly in the case of infected blood samples (as in the case of SPA-LDA).
| Chemometric analysis | Wavenumber selected (cm−1) |
|---|---|
| GA-LDA for DENV-3 in serum | 905, 926, 949, 984, 1047, 1055, 1176, 1196, 1277, 1311, 1321, 1396, 1448, 1554, 1603, 1753, 1761 |
| GA-LDA for DENV-3 in blood | 986, 1053, 1065, 1225, 1337, 1352, 1529, 1541, 1666, 1686, 1689 |
 | ||
| Fig. 6 Graph of the variables selected by GA-LDA, marked in the average spectrum of: (a) DENV-3 in serum and (b) DENV-3 in blood. | ||
Among the 17 wavenumbers selected by GA-LDA that best discriminate infected serum samples, we can highlight changes related to protein structures (amide III, ≈1311 cm−1, amide II, ≈1554 cm−1), COO− symmetric stretch in fatty acids (≈1396 cm−1) and lipid (≈1753 cm−1) vibrations. Among the 11 wavenumbers selected by GA-LDA in the case of infected blood samples, those that appear to be of major biological interest are related to RNA (≈1238 cm−1), protein structures (amide III, ≈1329 cm−1, amide I, ≈1661 cm−1) and lipid (≈1743 cm−1) vibrations. The changes associated with RNA make sense, since even within a single serotype there are minimal differences found between the RNA of one viral particle and another.
The good segregations shown for SPA-LDA and for GA-LDA give us an indication that these chemometric models will provide good results for figures of merit [sensitivity, specificity, PPV, NPV, YOU, LR(+) and LR(−)].
The performances of the method were evaluated through validation measures. Tables 3 and 4 presents the performance measures for PCA-LDA, SPA-LDA and GA-LDA for serum and blood samples with different concentrations of DENV-3, respectively. As can be seen, the models made some errors for the serum samples, however they obtained an excellent classification performance for the blood samples (in this case, PCA-LDA, SPA-LDA and GA-LDA obtained 100% sensitivity and specificity), demonstrating that ATR-FTIR spectroscopy together with classification techniques has the potential to quantitatively discriminate the dengue virus (in this case DENV-3) in serum and blood, and may, in the near future, assist in the more detailed diagnosis and correct treatment of patients.
| DENV-3 in serum | |||
|---|---|---|---|
| Stage performance features | PCA-LDA | SPA-LDA | GA-LDA |
| 1 × 103 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 50.0 |
| Specificity (%) | 100.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 100.0 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 66.6 |
| Youden index (YOU) | 100.0 | 100.0 | 50.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 1 × 102 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 100.0 |
| Specificity (%) | 100.0 | 100.0 | 50.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 66.6 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 100.0 |
| Youden index (YOU) | 100.0 | 100.0 | 50.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 2.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 1 PFU mL−1 | |||
| Sensitivity (%) | 0.0 | 50.0 | 50.0 |
| Specificity (%) | 0.0 | 50.0 | 100.0 |
| Positive predictive values (PPV) | 0.0 | 50.0 | 100.0 |
| Negative predictive values (NPV) | 0.05 | 0.0 | 66.6 |
| Youden index (YOU) | 100.0 | 0.0 | 50.0 |
| Positive likelihood ratios (LR+) | 0.0 | 1.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 1.0 | 0.5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 0.1 PFU mL−1 | |||
| Sensitivity (%) | 50.0 | 100.0 | 0.0 |
| Specificity (%) | 50.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 50.0 | 100.0 | 0.0 |
| Negative predictive values (NPV) | 50.0 | 100.0 | 50.0 |
| Youden index (YOU) | 0.0 | 100.0 | 0.0 |
| Positive likelihood ratios (LR+) | 1.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 1.0 | 0.0 | 1.0 |
| DENV-3 in blood | |||
|---|---|---|---|
| Stage performance features | PCA-LDA | SPA-LDA | GA-LDA |
| 1 × 103 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 100.0 |
| Specificity (%) | 100.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 100.0 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 100.0 |
| Youden index (YOU) | 100.0 | 100.0 | 100.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 1 × 102 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 100.0 |
| Specificity (%) | 100.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 100.0 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 100.0 |
| Youden index (YOU) | 100.0 | 100.0 | 100.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 10 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 100.0 |
| Specificity (%) | 100.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 100.0 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 100.0 |
| Youden index (YOU) | 100.0 | 100.0 | 100.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 0.1 PFU mL−1 | |||
| Sensitivity (%) | 100.0 | 100.0 | 100.0 |
| Specificity (%) | 100.0 | 100.0 | 100.0 |
| Positive predictive values (PPV) | 100.0 | 100.0 | 100.0 |
| Negative predictive values (NPV) | 100.0 | 100.0 | 100.0 |
| Youden index (YOU) | 100.0 | 100.0 | 100.0 |
| Positive likelihood ratios (LR+) | 0.0 | 0.0 | 0.0 |
| Negative likelihood ratios (LR−) | 0.0 | 0.0 | 0.0 |
Conclusion
The spectra acquisition of each infectious agent can be identified to avoid cross-reactions in cases of co-infections or co-morbidities. Serum and blood-based vibrational spectroscopy has been applied in studies with various diseases, but it has never been used in the quantitative classification of dengue virus (determination of dengue viral load). The importance of this determination is related to the fact that the viral load is associated with disease time, and, in most cases, is directly related to the severity of the disease. A patient with a high viral load needs a more urgent treatment. Thus, a rapid and sensitive technique for quantitative determination of the virus may assist in the correct treatment of patients or inform those patients who do not need treatment. The results of this study suggest that IR spectroscopy with multivariate classification techniques (PCA-LDA, SPA-LDA and GA-LDA, in this case) can be applied in this clinical perspective. Principal component-based dimensional reduction techniques (PCA) and variable selection (SPA and GA) combined with LDA were performed in an attempt to extract information from the ATR-FTIR spectra that best differentiate the DENV-3 concentrations. In comparing the DENV-3 samples in serum and DENV-3 in blood, the implemented classification models obtained better results for blood samples, with 100% sensitivity and specificity values for PCA-LDA, SPA-LDA and GA-LDA. Although the results have been encouraging, it is necessary to carry out studies with a larger number of samples for greater reliability. However, this study shows that further research is worth exploring in this perspective and suggests that in the near future we will be able to rely on portable spectroscopic instruments that emit specific radiation and simultaneous multivariate analyzes in the clinical and virological study environments.Acknowledgements
Marfran C. D. Santos would like to thank CAPES for its financial support and the PPGQ-UFRN for its scientific support. This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (grant No. 305962/2014-0 and 400328/2014-3).References
- Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition, World Health Organization, Geneva, 2009 Search PubMed.
- D. J. Gubler, Comp. Immunol. Microbiol. Infect. Dis., 2004, 27, 319–330 CrossRef CAS PubMed.
- I. Kurane, Comp. Immunol. Microbiol. Infect. Dis., 2007, 30, 329–340 CrossRef PubMed.
- M. G. Guzman, S. B. Halstead, H. Artsob, P. Buchy, J. Farrar, D. J. Gubler, E. Hunsperger, A. Kroeger, H. S. Margolis, E. Martínez, M. B. Nathan, J. L. Pelegrino, C. Simmons, S. Yoksan and R. W. Peeling, Nat. Rev. Microbiol., 2010, 8, S7–S16 CrossRef CAS PubMed.
- S. B. Halstead and S. N. Cohen, Microbiol. Mol. Biol. Rev., 2015, 79, 281–291 CrossRef PubMed.
- S. R. Sudulagunta, M. B. Sodalagunta, M. Sepehrar, S. K. B. Raja, A. S. Nataraju, M. Kumbhat, D. Sathyanarayana, S. Gummadi and H. Kumar, Oxford Med. Case Reports, 2016, vol. 11, pp. 269–272 Search PubMed.
- M. G. Guzman, M. Alvarez and S. B. Halstead, Arch. Virol., 2013, 158, 1445–1459 CrossRef CAS PubMed.
- R. W. Peeling, H. Artsob, J. L. Pelegrino, P. Buchy, M. J. Cardosa, S. Devi, D. A. Enria, J. Farrar, D. J. Gubler, M. G. Guzman, S. B. Halstead, E. Hunsperger, S. Kliks, H. S. Margolis, C. M. Nathanson, V. C. Nguyen, N. Rizzo, S. Vázquez and S. Yoksan, Nat. Rev. Microbiol., 2010, 8, S30–S37 CrossRef CAS PubMed.
- M. G. Guzmán and G. Kourí, Int. J. Infect. Dis., 2004, 8, 69–80 CrossRef.
- P. Heraud and M. J. Tobin, Stem Cell Res., 2009, 3, 12–14 CrossRef PubMed.
- F. Lee-Montiel, K. Reynolds and M. Riley, J. Biol. Eng., 2011, 5, 1–13 CrossRef PubMed.
- M. M. G. Godoy, E. P. A. Lopes, R. O. Silva, F. Hallwass, L. C. A. Koury, I. M. Moura, S. M. C. Gonçalves and A. M. Simas, J. Viral Hepatitis, 2010, 17, 854–858 CrossRef CAS PubMed.
- A. Sakudo, Y. Suganuma, R. Sakima and K. Ikuta, Clin. Chim. Acta, 2012, 413, 467–472 CrossRef CAS PubMed.
- J. G. Kelly, J. Trevisan, A. D. Scott, P. L. Carmichael, H. M. Pollock, P. L. Martin-Hirsch and F. L. Martin, J. Proteome Res., 2011, 10, 1437–1448 CrossRef CAS PubMed.
- B. E. Obinaju and F. L. Martin, Environ. Pollut., 2013, 183, 46–53 CrossRef CAS PubMed.
- G. Theophilou, K. M. G. Lima, P. L. Martin-Hirsch, H. F. Stringfellow and F. L. Martin, Analyst, 2016, 141, 585–594 RSC.
- A. S. Marques, M. C. N. de Melo, T. A. Cidral and K. M. G. de Lima, J. Microbiol. Methods, 2013, 98, 26–30 CrossRef PubMed.
- A. S. Marques, E. P. Moraes, M. A. A. Júnior, A. D. Moura, V. F. A. Neto, R. M. Neto and K. M. G. Lima, Talanta, 2015, 134, 126–131 CrossRef CAS PubMed.
- L. Cui, H. J. Butler, P. L. Martin-hirsch and F. L. Martin, Anal. Methods, 2016, 1–7 Search PubMed.
- A. S. Marques, M. C. N. Melo, T. A. Cidral and K. M. G. Lima, J. Microbiol. Methods, 2014, 98, 26–30 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Bioinformatics, 2005, 21, 860–868 CrossRef CAS PubMed.
- K. M. G. Lima, K. Gajjar, G. Valasoulis, M. Nasioutziki, M. Kyrgiou, P. Karakitsos, E. Paraskevaidis, P. L. Martin Hirsch and F. L. Martin, Anal. Methods, 2014, 6, 9643–9652 RSC.
- R. W. Kennard and L. A. Stone, Technometrics, 1969, 11, 137–148 CrossRef.
| This journal is © The Royal Society of Chemistry 2017 |



















