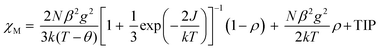DOI:
10.1039/C5QI00088B
(Research Article)
Inorg. Chem. Front., 2015,
2, 763-770
Synthesis, structure, and magnetic properties of dicopper and tricobalt complexes based on N-(2-pyridylmethyl)iminodiethanol†
Received
30th May 2015
, Accepted 26th June 2015
First published on 29th June 2015
Abstract
Dinuclear [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) and trinuclear [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2) coordination complexes have been prepared by the reaction of the tetradentate H2pmide ligand and Cu(NO3)2·3H2O or Co(CH3CO2)2·4H2O/NaNCS in organic solvent (H2pmide = N-(2-pyridylmethyl)iminodiethanol). In 1, one copper(II) ion is bonded with a H2pmide, and the other is coordinated with a mono-deprotonated Hpmide−. The copper(II) ions are linked through an ethoxo group of the Hpmide− ligand and two oxygen atoms of a nitrate ion, resulting in an asymmetric dinuclear structure. In 2, two terminal cobalt(III) units are coordinated with pmide2−, NCS−, and CH3CO2−. The terminal units are each connected to a central cobalt(II) cation through the two oxygen atoms of pmide2− and one bridged acetate ion, giving rise to a mixed-valence trinuclear cobalt complex formulated as CoIII–CoII–CoIII. Complex 1 shows a strong antiferromagnetic interaction through the ethoxo and nitrato groups between the copper(II) ions [g = 2.070, J = –88.1 cm−1 (–61.2 K)]. However, complex 2 behaves as a paramagnetic cobalt(II) monomer, due to the diamagnetic cobalt(III) ions in the terminal units [g = 2.526, |D| = 51.8 cm−1 (36.0 K)].
Introduction
Transition metal complexes incorporating tetradentate ligands have attracted much interest in the area of coordination chemistry, because they show significant structures, catalytic activities, and magnetic properties.1 One of the tetradentate ligands that have pyridine, amine, and hydroxyl groups is N-(2-pyridylmethyl)iminodiethanol (H2pmide), an asymmetrical tripodal N2O2-type ligand. Although the ligand has been used in the preparation of several transition metal complexes, H2pmide based metal compounds have been very limited. The H2pmide ligand acts as a capping ligand as well as a bridging ligand through the deprotonation of the hydroxyl groups. For example, [ReVO(pmide)Cl] and [VO2(Hpmide)] were prepared as monomeric complexes and characterized by various spectroscopic methods.2,3 As multinuclear coordination compounds, [{Ni(Hpmide)}2Ni(CH3CO2)2(HCO2)2] and [Mn8O4(Hpmide)4(EtCO2)6]·(ClO4)2 have been reported.4,5 The former as a trimer is composed of two terminal subunits, {Ni(Hpmide)}+ and a central nickel(II) cation. The terminal building units are each joined to a central nickel(II) cation via the μ2-oxygen of Hpmide−, one μ1-bridged acetate, and one μ2-bridged formate. However, no magnetic study of the trimer was carried out.4 The latter as an octamer possesses four manganese(II) and four manganese(III) ions and consists of three face-sharing cubes with manganese cations on alternate corners. The Mn8 complex acts as a single-molecule-magnet and shows a spin-frustration effect in DC magnetic susceptibility.5 Very recently, we reported on the structure, catalytic activity, and magnetic properties of bis-ethoxy-bridged dinuclear iron(III) complexes with Hpmide− or pmide2−, i.e., [(Hpmide)Fe(NO3)]2(NO3)2·2CH3OH and [(pmide)Fe(N3)]2.6 In these complexes, the H2pmide ligand is mono- and di-deprotonated depending on anions (i.e., NO3−, N3−). As a result the complexes are shown in different conformations from each other and their magnetic interactions are dependent on the different environments around the iron(III) ions. Thus, in an effort to explore the role of anions and transition metal ions and prepare new coordination complexes based on the H2pmide ligand, we have synthesized two coordination complexes composed of copper(II)/Co(II/III) and H2pmide as well as thiocyanate ions. Herein, we report the synthesis, crystal structure, and magnetic properties of oxygen-bridged dicopper(II) and tricobalt(II/III) complexes, [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) and [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2), respectively (Scheme 1).
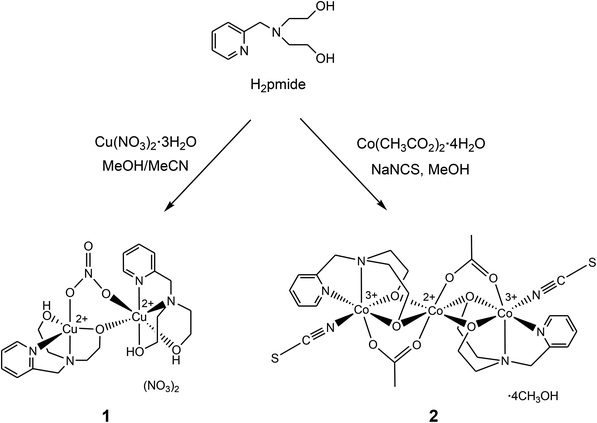 |
| | Scheme 1 Synthesis of complexes 1 and 2. | |
Experimental section
General
All the chemicals used in the synthesis were of reagent grade and used without further purification. N-(2-Pyridylmethyl)iminodiethanol (H2pmide) was prepared according to the literature procedure.5 Infrared spectra were recorded with a Thermo Fisher Scientific IR200 spectrophotometer (±1 cm−1) using a KBr disk. UV/Vis absorption spectra were recorded with a SCINCO S-2100 spectrophotometer. Elemental analyses were carried out using a Fissons/Carlo Erba EA1108 instrument in air or N2 gas. Cyclic voltammograms (CV) were obtained using an EPSILON electrochemistry analyzer in DMF [0.1 M [N(n-Bu)4]BF4 supporting electrolyte] at a scan rate of 100 mV s−1 using a Pt working electrode vs. an Ag/Ag+ electrode. Ferrocenium/ferrocene (Fc+/Fc) was used to calibrate the Ag/Ag+ electrode. Magnetic susceptibilities were measured in an applied field of 5000 Oe between 2 and 300 K on a Quantum Design MPMS superconducting quantum interference device (SQUID) magnetometer. Diamagnetic corrections were made [330.7 (1) and 400.2 × 10−6 (2) emu mol−1] by using Pascal's constants.
Syntheses
[Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1).
To a methanol solution (4 mL) of Cu(NO3)2·3H2O (145 mg, 0.60 mmol) was added dropwise an acetonitrile solution (4 mL) of H2pmide (79 mg, 0.40 mmol). The color became blue, and the solution was stirred for 30 min at room temperature. Blue crystals of 1 were obtained by diffusion of diethyl ether into the blue solution for several days, and were collected by filtration and washed with diethyl ether and dried in air. Yield: 73 mg (52%). Anal Calcd for C20H31Cu2N7O13: C, 34.09; H, 4.43; N, 13.92. Found: C, 33.81; H, 4.44; N, 14.04. FT-IR (KBr, cm−1): 3400, 3250, 3079, 2959, 2925, 2868, 1610, 1445, 1384, 1294, 1067, 917, 770.
[(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2).
To a methanol solution (4 mL) of Co(OAc)2·4H2O (150 mg, 0.60 mmol) were added dropwise a methanol solution (2 mL) of H2pmide (79 mg, 0.40 mmol) and a methanol solution (3 mL) of sodium thiocyanate (97 mg, 1.2 mmol). The solution was stirred for 30 min at room temperature. The red crystals of 2 were obtained by diffusion of diethyl ether into the dark red solution for several days, and were collected by filtration and washed with diethyl ether and dried in air. Yield: 96 mg (52%). Anal Calcd for C30H50Co3N6O12S2: C, 38.84; H, 5.43; N, 9.06; S, 6.91. Found: C, 38.67; H, 5.31; N, 9.08; S, 7.12. FT-IR (KBr, cm−1): 3433, 3024, 2925, 2108, 1585, 1430, 1050, 560. UV/Vis (DMF): λmax, nm (ε, M−1 cm−1) = 378 (2400), 531 (780).
X-ray crystallographic data collection and refinement
A crystal of 1 was mounted on a Hampton cryoloop with paratone-N oil. Intensity data for 1 were collected with a Bruker APEX CCD-based diffractometer (Korea Basic Science Institute, Chonju Branch) and using Mo Kα radiation (λ = 0.71073 Å, graphite monochromator) at 200(2) K. The raw data were processed to give structure factors using the Bruker SAINT program and corrected for Lorentz and polarization effects.7 The intensity data of 1 were corrected for absorption using the SADABS program with multi-scan data.8 A crystal of 2 was coated with paratone-N oil and the diffraction data were measured at 100(2) K with synchrotron radiation (λ = 0.70001 Å) on an ADSC Quantum-210 detector at 2D SMC with a silicon (111) double crystal monochromator (DCM) at the Pohang Accelerator Laboratory, Korea. The ADSC Q210 ADX program9 was used for data collection (detector distance is 63 mm, omega scan; Δω = 1°, exposure time is 1 s per frame) and HKL3000sm (Ver. 703r)10 was used for cell refinement, reduction and absorption correction. The crystal structures of 1 and 2 were solved by direct methods,11 and refined by full-matrix least-squares refinement using the SHELXL-2014 computer program.12 The positions of all non-hydrogen atoms were refined with anisotropic displacement factors. All hydrogen atoms were placed using a riding model, and their positions were constrained relative to their parent atoms using the appropriate HFIX command in SHELXL-2014. In 1, The C20 atom of the H2pmide ligand was refined as disordered over two sets of sites (C20a and C20b) with refined occupancies of 0.535(1) and 0.465(1), respectively. In 2, the final refinement was performed with the modification of the structure factors for contribution of the disordered solvent electron densities using the SQUEEZE option of the PLATON program.13 The crystallographic data and the result of refinements of 1–2 are summarized in Table 1.
Table 1 Crystallographic data and structure refinement for 1 and 2
| Compound |
1
|
2
|
|
R
1 = ∑||Fo| − |Fc||/∑|Fo|, wR2 = [∑w(Fo2 − Fc2)2/∑w(Fo2)2]1/2.
|
| Empirical formula |
C20H31Cu2N7O13 |
C30H50Co3N6O12S2 |
| Formula weight |
704.60 |
927.67 |
| Crystal system |
Monoclinic |
Monoclinic |
| Space group |
P21/c |
C2/c |
| Color |
Blue |
Purple |
| Crystal size, mm3 |
0.34 × 0.20 × 0.14 |
0.10 × 0.04 × 0.04 |
|
a, Å |
17.4759(12) |
26.164(5) |
|
b, Å |
9.7067(6) |
10.106(2) |
|
c, Å |
16.4929(11) |
19.468(4) |
|
V, Å3 |
2795.2(3) |
4174.2(19) |
|
Z
|
4 |
4 |
|
d
calc, g cm−3 |
1.674 |
1.476 |
|
λ, Å |
0.71073 |
0.70001 |
|
T, K |
200(2) |
100(2) |
|
μ, mm−1 |
1.598 |
1.289 |
|
F(000) |
1448 |
1924 |
| Reflections collected |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103 103 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 739 739 |
| Independent reflections |
6931 |
5373 |
| Reflections with I > 2σ(I) |
4127 |
4140 |
| Goodness-of-fit on F2 |
1.120 |
1.073 |
| Final R indices [I > 2σ(I)]a |
R
1 = 0.0545 |
R
1 = 0.0711 |
| wR2 = 0.1157 |
wR2 = 0.2220 |
| CCDC no. |
1402755
|
1402754
|
Results and discussion
Synthesis and characterization
From a mixed methanol/acetonitrile solution of H2pmide and Cu(NO3)2·3H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) at ambient temperature in the presence of air, a blue precipitate of [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) was obtained in moderate yield (Scheme 1). Unexpectedly, one H2pmide is intact without deprotonation, and the other is mono-deprotonated in 1. Thus, complex 1 is formed as an unsymmetric dinuclear copper(II) species through a deprotonated ethoxo group. The IR spectrum (KBr pellet) of 1 displayed a strong band of the nitrate ions at 1384 cm−1 and broad bands of hydroxyl groups from 3400 to 3250 cm−1 and CH peaks corresponding to the pyridine and ethoxyl groups at 3079, 2959, and 2925 cm−1.14 The reaction of 1 equiv. of H2pmide and Co(OAc)2·4H2O and 2 equiv. of NaSCN in methanol solution under aerobic conditions affords the trinuclear complex [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2) in moderate yield (Scheme 1). Interestingly, complex 2 is obtained as a mixed-valence trimeric CoIII–CoII–CoIII species. As shown in the crystal structure of 2 (vide infra), cobalt(II) and cobalt(III) ions in 2 are connected by oxygen atoms of ethoxo and acetato ligands and not the sought thiocyanato-bridged trinuclear complex. The IR spectrum (KBr pellet) of 2 showed very sharp and strong peaks at 2108 and 1585 cm−1 which are assigned to coordinated thiocyanate ions and acetate ions, respectively.14 The composition of 1–2 was also identified by elemental analyses, infrared spectroscopy, and single crystal X-ray diffraction.
3) at ambient temperature in the presence of air, a blue precipitate of [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) was obtained in moderate yield (Scheme 1). Unexpectedly, one H2pmide is intact without deprotonation, and the other is mono-deprotonated in 1. Thus, complex 1 is formed as an unsymmetric dinuclear copper(II) species through a deprotonated ethoxo group. The IR spectrum (KBr pellet) of 1 displayed a strong band of the nitrate ions at 1384 cm−1 and broad bands of hydroxyl groups from 3400 to 3250 cm−1 and CH peaks corresponding to the pyridine and ethoxyl groups at 3079, 2959, and 2925 cm−1.14 The reaction of 1 equiv. of H2pmide and Co(OAc)2·4H2O and 2 equiv. of NaSCN in methanol solution under aerobic conditions affords the trinuclear complex [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2) in moderate yield (Scheme 1). Interestingly, complex 2 is obtained as a mixed-valence trimeric CoIII–CoII–CoIII species. As shown in the crystal structure of 2 (vide infra), cobalt(II) and cobalt(III) ions in 2 are connected by oxygen atoms of ethoxo and acetato ligands and not the sought thiocyanato-bridged trinuclear complex. The IR spectrum (KBr pellet) of 2 showed very sharp and strong peaks at 2108 and 1585 cm−1 which are assigned to coordinated thiocyanate ions and acetate ions, respectively.14 The composition of 1–2 was also identified by elemental analyses, infrared spectroscopy, and single crystal X-ray diffraction.
Description of crystal structures
Structure of 1.
Complex 1 crystallizes in the monoclinic P21/c space group, and the unit cell includes four dinuclear units. The ORTEP drawing of 1 is shown in Fig. 1. The selected bond lengths and angles are listed in Table 2. The asymmetric unit of 1 consists of two crystallographically independent copper(II) cations (Cu1 and Cu2) and three nitrate anions. In the structure of 1, each copper(II) ion has a different coordination environment. The Cu1 atom is coordinated with two nitrogen atoms and two oxygen atoms of a mono-deprotonated Hpmide− ligand and an oxygen atom of the nitrate ion. The Cu1–NHpmide and Cu1–OHpmide bond distances are in the ranges of 1.983(4)–2.017(4) and 1.928(3)–2.412(4) Å, respectively. The Cu1–Onitrate bond distance is 1.983(4) Å. The Cu1 atom is 0.117(1) Å out of the square-coordination N2O2 plane toward the O1 atom. Additionally, the structural index parameter for the coordination geometry in the five-coordinated complex introduced by Reedijk,15τ = (β − α)/60, is τ = 0.16 (for Cu1), where α and β represent two basal angles (β > α), and the parameters for an ideal square pyramid (SP) and trigonal bipyramid (TBP) are τ = 0 (α = β = 180°) and τ = 1 (α = 120°, β = 180°), respectively. Thus, the coordination geometry around Cu1 in 1 indicates a geometry close to SP and can be described as a distorted square pyramid. The Cu2 atom is bonded to two nitrogens and two oxygens of the H2pmide ligand and two oxygen atoms from nitrate and Hpmide− coordinated to Cu1, resulting in a tetragonally distorted octahedral geometry. The bond lengths around the Cu2 atom in the equatorial plane are in the range of 1.921(3)–2.016(4) Å, while the axial Cu2–Ohydroxyl and Cu2–Onitrate bond distances are 2.328(4) and 2.612(4) Å, respectively. The bond lengths axially are somewhat longer than those of square planar, due to the Jahn–Teller distortion.16 Each copper(II) ion is bridged by an ethoxo group of the Hpmide− ligand belonging to Cu1 and nitrate ions, resulting in an asymmetric dinuclear copper(II) complex.17 The angle of Cu1–O2–Cu2 is 119.7(2)°. The bite distance and angle of the five-membered chelate ring are in the ranges of 2.677(1)–2.890(1) Å and 81.84(19)–86.57(16)°, respectively. The shortest Cu⋯Cu contact within the dimer is 3.167(1) Å and the shortest Cu⋯Cu distance between the dimers is 7.190(1) Å. Furthermore, the hydroxyl oxygen atoms of the H2pmide and Hpmide− ligands formed strong hydrogen bonds with oxygen atoms of uncoordinated nitrate ions (O1⋯O12(x, y + 1, z) 2.879(7) Å, ∠O1H1O12 159.5°, O3⋯O8(−x + 1, y + 1/2, −z + 1/2) 2.879(7) Å, ∠O3H3O8 168.4°, O4⋯O11(x, −y + 3/2, z + 1/2) 2.706(8) Å, ∠O4H4O11 154.8°). Due to the hydrogen bonding interactions, 1 is extended from a one-dimensional supramolecular structure along the c-axis (Fig. 2).18
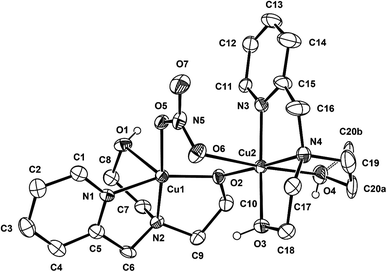 |
| | Fig. 1 ORTEP view of 1. The atoms are represented by 30% probable thermal ellipsoid. Hydrogen atoms and uncoordinated anions are omitted for clarity except for the hydrogen atom of the hydroxyl groups. | |
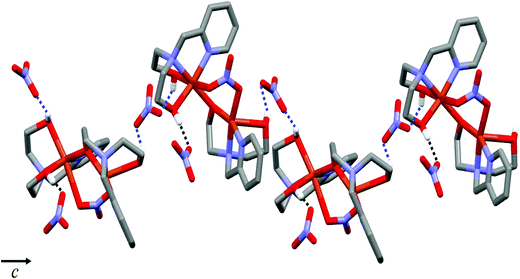 |
| | Fig. 2 One-dimensional structure of 1via hydrogen bonds. Hydrogen bonding interactions are shown as dashed lines. | |
Table 2 Selected bond distances (Å) and angles (°) for 1
| Cu1–O1 |
2.412(4) |
Cu1–N1 |
1.983(4) |
| Cu1–O2 |
1.928(3) |
Cu1–N2 |
2.017(4) |
| Cu1–O5 |
1.983(4) |
Cu2–O2 |
1.921(3) |
| Cu2–O3 |
2.007(4) |
Cu2–N3 |
1.999(4) |
| Cu2–O4 |
2.328(4) |
Cu2–N4 |
2.016(4) |
| Cu2–O6 |
2.612(4) |
|
|
| |
| O1–Cu1–O2 |
113.26(15) |
O2–Cu1–O5 |
96.56(15) |
| O1–Cu1–O5 |
88.17(14) |
O2–Cu1–N2 |
86.57(16) |
| O1–Cu1–N1 |
83.60(16) |
O5–Cu1–N2 |
169.02(16) |
| O1–Cu1–N2 |
80.94(15) |
N1–Cu1–O5 |
95.80(16) |
| O2–Cu1–N1 |
159.34(17) |
N1–Cu1–N2 |
84.41(17) |
| O2–Cu2–O3 |
95.84(16) |
O3–Cu2–N3 |
164.41(17) |
| O2–Cu2–O4 |
103.36(16) |
O3–Cu2–N4 |
83.42(17) |
| O2–Cu2–N3 |
95.97(17) |
O4–Cu2–N3 |
92.41(17) |
| O2–Cu2–N4 |
174.80(18) |
O4–Cu2–N4 |
81.84(19) |
| O3–Cu2–O4 |
94.72(17) |
N3–Cu2–N4 |
83.89(18) |
Structure of 2.
Compound 2 crystallizes in the monoclinic C2/c space group, and the unit cell includes four trinuclear complexes, and the ORTEP drawing of 2 is shown in Fig. 3. The selected bond lengths and angles are listed in Table 3. The core structure of 2 is composed of one mixed-valence CoIIICoIICoIII trimer and four methanol molecules. The central cobalt(II) ion, Co2, is bonded to two oxygen atoms of acetate anions (av. Co2–Oacetato = 2.075(3) Å) and four oxygen atoms of two deprotonated pmide2− ligands (av. Co2–Opmide = 2.092(2) Å). On the other hand, the terminal cobalt(III) ions are bonded to two nitrogen atoms and two oxygen atoms of the pmide2− ligand (av. Co1–Npmide = 1.925(2) and Co1–Opmide = 1.985(2) Å) and one nitrogen atom of the thiocyanate ion (Co1–Nthiocyanto = 1.933(4) Å) and one oxygen atom from the acetate anion (Co1–Oacetato = 1.910(3) Å). All cobalt ions adopt distorted octahedral geometries. The bond distances relating to the Co2 ion are in good agreement with those of low-spin cobalt(III) ions reported previously,19 while the bond lengths around the Co1 ion are somewhat longer than those of the Co2 ion and indicate a high-spin cobalt(II) ion.20 The H2pmide ligands in 2 are fully deprotonated to give pmide2−. The distorted octahedral coordination sphere of the central cobalt (Co2) involves an O6 donor set including four Oalkoxo (pmide2−) and two O (acetato) donors. The octahedral coordination sphere of the terminal cobalt (Co1) shows an N3O3 donor set including two N (pmide2−), one Nthiocyanato, two Oalkoxo (pmide2−), and one O (acetato) donors. The CoII⋯CoIII distance within the trimeric cation is 2.9705(15) Å, shorter than those (3.155 and 3.551(4) Å) found in the case of CoII⋯CoII systems.21 The CoIII⋯CoIII (−x, −y + 1, −z + 1) distance is 6.0090(11) Å and the shortest Co⋯Co distance between the trimers is 7.028(2) Å. The bond angles relating to the terminal cobalt (Co1) and the central cobalt (Co2) lie in the range from 85.10(13) to 171.34(13)° and from 76.42(12) to 180.0°, respectively. Due to the bond angles, the central cobalt (Co2) is much closer to octahedral geometry than terminal cobalt (Co1). In order to determine the oxidation states of cobalt ions in 2 using the bond lengths of Co–N/O, we have used the bond valence sum (BVS) method. Bond valences (s) are obtained using the equation s = exp[(ro − r)/B], where B = 0.37 Å and ro is used from published tables.22 The BVS for the cobalt ion is then the sum of bond valences for each bond made to that cobalt ion. The BVS calculations give the valence sums of 3.07 for Co1 and 2.03 for Co2. The results indicate that the Co1 and Co2 ions are trivalent and divalent, respectively. That is, this strongly demonstrates the +2 oxidation state for the central cobalt ion (Co2) and the +3 oxidation states for terminal cobalt ions (Co1), i.e., CoIII(S = 0, LS)–CoII(S = 3/2, HS)–CoIII(S = 0, LS).
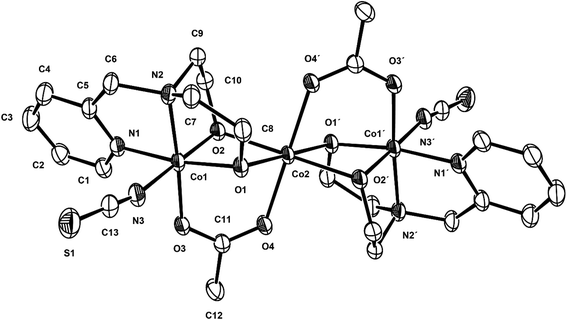 |
| | Fig. 3 ORTEP view of 2. The atoms are represented by 30% probable thermal ellipsoid. Hydrogen atoms and solvent molecules are omitted for clarity. | |
Table 3 Selected bond distances (Å) and angles (°) for 2
| Symmetry transformations used to generate equivalent atoms: (i) #1 −x + 1, −y + 1, −z + 1. |
| Co1–O1 |
1.905(3) |
Co1–N1 |
1.919(3) |
| Co1–O2 |
1.884(3) |
Co1–N2 |
1.930(3) |
| Co1–O3 |
1.910(3) |
Co1–N3 |
1.933(4) |
| Co2–O1 |
2.105(3) |
Co2–O4 |
2.075(3) |
| Co2–O2 |
2.079(3) |
Co1⋯Co2 |
2.9705(15) |
| |
| O1–Co1–O2 |
86.17(13) |
O2–Co1–N3 |
177.65(14) |
| O1–Co1–O3 |
98.09(11) |
O2–Co1–O3 |
92.05(12) |
| O1–Co1–N1 |
171.34(13) |
O3–Co1–N1 |
90.37(12) |
| O1–Co1–N2 |
86.45(12) |
O3–Co1–N2 |
175.46(13) |
| O1–Co1–N3 |
91.57(15) |
O3–Co1–N3 |
88.93(13) |
| O2–Co1–N1 |
91.69(14) |
N1–Co1–N2 |
85.10(13) |
| O2–Co1–N2 |
88.36(13) |
N1–Co1–N3 |
90.45(15) |
| O1–Co2–O2 |
76.42(12) |
O2–Co2–O1i |
103.58(12) |
| O1–Co2–O4 |
86.05(11) |
O4–Co2–O2i |
94.46(11) |
| O2–Co2–O4 |
85.54(11) |
O4i–Co2–O1 |
93.95(11) |
Magnetic properties of 1 and 2
Variable-temperature 2–300 K magnetic susceptibility, χ, measurements on solid samples of 1 and 2 were performed on a SQUID magnetometer (external field 5000 Oe) and are shown in Fig. 4 and 5, respectively. For 1, the effective magnetic moment μeff at 300 K is 2.23 μB/Cu2. This value is less than the spin-only value of 2.45 μB/Cu2 expected for two independent CuII spins (g = 2, S = 1/2), indicative of strong antiferromagnetic interaction between the copper(II) ions. μeff(T) decreases with decreasing temperature to 0.40 μB at 2 K, consistent with the presence of an antiferromagnetic coupling between the copper(II) ions (Fig. 4). χ(T) for 1 was fit to an analytical expression, eqn (1), derived by Bleaney–Bowers for a coupled S = 1/2 dinuclear spin model based on the Hamiltonian H = –2JS1·S2.23| | 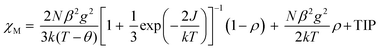 | (1) |
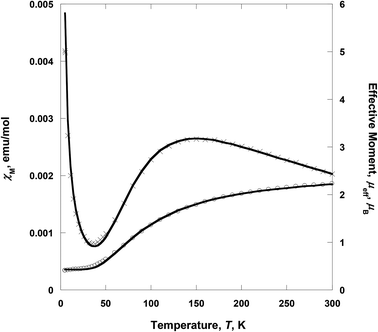 |
| | Fig. 4 Temperature dependence of χM (×) and μeff (○) for 1. The solid lines are the best-fit curves. | |
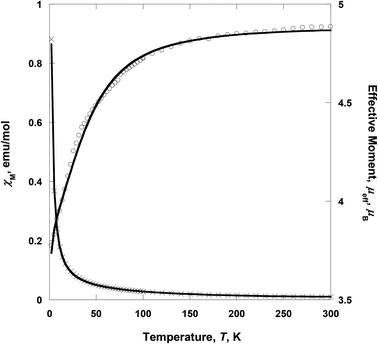 |
| | Fig. 5 Temperature dependence of χM (×) and μeff (○) for 2. The solid lines are the best-fit curves. | |
The best fit to eqn (1) had J = −88.1 cm−1, g = 2.070, the spin impurity, ρ = 0.024, and θ = −1.98 K (TIP = 200 × 10−6 emu mol−1). It should be noted that the J value of 1 indicates that the ethoxo and nitrato groups mediate the antiferromagnetic coupling between the copper(II) ions separated by ca. 3.329 Å. In the dinuclear system of 1, the crystal structure shows that Cu1 is square pyramidal but that of Cu2 is octahedral.17a The dinuclear unit is also connected through two different ligands, i.e., ethoxo and nitrato groups. Thus, the ethoxo group can interact with the unpaired electrons in the dx2−y2 orbitals on Cu1 and Cu2 ions, respectively. In addition, the nitrato ligand is able to interact with the unpaired electron in the dx2−y2 orbital on each copper(II) ion by direct orbital overlap. However, in the case of the nitrato group, the torsion angle of Cu1O5O6Cu2 is 64.9° and the bond distances of Cu1–O5 and Cu2–O6 are somewhat longer than those of Cu1–O2 and Cu2–O2. Due to these geometric factors, efficient coupling between the copper(II) ions can occur through the Cu–O–Cu bond, not the Cu–ONO–Cu one. Thus, the Cu–O–Cu bond contributes to the magnetic interaction more than the Cu–ONO–Cu one. Even though 1 has different linkers and bonding modes between the copper(II) ions, the magnetic properties show a strong antiferromagnetic coupling. The strong antiferromagnetic coupling can be attributed to the Cu–O–Cu angle (119.7°) and the dx2−y2 orbital interaction. Furthermore, the coupling constant of 1 is comparable to −88.2 and −90.4 cm−1 for [Cu2(dpyam)4(μ-CO3)][BF4]2·4H2O and [Cu2(dpyam)4(μ-CO3)][ClO4]2·4H2O, respectively, although the bridging modes are slightly different (dpyam = di-2-pyridylamine).24,25
Complex 2 consists of two low-spin CoIII as wingtip ions and one high-spin CoII as the central ion, i.e., CoIII(LS, S = 0)–CoII(HS, S = 3/2)–CoIII(LS, S = 0). The wingtip cobalt(III) ions are diamagnetic (S = 0), whereas the central cobalt(II) ion is paramagnetic (S = 3/2). At room temperature, the effective moment, μeff [= (8χT)1/2], value is 4.72 μB, suggesting that the cobalt(II) ion is in a high-spin state. This value is larger than the spin-only value (3.87 μB) for a high-spin CoII, indicative of the strong spin–orbit coupling.26 On lowering the temperature, μeff(T) decreases gradually with decreasing the temperature to 3.68 μB at 2 K (Fig. 5). The decrease of the effective magnetic moment at low-temperature (<100 K) was most likely due to the strong spin–orbit coupling of the CoII ion. Thus the spin–orbit coupling highly contributes to the zero-field splitting (ZFS). χ(T) for 2 was simulated using the julX program package (ver. 1.4.1).27 The simulations are based on the usual spin-Hamiltonian operator for the cobalt trimer (S = 0, S = 3/2, S = 0):
| | | H = gβS·B + D[Sz2 − S(S + 1)/3 + (E/D)(Sx2 − Sy2)] | (2) |
where
S is the total spin multiplet (3/2),
g is the average electronic
g value, and
D is the axial zero-field splitting parameter,
E/
D is the rhombicity of the zero-field splitting. The best fit from the data had
g = 2.526, |
D| = 51.8 cm
−1, and the spin impurity,
ρ = 0.005 (TIP = 100 × 10
−6 emu mol
−1).
27 The zero-field splitting parameter between ±1/2 and ±3/2 Kramers doublets was comparable to those for [(L1)
4Co
3(H
2O)
2](NO
3)
4·6H
2O (|
D| = 48.9 cm
−1, L1 =
N-(pyridine-2-ylmethylene)pyrazine-2-carbohydrazide)
28 and [CoL2Cl
2] (|
D| = 46.8 cm
−1, L2 = 4-dodecynyl-2,6-dipyrazolyl pyridine).
29 Additionally, above 2 K,
χM−1(
T) can be fit to the Curie–Weiss expression
χM =
C/(
T −
θ) with
θ = −9.29 K (
C = 3.068 emu K mol
−1) for
2.
30 This value can be also related to the large
D value and/or intermolecular interactions.
Conclusions
We have synthesized an unsymmetrical dinuclear copper complex (1) and a mixed-valence trinuclear cobalt compound (2) using H2pmide and copper/cobalt ions. In 1, each copper(II) ion has two different tetradentate ligands, i.e., H2pmide and Hpmide−, which give rise to an asymmetric structure. Complex 1 exhibits strong antiferromagnetic coupling due to the superexchange via the bridged ethoxy group of Hpmide−. In 2, the Co(II) and Co(III) ions are coordinated via two ethoxo groups and one μ1,3-acetato ligand. The mixed-valence trinuclear cobalt complex, CoIII(S = 0)–CoII(S = 3/2)–CoIII(S = 0), has one paramagnetic cobalt(II) ion and two diamagnetic cobalt(III) ions. Thus, the trimer behaves as a monomeric paramagnetic species and shows large zero-field splitting due to the high spin cobalt(II) ion.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2013R1A1A2007314 and 2014R1A1A2058815). X-ray crystallography with the PLS-II 2D-SMC beamline was supported by MSIP and POSTECH.
Notes and references
-
(a) R. Zong and R. P. Thummel, J. Am. Chem. Soc., 2004, 126, 10800 CrossRef CAS PubMed;
(b) N. Takeda, D. Watanabe, T. Nakamura and M. Unno, Organometallics, 2010, 29, 2839 CrossRef CAS;
(c) R. J. Cross, L. J. Farrugia, P. D. Newman, R. D. Peacock and D. Stirling, Inorg. Chem., 1999, 38, 1186 CrossRef CAS PubMed;
(d) T. Yagyu, K. Yano, T. Kimata and K. Jitsukawa, Organometallics, 2009, 28, 2342 CrossRef CAS;
(e) T. Ohkawara, K. Suzuki, K. Nakano, S. Mori and K. Nozaki, J. Am. Chem. Soc., 2014, 136, 10728 CrossRef CAS PubMed;
(f) J. R. Khusnutdinova, J. Luo, N. P. Rath and L. M. Mirica, Inorg. Chem., 2013, 52, 3920 CrossRef CAS PubMed;
(g) K. S. Min, A. G. DiPasquale, A. L. Rheingold, H. S. White and J. S. Miller, J. Am. Chem. Soc., 2009, 131, 6229 CrossRef CAS PubMed;
(h) A. R. Jeong, J. W. Shin, B. G. Kim and K. S. Min, Bull. Korean Chem. Soc., 2014, 35, 273 CrossRef CAS.
- A. Mondal, S. Sarkar, D. Chopra, T. N. Guru Row and K. K. Rajak, Dalton Trans., 2004, 3244 RSC.
- D. C. Crans, A. D. Keramidas, S. S. Amin, O. P. Anderson and S. M. Miller, J. Chem. Soc., Dalton Trans., 1997, 2799 RSC.
- R. W. Saalfrank, I. Bernt and F. Hampel, Chem. – Eur. J., 2001, 7, 2770 CrossRef CAS.
- C.-C. Wu, S. Datta, W. Wernsdorfer, G.-H. Lee, S. Hill and E.-C. Yang, Dalton Trans., 2010, 39, 10160 RSC.
- J. W. Shin, J. M. Bae, C. Kim and K. S. Min, Dalton Trans., 2014, 43, 3999 RSC.
-
Saint Plus
, Version 6.02, Bruker Analytical X-ray, Madison, WI, 1999 Search PubMed.
-
SADABS
, Version 2.03, Bruker AXS Inc., Madison, WI, 2000 Search PubMed.
-
A. J. Arvai and C. Nielsen, ADSC Quantum-210 ADX Program, Area Detector System Corporation, Poway, CA, USA, 1983 Search PubMed.
-
Z. Otwinowski and W. Minor, in Methods in Enzymology, ed. C. W. Carter Jr. and R. M. Sweet, Academic Press, New York, 1997, vol. 276, part A, p. 307 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Fundam. Crystallogr., 1990, 46, 467 CrossRef.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2015, 71, 3 CrossRef PubMed.
- A. L. Spek, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2015, 71, 9 CrossRef CAS PubMed.
-
K. Nakamoto, Infrared and Raman Spectra of Inorganic and Coordination Compounds, John Wiley and Sons, Inc., New Jersey, 6th edn, 2009, Part B, pp. 64–126 Search PubMed.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, J. Chem. Soc., Dalton Trans., 1984, 1349 RSC.
-
(a) D. Reinen, Coord. Chem. Rev., 2014, 272, 30 CrossRef CAS PubMed;
(b) M. J. Gajewska, W.–M. Ching, Y.–S. Wen and C.–H. Hung, Dalton Trans., 2014, 43, 14726 RSC.
-
(a) T. N. Sorrell, C. J. O'Connor, O. P. Anderson and J. H. Reibenspies, J. Am. Chem. Soc., 1985, 107, 4199 CrossRef CAS;
(b) V. McKee, M. Zvagulis, J. V. Dagdigian, M. G. Patch and C. A. Reed, J. Am. Chem. Soc., 1984, 106, 4765 CrossRef CAS.
-
(a)
J. W. Steed and J. L. Atwood, Supramolecular Chemistry, John Wiley & Sons, Ltd, Chichester, 2nd edn, 2009 Search PubMed;
(b)
G. A. Jeffrey, An Introduction to Hydrogen Bonding, Oxford University Press, New York, 1997 Search PubMed.
- J. W. Shin, S. R. Rowthu, M. Y. Hyun, Y. J. Song, C. Kim, B. G. Kim and K. S. Min, Dalton Trans., 2011, 40, 5762 RSC.
-
(a) L. Antolini, A. C. Fabretti, D. Gatteschi, A. Giusti and R. Sessolilb, Inorg. Chem., 1991, 30, 4858 CrossRef CAS;
(b) Y.–L. Zhang, S.–P. Chen and S.–L. Gao, Z. Anorg. Allg. Chem., 2009, 635, 537 CrossRef CAS PubMed.
-
(a) W.–K. Dong, G. Li, Z.–K. Wang and X.–Y. Dong, Spectrochim. Acta, Part A, 2014, 133, 340 CrossRef CAS PubMed;
(b) M. Diop, P. Aly–Gaye, F. Bouyang–Tamboura, M. Gaye, P. Pérez–Lourido, L. Valencia and G. Castro, Z. Anorg. Allg. Chem., 2014, 640, 1392 CrossRef CAS PubMed.
-
(a) W. Liu and H. H. Thorp, Inorg. Chem., 1993, 32, 4102 CrossRef CAS;
(b) I. D. Brown and D. Altermatt, Acta Crystallogr., Sect. B: Struct. Sci., 1985, 41, 244 CrossRef;
(c) N. E. Brese and M. O'Keeffe, Acta Crystallogr., Sect. B: Struct. Sci., 1991, 47, 192 CrossRef;
(d) J. Zachara, Inorg. Chem., 2007, 46, 9760 CrossRef CAS PubMed.
-
O. Kahn, Molecular Magnetism, VCH, New York, 1993, pp. 103–134 Search PubMed.
- S. Youngme, C. Pakawatchai, W. Somjitsripunya, S. Chantrapromma, K. Chinnakali and H.-K. Fun, Inorg. Chim. Acta, 2000, 303, 181 CrossRef CAS.
- S. Youngme, N. Chaichit, P. Kongsaeree, G. A. Albada and J. Reedijk, Inorg. Chim. Acta, 2001, 324, 232 CrossRef CAS.
-
(a) L.– C. Wu, T.–C. Weng, I.–J. Hsu, Y.–H. Liu, G.–H. Lee, J.–F. Lee and Y. Wang, Inorg. Chem., 2013, 52, 11023 CrossRef CAS PubMed;
(b) M. Rat, R. A. de Sousa, A. Tomas, Y. Frapart, J. P. Tuchagues and I. Artaud, Eur. J. Inorg. Chem., 2003, 759 CrossRef CAS PubMed.
-
E. Bill, julX program, version 1.4.1, Max Planck Institute for Bioinorganic Chemistry, Mülheim an der Ruhr, Germany, 2008 Search PubMed.
- D. Wu, X. Zhang, P. Juang, W. Huang, M. Ruan and Z. W. Ouyang, Inorg. Chem., 2013, 52, 10976 CrossRef CAS PubMed.
- C. Rajnák, J. Titiš, I. Šalitroš, R. Boča, O. Fuhr and M. Ruben, Polyhedron, 2013, 65, 122 CrossRef PubMed.
- E. Shurdha, C. E. Moore, A. L. Rheingold and J. S. Miller, Inorg. Chem., 2011, 50, 10546 CrossRef CAS PubMed.
|
| This journal is © the Partner Organisations 2015 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103
103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 739
739![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) at ambient temperature in the presence of air, a blue precipitate of [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) was obtained in moderate yield (Scheme 1). Unexpectedly, one H2pmide is intact without deprotonation, and the other is mono-deprotonated in 1. Thus, complex 1 is formed as an unsymmetric dinuclear copper(II) species through a deprotonated ethoxo group. The IR spectrum (KBr pellet) of 1 displayed a strong band of the nitrate ions at 1384 cm−1 and broad bands of hydroxyl groups from 3400 to 3250 cm−1 and CH peaks corresponding to the pyridine and ethoxyl groups at 3079, 2959, and 2925 cm−1.14 The reaction of 1 equiv. of H2pmide and Co(OAc)2·4H2O and 2 equiv. of NaSCN in methanol solution under aerobic conditions affords the trinuclear complex [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2) in moderate yield (Scheme 1). Interestingly, complex 2 is obtained as a mixed-valence trimeric CoIII–CoII–CoIII species. As shown in the crystal structure of 2 (vide infra), cobalt(II) and cobalt(III) ions in 2 are connected by oxygen atoms of ethoxo and acetato ligands and not the sought thiocyanato-bridged trinuclear complex. The IR spectrum (KBr pellet) of 2 showed very sharp and strong peaks at 2108 and 1585 cm−1 which are assigned to coordinated thiocyanate ions and acetate ions, respectively.14 The composition of 1–2 was also identified by elemental analyses, infrared spectroscopy, and single crystal X-ray diffraction.
3) at ambient temperature in the presence of air, a blue precipitate of [Cu(H2pmide)(NO3)Cu(Hpmide)](NO3)2 (1) was obtained in moderate yield (Scheme 1). Unexpectedly, one H2pmide is intact without deprotonation, and the other is mono-deprotonated in 1. Thus, complex 1 is formed as an unsymmetric dinuclear copper(II) species through a deprotonated ethoxo group. The IR spectrum (KBr pellet) of 1 displayed a strong band of the nitrate ions at 1384 cm−1 and broad bands of hydroxyl groups from 3400 to 3250 cm−1 and CH peaks corresponding to the pyridine and ethoxyl groups at 3079, 2959, and 2925 cm−1.14 The reaction of 1 equiv. of H2pmide and Co(OAc)2·4H2O and 2 equiv. of NaSCN in methanol solution under aerobic conditions affords the trinuclear complex [(pmide)2Co3(CH3CO2)2(NCS)2]·4CH3OH (2) in moderate yield (Scheme 1). Interestingly, complex 2 is obtained as a mixed-valence trimeric CoIII–CoII–CoIII species. As shown in the crystal structure of 2 (vide infra), cobalt(II) and cobalt(III) ions in 2 are connected by oxygen atoms of ethoxo and acetato ligands and not the sought thiocyanato-bridged trinuclear complex. The IR spectrum (KBr pellet) of 2 showed very sharp and strong peaks at 2108 and 1585 cm−1 which are assigned to coordinated thiocyanate ions and acetate ions, respectively.14 The composition of 1–2 was also identified by elemental analyses, infrared spectroscopy, and single crystal X-ray diffraction.