An aptamer array for discriminating tetracycline antibiotics based on binding-enhanced intrinsic fluorescence†
Yichen
Zhao
,
Biwen
Gao
,
Yijing
Chen
and
Juewen
Liu
 *
*
Department of Chemistry, Waterloo Institute for Nanotechnology, University of Waterloo, Waterloo, Ontario N2L 3G1, Canada. E-mail: liujw@uwaterloo.ca
First published on 6th March 2023
Abstract
Tetracyclines are a class of antibiotics with a similar four-ringed structure. Due to this structural similarity, they are not easily differentiated from each other. We recently selected aptamers using oxytetracycline as a target and focused on an aptamer named OTC5, which has similar affinities for oxytetracycline (OTC), tetracycline (TC), and doxycycline (DOX). Tetracyclines exhibit an intrinsic fluorescence that is enhanced upon aptamer binding, allowing convenient binding assays and label-free detection. In this study, we analyzed the top 100 sequences from the previous selection library. Three other sequences were found to differentiate between different tetracyclines (OTC, DOX, and TC) by the selective enhancement of their intrinsic fluorescence. Among them, the OTC43 aptamer was more selective for OTC with a limit of detection (LOD) of 0.7 nM OTC, OTC22 was more selective for DOX (LOD 0.4 nM), and OTC2 was more selective for TC (0.3 nM). Using these three aptamers to form a sensor array, principal component analysis was able to discriminate between the three tetracyclines from each other and from the other molecules. This group of aptamers could be useful as probes for the detection of tetracycline antibiotics.
Introduction
Tetracyclines are a group of antibiotics with very similar structures.1 Historically, oxytetracycline (OTC) and tetracycline (TC) were among the first discovered antibiotics but they had obvious side effects such as staining of the teeth and bones. As a result, they are now mainly used in animals.2 Doxycycline (DOX) was discovered later and remains a very useful antibiotic to date. The detection of tetracyclines is an important analytical task for environmental monitoring and food safety.3–5Molecular probes to recognize them individually are highly desirable. However, their subtle structural differences present significant analytical challenges. Antibodies against tetracyclines have been developed. In one study, the antibodies were unable to distinguish TC and chlortetracycline.6 In another case, a highly selective chlortetracycline antibody was reported.7 Antibodies for DOX were developed and used for the sensing of seven different tetracyclines in milk with cross-reactivity from 47–102%.8 Similarly, antibodies have also been used for TC detection in honey, although cross reactivity was seen against OTC, rolitetracycline, methacycline, and chlortetracycline.9 Antibody-based assays for small molecular targets are often performed in a competitive format and monoclonal antibodies are very expensive.
Aptamers are nucleic acid based ligands that can selectively bind target molecules.10–15 While aptamers for tetracyclines have been previously reported, they were all isolated using the target-immobilization method,16,17 which may limit the orientation of aptamer binding due to steric hindrance. Library-immobilized aptamer selection allows the use of unmodified targets.18–24 Many new aptamers with low Kd values have been selected.18–20,25–34 We recently used the DNA library immobilization method to select aptamers that can bind OTC.35 Compared to the other aptamer selections performed in our lab, the OTC selection resulted in a much more diverse sequence distribution. Most of our targets (e.g., theophylline and uric acid) had only one or two major families,31,36 but the OTC selection resulted in at least ten families. In our initial work, we only analyzed the top 15 most abundant sequences and tested two of the families. Among them, the OTC5 aptamer has a similar high affinity to OTC, TC and DOX.35
Because tetracyclines are quite complex molecules, it is possible that aptamers can bind in various ways, which may explain the sequence diversity in the final library. While the OTC5 aptamer is a general binder, we hypothesized that we might be able to find other sequences that can discriminate between these tetracyclines and form a sensor array to detect them individually and collectively.
Tetracyclines have intrinsic fluorescence, and aptamer binding may increase the fluorescence intensity, allowing for label-free detection.37,38 In this work, we first performed fluorescent binding assays to identify aptamers with distinct responses to the three tetracyclines. Subsequently, a subset of these aptamers were used to form a sensor array to detect tetracyclines with ultrahigh sensitivity and specificity.
Materials and methods
Chemicals
All DNA samples were purchased from Integrated DNA Technologies (Coralville, IA). TC, OTC, DOX, and other chemicals used for the selectivity tests were purchased from Sigma-Aldrich. All buffers were from Mandel Scientific (Guelph, ON, Canada). Solutions of OTC, TC and DOX were freshly prepared in a buffer containing 10 mM pH 6.0 MES, 50 mM NaCl, and 2 mM MgCl2.Fluorescence spectroscopy
The experiments were performed on a Tecan Spark microplate reader with an excitation wavelength of 370 nm and an emission wavelength of 535 nm. For titrations, 100 nM OTC, TC or DOX was dissolved in the buffer. The ten aptamers were titrated such that the final volume change was maintained at less than 10% of the initial 100 μL volume. The solution was mixed well after each titration and allowed to equilibrate for 1 min before reading. Similar methods were used for the measurement of antibiotic concentration-dependent fluorescence with 0 or 2 μM OTC2 aptamer. These procedures were carried out for DOX with the OTC22 aptamer and OTC with the OTC43 aptamer.Selectivity of the sensors
To test the selectivity of the sensors against a series of different molecules (fluorescein, dopamine, quinine, chloramphenicol, kanamycin, ampicillin, and penicillin), 100 nM of each molecule was dissolved in buffer. The fluorescence intensities of these molecules were measured before and after adding 2 μM OTC2, OTC22, or OTC43 aptamers.Sensor array
To test if our proposed OTC sensor array could discriminate between the different tetracyclines, the fluorescence difference of 50 nM antibiotics in the presence of 0 or 2 μM OTC2, OTC22 or OTC43 aptamers were run in six replicates and the data was analyzed using principal component analysis (PCA, R software). Different OTC concentrations were also tested in a separate study.Results and discussion
Ten aptamers for tetracyclines
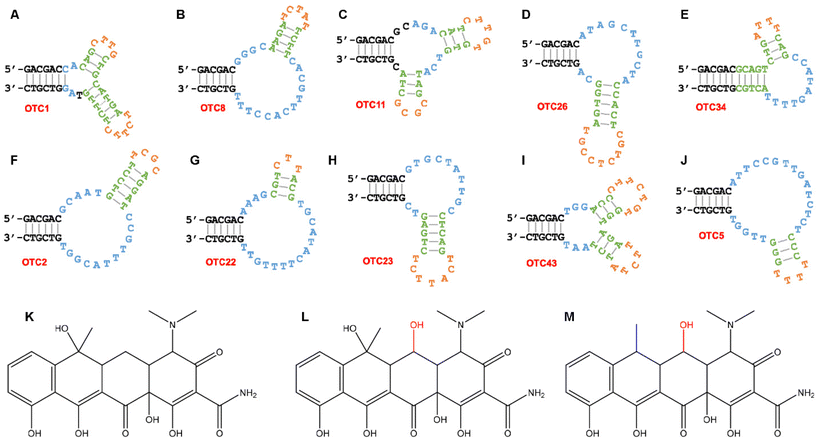
The use of OTC as a target for aptamer selection has been reported previously by our lab.35 In this work, we aimed to analyze more sequences to understand the binding of different aptamers. To be comprehensive, we analyzed the top 100 most abundant sequences, which collectively represented >65% of the final library (round 20). Using Clustal Omega and manual analysis,39 we divided these 100 sequences into 10 families along with eight orphan sequences (Fig. S1†). We then chose one sequence from each family. The predicted secondary structures of the ten sequences are shown in Fig. 1. All of the aptamers have bulged two-way or three-way junction structures. The internal hairpin regions are colored green/orange, whereas the sequences in the potential binding loops are shown in blue. We previously studied the OTC5 aptamer (Fig. 1J), which can bind to all the three tetracyclines with Kd values of approximately 100 nM. Thus, this study focused on the other nine aptamers in Fig. 1. The structures of the three tetracycline antibiotics are shown in Fig. 1K–M and the goal is to use these aptamers to distinguish them.Aptamer binding induced fluorescence changes
Tetracyclines are fluorescent, and their fluorescence is enhanced upon aptamer binding.35,37,38 Therefore, we used their intrinsic fluorescence to study binding. Previous work has indicated that the optimal condition for fluorescence enhancement was with 2 mM Mg2+ at pH 6.0,40 and we used this condition for all the work here. Fig. 2A shows the fluorescence spectra of 100 nM OTC after the addition of 2 μM OTC2 aptamer. A large fluorescence enhancement was observed. Such enhancement can also be readily observed by the naked eye with the excitation of a handheld UV lamp in a darkroom (Fig. 2B). The enhanced fluorescence can be attributed to the aptamer providing a hydrophobic binding pocket to decrease other pathways of relaxation of excited OTC. Binding curves were obtained by gradually titrating the aptamers (Fig. 3). These curves all have a similar y-axis scale (except for Fig. 3B) to highlight the different extents of fluorescence enhancement in different aptamers. The aptamers can be divided into two groups based on the amount of fluorescence enhancement. For biosensor applications, the group in the bottom row is more interesting due to a much larger fluorescence enhancement. The same data were also plotted with a smaller y-axis scale in Fig. S2† to show all the trends of fluorescence enhancement. Outside the 10 families, we also tested an orphan sequence, OTC7, and barely any fluorescence change was observed (Fig. S3†). Therefore, OTC7 served as a negative control to show that not all DNA sequences can enhance the fluorescence of the tetracyclines. The folds of fluorescence enhancement were plotted in Fig. 2C. Based on these data, five aptamers stood out due to their exceptionally high fluorescence enhancement (OTC2, OTC22, OTC23, OTC43 and the previously studied OTC5).Based on the subtle structural differences of the tetracyclines (Fig. 1K–M), we can deduce the binding orientation of different aptamers. One major type of the aptamers has the highest affinity for OTC, followed by TC, and barely binds to DOX (e.g. OTC1, OTC11, OTC26 and OTC43). These aptamers are likely to bind to the common structures of OTC and TC. The other type has the highest affinity for DOX (for example OTC8 and OTC22). OTC2 and OTC5, on the other hand, likely bind in the manner indicated in the top panel of Fig. 2E, and all three tetracyclines can be bound. The proposed ways of binding OTC by three representative aptamers are shown in Fig. 2E.
We did not observe absolute selectivity for any of the tetracyclines. In a previous study, an RNA aptamer with excellent affinity and specificity for DOX was reported.38 However, that was a much longer sequence and even the truncated aptamer contained 74-nt. Our DNA aptamers were only approximately 42-nt, and the binding contact points could be less. It might be possible to use a much longer random library to achieve even better selectivity, but that would increase the cost of synthesis.
The main goal of this study was to distinguish these three tetracyclines, for which we needed to have probes that have differential responses to them. Therefore, we chose OTC2, OTC22 and OTC43, since they each responded the most to a different tetracycline. We also plotted the Kd values of these three aptamers (Fig. 2D), which also showed the relative selectivity for the three tetracyclines. For the remainder of the study, we focused on these three aptamers.
Highly sensitive detection
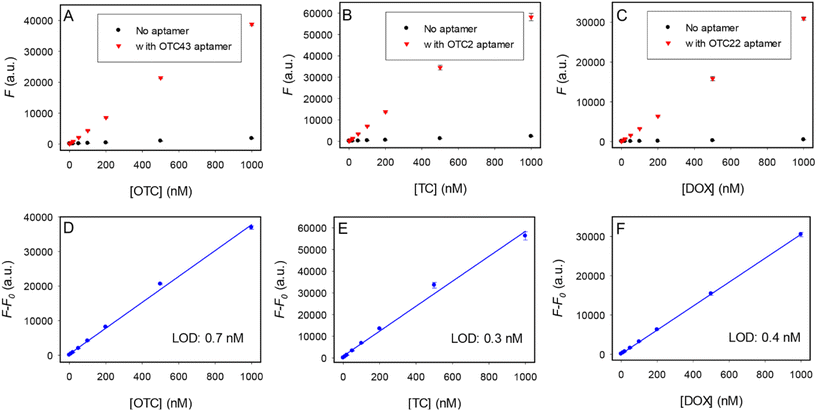
Among the three with the highest fluorescence enhancement, OTC43 had the best selectivity for OTC, OTC22 had the best selectivity for DOX, and OTC2 had the highest response for TC, although the difference was quite small for OTC2. We then tested the sensitivity of label-free detection using the respective best aptamers (Fig. 4). The assays followed the label-free detection method of first measuring the fluorescence of a target tetracycline followed by the addition of an aptamer (Fig. 4A–C). The quantification was based on the difference between these two (Fig. 4D–F).35 The responses were all linear for target molecules up to 1 μM. The detection limits were calculated to be 0.7 nM OTC, 0.3 nM TC, and 0.4 nM DOX.A sensor array for discriminating the three antibiotics
After confirming excellent sensitivity, we measured the selectivity of the three aptamers by calculating the fluorescence enhancement upon adding the aptamers. As shown in Fig. 5A, only the three tetracyclines showed increased fluorescence upon addition of the aptamers. Other molecules such as fluorescein and quinine, although fluorescent, did not show a fluorescence change by the aptamers. The remaining antibiotics were not fluorescent and also did not show any fluorescence changes by the aptamers. Therefore, all these three aptamers are highly selective for the tetracyclines.Based on the data in Fig. 5A, we used principal component analysis (PCA), which is a powerful data treatment method to maximally differentiate target molecules based on sensor array data.41–44 As shown in Fig. 5B, with 50 nM antibiotics, TC, DOX, and OTC were well separated, and the rest of the antibiotics and other molecules were clustered in another region. In addition, we measured the responses to different concentrations of OTC (Fig. 5C). Because the second principal component (PC2) was much smaller, the first principal component (PC1) could be used to quantify the responses (Fig. 5D). The linear range of detection for OTC was found to be from 10 nM to 200 nM.
Discussion
In this work, we reported a range of new aptamers that can bind to various tetracycline antibiotics. Some aptamers could bind to all three tetracyclines with a similar affinity, whereas some other aptamers showed selectivity for only one or two of them. This selectivity may reflect the manner in which these aptamers bind to the antibiotics. Most aptamers with low binding-induced fluorescence enhancement also have a weak binding affinity.Although aptamer binding induced fluorescence enhancement of tetracyclines has been well studied in RNA aptamers,37,38 this property was not explored in DNA aptamers until our recent publications.35,40 In the current work, we demonstrated at least ten different aptamers that all showed such fluorescence enhancement. Therefore, this fluorescence enhancement could be a convenient binding assay for DNA aptamers against tetracyclines. Whether these ten aptamers bind the tetracyclines independently or if some of the aptamers belong to the same family will need to be answered in future studies; however, it is quite clear that there are multiple different ways for aptamers to bind to tetracyclines based on their relative fluorescence enhancements to different targets. These aptamers can serve as probes for the design of biosensors to detect tetracyclines.
Conclusions
In this work, we analyzed the top 100 most abundant sequences in a previous aptamer selection library using OTC as the target. Using the enhancement of the intrinsic fluorescence of the tetracyclines for binding assays, three aptamers were able to differentiate between three different tetracyclines and their binding led to a more than 10-fold increase in fluorescence intensity. OTC43 was found to be specific to OTC with a LOD of 0.7 nM, OTC22 was found to be specific to DOX with a LOD of 0.4 nM, and OTC2 was moderately specific to TC with a LOD of 0.3 nM under optimal buffer conditions. PCA showed that these new sequences could differentiate between the three tetracyclines as well as other antibiotics and fluorescent molecules. We hypothesize that this selectivity could be due to the way each aptamer binds to a different edge of the tetracyclines. This label-free, highly selective aptamer sensor array can be readily paired with other techniques to form improved sensing platforms for different tetracycline antibiotics, especially for portable biosensors.45–47Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding for this work was from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Strathclyde/Waterloo Joint Transatlantic Funding.References
- I. Chopra and M. Roberts, Microbiol. Mol. Biol. Rev., 2001, 65, 232–260 CrossRef CAS.
- F. Granados-Chinchilla and C. Rodríguez, J. Anal. Methods Chem., 2017, 2017, 1315497 Search PubMed.
- X. Tao, Y. Peng and J. Liu, J. Food Drug Anal., 2020, 28, 575–594 CAS.
- L. Hermouche, M. Bendany, K. Abbi, Y. El Hamdouni, N. Labjar, M. El Mahi, E. m. Lotfi, M. Dalimi, D. Dhiba and S. El Hajjaji, Int. J. Environ. Anal. Chem., 2021, 1–23 CrossRef.
- S. P. Karthick and R. K. V, Int. J. Pharm. Pharm., 2014, 6, 48–51 Search PubMed.
- Y. Zhang, S. Lu, W. Liu, C. Zhao and R. Xi, J. Agric. Food Chem., 2007, 55, 211–218 CrossRef CAS PubMed.
- P. Poungmalai, A. Buakeaw, S. Puthong and N. Khongchareonporn, Food Agric. Immunol., 2021, 32, 163–173 CrossRef CAS.
- F. Gao, G. X. Zhao, H. C. Zhang, P. Wang and J. P. Wang, J. Environ. Sci. Health, Part B, 2013, 48, 92–100 CrossRef CAS.
- N. Pastor-Navarro, S. Morais, Á. Maquieira and R. Puchades, Anal. Chim. Acta, 2007, 594, 211–218 CrossRef CAS PubMed.
- Y. Wu, I. Belmonte, K. S. Sykes, Y. Xiao and R. J. White, Anal. Chem., 2019, 91, 15335–15344 CrossRef CAS PubMed.
- L. Wu, Y. Wang, X. Xu, Y. Liu, B. Lin, M. Zhang, J. Zhang, S. Wan, C. Yang and W. Tan, Chem. Rev., 2021, 121, 12035–12105 CrossRef CAS PubMed.
- E. M. McConnell, J. Nguyen and Y. Li, Front. Chem., 2020, 8, 434 CrossRef CAS.
- H. Yu, O. Alkhamis, J. Canoura, Y. Liu and Y. Xiao, Angew. Chem., Int. Ed., 2021, 60, 16800–16823 CrossRef CAS PubMed.
- M. Debiais, A. Lelievre, M. Smietana and S. Müller, Nucleic Acids Res., 2020, 48, 3400–3422 CrossRef CAS PubMed.
- L. He, R. Huang, P. Xiao, Y. Liu, L. Jin, H. Liu, S. Li, Y. Deng, Z. Chen, Z. Li and N. He, Chin. Chem. Lett., 2021, 32, 1593–1602 CrossRef CAS.
- J. H. Niazi, S. J. Lee, Y. S. Kim and M. B. Gu, Bioorg. Med. Chem., 2008, 16, 1254–1261 CrossRef CAS.
- J. H. Niazi, S. J. Lee and M. B. Gu, Bioorg. Med. Chem., 2008, 16, 7245–7253 CrossRef CAS.
- R. Nutiu and Y. Li, Angew. Chem., Int. Ed., 2005, 44, 1061–1065 CrossRef CAS PubMed.
- N. Nakatsuka, K.-A. Yang, J. M. Abendroth, K. M. Cheung, X. Xu, H. Yang, C. Zhao, B. Zhu, Y. S. Rim, Y. Yang, P. S. Weiss, M. N. Stojanović and A. M. Andrews, Science, 2018, 362, 319–324 CrossRef CAS PubMed.
- K.-A. Yang, M. Barbu, M. Halim, P. Pallavi, B. Kim, D. M. Kolpashchikov, S. Pecic, S. Taylor, T. S. Worgall and M. N. Stojanovic, Nat. Chem., 2014, 6, 1003–1008 CrossRef CAS PubMed.
- H. Yu, Y. Luo, O. Alkhamis, J. Canoura, B. Yu and Y. Xiao, Anal. Chem., 2021, 93, 3172–3180 CrossRef CAS PubMed.
- H. X. Yu, W. J. Yang, O. Alkhamis, J. Canoura, K. A. Yang and Y. Xiao, Nucleic Acids Res., 2018, 46, e43 CrossRef PubMed.
- C. Lyu, I. M. Khan and Z. Wang, Talanta, 2021, 229, 122274 CrossRef.
- M. Rajendran and A. D. Ellington, Nucleic Acids Res., 2003, 31, 5700–5713 CrossRef CAS PubMed.
- K.-A. Yang, H. Chun, Y. Zhang, S. Pecic, N. Nakatsuka, A. M. Andrews, T. S. Worgall and M. N. Stojanovic, ACS Chem. Biol., 2017, 12, 3103–3112 CrossRef CAS PubMed.
- W. J. Yang, H. X. Yu, O. Alkhamis, Y. Z. Liu, J. Canoura, F. F. Fu and Y. Xiao, Nucleic Acids Res., 2019, 47, e71 CrossRef CAS.
- H. L. Gao, J. X. Zhao, Y. Huang, X. Cheng, S. Wang, Y. Han, Y. Xiao and X. H. Lou, Anal. Chem., 2019, 91, 14514–14521 CrossRef CAS.
- P.-J. J. Huang and J. Liu, Anal. Chem., 2022, 94, 3142–3149 CrossRef CAS.
- Y. Luo, J. Wang, L. Yang, T. Gao and R. Pei, Sens. Actuators, B, 2018, 276, 128–135 CrossRef CAS.
- H. Tian, N. Duan, S. Wu and Z. Wang, Anal. Chim. Acta, 2019, 1081, 168–175 CrossRef CAS.
- Y. Liu and J. Liu, Analysis Sensing, 2022, 2, e202200010 CAS.
- Y. Luo, Z. Jin, J. Wang, P. Ding and R. Pei, Analyst, 2021, 146, 1986–1995 RSC.
- P.-J. J. Huang and J. Liu, Angew. Chem., Int. Ed., 2023, 62, e202212879 Search PubMed.
- A. Boussebayle, F. Groher and B. Suess, Methods, 2019, 161, 10–15 CrossRef CAS PubMed.
- Y. Zhao, S. Ong, Y. Chen, P.-J. J. Huang and J. Liu, Anal. Chem., 2022, 94, 10175–10182 CrossRef CAS PubMed.
- P.-J. J. Huang and J. Liu, ACS Chem. Biol., 2022, 17, 2121–2129 CrossRef CAS PubMed.
- M. Müller, J. E. Weigand, O. Weichenrieder and B. Suess, Nucleic Acids Res., 2006, 34, 2607–2617 CrossRef PubMed.
- Z. J. Tickner, G. Zhong, K. R. Sheptack and M. Farzan, Biochemistry, 2020, 59, 3473–3486 CrossRef CAS PubMed.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Söding, J. D. Thompson and D. G. Higgins, Mol. Syst. Biol., 2011, 7, 539 CrossRef PubMed.
- Y. Zhao, B. Gao, P. Sun, J. Liu and J. Liu, Biosensors, 2022, 12, 717 CrossRef CAS.
- H. Haick and N. Tang, ACS Nano, 2021, 15, 3557–3567 CrossRef CAS.
- S.-F. Xue, X.-Y. Han, Z.-H. Chen, Q. Yan, Z.-Y. Lin, M. Zhang and G. Shi, Anal. Chem., 2018, 90, 10614–10620 CrossRef CAS PubMed.
- M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H. F. Bunz and V. M. Rotello, Nat. Chem., 2009, 1, 461–465 CrossRef CAS PubMed.
- M. S. Hizir, N. M. Robertson, M. Balcioglu, E. Alp, M. Rana and M. V. Yigit, Chem. Sci., 2017, 8, 5735–5745 RSC.
- J. Zhang, T. Lan and Y. Lu, Annu. Rev. Anal. Chem., 2022, 15, 151–171 CrossRef PubMed.
- R. Liu, Y. Hu, Y. He, T. Lan and J. Zhang, Chem. Sci., 2021, 12, 9022–9030 RSC.
- Y. Zhao, K. Yavari and J. Liu, TrAC, Trends Anal. Chem., 2022, 146, 116480 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00154g |
| This journal is © The Royal Society of Chemistry 2023 |