Recent advances in the discovery of plant-derived antimicrobial natural products to combat antimicrobial resistant pathogens: insights from 2018–2022
Sunmin
Woo
 a,
Lewis
Marquez
b,
William J.
Crandall
a,
Lewis
Marquez
b,
William J.
Crandall
 b,
Caitlin J.
Risener
b,
Caitlin J.
Risener
 b and
Cassandra L.
Quave
b and
Cassandra L.
Quave
 *ac
*ac
aCenter for the Study of Human Health, Emory University, USA
bMolecular and Systems Pharmacology Program, Laney Graduate School, Emory University, USA
cDepartment of Dermatology, Emory University School of Medicine, USA. E-mail: cquave@emory.edu
First published on 13th July 2023
Abstract
Covering: 2018 to 2022
Antimicrobial resistance (AMR) poses a significant global health threat. There is a rising demand for innovative drug scaffolds and new targets to combat multidrug-resistant bacteria. Before the advent of antibiotics, infections were treated with plants chosen from traditional medicine practices. Of Earth's 374![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 plant species, approximately 9% have been used medicinally, but most species remain to be investigated. This review illuminates discoveries of antimicrobial natural products from plants covering 2018 to 2022. It highlights plant-derived natural products with antibacterial, antivirulence, and antibiofilm activity documented in lab studies. Additionally, this review examines the development of novel derivatives from well-studied parent natural products, as natural product derivatives have often served as scaffolds for anti-infective agents.
000 plant species, approximately 9% have been used medicinally, but most species remain to be investigated. This review illuminates discoveries of antimicrobial natural products from plants covering 2018 to 2022. It highlights plant-derived natural products with antibacterial, antivirulence, and antibiofilm activity documented in lab studies. Additionally, this review examines the development of novel derivatives from well-studied parent natural products, as natural product derivatives have often served as scaffolds for anti-infective agents.
1. Introduction
Antimicrobial resistance (AMR) poses a significant public health threat in the modern era. As antibiotics lose their effectiveness due to the development of drug resistance, the treatment of recalcitrant infections becomes increasingly challenging, and in some cases, impossible. Researchers from the Centers for Disease Control and Prevention (CDC), World Health Organization (WHO), and public health institutions around the world continue to assert that remedial actions must be implemented globally to address the spread of AMR.1–3 According to the CDC, more than two million AMR infections occur annually in the United States, resulting in an estimated 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths; the medical cost for AMR treatment is over $4.7 billion.2 Although the degree of AMR varies greatly depending on the bacterial species and geographic region, AMR is prevalent across the globe. For example, in the European Union/European Economic Area (EU/EEA) alone, it is estimated that more than 670
000 deaths; the medical cost for AMR treatment is over $4.7 billion.2 Although the degree of AMR varies greatly depending on the bacterial species and geographic region, AMR is prevalent across the globe. For example, in the European Union/European Economic Area (EU/EEA) alone, it is estimated that more than 670![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 infections are caused by AMR each year and about 33
000 infections are caused by AMR each year and about 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths as a result of it.4 An assessment of 2019 global data found that an estimated 4.95 million deaths were associated with bacterial AMR, including 1.27 million deaths attributed to bacterial AMR.5 During the global COVID-19 pandemic, AMR rates have continued to rise. In the period from March to October 2020, nearly 80% of hospitalized patients for COVID-19 received antibiotics, which contributed to this AMR trend.6
000 deaths as a result of it.4 An assessment of 2019 global data found that an estimated 4.95 million deaths were associated with bacterial AMR, including 1.27 million deaths attributed to bacterial AMR.5 During the global COVID-19 pandemic, AMR rates have continued to rise. In the period from March to October 2020, nearly 80% of hospitalized patients for COVID-19 received antibiotics, which contributed to this AMR trend.6
In February 2017, the WHO published a list of pathogens in urgent need of new antibiotics and provided guidelines to focus researchers' research and development efforts on high-priority targets. Six pathogens, in particular, were issued with priority ratings and named the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.).7,8 To overcome the challenges of AMR, the development of rapid point-of-care diagnostic methods, discovery of new drug candidates, and rapid development of existing candidates are all necessary.
About half of the drugs registered by the FDA between 1981 and 2019 were natural products, mimetics, or derivatives, which was roughly double the share of all synthetic drugs (24.6%).9 Among these drugs, natural products continue to be an important source for the discovery and development of anti-infective drug candidates. Around 42.3% of all anti-infective agents are derived from natural products, their derivatives, and natural product mimics.9 Natural products are characterized by a great diversity of scaffolds and high structural complexity. Among plant-derived natural products, alkaloids, phenolic derivatives, and terpenoids are promising sources of antibacterial lead compounds that can help fill the drug development pipeline. In addition to their potential value as classic antibiotics targeting growth and survival, some plant-derived natural products also show promise as synergists, targeting bacterial virulence and pathogenesis pathways, or by sensitizing bacteria to established classes of antibiotics.10–12
Bacterial resistance to antimicrobials is enabled through a few core mechanisms, including inactivation or alteration of antibacterial molecules, modification of bacterial target sites, reduction of antibiotic penetration/accumulation, and formation of bacterial biofilms.13 Plant natural products can act as antibacterial agents through a variety of modes of action targeting these resistance mechanisms and other pathways necessary for bacterial survival. In this paper, we review recent findings from the last five years (Jan 2018–Sep 2022) on plant natural products that exhibit anti-infective properties against pathogenic bacteria, including growth inhibition, antivirulence, and antibiofilm activity. We also introduce traditional knowledge of medicinal plants used for the treatment of infectious diseases in the study of source material for plant natural product discovery in the context of ethnobotany—the study of local plants and their practical uses through accumulated knowledge of local culture and people.14 We present a summary of activities for plant-derived natural products exhibiting promising activity as antibiotic, antivirulence, and antibiofilm leads in Tables 1–3. This review will be of critical importance to the natural products research community, filling a gap in comprehensive reviews on plant-derived natural products for treating infection.
| Natural product compounds | Chemical class | Plant source | Antibacterial activity | Proposed mechanism | Other biological activity | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Bacterial target | Activity (MIC) | Derivative activity (MIC) | ||||||
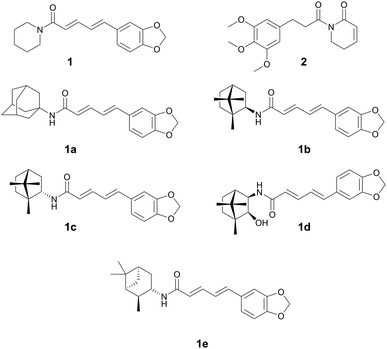
| Piperine (1) | Amide alkaloids | Piper spp., Piperaceae | Staphylococcus aureus | (1) 3.9–15.6 μg mL−1 | (1a–1e) 0.15–0.36 μM (M. tuberculosis) | — | Anti-tumor, antifungal. analgesic, and antidepressant | 30 |
| Piperlongumine (2) | Pseudomonas aeruginosa; Mycobacterium tuberculosis | (2) 3.9–31.2 μg mL−1 (P. aeruginosa) | Host targeted mechanism: increased mTORC1 signaling leading to increased bacterial clearance by macrophages | 31 | ||||
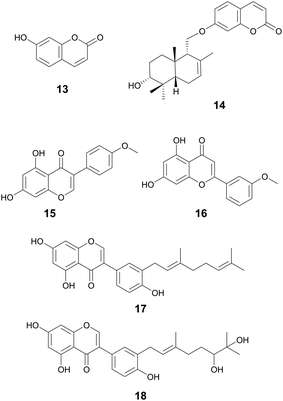
| Biochanin A (15) | Flavonoids | Fabaceae spp. | P. aeruginosa | 0.97 μg mL−1 | — | — | Antidiabetic, anticancer | 82 |
| Acacetin (16) | Flavonoids | Fabaceae spp. | P. aeruginosa | 3.90 μg mL−1 | — | — | MAO-A/B inhibitor | 82 |
| Flavonoids (isoflavonoids, flavones) | Flavonoids (17, 18) | Ficus auriculata Lour., Moraceae (17) | Various | 1.25–10 μM (17) | — | Potentially through nonspecific action upon the bacterial cell membrane (17) | α-Glucosidase inhibition IC50 50.5 ± 3.7 μM (17) | 84 |
| Myrsininone A (17) | Ficus auriculata Lour., Moraceae (18) | Various | 10–20 μM | |||||
| (18) | Anthraquinone (6) | Stereospermum fimbriatum (Wall. Ex G. Don) DC., Bignoniaceae (6) | Methicillin-resistant S. aureus (MRSA) | 6.25 μg mL−1 | Unknown (18–19) | 49 | ||
| (6) | ||||||||
| Carnosic acid and derivatives Carnosic acid (29) | Diterpenoids | Lepechinia meyenii (Walp.) Epling, Lamiaceae | Enterococcus faecalis, various MRSA strains | 15.6, 7.8–15.6 μg mL−1 (29) | Anti-MRSA activity; (30), 31.2 μg mL; (31) 3.9 μg mL−1 | Interaction with bacterial membrane phospholipids | Antioxidant and anti-adipogenic | 125 |
| Triterpenic derivatives (34–38) | Terpenoids | Various | S. aureus | (34–38) 0.15 μM | — | Disrupts S. aureus cell membrane and inhibits NADH oxidation | — | 139 |
| Oleanolic acid derivative (39) | Terpenoids | Various | S. aureus | 4 μg mL−1 (MRSA) | — | Deregulate peptidoglycan metabolism | Anti-chlamydial activity | 141 |
| Natural product | Compound class | Plant source | Antibacterial activity | Mechanism | Other biological activity | References | ||
|---|---|---|---|---|---|---|---|---|
| Bacterial target | Activity | Derivative activity | ||||||
| a Unspecified IC50, IC90, or activities of multiple assays listed as range. | ||||||||
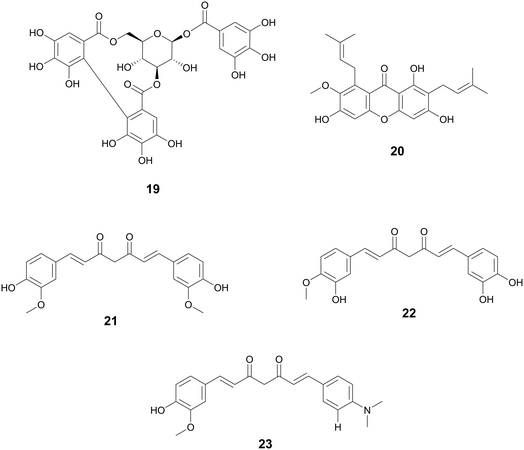
| Curcumin (21) | Diarylheptanoids | Streptococcus pneumoniae Nan A; Vibrio cholerae | 0.6–5.3 μM (IC50) | 0.2–1.9 μM (IC50) | Sialidase inhibitors | — | 161 | |
| (22) | ||||||||
| (23) | ||||||||
| Ligustchuanes (3) | Lignans | Ligusticum striatum DC., Apiaceae | Staphylococcus aureus | (4) 3–6 μMa | — | Inhibits α-hemolysin secretion | — | 162 |
| (4) | (3) 96 μMa | |||||||
| Paeonol (28) | Methoxybenzenes | Paeonia spp., Paeoniaceae, dried root bark | Salmonella enterica serovar Typhimurium | 95–190 μMa | — | T3SS inhibitor, reduces SipA translocation and expression of effector proteins | — | 163 |
| Salvianolic acid A (24) | Phenolic derivatives | Salvia miltiorrhiza Bunge, Lamiaceae | S. aureus | 5.75 μg mL−1 (IC50) | — | Inhibition of SrtA activity | — | 164 |
| Resveratrol oligomers (25) | Stilbenes | Vitis vinifera L., Vitaceae | Pseudomonas syringae pv. Tomato DC3000 | 25–100 μMa | — | Inhibition of T3SS through downregulation of hrp gene expression | — | 165 |
| (26) | ||||||||
| (27) | ||||||||
| Corilagin (19) | Tannins | Terminalia chebula Retz., Combretaceae | S. aureus | 6–10 μg mL−1a | — | Quorum sensing inhibitor through agrA, anti-α-hemolysin | Biofilm inhibition, | 166 |
| Pigment inhibition | ||||||||
| Decreased STX production | ||||||||
| Betulin (40) | Terpenoids | Betula spp., Betulaceae | Listeria monocytogenes | 4.5–36 μMa | — | Inhibition of exotoxin, LLO activity | — | 147 |
| Celastrol (41) | Terpenoids | Tripterygium spp., Celastraceae | S. aureus | 0.44–2.2 μMa | — | Decreased expression of virulence transcriptional regulators SigB, msaB, and stress adaptor yjbH | — | 167 |
| Triterpenoid acids (42) | Terpenoids | Schinus terebinthifolia Raddi, Anacardiaceae | S. aureus | 2–70 μM (IC50) | — | Quorum sensing inhibitors | Biofilm inhibition | 168 |
| (43) | ||||||||
| (44) | ||||||||
| Natural product compounds | Natural product class | Plant source | Antibacterial activity | Mechanism | Other biological activity | References | ||
|---|---|---|---|---|---|---|---|---|
| Bacterial target | Activity | Derivative activity | ||||||
| Umbelliferone (13) | Coumarins | Apiaceae family (13) | Staphylococcus epidermidis | 200 μg mL−1 MBIC (13) | Downregulation of genes key for biofilm formation via adhesion (icaD, aap, and pbh) | Anti-fungal, antioxidant, and antidiabetic activities | 73 and 74 | |
| Feselol (14) | Ferula narthex Boiss., Apiaceae (14) | Staphylococcus aureus and S. epidermidis | 87 μg mL−1 MBIC (14) (S. aureus); 174 μg mL−1 MBIC (14) (S. epidermidis) | |||||
| Phillygenin (5) | Lignans | Forsythia suspensa Vahl, Oleaceae | Helicobacter pylori | 100–150 μg mL−1 MBIC | — | — | Antioxidant, protection of HDL and LDL lipid peroxidation, and may also have anti-inflammatory activity | 43 |
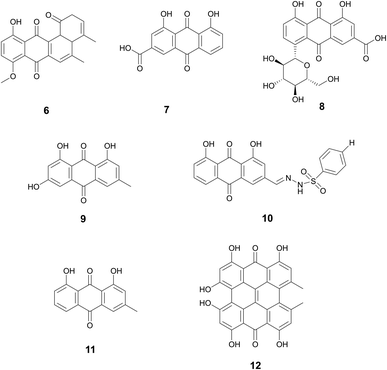
| Rhein (7) | Anthraquinones | Rheum palmatum L., Polygonaceae; Senna tora (L.) Roxb., Fabaceae; Reynoutria multiflora (Thunb.) Moldenke, Polygonaceae; and Aloe vera (L.) Burm.f.,Asphodelaceae | Streptococcus mutans | 60% inhibition at 10 μg mL−1, complete inhibition at 50 μg mL−1 (8) | — | (7) may downregulate the expression of brpA and luxS in S. mutans | Moderate bioactivity against human PTP1B (protein tyrosine phosphatase 1B) | 52 and 57 |
| Rhein-8-O-β-D-glucopyranoside (8) | MBEC50 of 6.31 μg mL−1 and MBEC90 > 50 μg mL−1 (7) | Promote the metabolic activity and purgative activity of sennoside A | ||||||
| Neuroprotection, bacterial growth inhibition, anticancer, anti-inflammatory, antiapoptotic | ||||||||
| Emodin (9) | Anthraquinones | Frangula alnus Mill., Rhamnaceae (9) | Methicillin resistant S. aureus (MRSA) ((9) | 0.05–3.125 μg mL−1 MBIC | 40% biofilm formation inhibition at 4 μg mL−1 (10) | Decrease in ica gene expression (9) | Antioxidant, anti-inflammatory, antiviral, anti-diabetic, and anticancer activity (9) | 62 and 66 |
| N′-((4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracen-2-yl) methylene) benzenesulfonohydrazide (10) | Rheum palmatum L., Polygonaceae; and Senna obtusifolia (L.) H.S.Irwin & Barneby, Fabaceae (cassia seed) (10) | Methicillin sensitive S. aureus (MSSA) (9) | 30% biofilm eradication at 12.5 μg mL−1 (9) | Dispersion of the biofilm into the planktonic culture (10) | ||||
| MRSA (10) | ||||||||
| Chrysophanol (11) | Anthraquinones | Rheum palmatum L., Polygonaceae | Streptococcus suis | MBIC: 0.124 μg mL−1 | — | Lack of interconnection between bacterial cells prevents biofilm formation | Cardioprotective, anticancer, neuroprotective, and anti-inflammation | 67 |
| Hypericin (12) | Anthraquinones | Hypericum perforatum L., Hypericaceae | MRSA | 60% inhibition at 8 μg mL−1 | — | Inhibits sarA expression in MRSA | Anticancer and antiviral | 69 |
| α-Mangostin (20) | Xanthones | Garcinia mangostana L., Clusiaceae | S. aureus | Attachment inhibition 75% at 4 μg mL | Decrease in gene expression of dnaK | Anti-inflammatory, neuroprotective, and anti-viral | 94 | |
| Formation inhibition: 2 μg mL−1 (MBIC) | ||||||||
| Eradication: | ||||||||
| 96% at 4 μg mL−1 | ||||||||
| Andrographolide: diterpene lactone (32) | Terpenoids | Helichrysum caespititium (DC.) Sond, Asteraceae (32) | Neisseria gonorrhoeae (32) | Attachment inhibition 81% and formation inhibition 32% at 60 μg mL−1 (32) | (32) Disturbs extracellular polysaccharides cohesiveness | 131 and 133 | ||
| Sulfonate (33) | Andrographis paniculata (Burm.f.) Wall, Acanthaceae (33) | S. aureus | Biofilm eradication at 25 mg mL−1 (33) | (33) Down-regulates gene expression of intercellular adhesion genes (icaA, icaD, and PIA) | ||||
| Oleanolic acid (45) | Terpenoids | Olea europaea L., Oleaceae | S. aureus | (45) No eradication at 50 μg mL−1 | (46) 45% eradication at 50 μg mL−1 | Penetration of bacterial cellular membranes causing damage as well | Anti-cancer anti-diabetic, hepatoprotective, anti-parasitic, anti-bacterial, and antioxidant activities | 157 |
| Oleanolic acid-HDA (46) | ||||||||
| Maslinic acid (47) | Terpenoids | Olea europaea L., Oleaceae | S. aureus | (47) 20% eradication at 50 μg mL−1 | (48) 30% eradication at 50 μg mL−1 | Penetration of bacterial cellular membranes causing damage as well | Cardioprotection, anticancer, antiparasitic, and anti-inflammatory | 157 |
| Maslinic acid-HDA (48) | ||||||||
2. Methods for review
Our previous comprehensive review of plant derived natural products with antibacterial activity examined the literature from January 1st, 2012 to September 3rd, 2019.10 This review seeks to build on that by examining the literature for plant-derived and plant-inspired antimicrobial compounds that met our rigorous inclusion criteria from articles published from January 1st, 2018 to September 30th, 2022. This review specifically focuses on recently reported natural products with antibacterial, antivirulence, or antibiofilm activity.Articles were searched on PubMed with specific key words for each type of activity. Antibacterial key terms searched were “(antibiotics) AND (natural product)” and “(antibiotics) AND (isolated) AND (natural product)”, “(natural product) AND (antibiotic) AND (novel)”, “(antibiotic) AND (natural product)”, “(plant) AND (antibiotic) AND (novel compound)”, and “(antibiotics) AND (extract)” and “(antivirulence) AND (plant)” on PubMed and Google Scholar; Reaxys key terms were “antibiotic, natural product ≥2018” and “Pharmacological data, article, antibiotic”. Antivirulence key terms searched were “(virulence) AND (natural product)” on PubMed and Google Scholar; Reaxys key terms were “antivirulence, natural product ≥2018” and “Pharmacological data, article, antivirulence”. Antibiofilm key terms searched were “(biofilm) AND (natural product)” and “(biofilm) AND (plant)” on PubMed and Google Scholar; Reaxys key terms were “biofilm, natural product ≥2018” and “Pharmacological data, article, biofilm”. As lead hits were selected, similar searches were performed including the compound name and “derivative”.
The initial search yielded thousands of results, prompting us to establish criteria to refine our selection. First, we selected papers to only include terrestrial plant-derived natural products with activity specifically against multidrug resistant bacteria. Next, we filtered our results to only include single compounds rather than essential oils or extracts. Additionally, we searched for novel products and allowed for derivatives from well-studied parent compounds (such as myricetin and quercetin), while excluding results that utilized nanoparticle technology. For antibiofilm results, we cross-checked with the 2020 Natural Product Reports review “Natural products as inspiration for the development of bacterial antibiofilm agents” by R. J. Melander et al. to prevent redundant reporting in the literature.15
An additional round of literature review on PubMed and Google Scholar was developed after identifying our lead compounds. Our first round was centered on reports of each compound's mechanism of action in bacteria, and then we transitioned to reviewing their bioactivity against other diseases. Finally, we searched records for ethnobotanical information for each plant. The resources for ethnobotanical information included review articles, textbook chapters, and primary field reports as available.
2.1 Data reporting
2.2 Chemical structures
Chemical structures reported in this review are based upon the published structure within each manuscript if available and PubChem for those not reported. Structural figures were produced using ChemDraw 21.0.0 by PerkinElmer Informatics.2.3 Plant nomenclature
Plant nomenclature is updated periodically, and it is important to differentiate the accepted plant names from common names. The full accepted name of each plant in this review was to include genus, species, author epithet, and family. Each plant mentioned in the literature was verified on World Flora Online (http://www.worldfloraonline.org) to ensure that the nomenclature is current and up to date.3. Plant-derived natural products with antimicrobial activity
3.1 Alkaloids
Alkaloids have been used in botanical medicine for over >3500 years, with the Ebers papyrus (∼1500 BCE) mentioning the first recorded medical use of the opium poppy.24 They are a diverse class of natural products originally documented as basic, organic nitrogen-containing secondary metabolites produced by plants, microbes, and animals. The class is broadened to now include most nitrogen-containing natural products of low molecular weight or their derivatives.25 The most prevalent class are indole alkaloids, with other classes including, tropane, quinoline, isoquinoline, pyridine, pyrrolidine, pyrrolizidine, and steroidal alkaloids. Many plant alkaloids and their derivatives are still prescribed today for a variety of medical reasons such as severe pain (oxycodone, morphine, fentanyl), as antimalarials (quinine), to treat hypotension (ephedrine), or used recreationally (caffeine, nicotine, cocaine). Medicinal chemists often explore the inclusion of nitrogen into the parent compound due to its remarkable ability to improve the pharmacological profile of new drug derivatives.26 Several plant natural product alkaloids, or their derivatives, with antibacterial activity are described below.Mgbeahuruike et al. found 1 and 2 to have antibacterial activity against Staphylococcus aureus and Pseudomonas aeruginosa.30 Philipova et al. synthesized a series of piperine derivatives with different cyclic amines.31 These piperine derivatives 1a–1e were found to have potent anti-bacterial activity against Mycobacterium tuberculosis with MICs between 0.15–0.36 μM. 1e was the most promising compound with an MIC of 0.18 μM and cytotoxicity IC50 of 249.8 μM (vs. HEK293) and a selectivity index of >1300. The authors also conducted quantitative structure–activity relationship studies and found that quaternary carbons were necessary for anti-tuberculosis activity.
While the mechanism of action (MOA) for these compounds has not been identified, it has been theorized that piperine's antibacterial activity occurs through an increase in mTORC1 signaling in the host. A study reported that mTORC signaling led to an increase in the bacterial phagocytic ability of peritoneal macrophages in mice and the subsequent increase in bacterial clearance (Fig. 1).32
3.2 Phenolics
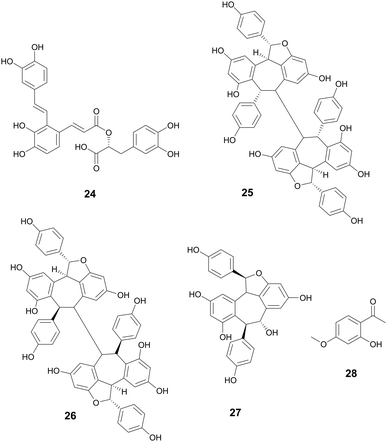
The oldest historical record of plant phenolic compounds used as medicine comes from the Ebers papyrus (∼1500 BCE), where ancient Sumerians and Egyptians recorded the uses of what is now known as willow for treating pain and fever.33 Thousands of years later, salicin and salicylic acid were isolated and later derivatized to acetylsalicylic acid. Better known by its generic name, Aspirin, it has since become the most used drug in the world,33 providing one of the ultimate examples of ethnobotanical records guiding the discovery of a new drug. There are over 8000 known plant phenolic compounds,34 making it the largest class of plant secondary metabolites.35 They are seemingly ubiquitously expressed in plants and play many roles, from defense and signaling to pigment biosynthesis and regulation growth.35 Phenolics are classified as having at least one hydroxyl group directly bonded to a mono or polycyclic aromatic hydrocarbon and are biosynthesized through shikimate or phenylpropanoid pathway.36 The phenolic compounds highlighted below are those that exhibit antibacterial properties.Phillygenin is a Phillyrin aglycone lignan.43 Li et al. reported phillygenin (5) isolated from Forsythia suspensa Vahl, Oleaceae, to prevent biofilm formation at 100–150 μg mL−1 in Helicobacter pylori. Additional studies on 5 show it can act as an antioxidant, protect high density lipoprotein and low density lipoprotein lipid peroxidation, and may also have anti-inflammatory activity.44–46 The fruit from F. suspensa has a rich history in Chinese traditional medicine, with uses including the treatment of gonorrhea, inflammation, cough, ulcers, and parasites (Fig. 2).47
Rhein (7), rhein-8-O-β-D-glucopyranoside (8), emodin (9), N′-((4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracen-2-yl)methylene) benzenesulfonohydrazide (an aloe emodin-conjugated sulfonyl hydrazone) (10), chrysophanol (11), and hypericin (12) are all examples of this diverse and rich class of biofilm inhibitors.
Anti-biofilm activity of anthraquinone rhein (7), which can be isolated from traditional medicinal plants Rheum palmatum L. (Polygonaceae), Senna tora (L.) Roxb. (Fabaceae), Reynoutria multiflora (Thunb.) Moldenke (Polygonaceae), and Aloe vera (L.) Burm.f. (Asphodelaceae), was discovered by Folliero et al.52 This study identified 7 to eradicate biofilm formation against Streptococcus mutans with an MBEC50 of 6.31 μg mL−1 and MBEC90 > 50 μg mL−1. At 6.25 μg mL−1, inhibition of biofilm formation and decreased thickness of the biofilm was observed. In addition to its antibiofilm activity, studies have shown that rhein also demonstrates growth inhibition of various bacteria strains.53–55 Rhein is also neuro and hepatoprotective, as well as anti-cancer and anti-inflammatory.56
A related compound, (8) was also reported to have anti-biofilm properties.57 Zhang et al. studied 8, which is isolated from the roots of Aucklandia costus Falc. (Asteraceae) and the roots of Rheum palmatum. A dose-dependent effect of 8 was seen against S. mutans, with 60% inhibition of biofilm formation at 10 μg mL−1 of 8 and complete inhibition at 50 μg mL−1. Zhang et al. also observed a decrease in the expression of the genes brpA and luxS, which engage in biofilm and may be one MOA through which 8 inhibits biofilm formation. Studies have reported that 8 inhibited the activity of PTP1B, prevents apoptosis, and promotes the purgative activity of sennoside A.58–60
Rhein and 8 are isolated primarily from Rheum palmatum, which has a rich history in traditional Chinese medicine.61 Traditional use includes it in preparations to treat fever, and eliminate blood stasis, burns, and cancer.61
Đukanović et al. demonstrated the antibiofilm of emodin (9) isolated from Frangula alnus Mill., Rhamnaceae bark activity against methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains.62 Significant biofilm formation inhibition was observed in both strains at concentrations as low as 0.05 μg mL−1 with the highest concentration tested at 3.125 μg mL−1. Emodin was also evaluated on mature biofilms to determine its potential to eradicate biofilm. Methicillin-resistant strains demonstrated 30% biofilm eradication at a concentration of 12.5 μg mL−1. Additionally, this study found some strains displayed a decrease in ica genes, which polysaccharide intercellular adhesin is dependent upon and plays a key role in biofilm formation.63 Emodin also demonstrates antioxidant, anti-inflammatory, antiviral, anti-diabetic, and anticancer.64F. alnus has traditional medicinal use in Europe with the bark used as a laxative or to induce vomiting.65
A similar compound derived from aloe emodin, aloe emodin-conjugated sulfonyl hydrazone 5a (10), reportedly demonstrates anti-biofilm properties as well.66 Aloe emodin is a key component in much of Chinese traditional medicine and can be derived from Rheum palmatum and Senna obtusifolia (L.) H.S.Irwin & Barneby, Fabaceae (cassia seed). Significant biofilm reduction (40%) by this compound was observed at 4 μg mL−1 in methicillin-resistant S. aureus. In addition to biofilm inhibition, the compound also demonstrated dispersion of the biofilm into the planktonic culture which led to decreased resistance. The method of action in which this specific emodin derivative acts as a biofilm inhibitor remains to be explored.
Another phenolic compound found within Rheum palmatum that also demonstrates antibiofilm activity is chrysophanol (11).67 This study reports inhibition of biofilm formation (around 60%) in Staphylococcus suis at concentrations as low as 0.112 μg mL−1. SEM results demonstrated a single molecule state of the bacteria cells with a lack of interconnection to create a biofilm, however, the exact mechanism of antibiofilm action needs to be further explored. Studies report chrysophanol also has uses in treating cardiovascular disease, treating cancer, providing neuroprotection, and preventing inflammation.68
Hypericin (12) derived from Hypericum perforatum L., Hypericaceae was reported by Wang et al. to inhibit biofilm formation (∼60%) in S. aureus at 8 μg mL−1.69 This study reports that hypericin decreases gene expression of sarA, a key gene for biofilm formation, but still requires further mechanism of action studies. Additional studies report hypericin has anticancer activity and antiviral activity.70,71Hypericum perforatum has a history dating back the ancient Greek physicians for wound healing, promoting diuresis, and eliminating parasites.72 As its use spread throughout Europe in the 1500s, it was also used to treat anxiety and depression (Fig. 3).72
Myrsininone A (17) and 18 are also both isoflavones, isolated from the fruits of Ficus auriculata Lour. from the Moraceae family. Myrsininone A has been reported to have anti-diabetic activity and has been found to inhibit α-glucosidase.83 Both 17 and 18 displayed potent activity against various bacterial strains with MICs between 1.25–10 μM and 10–20 μM, respectively.84F. auriculata is native to Asia and the Himalayan regions. Leaves of this plant have been used as a paste for the treatment of wounds and the fruits have been used for hepatic and neurodegenerative diseases.85
While the mechanism of action for the antibacterial activity of many flavonoids remains unknown and many targets and MOAs have been proposed, it has been postulated that they may act through nonspecific action upon the bacterial cell membrane. Flavonoids have been found to permeate the cell membrane,86 disrupt cellular membrane integrity,87 and a relationship has been shown between their lipophilicity and antibacterial activity (Fig. 4).88
Turmeric has been used in Vedic culture for around 4000 years as a culinary spice, for cosmetics, and as medicine.104 Ayurvedic medicinal practices use turmeric to treat respiratory conditions, anorexia, rheumatism, runny nose, cough, sprains, and swelling.104,105 It has also been used to as an antibacterial agent for the treatment of burns and cuts in many South Asian countries (Fig. 5).104
Hopeaphenol, isohopephenol, and ampelopsin A (25–27) are oligomeric stilbenoids isolated from the roots of Vitis vinifera L., Vitaceae. Their biosynthesis is derived from the oligomerization of resveratrol, of which dimers through octamers have been found.110,111 Besides V. vinifera, resveratrol oligomers have been isolated from various other Vitis species and plant families such as Cyperaceae, Dipterocarpaceae, Fabiaceae, Gnetaceae, and Paeoniaceae.112–114 Resveratrol oligomers have been found to interfere with the type III secretion system (T3SS) of various human and plant pathogens.115,116 The T3SS system is a needle-like structure in Gram-negative bacteria that allows the bacteria to penetrate host cell membranes and transfer various substrates.18 In Pseudomonas syringae pv. Tomato DC3000, these oligomers exhibit antivirulence through the decreased expression of various hrp gene clusters of the T3SS including hrpA, hrpL, and hop1,116 while resveratrol itself had no effect. Resveratrol can be produced at increasing amounts at sites of plant infection, allowing also for the biosynthesis of oligomers with more potent activity.110 Various resveratrol oligomers have been shown to have antibacterial, antioxidant, anti-neurodegenerative, anti-tumor, as well as heart and liver protective effects.111 Many plant foods and medicines contain resveratrol117 and various oligomers, with TCM preparations displaying numerous decoctions and granules in which resveratrol has been found.118 These traditional preparations focus on treating lung diseases such as pneumonia, chronic obstructive pulmonary disease, asthma, and idiopathic pulmonary fibrosis and asthma.
3.3 Terpenoids
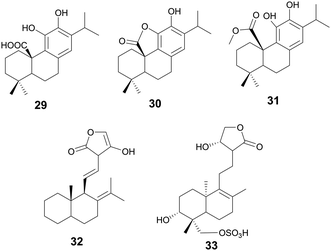
Terpenoids, a class of organic compounds comprised of an isoprene unit, have been used in medicine for thousands of years. Roughly 2000 years ago, the use of Artemisia annua L. for fever reduction was recorded in a text called “52 Prescriptions”, recovered from the Mawangdui Han dynasty tomb.124 More than a thousand years later, the active compound Artemisinin (a sesquiterpene lactone) was discovered and in 2015, Tu Youyou was awarded the Nobel Prize for her contributions. Terpenoids play broad roles in plant growth and development and plant odors. Another example of is that of paclitaxel, a tetracyclic diterpenoid isolated from the Pacific Yew tree used successfully in the clinic as a chemotherapeutic. Outlined below are terpenoids found within the search criteria as possessing various antibacterial properties.Compound 32 is a diterpene lactone andrographolide isolated from Helichrysum caespititium (DC.) Sond., Asteraceae, with antibiofilm properties against Neisseria gonorrhoeae.131 When tested at 60 μg mL−1, 32 inhibited biofilm cell attachment by 81%, but only inhibited biofilm formation by 32%. The potential mechanism of action may be a disruption of the EPS cohesiveness. H. caespititium has many known uses in traditional medicine across South and Central Africa, individually against respiratory infections, and as a component of traditional medicinal mixtures for various other diseases.132
Andrographolide sulfonate (33) isolated from Andrographis paniculata (Burm.f.) Wall., Acanthaceae, demonstrates antibiofilm activity against MRSA at a test concentration of 25 mg mL−1.133 This concentration is considered very high, but the minimum inhibitory concentration of bacterial growth for this compound is 50 μg mL−1. This highlights the potential for derivatives to work as antibiofilm inhibitors. Additionally, this study observed downregulation of genes associated with intracellular adhesion (icaA, icaD, and PIA), key to biofilm formation, in the presence of the compound. Andrographolide sulfonates are part of injections based on TCM and are currently approved in China to treat respiratory infections, tonsillitis, and numerous other infections.134A. paniculata has uses in traditional Chinese medicine to treat snake bites, sore throats, and to treat colds and fevers (Fig. 7).135
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 natural triterpenes known.136 An annual review by Connolly and Hill catalogues the latest triterpenes isolated from nature.137 Pentacyclic triterpenes are a class of triterpenes characterized by their 5-membered rings and include the compounds ursane, lupane, and oleanane.138
000 natural triterpenes known.136 An annual review by Connolly and Hill catalogues the latest triterpenes isolated from nature.137 Pentacyclic triterpenes are a class of triterpenes characterized by their 5-membered rings and include the compounds ursane, lupane, and oleanane.138
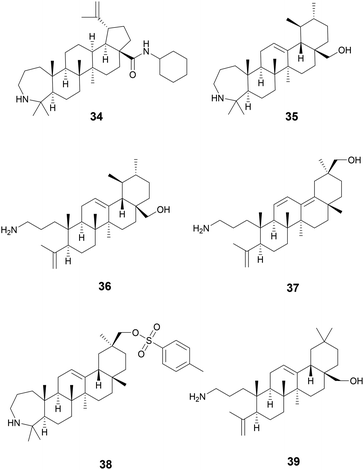
Starting with various pentacyclic triterpene skeletons, Kazakova et al. (2021) synthesized multiple pentacyclic triterpenoid derivatives (34–38) and studied their efficacy against the ESKAPE pathogens.139 These derivatives were found to have MICs of 0.15 μM against S. aureus. The MOA for growth inhibition by these pentacyclic triterpenoid derivatives is currently unknown. But, pentacyclic triterpenes have been reported to disrupt the bacterial cell membrane and inhibit NADH oxidation in S. aureus.140
In a separate study by Kazakova et al. (2021), the authors tested the antibacterial activity of different oleanolic acid derivatives.14139 is a derivative of oleanolic acid, a compound found in over 1600 plant species and found in many members of the Oleaceae family.14239 was found to have anti-MRSA activity and also displayed anti-chlamydial activity. The MOA for 39 was not reported, but a previous study has reported that oleanolic acid and related derivatives have been found to deregulate peptidoglycan metabolism in S. aureus (Fig. 8).143
Betulin (40) and celastrol (41) are two triterpenoids also possessing antivirulence activity. Betulin is a pentacyclic triterpenoid of lupane structure and contains a hydroxy and hydroxymethyl group. It can be found in a wide variety of Betula species (Betulaceae family) and is commonly isolated form birch bark which can contain around 20–30% betulin by dry weight.144 Many pharmacopoeias around the world have traditional medicinal application of plants of Betula species.145 In North America, gray birch bark (Betula populifolia Marshall, Betulaceae) was used by the Maliseet and Mi'kmaq to treat infected wounds.146
Experiments using Listeria monocytogenes show that 40 was able to inhibit listeriolysin O (LLO) hemolysis likely by disrupting LLO oligomerization in concentrations as low as 4.5 μM. Additionally, bacterial burden in livers and kidneys of mice infected with L. monocytogenes was significantly reduced when administered at 50 mg kg−1.147
Celestrol, also known as tripterine, (41) is a pentacyclic quinone methide triterpene and is present in high abundances in the roots of Tripterygium wilfordii Hook.f., Celastraceae.148T. wilfordii has been used for more than two thousand years in TCM149 to treat various inflammatory disorders namely rheumatoid arthritis150 and has also been shown to possess anticancer properties.151 In S. aureus, celastrol inhibits a virulence factor called carotenoid staphyloxanthin (STX) which is essential for S. aureus to disrupt reactive oxygen species produced by host macrophages and neutrophils, thus reducing bacterial clearance by the innate immune system.152
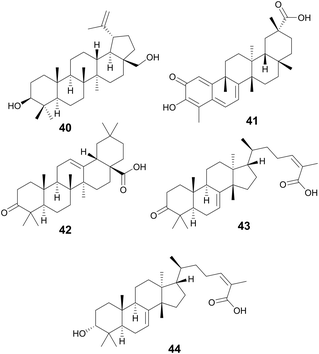
Compounds 42–44 are triterpenoid acids, classified by having 30 carbons forming 6 isoprene units in their skeleton. Triterpenoids can be linear or cyclic, with 43 and 44 showing tetracyclic tirucallane-like structure and 42 with a pentacyclic oleanolic acid-like structure. Isolated from the fruits of Schinus terebinthifolia Raddi, Anacardiaceae, these compounds exhibit antivirulence in MRSA through quorum sensing inhibition of the accessory gene regulator (agr) system and broader antivirulence effects through targeting the sae-two component system.153 The agr signaling system has been found to be crucial for MRSA skin infection and leads to the production of toxins and exoenzymes.154 Compounds 43 and 44 exhibited the best activity with IC50 of the agr system ranging between 2–9 μM, decreasing δ-toxin, while exhibiting little to no growth inhibition at these concentrations. A traditional medicine in Brazil, its indigenous habitat,155 various parts of the plant have been used to treat wounds and ulcers of the skin, tumor, arthritis, infections of the urinary and respiratory tracts, and contusions (Fig. 9).155,156
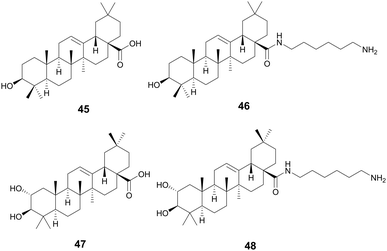
Oleanolic (45) and maslinic acid (47) were isolated from the waste of olive oil, Olea europaea L., Oleaceae, and two promising C-28 amide derivatives were synthesized: oleanolic acid-HDA (46) and maslinic acid-HDA (48).157 These derivatives from oleanolic acid (45) and maslinic acid (47) demonstrated potential biofilm inhibition properties. At 50 μg mL−1 oleanolic acid (45) did not demonstrate notable destructive activity, but its derivatives 46–48 were able to remove 99% of the pre-existing biofilm grown on catheters. 46 and 48 were also tested against a continuous biofilm after 4 days, and after 24 hours demonstrated a decrease in biofilm (30% for 46 and 45% for 48) and a decrease in the thickness (∼10 μm). These derivatives potentially penetrate the bacterial cellular membranes causing damage, indicating a potential MOA. Oleanolic acid (45) is also reported to have anti-cancer anti-diabetic, hepatoprotective, anti-parasitic, anti-bacterial, and antioxidant activities.158 Maslinic acid (47) also has a range of additional activities including cardio protection, anticancer, antiparasitic, and anti-inflammatory.159O. europaea can be found in medicinal practices throughout the globe including Japan, the United States, Brazil, and the Mediterranean as a laxative, anti-asthmatic, antibacterial, and anti-inflammatory (Fig. 10).160
4. Conclusion and perspectives
Antibiotic resistance (AMR) continues to be a widespread worldwide issue. The recent manifestation of COVID-19 has exacerbated the rise of antibiotic resistance as more individuals are exposed to nosocomial MDR bacterial infections and an increase in patients on antibiotics that developed pneumonia as a co-infection. Some predict that the pace of antibiotic development will not be able to keep up with the pace of bacterial resistance, leading to an antibiotic crisis and potentially the next pandemic.169 Natural products are an incredible tool in urgent drug discovery efforts and are the basis of the field of antimicrobial development. Alexander Fleming accidently discovered one of the most famous natural products, penicillin, in the early days of anti-infective research. Selman Waksman, who coined the term “antibiotic”, took an interest in the mechanisms behind penicillin's activity, the ability of microorganisms to produce antimicrobial agents, and developed the first screening protocols that led to the isolation of 15 antibiotics. Since the 1940's, the advancement of protocols and technologies available to mine, extract, produce, characterize, and manipulate natural products should have led to additional novel antibiotic development, but there is a stall in novel antibiotic classes brought to the market for clinical use. Furthering the field of natural product research has great potential to fill this gap in medicine.In this review, we highlight research results from 2018–2022 on plant-derived natural products that demonstrate anti-infective characteristics against pathogenic bacteria, including growth inhibition, antivirulence and antibiofilm activity. Although various structures and natural sources with anti-infective activity were reported, under the inclusion criteria, only three main classes of compounds were discussed in this review, in which a large diversity of phenolic derivatives was observed. Alkaloids, phenolic derivatives, and terpenoids are extremely diverse chemical classes and are among the most abundant types of compounds found in plants. Some novelty arose from the synthesis of derivatives, re-emphasizing complementarity of medicinal chemistry in natural products drug discovery. A significant portion of the discussed compounds were of known structure, with novel biological activities as antibacterial agents.
5. Potential clinical relevance of the reported antimicrobial compounds
Natural product research involves many difficulties in the process from plant collection to isolation and structural identification. In particular, appropriate reporting criteria for plant materials must include adequate documentation of the plant collection (e.g., herbarium vouchers), species authentication, sample preparation, and validation of extraction methodology. Articles in which plant material were not validated or properly reported were not included in this review work. In addition, inconsistencies in the reporting of experimental results creates an issue for data comparability. For experimental design and data interpretation, publications such as the CLSI (Clinical and Laboratory Standards Institute) are an excellent resource for many antibacterial studies but are limited in their scope for biofilm and antivirulence assays. An additional disadvantage is the inconsistent definition of relevant concentrations for all studies (i.e., studies reporting novel compounds with activity in the μg mL−1 range vs. mg mL−1). Compounds that require significant starting material may not be great candidates for drug discovery efforts alone but can provide a basis for modification through medicinal chemistry. Further additions and definitions in the field can ensure uniform research and reproducibility, as well as ensuring the reported compounds have the potential to be potent preclinical drug candidates.Our search criteria were stringent, thus leaving out additional natural products that may have potential in the clinic. An approach not included under the criteria of this review were natural products aided by the delivery of nanoparticles whether in vivo or examining in vitro cell permeability. Many natural products lack the physiological and chemical properties needed to make a drug with good bioavailability and pharmacokinetic properties. Thus, methods of better delivery could provide an avenue in which natural products with promising in vitro biological activity could play a role in antibacterial drug development.
Notably, as of May 2023, there are currently no FDA approved antibacterial drugs that are botanical natural product. Preclinical animal studies for the compounds listed in this review are rare. The following preclinical animal studies were found for piperine and curcumin,170 and for celastrol.171 All other compounds listed in this review had no registered preclinical animal studies at the registry databases we searched (https://www.animalstudyregistry.org, https://preclinicaltrials.eu/). We found more success searching for these compounds at the NIH's clinical trial database (https://clinicaltrials.gov/). The compounds found (1, 3, 17, 22, 23, 27, 28, 29, 33, 39, 41, 42, 44, 45 and 47) with active/completed clinical trials, were all unrelated to antimicrobial studies. But, outside of well-studied commonly described natural products (piperine, curcumin, hypericin, salvianolic acid A) there were very few (≤5 clinical trials) for these compounds. Bioavailability and/or pharmacokinetic clinical trials were completed or being conducted for six compounds (1, 17, 23, 29, 41 and 42). The trials examining the pharmacokinetic properties of these compounds are a promising first step. Characterizing the safety profiles of these compounds in human subjects may facilitate future studies examining the antimicrobial properties of these botanical natural products.
While a few studies in this review merely acknowledge the traditional use of these plants in their discussion, over half (28/48) of the reported compounds have a history of traditional uses (compounds 6–8, 17–23, 27, 30–37, 39, 40, 42–48). While not always explicitly stated, many studies on plant-derived bioactive natural products are based on plants with strong documentation for traditional medicinal applications in ethnobotanical literature. Herein, many of the studies reviewed sought out unidentified or derivative compounds (18, 19, 39, 46, 48) from traditional medicinal plants, or to further explore the bioactivity of previously identified bioactive compounds (27, 33, 34, 36, 39, 42–48). The studies of natural products and their derivatives with bioactivity can contribute to the search for new drug candidates.
Plant-derived natural products can inspire the development of the next generation of antimicrobials. In addition, ethnobotanical knowledge concerning medicinal plant uses can be a valuable starting point for researchers. In the future, active collaboration is needed not only by natural products and microbiology researchers but also by scientists from the fields of medicinal chemistry and ethnobotany to discover and develop the next generation of anti-infective agents.
6. Author contributions
All authors participated in drafting and editing the manuscript.7. Conflicts of interest
CLQ is a founder and CEO/CSO of PhytoTEK LLC, a company developing plant-derived antibiofilm compounds. CLQ is a founder and CSO of Verdant Scientific LLC, a company developing plant-derived antivirulence compounds. CLQ is an Editor for the Natural Product Reports Special Issue “Botanical Natural Products in Drug Discovery, Past, Present and Future”.8. Acknowledgements
This work was supported by the National Institutes of Health, National Center for Complementary and Integrative Health (R01AT011990 to CLQ; F31AT011988 to CJR; F31AT012140 to WC) and the Jones Center at Ichauway (to LM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.9. References
- WHO, Global Action Plan on Antimicrobial Resistance, World Heatlh Organization, 2015 Search PubMed.
- CDC, Journal, 2019 Search PubMed.
- J. O'Neill, Tackling Drug-Resistant Infections Globally: Final Report and Recommendations, 2016 Search PubMed.
- WHO, Antimicrobial resistance surveillance in Europe 2022 – 2020 data, World Heatlh Organization, 2022 Search PubMed.
- Antimicrobial Resistance Collaborators, Lancet, 2022, 399, 629–655 CrossRef PubMed.
- CDC, COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022, Centers for Disease Control and Prevention, 2022 Search PubMed.
- L. B. Rice, J. Infect. Dis., 2008, 197, 1079–1081 CrossRef PubMed.
- WHO, Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis, World Health Organization, 2017 Search PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- G. Porras, F. Chassagne, J. T. Lyles, L. Marquez, M. Dettweiler, A. M. Salam, T. Samarakoon, S. Shabih, D. R. Farrokhi and C. L. Quave, Chem. Rev., 2021, 121, 3495–3560 CrossRef CAS PubMed.
- C. M. J. Wesseling and N. I. Martin, ACS Infect. Dis., 2022, 8, 1731–1757 CrossRef CAS PubMed.
- R. P. Singh, MedChemComm, 2015, 6, 259–272 RSC.
- D. M. P. De Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Clin. Microbiol. Rev., 2020, 33(3), e00181 CrossRef CAS PubMed.
- R. E. Schultes and S. von Reis, Ethnobotany: Evolution of a Discipline, Timber Press, 2008 Search PubMed.
- R. J. Melander, A. K. Basak and C. Melander, Nat. Prod. Rep., 2020, 37, 1454–1477 RSC.
- W. Levinson, P. Chin-Hong, E. A. Joyce, J. Nussbaum and B. Schwartz, in Review of Medical Microbiology & Immunology: A Guide to Clinical Infectious Diseases, McGraw Hill, New York, NY, vol. 16e, 2020 Search PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef PubMed.
- S. W. Dickey, G. Y. C. Cheung and M. Otto, Nat. Rev. Drug Discovery, 2017, 16, 457–471 CrossRef CAS PubMed.
- A. Casadevall and L. A. Pirofski, Infect. Immun., 1999, 67, 3703–3713 CrossRef CAS PubMed.
- R. M. Donlan, Emerging Infect. Dis., 2002, 8, 881–890 CrossRef PubMed.
- M. H. Muhammad, A. L. Idris, X. Fan, Y. Guo, Y. Yu, X. Jin, J. Qiu, X. Guan and T. Huang, Front. Microbiol., 2020, 11, 928 CrossRef PubMed.
- G. A. O’Toole, J. Visualized Exp., 2011, 47, e2437 Search PubMed.
- J. Azeredo, N. F. Azevedo, R. Briandet, N. Cerca, T. Coenye, A. R. Costa, M. Desvaux, G. Di Bonaventura, M. Hébraud, Z. Jaglic, M. Kačániová, S. Knøchel, A. Lourenço, F. Mergulhão, R. L. Meyer, G. Nychas, M. Simões, O. Tresse and C. Sternberg, Crit. Rev. Microbiol., 2017, 43, 313–351 CrossRef CAS PubMed.
- N. H. Aboelsoud, J. Med. Plants Res., 2010, 4, 082–086 Search PubMed.
- M. Heinrich, J. Mah and V. Amirkia, Molecules, 2021, 26, 1836 CrossRef CAS PubMed.
- L. D. Pennington and D. T. Moustakas, J. Med. Chem., 2017, 60, 3552–3579 CrossRef CAS PubMed.
- P. Biswas, U. Anand, S. C. Saha, N. Kant, T. Mishra, H. Masih, A. Bar, D. K. Pandey, N. K. Jha, M. Majumder, N. Das, V. S. Gadekar, M. S. Shekhawat, M. Kumar, Radha, J. Proćków, J. M. P. d. l. Lastra and A. Dey, J. Cell. Mol. Med., 2022, 26, 3083–3119 CrossRef PubMed.
- A. A. Durant-Archibold, A. I. Santana and M. P. Gupta, J. Ethnopharmacol., 2018, 217, 63–82 CrossRef PubMed.
- I.-U. Haq, M. Imran, M. Nadeem, T. Tufail, T. A. Gondal and M. S. Mubarak, Phytother. Res., 2021, 35, 680–700 CrossRef CAS PubMed.
- E. E. Mgbeahuruike, M. Stålnacke, H. Vuorela and Y. Holm, Antibiotics, 2019, 8, 55 CrossRef CAS PubMed.
- I. Philipova, V. Valcheva, R. Mihaylova, M. Mateeva, I. Doytchinova and G. Stavrakov, Chem. Biol. Drug Des., 2018, 91, 763–768 CrossRef CAS PubMed.
- H. Pan, L. H. Xu, M. Y. Huang, Q. B. Zha, G. X. Zhao, X. F. Hou, Z. J. Shi, Q. R. Lin, D. Y. Ouyang and X. H. He, Oncotarget, 2015, 6, 32468–32483 CrossRef PubMed.
- M. J. R. Desborough and D. M. Keeling, Br. J. Haematol., 2017, 177, 674–683 CrossRef PubMed.
- Y. Zhang, P. Cai, G. Cheng and Y. Zhang, Nat. Prod. Commun., 2022, 17, 1934578X211069721 CAS.
- J. Dai and R. J. Mumper, Molecules, 2010, 15, 7313–7352 CrossRef CAS PubMed.
- A. Sharma, B. Shahzad, A. Rehman, R. Bhardwaj, M. Landi and B. Zheng, Molecules, 2019, 24, 2452 CrossRef CAS PubMed.
- S.-J. Wan, H.-G. Ren, J.-M. Jiang, G. Xu, Y. Xu, S.-M. Chen, G. Chen, D. Zheng, M. Yuan, H. Zhang and H.-X. Xu, Front. Chem., 2022, 10, 877469 CrossRef CAS PubMed.
- R. B. Teponno, S. Kusari and M. Spiteller, Nat. Prod. Rep., 2016, 33, 1044–1092 RSC.
- H. A. Abbas, H. Atallah, M. A. El-Sayed and A. M. El-Ganiny, Arch. Microbiol., 2020, 202, 2751–2760 CrossRef CAS PubMed.
- Z. Chen, C. Zhang, F. Gao, Q. Fu, C. Fu, Y. He and J. Zhang, Food Chem. Toxicol., 2018, 119, 309–325 CrossRef CAS PubMed.
- Z. Zhao, P. Guo and E. Brand, Chin. Med., 2018, 13, 18 CrossRef PubMed.
- X. Zhang, B. Han, Z. Feng, J. Jiang, Y. Yang and P. Zhang, Acta Pharm. Sin. B, 2018, 8, 818–824 CrossRef PubMed.
- W. Liu, G. Chu, N. Chang, X. Ma, M. Jiang and G. Bai, RSC Adv., 2017, 7, 40418–40426 RSC.
- C. C. Chen, H. Y. Chen, M. S. Shiao, Y. L. Lin, Y. H. Kuo and J. C. Ou, Planta Med., 1999, 65, 709–711 CrossRef CAS PubMed.
- M.-J. Chang, T. M. Hung, B.-S. Min, J.-C. Kim, M. H. Woo, J. S. Choi, H. K. Lee and K. Bae, Biosci., Biotechnol., Biochem., 2008, 72, 2750–2755 CrossRef CAS PubMed.
- H. Lim, J. G. Lee, S. H. Lee, Y. S. Kim and H. P. Kim, J. Ethnopharmacol., 2008, 118, 113–117 CrossRef CAS PubMed.
- Z. Wang, Q. Xia, X. Liu, W. Liu, W. Huang, X. Mei, J. Luo, M. Shan, R. Lin, D. Zou and Z. Ma, J. Ethnopharmacol., 2018, 210, 318–339 CrossRef CAS PubMed.
- E. M. Malik and C. E. Müller, Med. Res. Rev., 2016, 36, 705–748 CrossRef CAS PubMed.
- A. F. Izyani Awang, Q. U. Ahmed, S. A. A. Shah, J. M. Jaffri, K. Ghafoor, A. B. M. H. Uddin, S. Ferdosh and M. Z. Islam Sarker, Nat. Prod. Res., 2020, 34, 629–637 CrossRef CAS PubMed.
- U.S. Department of Agriculture Agricultural Research Service, Dr Duke's Phytochemical and Ethnobotanical Databases, https://phytochem.nal.usda.gov/) Search PubMed.
- M. Y. Fosso, K. Y. Chan, R. Gregory and C.-W. T. Chang, ACS Comb. Sci., 2012, 14, 231–235 CrossRef CAS PubMed.
- V. Folliero, F. Dell'Annunziata, E. Roscetto, A. Amato, R. Gasparro, C. Zannella, V. Casolaro, A. De Filippis, M. R. Catania, G. Franci and M. Galdiero, Microbiol. Res., 2022, 261, 127062 CrossRef CAS PubMed.
- A. T. Nguyen and K.-y. Kim, J. Med. Microbiol., 2020, 69, 689–696 CrossRef CAS PubMed.
- L. K. Müller-Heupt, N. Vierengel, J. Groß, T. Opatz, J. Deschner and F. D. von Loewenich, Antibiotics, 2022, 11, 186 CrossRef PubMed.
- Q. Li, Y. Huang, X. Liu, J. Gan, H. Chen and C. G. Yang, J. Biol. Chem., 2016, 291, 11083–11093 CrossRef CAS PubMed.
- Y.-X. Zhou, W. Xia, W. Yue, C. Peng, K. Rahman and H. Zhang, J. Evidence-Based Complementary Altern. Med., 2015, 2015, 578107 Search PubMed.
- Y. Zhang, Y. Zhu, Y. Zuo, C. Tang, F. Zhou, X. Cui and L. Wang, Curr. Microbiol., 2021, 78, 323–328 CrossRef CAS PubMed.
- S. Li, T. Y. An, J. Li, Q. Shen, F. C. Lou and L. H. Hu, J. Asian Nat. Prod. Res., 2006, 8, 281–286 CrossRef CAS PubMed.
- L. S. Zhang, J. Li and L. Jia-Ping, Exp. Ther. Med., 2020, 19, 2871–2878 CAS.
- K. Takayama, H. Tsutsumi, T. Ishizu and N. Okamura, Biol. Pharm. Bull., 2012, 35, 2204–2208 CrossRef CAS PubMed.
- P. Xiao, L. He and L. Wang, J. Ethnopharmacol., 1984, 10, 275–293 CrossRef PubMed.
- S. Đukanović, T. Ganić, B. Lončarević, S. Cvetković, B. Nikolić, D. Tenji, D. Randjelović and D. Mitić-Ćulafić, J. Appl. Microbiol., 2022, 132, 1840–1855 CrossRef PubMed.
- K. Schilcher and A. R. Horswill, Microbiol. Mol. Biol. Rev., 2020, 84(3), e00026 CrossRef PubMed.
- J. Sharifi-Rad, J. Herrera-Bravo, S. Kamiloglu, K. Petroni, A. P. Mishra, M. Monserrat-Mesquida, A. Sureda, M. Martorell, D. S. Aidarbekovna, Z. Yessimsiitova, A. Ydyrys, C. Hano, D. Calina and W. C. Cho, Biomed. Pharmacother., 2022, 154, 113555 CrossRef CAS PubMed.
- C. Earwood, Folk Life, 1999, 38, 22–31 CrossRef.
- Z. Deng, R. R. Y. Bheemanaboina, Y. Luo and C. H. Zhou, Bioorg. Chem., 2022, 127, 106035 CrossRef CAS PubMed.
- J.-W. Bai, X.-R. Chen, Y. Tang, W.-Q. Cui, D.-L. Li, B.-O. God'spower and Y. Yang, RSC Adv., 2019, 9, 28996–29004 RSC.
- S. Su, J. Wu, Y. Gao, Y. Luo, D. Yang and P. Wang, Biomed. Pharmacother., 2020, 125, 110002 CrossRef CAS PubMed.
- G. Wang, L. Li, X. Wang, X. Li, Y. Zhang, J. Yu, J. Jiang, X. You and Y. Q. Xiong, Acta Pharm. Sin. B, 2019, 9, 1174–1182 CrossRef PubMed.
- N. Choudhary, T. E. Collignon, D. Tewari and A. Bishayee, Phytomedicine, 2022, 105, 154356 CrossRef CAS PubMed.
- F. F. Mohamed, D. Anhlan, M. Schöfbänker, A. Schreiber, N. Classen, A. Hensel, G. Hempel, W. Scholz, J. Kühn, E. R. Hrincius and S. Ludwig, Pharmaceuticals, 2022, 15, 530 CrossRef CAS PubMed.
- K. M. Klemow, A. Bartlow, J. Crawford, N. Kocher, J. Shah and M. Ritsick, in Herbal Medicine: Biomolecular and Clinical Aspects, ed. I. F. F. Benzie and S. Wachtel-Galor, CRC Press/Taylor & Francis, Boca Raton (FL), 2011 Search PubMed.
- T. K. Swetha, M. Pooranachithra, G. A. Subramenium, V. Divya, K. Balamurugan and S. K. Pandian, Front. Cell. Infect. Microbiol., 2019, 9, 357 CrossRef CAS PubMed.
- A. Amin, M. Hanif, K. Abbas, M. Ramzan, A. Rasheed, A. Zaman and L. Pieters, Microb. Pathog., 2020, 144, 104184 CrossRef CAS PubMed.
- O. Mazimba, Bull. Fac. Pharm., 2017, 55, 223–232 Search PubMed.
- M. S. Amiri and M. R. Joharchi, Avicenna J. Phytomed., 2016, 6, 621–635 Search PubMed.
- P. Mahendra and S. Bisht, Pharmacogn. Rev., 2012, 6, 141–146 CrossRef PubMed.
- M. C. Dias, D. C. G. A. Pinto and A. M. S. Silva, Molecules, 2021, 26, 5377 CrossRef CAS PubMed.
- M. J. Oza and Y. A. Kulkarni, Biomed. Pharmacother., 2018, 107, 1119–1127 CrossRef CAS PubMed.
- V. Desai, A. Jain, H. Shaghaghi, R. Summer, J. C. K. Lai and A. Bhushan, Anticancer Res., 2019, 39, 57 CrossRef CAS PubMed.
- N. D. Chaurasiya, V. Gogineni, K. M. Elokely, F. León, M. J. Núñez, M. L. Klein, L. A. Walker, S. J. Cutler and B. L. Tekwani, J. Nat. Prod., 2016, 79, 2538–2544 CrossRef CAS PubMed.
- R. Pachaiappan, T. P. Rajamuthu, A. Sarkar, P. Natrajan, N. Krishnan, M. Sakthivelu, P. Velusamy, P. Ramasamy and S. C. B. Gopinath, Process Biochem., 2022, 116, 84–93 CrossRef CAS.
- Z.-F. Shi, C. Lei, B.-W. Yu, H.-Y. Wang and A.-J. Hou, Chem. Biodiversity, 2016, 13, 445–450 CrossRef CAS PubMed.
- T.-M. Shao, H.-X. Liao, X.-B. Li, G.-Y. Chen, X.-P. Song and C.-R. Han, Nat. Prod. Res., 2022, 36, 1191–1196 CrossRef CAS PubMed.
- G. Tamta, N. Mehra and S. Tandon, J. Drug Delivery Ther., 2021, 11, 163–169 CrossRef CAS.
- G. Yuan, Y. Guan, H. Yi, S. Lai, Y. Sun and S. Cao, Sci. Rep., 2021, 11, 10471 CrossRef CAS PubMed.
- C. Babii, L. G. Bahrin, A.-N. Neagu, I. Gostin, M. Mihasan, L. M. Birsa and M. Stefan, J. Appl. Microbiol., 2016, 120, 630–637 CrossRef CAS PubMed.
- J. Echeverría, J. Opazo, L. Mendoza, A. Urzúa and M. Wilkens, Molecules, 2017, 22, 608 CrossRef PubMed.
- O. T. Schmidt and R. Lademann, Justus Liebigs Ann. Chem., 1951, 571, 232–237 CrossRef CAS.
- A. Olmedo-Juárez, T. I. Briones-Robles, A. Zaragoza-Bastida, A. Zamilpa, D. Ojeda-Ramírez, P. Mendoza de Gives, J. Olivares-Pérez and N. Rivero-Perez, Microb. Pathog., 2019, 136, 103660 CrossRef PubMed.
- X. Li, Y. Deng, Z. Zheng, W. Huang, L. Chen, Q. Tong and Y. Ming, Biomed. Pharmacother., 2018, 99, 43–50 CrossRef CAS PubMed.
- B. Sarin, N. Verma, J. P. Martín and A. Mohanty, Sci. World J., 2014, 2014, 839172 Search PubMed.
- Z. Feng, X. Lu, L. Gan, Q. Zhang and L. Lin, Molecules, 2020, 25, 598 CrossRef CAS PubMed.
- L. Felix, B. Mishra, R. Khader, N. Ganesan and E. Mylonakis, Front. Cell. Infect. Microbiol., 2022, 12, 898794 CrossRef CAS PubMed.
- V. K. Singh, M. Syring, A. Singh, K. Singhal, A. Dalecki and T. Johansson, Int. J. Med. Microbiol., 2012, 302, 242–252 CrossRef CAS PubMed.
- F. Gutierrez-Orozco, C. Chitchumroonchokchai, G. B. Lesinski, S. Suksamrarn and M. L. Failla, J. Agric. Food Chem., 2013, 61, 3891–3900 CrossRef CAS PubMed.
- P. Parekh, N. Sharma, M. Sharma, A. Gadepalli, A. A. Sayyed, S. Chatterjee, A. Kate and A. Khairnar, Metab. Brain Dis., 2022, 37, 2853–2870 CrossRef CAS PubMed.
- M. Tarasuk, P. Songprakhon, T. Chieochansin, K. Choomee, K. Na-Bangchang and P. T. Yenchitsomanus, Sci. Rep., 2022, 12, 16088 CrossRef CAS PubMed.
- J. Pedraza-Chaverri, N. Cárdenas-Rodríguez, M. Orozco-Ibarra and J. M. Pérez-Rojas, Food Chem. Toxicol., 2008, 46, 3227–3239 CrossRef CAS PubMed.
- J. Sharifi-Rad, Y. E. Rayess, A. A. Rizk, C. Sadaka, R. Zgheib, W. Zam, S. Sestito, S. Rapposelli, K. Neffe-Skocińska, D. Zielińska, B. Salehi, W. N. Setzer, N. S. Dosoky, Y. Taheri, M. El Beyrouthy, M. Martorell, E. A. Ostrander, H. A. R. Suleria, W. C. Cho, A. Maroyi and N. Martins, Front. Pharmacol., 2020, 11, 01021 CrossRef CAS PubMed.
- S. A. Noureddin, R. M. El-Shishtawy and K. O. Al-Footy, Eur. J. Med. Chem., 2019, 182, 111631 CrossRef CAS PubMed.
- L. Yang, H. Connaris, J. A. Potter and G. L. Taylor, BMC Struct. Biol., 2015, 15, 15 CrossRef PubMed.
- A. Vogel and J. Pelletier, J. Pharm., 1815, 1, 289–300 Search PubMed.
- A. B. Prasad S, in Turmeric, the Golden Spice: From Traditional Medicine to Modern Medicine, ed. W.-G. S. Benzie IFF, CRC Press/Taylor & Francis, Boca Raton (FL), 2nd edn, 2011, ch. 13 Search PubMed.
- C. C. Araújo and L. L. Leon, Mem. Inst. Oswaldo Cruz, 2001, 96, 723–728 CrossRef PubMed.
- L. Lian-Niang, T. Rui and C. Wei-Ming, Planta Med., 1984, 50, 227–228 CrossRef CAS PubMed.
- D. Mu, Y. Luan, L. Wang, Z. Gao, P. Yang, S. Jing, Y. Wang, H. Xiang, T. Wang and D. Wang, Emerging Microbes Infect., 2020, 9, 169–179 CrossRef CAS PubMed.
- L. Wang, R. Ma, C. Liu, H. Liu, R. Zhu, S. Guo, M. Tang, Y. Li, J. Niu, M. Fu, S. Gao and D. Zhang, Curr. Pharm. Des., 2017, 23, 1077–1097 CrossRef CAS PubMed.
- C.-Y. Su, Q.-L. Ming, K. Rahman, T. Han and L.-P. Qin, Chin. J. Nat. Med., 2015, 13, 163–182 CAS.
- M. H. Keylor, B. S. Matsuura and C. R. J. Stephenson, Chem. Rev., 2015, 115, 8976–9027 CrossRef CAS PubMed.
- J. Shen, Q. Zhou, P. Li, Z. Wang, S. Liu, C. He, C. Zhang and P. Xiao, Molecules, 2017, 22, 2050 CrossRef PubMed.
- J. E. Kang, N. Yoo, B. J. Jeon, B. S. Kim and E.-H. Chung, Front. Plant Sci., 2022, 13, 885625 CrossRef PubMed.
- J. Kawabata, E. Fukushi, M. Hara and J. Mizutani, Magn. Reson. Chem., 1992, 30, 6–10 CrossRef CAS.
- P. Coggon, T. King and S. Wallwork, Chem. Commun., 1966, 439–440 RSC.
- C. E. Zetterström, J. Hasselgren, O. Salin, R. A. Davis, R. J. Quinn, C. Sundin and M. Elofsson, PLoS One, 2013, 8, e81969 CrossRef PubMed.
- J. E. Kang, B. J. Jeon, M. Y. Park, H. J. Yang, J. Kwon, D. H. Lee and B. S. Kim, Pest Manage. Sci., 2020, 76, 2294–2303 CrossRef CAS PubMed.
- J. Burns, T. Yokota, H. Ashihara, M. E. J. Lean and A. Crozier, J. Agric. Food Chem., 2002, 50, 3337–3340 CrossRef CAS PubMed.
- B. N. Ma and X. J. Li, Chin. Herb. Med., 2020, 12, 349–358 CrossRef PubMed.
- H. Yang, R. Zhang, C. Jia, M. Chen, W. Yin, L. Wei and H. Jiao, Pharmaceut. Biol., 2019, 57, 22–28 Search PubMed.
- W.-C. Chuang, W.-C. Lin, S.-J. Sheu, S.-H. Chiou, H.-C. Chang and Y.-P. Chen, Planta Med., 1996, 62, 347–351 CrossRef CAS PubMed.
- S. Ducki, J. A. Hadfield, N. J. Lawrence, X. Zhang and A. T. McGown, Planta Med., 1995, 61, 586–587 CrossRef CAS PubMed.
- Q. Lv, S. Li, H. Wei, Z. Wen, Y. Wang, T. Tang, J. Wang, L. Xia and X. Deng, Appl. Microbiol. Biotechnol., 2020, 104, 1673–1682 CrossRef CAS PubMed.
- M. Zhao and S. P. Wu, S. Afr. J. Bot., 2019, 124, 556–563 CrossRef CAS.
- A. C. R. g. Qinghaosu, Chin. Med. J., 1979, 92, 811–816 Search PubMed.
- M. F. Chabán, C. Karagianni, M. B. Joray, D. Toumpa, C. Sola, M. I. Crespo, S. M. Palacios, C. M. Athanassopoulos and M. C. Carpinella, J. Ethnopharmacol., 2019, 239, 111930 CrossRef PubMed.
- M.-Y. Park and M.-K. Sung, J. Sci. Food Agric., 2015, 95, 828–835 CrossRef CAS PubMed.
- M. A. González, Nat. Prod. Rep., 2015, 32, 684–704 RSC.
- S. Manner, M. Vahermo, M. E. Skogman, S. Krogerus, P. M. Vuorela, J. Yli-Kauhaluoma, A. Fallarero and V. M. Moreira, Eur. J. Med. Chem., 2015, 102, 68–79 CrossRef CAS PubMed.
- F. J. Aranda and J. VillalaıÍn, Biochim. Biophys. Acta Biomembr., 1997, 1327, 171–180 CrossRef CAS PubMed.
- R. W. Bussmann and D. Sharon, J. Ethnobiol. Ethnomed., 2006, 2, 47 CrossRef PubMed.
- K. Bassey, P. Mamabolo and S. Cosa, Biology, 2021, 10, 1224 CrossRef CAS PubMed.
- A. Maroyi, J. Appl. Pharm. Sci., 2020, 10, 142–150 CrossRef CAS.
- L. Zhang, B. Wen, M. Bao, Y. Cheng, T. Mahmood, W. Yang, Q. Chen, L. Lv, L. Li, J. Yi, N. Xie, C. Lu and Y. Tan, Front. Pharmacol., 2021, 12, 720685 CrossRef CAS PubMed.
- Y. Zhao, P. Huang, Z. Chen, S. W. Zheng, J. Y. Yu and C. Shi, J. Huazhong Univ. Sci. Technol., Med. Sci., 2017, 37, 293–299 CrossRef CAS PubMed.
- B. Zeng, A. Wei, Q. Zhou, M. Yuan, K. Lei, Y. Liu, J. Song, L. Guo and Q. Ye, Phytother. Res., 2022, 36, 336–364 CrossRef CAS PubMed.
- H. Sheng and H. Sun, Nat. Prod. Rep., 2011, 28, 543–593 RSC.
- R. A. Hill and J. D. Connolly, Nat. Prod. Rep., 2020, 37, 962–998 RSC.
- R. Xu, G. C. Fazio and S. P. T. Matsuda, Phytochemistry, 2004, 65, 261–291 CrossRef CAS PubMed.
- O. Kazakova, E. Tret’yakova and D. Baev, J. Antibiot., 2021, 74, 559–573 CrossRef CAS PubMed.
- L. de León, M. R. López and L. Moujir, Microbiol. Res., 2010, 165, 617–626 CrossRef PubMed.
- O. Kazakova, L. Rubanik, I. Smirnova, N. Poleschuk, A. Petrova, Y. Kapustsina, I. Baikova, E. Tret’yakova and E. Khusnutdinova, Med. Chem. Res., 2021, 30, 1408–1418 CrossRef CAS.
- J. Pollier and A. Goossens, Phytochemistry, 2012, 77, 10–15 CrossRef CAS PubMed.
- L. Huang, H. Luo, Q. Li, D. Wang, J. Zhang, X. Hao and X. Yang, Eur. J. Med. Chem., 2015, 95, 64–75 CrossRef CAS PubMed.
- P. Šiman, A. Filipová, A. Tichá, M. Niang, A. Bezrouk and R. Havelek, PLoS One, 2016, 11, e0154933 CrossRef PubMed.
- S. Rastogi, M. M. Pandey and A. Kumar Singh Rawat, J. Ethnopharmacol., 2015, 159, 62–83 CrossRef CAS PubMed.
- D. E. Moerman, Native American Ethnobotany, Timber Press, Portland, OR, 1998 Search PubMed.
- G. Lu, L. Xu, P. Zhang, X. Dou, J. Yu and H. Feng, Fitoterapia, 2019, 139, 104409 CrossRef CAS PubMed.
- G. Sethi, K. S. Ahn, M. K. Pandey and B. B. Aggarwal, Blood, 2007, 109, 2727–2735 CrossRef CAS PubMed.
- L. Wang, Z. Wang, Z. Yang, K. Yang and H. Yang, Front. Mol. Biosci., 2021, 8, 664416 CrossRef CAS PubMed.
- J. Bao and S.-M. Dai, Rheumatol. Int., 2011, 31, 1123–1129 CrossRef PubMed.
- R. Kannaiyan, M. K. Shanmugam and G. Sethi, Cancer Lett., 2011, 303, 9–20 CrossRef CAS PubMed.
- F. A.-z. A. Yehia, N. Yousef and M. Askoura, BMC Microbiol., 2022, 22, 106 CrossRef CAS PubMed.
- H. Tang, G. Porras, M. M. Brown, F. Chassagne, J. T. Lyles, J. Bacsa, A. R. Horswill and C. L. Quave, Sci. Rep., 2020, 10, 8046 CrossRef CAS PubMed.
- G. Y. C. Cheung, R. Wang, B. A. Khan, D. E. Sturdevant and M. Otto, Infect. Immun., 2011, 79, 1927–1935 CrossRef CAS PubMed.
- J. F. Morton, Econ. Bot., 1978, 32, 353–359 CrossRef CAS.
- L. E. Fedel-Miyasato, A. S. Formagio, S. A. Auharek, C. A. Kassuya, S. D. Navarro, A. L. Cunha-Laura, A. C. Monreal, M. C. Vieira and R. J. Oliveira, Genet. Mol. Res., 2014, 13, 3411–3425 CrossRef CAS PubMed.
- N. Blanco-Cabra, K. Vega-Granados, L. Moya-Andérico, M. Vukomanovic, A. Parra, L. Álvarez de Cienfuegos and E. Torrents, ACS Infect. Dis., 2019, 5, 1581–1589 CrossRef CAS PubMed.
- T. B. Ayeleso, M. G. Matumba and E. Mukwevho, Molecules, 2017, 22, 1915 CrossRef PubMed.
- G. Lozano-Mena, M. Sánchez-González, M. E. Juan and J. M. Planas, Molecules, 2014, 19, 11538–11559 CrossRef PubMed.
- M. A. Hashmi, A. Khan, M. Hanif, U. Farooq and S. Perveen, J. Evidence-Based Complementary Altern. Med., 2015, 2015, 541591 Search PubMed.
- B. R. Kim, J.-Y. Park, H. J. Jeong, H.-J. Kwon, S.-J. Park, I.-C. Lee, Y. B. Ryu and W. S. Lee, J. Enzyme Inhib. Med. Chem., 2018, 33, 1256–1265 CrossRef CAS PubMed.
- S.-J. Wan, H.-G. Ren, J.-M. Jiang, G. Xu, Y. Xu, S.-M. Chen, G. Chen, D. Zheng, M. Yuan, H. Zhang and H.-X. Xu, Front. Chem., 2022, 10, 877469 CrossRef CAS PubMed.
- Q. Lv, S. Li, H. Wei, Z. Wen, Y. Wang, T. Tang, J. Wang, L. Xia and X. Deng, Appl. Microbiol. Biotechnol., 2020, 104, 1673–1682 CrossRef CAS PubMed.
- D. Mu, Y. Luan, L. Wang, Z. Gao, P. Yang, S. Jing, Y. Wang, H. Xiang, T. Wang and D. Wang, Emerging Microbes Infect., 2020, 9, 169–179 CrossRef CAS PubMed.
- J. E. Kang, B. J. Jeon, M. Y. Park, H. J. Yang, J. Kwon, D. H. Lee and B. S. Kim, Pest Manage. Sci., 2020, 76, 2294–2303 CrossRef CAS PubMed.
- K. Li, X. Han, R. Li, Z. Xu, T. Pan, J. Liu, B. Li, S. Wang, Y. Diao and X. Liu, J. Food Sci., 2019, 84, 1721–1729 CrossRef CAS PubMed.
- F. A.-z. A. Yehia, N. Yousef and M. Askoura, BMC Microbiol., 2022, 22, 106 CrossRef CAS PubMed.
- H. Tang, G. Porras, M. M. Brown, F. Chassagne, J. T. Lyles, J. Bacsa, A. R. Horswill and C. L. Quave, Sci. Rep., 2020, 10, 8046 CrossRef CAS PubMed.
- C. Wong, New Sci, 2023, 257, 14 CrossRef PubMed.
- The effect of orally ingested Curcumin and Piperine on the rate of orthodontic tooth movement and the extent of root resorption in a rodent model – A pilot study, https://preclinicaltrials.eu/database/view-protocol/197, accessed 04-24, 2023 Search PubMed.
- Celastrol attenuates the remodeling of pulmonary vascular and right ventricular in monocrotaline-induced pulmonary arterial hypertension in rat, https://search.animalstudyregistry.org/asr_viewer/documents.getPDF.action?doi=10.17590/asr.0000266, accessed 04-24, 2023 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |