AFM force-clamp spectroscopy captures the nanomechanics of the Tad pilus retraction
Johann
Mignolet
 ,
Marion
Mathelié-Guinlet
,
Marion
Mathelié-Guinlet
 ,
Albertus
Viljoen
,
Albertus
Viljoen
 and
Yves F.
Dufrêne
and
Yves F.
Dufrêne
 *
*
Louvain Institute of Biomolecular Science and Technology, UCLouvain, Croix du Sud, 4-5, bte, L7.07.07, Louvain-la-Neuve B-1348, Belgium. E-mail: yves.dufrene@uclouvain.be
First published on 13th May 2021
Abstract
Motorization of bacterial pili is key to generate traction forces to achieve cellular function. The Tad (or Type IVc) pilus from Caulobacter crescentus is a widespread motorized nanomachine crucial for bacterial survival, evolution and virulence. An unusual bifunctional ATPase motor drives Tad pilus retraction, which helps the bacteria to land on target surfaces. Here, we use a novel platform combining a fluorescence-based screening of piliated bacteria and atomic force microscopy (AFM) force-clamp spectroscopy, to monitor over time (30 s) the nanomechanics and dynamics of the Tad nanofilament retraction under a high constant tension (300 pN). We observe striking transient variations of force and height originating from two phenomena: active pilus retraction and passive hydrophobic interactions between the pilus and the hydrophobic substrate. That the Tad pilus is able to retract under high tensile loading – at a velocity of ∼150 nm s−1 – indicates that this nanomachine is stronger than previously anticipated. Our findings show that pilus retraction and hydrophobic interactions work together to mediate bacterial cell landing and surface adhesion. The motorized pilus retraction actively triggers the cell to approach the substrate. At short distances, passive hydrophobic interactions accelerate the approach phenomenon and promote strong cell-substrate adhesion. This mechanism could provide a strategy to save ATP-based energy by the retraction ATPase. Our force-clamp AFM methodology offers promise to decipher the physics of bacterial nanomotors with high sensitivity and temporal resolution.
New conceptsBacterial pili are flexible and dynamic nanofilaments that fulfil a wealth of cellular functions. The atypical Tad pilus from Caulobacter crescentus can retract through an active motorized process, which is critical for bacterial survival, evolution and virulence. Currently, we know little about the nanophysics of Tad pilus retraction, due to the complexity of the cell cycle and to the lack of appropriate ultrasensitive force probes. In this study, we investigate the nanomechanics and dynamics of Tad pilus retraction, using a platform combining a fluorescence-based piliated cell discrimination assay with atomic force microscopy (AFM) force-clamp spectroscopy. We discover that applying a constant tensile load (300 pN) to single pili connected to hydrophobic substrates leads to two types of transient variations in force and height, originating from pilus retraction and from hydrophobic binding. These findings support a model whereby pilus retraction and hydrophobic interactions work in concert to promote bacterial cell landing on surfaces. Our experiments emphasize the power of force-clamp AFM to understand the nanophysics and dynamics of motorized bacterial pili. In nanomedicine, our methodology may provide a means to screen for small molecules that can hinder pilus retraction in bacterial pathogens, thereby helping to prevent or treat infections. |
Introduction
Pili are surface-sensor nanofibers that decorate bacterial cell envelopes and act as grappling cables, promoting reversible adhesion to various surfaces which is crucial for the life cycle, gene transfer, virulence and biofilm formation.1,2 While the overall 3D-structure of pilin subunits and their organization inside the pilus filament are responsible for the gripping properties, pilus elongation and shrinking provide dynamics that expand its functional properties.2–7 In the widespread type IV pili (T4P), the filament length is dictated by the alternation of polymerization and depolymerization of pilin subunits.2 These cycles of pilus extension and retraction generate bacterial motion via traction forces in Pseudomonas aeruginosa and Neisseria gonorrhoeae,8,9 but are also essential for endothelial cell invasion by Neisseria meningitidis.10 In type IVa (T4aP) and some type IVb pili (T4bP), two canonical, dedicated ATP-driven motor proteins (ATPases) facilitate the unidirectional assembly and disassembly of pilin subunits.11–13 Optical tweezer experiments showed that the T4aP of N. gonorrhoeae powered by the PilT ATPase can generate a retraction force exceeding 100 pN.8,14 T4aP retraction forces allow the bacteria to establish fast and robust physical connection with host cells despite shear conditions.15,16 Moreover, these T4aP pili can aggregate in bundles to develop a cooperative retraction force in the nanonewton range.16 By contrast, the type IVc pilus (T4cP), also called Tad (Tight adherence) pilus, lacks unidirectional motors for elongation and retraction and relies on a bifunctional ATPase that carries out the two antagonistic processes.17 The Tad pilus has a predicted atypical structure made of individual pilin subunits composed of 2 very short α-helices.18 Akin to T4a/bP, Tad pilins are predicted to be made of hydrophobic α-helices that are intercalated tightly within the core of the pilus rod giving rise to a solid structure. Despite recent progress in understanding the biological and physical properties of the atypical Tad pilus from Caulobacter crescentus,17–22 we know little about the pilus nanomechanics and dynamics, which is likely to be due to the complex spatiotemporal orchestration of the Tad pilus biogenesis along the cell cycle.23,24Detailed knowledge of the nanophysics of Tad pilus retraction would greatly benefit from the tools of cellular and molecular mechanobiology, a growing field aiming at understanding how biosystems sense and respond to force.25–27 Here, we investigate the retraction of the “two-in-one” Tad pilus motor. We use the piliated bacterium Caulobacter crescentus that specifically assembles a single pilus filament at one pole of motile cells23,28 (Fig. 1a). We probe pilus retraction using atomic force microscopy (AFM)-based force-clamp spectroscopy, in which force is applied and kept at a constant level by continuously adapting the position of the piezoelectric device.25 This modality has been used to measure conformational changes and unfolding of single biomolecules at constant applied force29–31 and to quantify the lifetime of receptor–ligand interactions.32,33 In the present study, we use force-clamp AFM to study the nanomechanics and dynamics of single Tad pili under constant tension (300 pN) over extended periods of time (30 s). The results reveal transient changes in height and force traces, indicating that force-clamp spectroscopy detects Tad pilus retraction even when subjected to high tensile load. Pilus retraction together with hydrophobic interactions mediate bacterial cell landing on surfaces and favor firm cell-surface adhesion.
Results
Force-clamp AFM as a tool to monitor pilus retraction
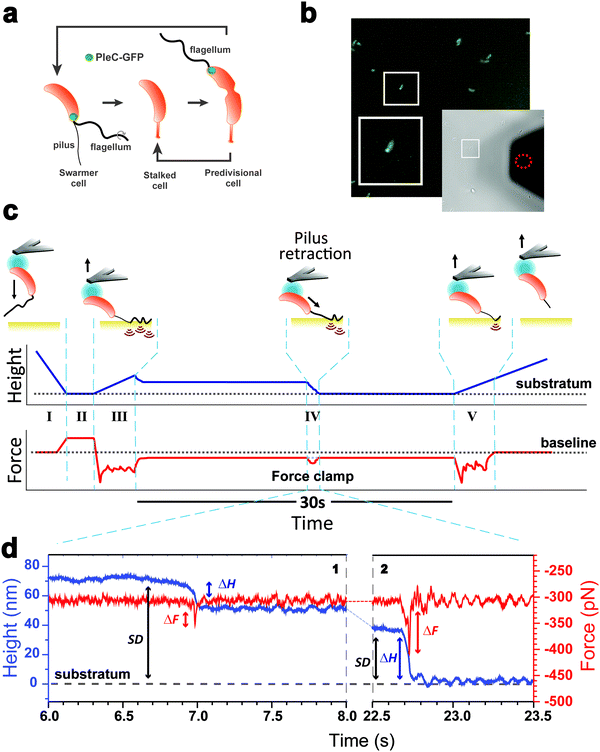
As piliated cells represent no more than 30% of the planktonic population18 (see cell cycle in Fig. 1a), we combined force-clamp AFM with a fluorescence-based cell discrimination assay to selectively probe single piliated cells (Fig. 1b). This allowed us to maintain an adhering pilus under constant tension while tracking its mechanical response over time. An AFM colloidal probe bearing a living piliated cell was brought into contact with a hydrophobic substrate, and then the cell was retracted from the surface with a force (F) of 300 pN, while the pilus was still contacting the surface (Fig. 1c). The force applied on the cell probe was kept constant over time (t = 30 s) by continuously adjusting the position (Z) of the piezoelectric device using an active feedback loop. We also recorded variations in height (ΔH) which are related to changes in Z piezo position (ΔZ) with respect to a zero baseline defined as the substrate surface.Our force-clamp experiments involved five steps (Fig. 1c): (I) an approach phase to initiate contact between the piliated cell and the hydrophobic substrate (F abruptly increases at contact); (II) a pause of 5 s at constant F to maximize the number of pilus-substrate contact points; (III) a first withdraw phase to separate the cell body from the surface and increase the separation distance (SD), while the pilus remains in contact but is subjected to increased mechanical tension; (IV) a 30 s force-clamp phase with a constant force of 300 pN, where we anticipate that the pilus will retract, causing changes in tensile force (ΔF). (V) Then the cell probe is withdrawn until the pilus detaches from the surface, resulting in strong multipeak forces associated with the rupture of multiple hydrophobic contact points.22
Mechanics and dynamics of the Tad pilus under constant force
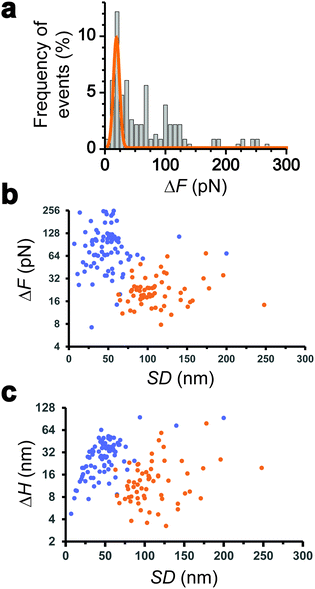
We collected more than 230 force-clamp recordings from 20 independent piliated cells. The clamp phase frequently featured striking signatures (Fig. 1d; 39% of the 230 force-clamp traces), i.e. transient increases in F absolute values, meaning that the pili were subjected to stronger tension, which were correlated with decreases in H, associated with an approach of the cell towards the substrate. We suggest that, during its retraction and shortening, the pilus was transiently loaded with a force exceeding the clamping force, thereby activating the feedback loop to compensate by a decrease in H.We observed a broad distribution of transient ΔF events (n = 147) events from 230 curves from 20 cells (Fig. 2a), with a sharp maximum at 20 pN (Gaussian fit), followed by a large spread of values, mostly ranging from 30 to 150 pN. This suggests that there were two groups of ΔF values, presumably reflecting two different phenomena. Supporting this view, plotting ΔF and ΔH as a function of the cell-substrate separation distance (SD) (Fig. 2b and c) revealed two populations of data. A first data cloud (“type 1” in orange; 27% occurrence; n = 61 events from 230 curves) featured very sharp events showing low variations in force (low ΔF) and height (low ΔH of 15 ± 13 nm), occurring far from the substrate (long SD of 111 ± 34 nm). The second cloud (“type 2” in blue; 37% occurrence; n = 86 events) displayed less sharp events with high ΔF, high ΔH (31 ± 17 nm), and short SD (49 ± 26 nm) values (Fig. 2b and c).
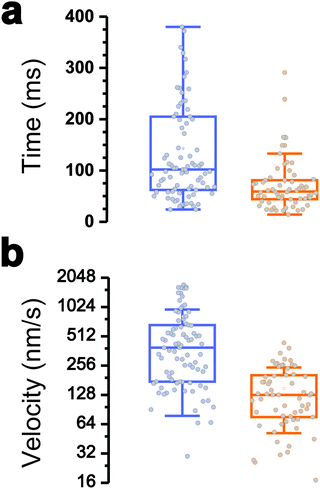
Interestingly, the dynamics of the two different behaviors were substantially different (Fig. 3a and b). The duration of type 1 events was clearly shorter than that of type 2 events (77 ± 72 ms and 143 ± 119 ms, respectively). There was also a major difference in velocities, as estimated from the H vs. t curves (148 ± 97 nm s−1 and 521 ± 443 nm s−1). Finally, a large fraction of the traces featuring type 1 events also showed type 2 events (84% of a total of 61 curves), with type 1 always preceding type 2, suggesting a time-dependent two-step process.
All together these results lead us to believe that type 1 signatures result from genuine Tad pilus retraction, followed by type 2 signatures representing short-range hydrophobic interactions between the pilus (and perhaps the cell body as well) and the hydrophobic substrate, as they occur near the surface and with fast dynamics. The later interpretation is supported by the high hydrophobicity of the pilins and by the strong hydrophobic adhesion of the pilus.
Retraction is abrogated in bacteria with static Tad pili
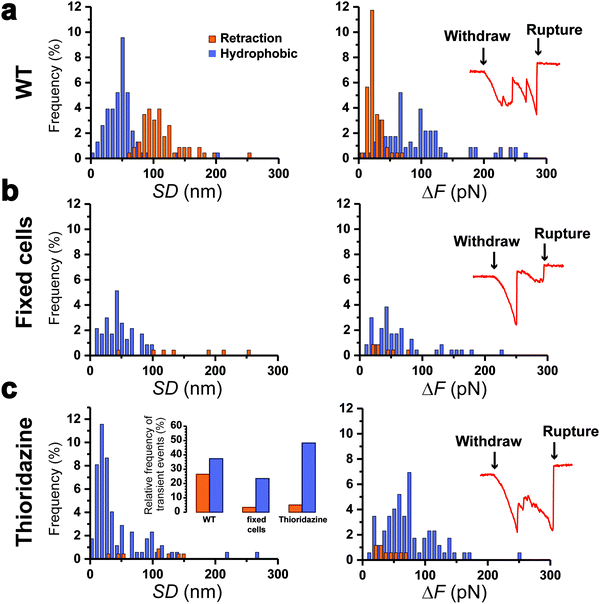
To further support our results, control experiments were performed in conditions where pilus retraction is impaired. Since the retraction ATPase is also involved in pilus assembly, existing motorization mutants that affect pilus retraction have partial loss-of-function effects17 and are thus inappropriate controls. This prompted us to hinder pilus retraction using chemicals. We treated Caulobacter cells with paraformaldehyde (PFA) prior to the piliated cell discrimination procedure, or added thioridazine during the force experiments. PFA crosslinks the pilin subunits between them, while thioridazine dissipates the sodium-based electrochemical gradient essential for ATP synthesis.10 We expect that these treatments should alter the processivity of the ATPase, through substrate incompatibility and ATP depletion, and therefore abolish pilus retraction. We found that this was indeed the case, as histograms of the distributions of SD and ΔF values (Fig. 4a–c) demonstrated that type 1 signatures were almost abolished on treated cells: 27% for the WT vs. 3 and 5% for PFA- and thioridazine-treated cells, respectively. Type 2 signatures however still occurred at relatively high frequency (Fig. 4c, left inset). To confirm that the two treatments did not alter the adhesion of Tad pili, we inspected the rupture events after the clamp phase (Fig. 4, right insets). We observed that during the last withdraw phase, the two types of treated cells produced typical adhesion events similar to those of the WT,22 meaning that pili were not dismantled by the chemical treatments. These results show that making the Tad pilus static abrogates type 1 events, while type 2 events do not depend on pilus motorization. This leads us to conclude that type 1 signatures are due to an active pilus retraction, whereas type 2 signatures arise from passive molecular interactions.Discussion
Pili are flexible and dynamic appendages that play multiple functional roles.4,13 One of these is to mediate surface colonization, which for some pili is activated by motorized elongation-retraction processes. Knowledge of the retraction mechanisms of these nanofilaments is critical to understand their functions. We have shown the power of force-clamp AFM to capture the nanomechanics and dynamics of motorized bacterial pili with high temporal resolution and force sensitivity. This is the first time that AFM force-clamp spectroscopy is implemented to study retraction events in motorized bacterial pili.Force-clamp traces recorded under a constant applied load of 300 pN between piliated C. crescentus cells and hydrophobic substrates feature two types of transient variations in force (ΔF) and height (ΔH). Type 1 signatures have low ΔF and ΔH but a long cell-substrate separation distance (SD). By contrast, type 2 signatures feature high ΔF and ΔH but short SD. Control experiments show that these behaviors originate, respectively, from Tad pilus active retraction and from passive hydrophobic interactions between pilus and substrate. This notion is consistent with (i) the strong hydrophobic properties of Tad pilins18 and (ii) the hydrophobic adhesion of the pilus, recently revealed using classical force-ramp AFM spectroscopy, where F is continuously increased at a constant velocity until rupture of the pilus-substrate connection.22 With this force-ramp mode, it was found that the Tad Pilus can sustain high pulling forces, up to 600 pN, explaining why we could here readily apply a high clamping force of 300 pN.
The retraction dynamics of T4aP has traditionally been studied using optical tweezers where a dielectric bead within an optical trap is pulled by a retracting pilus.8,14,15,34,35 Tensile load was minimal at the onset of retraction and progressively increased as the retracting pilus pulled on the bead in the optical trap. Pili therefore retracted at their optimal velocity (∼1000 nm s−1 for N. gonorrhoeae and ∼2000 nm s−1 for Myxococcus xanthus T4aP) until a stalling force (often called the maximal retractile force) was reached where retraction effectively came to a stop (∼100 pN and ∼150 pN for N. gonorrhoeae and M. xanthus T4aP).14,15,34 Force-clamp based optical tweezer studies then demonstrated that retraction frequency, duration and velocity were decreased for both species when pili were clamped at forces close to their respective stalling forces.34,35 Here, an AFM-based force-clamp set up is used to probe the C. crescentus Tad pilus retraction under tensile load. We are able to access a high applied force (300 pN) and still see retraction signatures at relatively high frequency. Tad pilus retraction occurs at a velocity of ∼150 nm s−1, which is in the range of values obtained by micropillar and optical tweezer experiments.19,20 It is remarkable that the Tad nanofilament can retract under an external load as high as 300 pN. This is likely to be of biological relevance as it will boost surface adhesion and colonization, key elements during cell cycle and virulence, in dynamic environments subjected to high shear stress, such as rivers or the blood flow.
These observations show that force-clamp AFM is capable of specifically detecting transient events corresponding to active pilus retraction and passive molecular interactions, to estimate ΔF and ΔH while correlating them with changes in SD values, and finally to assess retraction velocity. As retraction is detected under a high constant load, this implies that the force generated by the Tad retraction motor is much stronger than previously anticipated.19,20 During retraction, the pilus typically shortens by 15 nm (ΔH). If T4cP and T4aP fibers share a similar helical structure with a 4 nm pitch,6,36 this will mean that the Tad ATPase motor could process at least 4 helix turns in one retraction cycle before stopping. The reason why the pilus initiates retraction and then pauses is governed by external forces imposed by our force-clamp set-up parameters. However, this phenomenon could have a biological role, suggesting that the landing decision implemented by the cell body according to the pilus state or environmental cues is reversible. Therefore, motile swarmer cells will be able to reappraise the situation, especially in high shear stress environments, before they engage in firm attachment to a substrate and shift to a sedentary lifestyle.
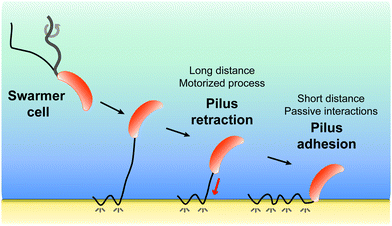
All together, our results favor a two-step model (Fig. 5) whereby pilus retraction and hydrophobic interactions act in concert to trigger fast bacterial cell landing on, and adhesion to, hydrophobic substrates. Tad pilus retraction actively induces the approach of the cell towards the surface. At short distances, hydrophobic interactions speed up the approach process and ensure tight cell-substrate connections leading to strong adhesion. We postulate that Caulobacter, and possibly other bacteria, may have evolved such a dual mechanism to compensate for the assembly of only one or two pili per cell, as opposed to the cooperative multiple pili in other bacteria, and to provide a strategy to save ATP-based energy consumed by the retraction ATPase.
In the future, studying biomolecular processes with fast kinetics will strongly benefit from advanced techniques with (sub)millisecond time resolution and force sensitivity. For instance, recently developed ultrafast optical trap force-clamp spectroscopy,37 which makes it possible to detect and study interactions on the sub-millisecond time scale, could be combined with AFM force spectroscopy to get a full picture of the physics and dynamics of the Tad pilus retraction.
Methods
Bacterial strains and growth conditions
Caulobacter crescentus NA1000 WT strain and a derivative were used in this study. Cells were grown at 30 °C in PYE. We added 1.5% agar into PYE plates, and tetracycline (1 μg ml−1) as required. Caulobacter WT was electroporated as previously described24 with pMR20-pleC-gfp (C-terminal translational fusion of PleC with the fluorescent protein GFP; low copy; tetR)38 to be subsequently used for the fluorescence-based swarmer cell discrimination. As required, cells were incubated with thioridazine (5 μM final concentration; Sigma Aldrich T9025-5G) or fixed with paraformaldehyde 3.7% (Alfa Aesar J61984) for 15 minutes and then rinsed in 1 ml of PBS.Preparation of hydrophobic substrates
Gold-coated glass coverslips were immersed overnight in an ethanol solution containing dodecanethiol (1 μl ml−1). Surfaces were then rinsed with ethanol and N2-blown.Colloidal probe preparation
Colloidal probes39 were obtained by attaching single silica microspheres (6.1 μm diameter; Bangs Laboratories) with a thin layer of UV-curable glue (NOA 63, Norland Edmund Optics) to triangular shaped tip-less cantilevers (NP-O10, Bruker) using a Nanowizard III atomic force microscope (JPK Instrument, Berlin, Germany). Cantilevers were then immersed for 1 h in Tris-buffered saline (TBS; tris, 50 mM; NaCl, 150 mM; pH 8.5) containing 4 mg ml−1 dopamine hydrochloride (Sigma-Aldrich), washed four times in PBS, and used directly for cell probe preparation.Swarmer cell selection protocol
Prior to force-clamp experiments, glass bottom Petri dishes (Wilco dish GWST3522) were coated with 20 μl of 1% poly-L-lysine for 20 min to promote Caulobacter reversible adherence. Excess poly-L-lysine was removed by washing and 20 μl of exponentially growing (OD660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm ∼ 0.5) bacteria were added in the Petri dish. After 40 min, medium excess was removed and Petri dishes were gently rinsed twice with 2 ml PBS. Finally, hydrophobic surfaces were mounted in the same Petri dish, in the near vicinity of adherent bacteria.
nm ∼ 0.5) bacteria were added in the Petri dish. After 40 min, medium excess was removed and Petri dishes were gently rinsed twice with 2 ml PBS. Finally, hydrophobic surfaces were mounted in the same Petri dish, in the near vicinity of adherent bacteria.
The colloidal probe was first calibrated with an empty glass Petri dish at low magnification (Plan N 40×/0.6 objective). The spring constant of cantilevers was determined by the thermal noise method, giving an average value of ∼0.08 N m−1. Next, bacteria were observed using an optical inverted fluorescence microscope (Zeiss AxioObserver Z1 equipped with a Hamamatsu Orca camera, ORCA Flash 4.0 C13440, with high magnification using an Alpha Plan-Apochromatic 100×/1.46 VIS-IR oil objective). A major technical challenge is that pili are assembled only in Caulobacter swarmer cells, which represent a small fraction of the whole population in batch culture. To circumvent this problem, we used a swarmer cell selection procedure relying on the PleC-GFP fluorescent fusion as previously described.22 PleC is a protein morphogen that localizes at the flagellated pole in piliated swarmer cells (smaller cell type), as well as in late pre-divisional cells (larger cell type). We were thus able to only attach single small cells with a polar fluorescent focus to the colloidal cantilevers. Bacteria were illuminated with GFP excitation light (470 nm ± 40) and the polydopamine-coated colloidal probe was put in contact for 10 s with a single piliated cell. The cell probe was then positioned over the hydrophobic gold-coated cover slip without de-wetting.
Force-clamp on discriminated swarmer cells
Force measurements were performed in PBS at room temperature using a NanoWizard 4 NanoScience AFM (JPK Instrument, Berlin, Germany). Force clamp maps were obtained by recording arrays of 8-by-8 force/distance–time curves on 10 μm by 10 μm areas with a constant cantilever approach-withdraw speed of 2000 nm s−1. Our experiments involved the following steps: (I) the piliated cell on the colloidal probe is brought into contact with the hydrophobic substrate with a maximum applied force of 250 pN; (II) the cell stays in contact for 5 s at constant height to increase the probability of pilus-substrate contact points; (III) the cantilever is moved away from the substrate with a specific distance in order to maximize the cell body-substrate separation distance (SD) but still maintain a pilus-substrate connection; (IV) the pilus-substrate interaction strength is clamped at a force of 300 pN owing to a controlled feedback loop that adjusts continuously the cell probe-substrate distance; (V) the pilus is finally disengaged with an ultimate withdraw phase over a 1 μm-course. The transient events occurring during the force-clamp phase were analyzed from distance–time and force–time traces to extract the separation distance (SD), the variation in height (ΔH) and the variation in force (ΔF). Force traces were smoothened with the Savitzky–Golay method (4th order). Data were analyzed using the Data Processing software from JPK Instruments V.6.1.142 (Berlin, Germany).Author contributions
J. M., M. M., A. V. and Y. D. conception and design. J. M. acquisition of data. J. M., M. M., A. V. and Y. D. analysis and interpretation of data, and writing of the manuscript.Conflicts of interest
The authors declare they have no conflict of interests.Acknowledgements
We are grateful to Prof. D. Alsteens and Dr F. Viela for inspiring discussions on force-clamp data and to Prof. P. H. Viollier and Dr Gaël Panis for providing the pleC-gfp reporter plasmid. Work at UCLouvain was supported by the Excellence of Science-EOS programme (Grant #30550343), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 693630), and the National Fund for Scientific Research (FNRS). Y. D. is Research Director at the FNRS.References
- J. L. Berry and V. Pelicic, FEMS Microbiol. Rev., 2015, 39, 134–154 CrossRef CAS PubMed.
- L. Craig, K. T. Forest and B. Maier, Nat. Rev. Microbiol., 2019, 17, 429–440 CrossRef CAS PubMed.
- L. Craig, M. E. Pique and J. A. Tainer, Nat. Rev. Microbiol., 2004, 2, 363–378 CrossRef CAS PubMed.
- T. Proft and E. N. Baker, Cell. Mol. Life Sci., 2009, 66, 613–635 CrossRef CAS PubMed.
- Y. Dufrene, A. Viljoen, J. Mignolet and M. Mathelie-Guinlet, Cell. Microbiol., 2021, e13324 Search PubMed.
- C. L. Giltner, Y. Nguyen and L. L. Burrows, Microbiol. Mol. Biol. Rev., 2012, 76, 740–772 CrossRef CAS PubMed.
- S. Melville and L. Craig, Microbiol. Mol. Biol. Rev., 2013, 77, 323–341 CrossRef CAS PubMed.
- A. J. Merz, M. So and M. P. Sheetz, Nature, 2000, 407, 98–102 CrossRef CAS PubMed.
- J. S. Mattick, Annu. Rev. Microbiol., 2002, 56, 289–314 CrossRef CAS PubMed.
- K. Denis, et al. , Nat. Microbiol., 2019, 4, 972–984 CrossRef CAS PubMed.
- Y. W. Chang, et al. , Science, 2016, 351, aad2001 CrossRef PubMed.
- M. Wolfgang, et al. , Mol. Microbiol., 1998, 29, 321–330 CrossRef CAS PubMed.
- R. Denise, S. S. Abby and E. P. C. Rocha, PLoS Biol., 2019, 17, e3000390 CrossRef PubMed.
- B. Maier, et al. , Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16012–16017 CrossRef CAS PubMed.
- B. Maier, M. Koomey and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10961–10966 CrossRef CAS PubMed.
- N. Biais, et al. , Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11358–11363 CrossRef CAS PubMed.
- C. K. Ellison, et al. , Sci. Adv., 2020, 5, eaay2591 CrossRef PubMed.
- L. Del Medico, et al. , Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9546–9553 CrossRef PubMed.
- C. K. Ellison, et al. , Science, 2017, 358, 535–538 CrossRef CAS PubMed.
- M. Sangermani, et al. , mBio, 2019, 10, e01237-19 CrossRef PubMed.
- R. A. Snyder, et al. , Proc. Natl. Acad. Sci. U. S. A., 2020, 17984–17991 CrossRef CAS PubMed.
- J. Mignolet, M. Mathelie-Guinlet, A. Viljoen and Y. F. Dufrene, Nano Lett., 2021, 21, 3075–3082 CrossRef CAS PubMed.
- J. Mignolet, G. Panis and P. H. Viollier, Curr. Opin. Microbiol., 2017, 42, 79–86 CrossRef PubMed.
- J. Mignolet, et al. , eLife, 2016, 5, e18647 CrossRef PubMed.
- B. Liu, W. Chen and C. Zhu, Annu. Rev. Phys. Chem., 2015, 66, 427–451 CrossRef CAS PubMed.
- M. Krieg, et al. , Nat. Rev. Phys., 2015, 1, 41–57 CrossRef.
- Y. F. Dufrene and A. Persat, Nat. Rev. Microbiol., 2020, 18, 227–240 CrossRef CAS PubMed.
- P. D. Curtis and Y. V. Brun, Microbiol. Mol. Biol. Rev., 2010, 74, 13–41 CrossRef CAS PubMed.
- A. F. Oberhauser, P. K. Hansma, M. Carrion-Vazquez and J. M. Fernandez, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 468–472 CrossRef CAS.
- J. Brujic, et al. , Biophys. J., 2007, 92, 2896–2903 CrossRef CAS PubMed.
- S. Garcia-Manyes, T. L. Kuo and J. M. Fernandez, J. Am. Chem. Soc., 2011, 133, 3104–3113 CrossRef CAS PubMed.
- B. T. Marshall, et al. , Nature, 2003, 423, 190–193 CrossRef CAS PubMed.
- M. Mathelie-Guinlet, et al. , Nat. Commun., 2020, 11, 5431 CrossRef PubMed.
- M. Clausen, V. Jakovljevic, L. Sogaard-Andersen and B. Maier, J. Bacteriol., 2009, 191, 4633–4638 CrossRef CAS PubMed.
- M. Clausen, M. Koomey and B. Maier, Biophys. J., 2009, 96, 1169–1177 CrossRef CAS PubMed.
- L. Craig and J. Li, Curr. Opin. Struct. Biol., 2008, 18, 267–277 CrossRef CAS PubMed.
- M. Capitanio, et al. , Nat. Methods, 2012, 9, 1013–1019 CrossRef CAS PubMed.
- R. T. Wheeler, J. W. Gober and L. Shapiro, Curr. Opin. Microbiol., 1998, 1, 636–642 CrossRef CAS PubMed.
- A. Beaussart, et al. , Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |