Open Access Article
Open Access ArticleAnti-inflammatory, antiallergic and COVID-19 protease inhibitory activities of phytochemicals from the Jordanian hawksbeard: identification, structure–activity relationships, molecular modeling and impact on its folk medicinal uses†
Sherif S. Ebada *abc,
Nariman A. Al-Jawabrib,
Fadia S. Youssef
*abc,
Nariman A. Al-Jawabrib,
Fadia S. Youssef ac,
Dina H. El-Kashefad,
Tim-Oliver Knedele,
Amgad Albohyf,
Michal Korinekghi,
Tsong-Long Hwang
ac,
Dina H. El-Kashefad,
Tim-Oliver Knedele,
Amgad Albohyf,
Michal Korinekghi,
Tsong-Long Hwang hij,
Bing-Hung Chengkl,
Guan-Hua Linm,
Chia-Yi Linm,
Sa'ed M. Aldalaienb,
Ahmad M. Disin,
Christoph Janiak
hij,
Bing-Hung Chengkl,
Guan-Hua Linm,
Chia-Yi Linm,
Sa'ed M. Aldalaienb,
Ahmad M. Disin,
Christoph Janiak e and
Peter Proksch*a
e and
Peter Proksch*a
aInstitute of Pharmaceutical Biology and Biotechnology, Heinrich-Heine University, D-40225 Düsseldorf, Germany. E-mail: ss_ebada@mutah.edu.jo; sherif_elsayed@pharma.asu.edu.eg; Peter.Proksch@hhu.de; Fax: +49-211-81-11923; Tel: +49-211-81-14163
bDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Mu'tah University, 61710 Al-Karak, Jordan
cDepartment of Pharmacognosy, Faculty of Pharmacy, Ain-Shams University, Abbasia, 11566 Cairo, Egypt
dDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt
eInstitut für Anorganische Chemie und Strukturchemie, Heinrich-Heine-Universität, D-40204 Düsseldorf, Germany
fDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, The British University in Egypt (BUE), Al-Sherouk City, Cairo-Suez Desert Road, 11837, Egypt
gGraduate Institute of Natural Products, College of Pharmacy, Kaohsiung Medical University, Kaohsiung, Taiwan
hDepartment of Biotechnology, College of Life Science, Kaohsiung Medical University, Kaohsiung, Taiwan
iGraduate Institute of Natural Products, College of Medicine, Chang Gung University, Taoyuan, Taiwan
jResearch Center for Chinese Herbal Medicine, Research Center for Food and Cosmetic Safety, Graduate Institute of Health Industry Technology, College of Human Ecology, Chang Gung University of Science and Technology, Taoyuan, Taiwan
kDepartment of Anesthesiology, Chang Gung Memorial Hospital, Taoyuan, Taiwan
lDepartment of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan
mThe Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung, Taiwan
nDepartment of Basic Pharmaceutical Sciences, Faculty of Pharmacy, Al-Isra University, 11622 Amman, Jordan
First published on 15th October 2020
Abstract
On Wednesday 11th March, 2020, the world health organization (WHO) announced novel coronavirus (COVID-19, also called SARS-CoV-2) as a pandemic. Due to time shortage and lack of either a vaccine and/or an effective treatment, many trials focused on testing natural products to find out potential lead candidates. In this field, an edible and folk medicinal Jordanian plant Crepis sancta (Asteraceae) was selected for this study. Phytochemical investigation of its enriched polyphenolic extract afforded four eudesmane sesquiterpenes (1–4) together with (6S,9R)-roseoside (5) and five different methylated flavonols (6–10). Structure elucidation of isolated compounds was unambiguously determined based on HRESIMS, X-ray crystallography, and exhaustive 1D and 2D NMR experiments. All isolated compounds were assessed for their in vitro anti-inflammatory, antiallergic and in silico COVID-19 main protease (Mpro) inhibitory activities. Among the tested compounds, compounds 5–10 revealed potent anti-inflammatory, antiallergic and COVID-19 protease inhibitory activities. Chrysosplenetin (10) is considered as a promising anti-inflammatory and antiallergic lead structure adding to the phytotherapeutic pipeline. Moreover, its inhibitory activity against SARS-CoV-2 Mpro, supported by docking and molecular dynamic studies, strengthens its potential as a lead structure paving the way toward finding out a natural remedy to treat and/or to control the current COVID-19 pandemic.
Introduction
Polymorphonuclear neutrophils (PMNs) are a major component of the innate immunity defending the human body against inadvertent entrance of xenobiotics and/or pathogens. PMNs perform their physiological function principally through the production of oxidative free radicals and non-oxidative proteases including neutrophil elastase (NE, a serine protease aka leukocyte elastase).1 The more PMNs infiltrated, the higher NE levels secreted in response accomplishing their defensive role by demolishing functional proteins of phagocytosed bacterial, fungal and/or viral pathogens. NE is an intracellular enzyme principally stored in azurophilic granules of PMNs. Beside its defensive role against pathogens, they were found to produce devastating effects on elastin-rich connective tissue particularly in the lungs.2 NE can also split and sever collagen, proteoglycans and other plasma proteins. Hence, its overexpression may result in deleterious effects on permeability barrier integrity between alveolar epithelial and endothelial cells leading to pathologic edematous symptoms in the inflamed lungs undergoing either acute lung injury (ALI), acute respiratory distress syndrome (ARDS) or chronic respiratory disorders such as asthma and chronic obstructive pulmonary disease (COPD).3 However, ARDS, ALI and COPD are among the leading causes of morbidity and mortality in the intensive care units (ICU) worldwide,4 some recurrent comorbidities have been reported for COPD with chronic disorders such as hypertension, diabetes, heart diseases, depression and pneumonia.5During the last two decades, two epidemics of zoonotic origins have been introduced into human community, namely, severe acute respiratory syndrome (SARS) commenced at Guangzhou province (China) in 2003 (ref. 6) and Middle East respiratory syndrome (MERS) emerged in Saudi Arabia about 10 years later in 2012.7 In December 2019 at Wuhan city (China), early reports indicated a group of five patients primarily diagnosed with pneumonia of an unknown etiology featuring ARDS and one of them died later. About six weeks later, the World Health Organization (WHO) announced the onset of a potential coronavirus outbreak caused by SARS-CoV2 (COVID-19, named by WHO on February 11, 2020) and one month later on March 11, 2020, WHO raised alerts and announced this outbreak as a pandemic.8
Their causative pathogens belong to betacoronaviruses family and are of zoonotic origins. They all are believed to emerge from the horseshoe bats as a natural reservoir for their immense majority and recently moved through intermediate animal hosts, dromedary camels for MERS and Malayan pangolins for COVID-19, to humans that then adopted human–human transmission leading to epidemics among humans.9
Till preparing this manuscript, COVID-19 spread worldwide affecting about 37.1 million patients including more than one million deaths that obliged many governments to implement social distancing together with total lockdown strategies to defend against this pandemic.
Due to the current lack of an effective pharmaceutical and/or an approved vaccine, the major WHO recommendations focus on infection control measures, improving lifestyle and diet enrichment with more-healthy components. Based on the ongoing research efforts directed toward finding a plausible target for developing a specific antiviral agent, a very recent paper describing COVID-19 virus main protease (Mpro), a key CoV enzyme mediating viral replication and transcription, that was recognized as a very promising target for drug development.10
As a part of our ongoing research and building on the previous notions, we thought of isolating and probing plant natural products as NE and/or COVID-19 Mpro inhibitors which may help to ameliorate the clinical manifestations related to ARDS and to cease or even slow down viral replication and transcription granting additional time for the immune system to defend itself against COVID-19.
In this study, we explored an edible and a folk medicinal plant of the genus Crepis (aka hawksbeard including about 200 species) belonging to the tribe Cichorieae and family Asteraceae. It is widely eaten and/or used for herbal medicine purposes in different countries and territories within the circumference of the Mediterranean Basin including Italy, Greece and Jordan. The fresh leaves of many Crepis species including that discussed in this study, C. sancta, are eaten either fresh, boiled or browned in salads by the locals. Traditionally, its decoction has been used for treating different ailments related mainly to respiratory and gastrointestinal systems such as common cold, cough, abdominal colic, constipation, hepatitis along with hypertension and hyperglycemia.11
Based on searching the reported literature of the genus Crepis, its aerial parts were reported to be rich in both flavonoid aglycones and glycosides12 in addition to eudesmanes and guaianolides sesquiterpenes present as both aglycone and glycosidic forms.13 Regarding assessed bioactivities, the extracts of genus Crepis have been reported to possess antimicrobial, antiviral and antiproliferative activities together with antioxidant, analgesic, antiulcer and anti-inflammatory.12,13
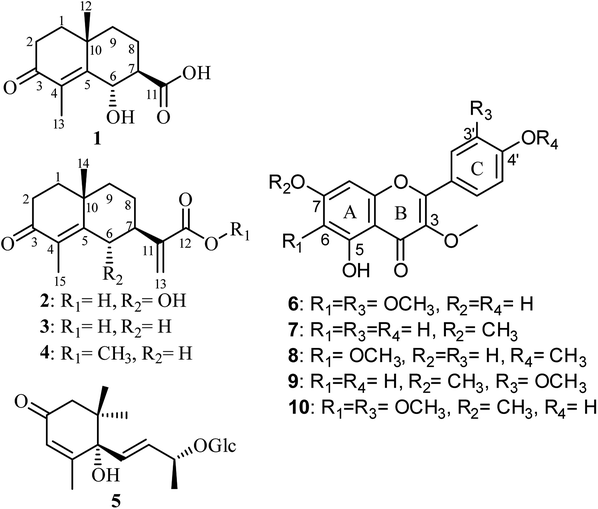
In this study, we report phytochemical exploration of polyphenolic-rich extract of C. sancta aerial parts affording ten different compounds belonging to eudesmane sesquiterpenes and methoxylated flavonoids. Structure elucidation of isolated compounds was unambiguously confirmed through HRESIMS, exhaustive 1D, 2D NMR spectroscopy and X-ray crystallography. We also report in this study results of in vitro anti-inflammatory and antiallergic activities of isolated compounds in addition to in silico COVID-19 main protease (Mpro) inhibitory activity assessment.
Materials and methods
General experimental procedures
Perkin-Elmer-241 MC polarimeter was used for determining optical rotation. LRESIMS and HRESIMS were determined on a LC-MS HP1100 Agilent Finnigan LCQ Deca XP Thermoquest and FTHRMS-Orbitrap (ThermoFinnigan) mass spectrometer, respectively. Chromatographic workups were conducted via column chromatography using different stationary phases such as silica gel 60 M (0.04–0.063 mm) and Sephadex LH20. For screening purposes, ready-made silica gel 60 F254 TLC plates (Merck, Darmstadt, Germany) were used. For visualizing TLC plates, UV light at 254 and 365 nm wavelengths were applied as non-destructive technique or after spraying with anisaldehyde reagent and heating. For analytical HPLC measurements, a Dionex Ultimate 3000 LC system equipped with a ready-made separation column (125 × 4 mm, L × ID), pre-packed with Eurospher-10C18 (Knauer, Germany) and coupled to a photodiode array (PDA) detector (UVD340S) was implemented, setting up the detection channels at 235, 254, 280 and 340 nm wavelengths. A standard gradient elution was applied using (MeOH, 0.01% formic acid in water): 0 min, 10% MeOH; 5 min, 10% MeOH; 35 min, 100% MeOH; 45 min, 100% MeOH, with a flow rate of 1 mL min−1. Preparative HPLC separations were accomplished using a RP-HPLC system of LaChrom-Merck Hitachi equipped with a pump L7100, UV detector L7400 and a column (300 × 8 mm, L × ID) prefilled with Euroshper-100C18 (Knauer, Germany) at a flow rate of 5.0 mL min. 1D (1H and 13C NMR) and 2D NMR (chemical shifts in ppm) spectra were determined on Bruker AVANCE DMX 600 (Switzerland) using methanol-d4 and DMSO-d6 solvents (Sigma-Aldrich, Germany).Plant material
Crepis sancta aerial parts were collected in November, 2017 at Al-Basirah region (Al-Tafilah Governorate, southern Jordan). The plant identity was authenticated by Prof. Dr Saleh A. Al-Qur'an at Department of Botany, Faculty of Science, Mu'tah University and a voucher specimen coded CSA-201711 was kept at Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Mu'tah University.Extraction, isolation and purification
The freshly collected aerial parts were kept in shade to dry, ground to afford a one-kg net weight that was then extracted by methanol (3 × 1 L). Methanol extract was then concentrated under vacuum till dryness affording a total residue (9.8 g). The total extract was then defatted via liquid–liquid fractionation procedure by being dispersed in 90% MeOH (500 mL) and shaken against n-hexane (1 L). The two immiscible liquid phases were then separated and the aqueous methanol layer was evaporated till dryness under reduced pressure resulting in a solid residue (5.9 g). Afterwards, the defatted residual solid was chromatographed through vacuum liquid chromatography (VLC) using silica gel stationary phase and implementing a gradient elution system using n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc and DCM
EtOAc and DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH with a 20% increment affording 12 fractions (CSV1-CSV12). All obtained fractions were subjected to TLC and analytical HPLC procedures. Fractions CSV2, CSV5-CSV8, CSV11 and CSV12 were chosen for further preparative HPLC purification procedure. Fractions CSV2 (58 mg) and CSV11 (63 mg), eluted with n-hexane
MeOH with a 20% increment affording 12 fractions (CSV1-CSV12). All obtained fractions were subjected to TLC and analytical HPLC procedures. Fractions CSV2, CSV5-CSV8, CSV11 and CSV12 were chosen for further preparative HPLC purification procedure. Fractions CSV2 (58 mg) and CSV11 (63 mg), eluted with n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (8:2) and with DCM
EtOAc (8:2) and with DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (2:8), were subjected to preparative HPLC yielding 4 (2.7 mg) and 3 (3.5 mg), respectively. Four fractions (CSV5-CSV8) were collected together and applied on column chromatography using Sephadex LH-20 as stationary phase and methanol as a mobile phase followed by preparative HPLC for final purification to yield 6 (4.5 mg), 7 (5.7 mg), 8 (6 mg), 9 (8 mg) and 10 (12 mg). Fraction CSV12 (82 mg) eluted by DCM
MeOH (2:8), were subjected to preparative HPLC yielding 4 (2.7 mg) and 3 (3.5 mg), respectively. Four fractions (CSV5-CSV8) were collected together and applied on column chromatography using Sephadex LH-20 as stationary phase and methanol as a mobile phase followed by preparative HPLC for final purification to yield 6 (4.5 mg), 7 (5.7 mg), 8 (6 mg), 9 (8 mg) and 10 (12 mg). Fraction CSV12 (82 mg) eluted by DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1:4) was further purified using preparative HPLC yielding 1 (1.3 mg), 2 (1.6 mg) and 5 (1.2 mg).
MeOH (1:4) was further purified using preparative HPLC yielding 1 (1.3 mg), 2 (1.6 mg) and 5 (1.2 mg).
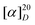 +97.6° (c 0.02, MeOH); UV (MeOH) λmax 248 nm; 1H NMR and 13C NMR see ESI Table S1;† HRESIMS m/z 239.1282 [M+H]+ (calcd for C13H19O4; m/z 239.1283) and m/z 237.1128 [M−H]− (calcd for C13H17O4; m/z 237.1127).
+97.6° (c 0.02, MeOH); UV (MeOH) λmax 248 nm; 1H NMR and 13C NMR see ESI Table S1;† HRESIMS m/z 239.1282 [M+H]+ (calcd for C13H19O4; m/z 239.1283) and m/z 237.1128 [M−H]− (calcd for C13H17O4; m/z 237.1127).Cell viability assay
The mucosal mast-cell derived rat basophilic leukemia cells (RBL-2H3) were obtained from Bioresource Collection and Research Center (Hsin-Chu, Taiwan). The cells were cultured in DMEM containing 10% FBS, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin in 10 cm cell culture dishes at 37 °C in a humidified chamber with 5% CO2 in air. The potential cytotoxic effects of the samples (10 to 100 μg mL−1 for the crude extract and 1 to 400 μM for the pure compounds) on RBL-2H3 cells were determined by the methylthiazole tetrazolium (MTT) assay as described previously.14Superoxide anion generation and elastase release assays by human neutrophils
The human neutrophils were obtained from venous blood of healthy adult volunteers (20–30 years old) following the reported procedure.15 Measurement of superoxide anion generation by the activated neutrophils was based on the reduction of ferricytochrome c as described before.15,16 Elastase release by activated neutrophils was determined using N-methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide as elastase substrate according to the reported method.16 The concentration was 1 to 10 μg mL−1 for the crude samples and 0.3 to 10 μM for the pure compounds. LY294002 was used as the positive control.Degranulation assay in mast cells
The level of degranulation in mast cells was evaluated based on A23187- and antigen-induced β-hexosaminidase release in RBL-2H3 cells according to a reported method with some modifications.14 Briefly, the cells were seeded in a 96-well plate (2 × 104 cells per well, for the A23187-induced assay) or a 48-well plate (3 × 104 cells per well, for the antigen-induced assay) overnight. At the same time, the cells for the antigen-induced assay were sensitized with anti-DNP IgE (0.5 μg mL−1; Sigma). RBL-2H3 cells were treated with the samples (0.5, 5 and 50 μM) for 30 min in Tyrode's buffer with maximal DMSO dose of 0.5%. For the A23187-induced assay, the cells were activated by adding A23187 (final concentration 0.5 μM), while cells for antigen-induced assay were activated by the addition of DNP-BSA (final concentration 100 ng mL−1) for 30 min. Azelastine served as a positive control. The amount of β-hexosaminidase was detected using the method utilizing p-NAG as a substrate according to the procedure described before.14X-ray crystallographic analysis
Crystals of chrysosplenetin (10) have been obtained by crystallization from pure methanol.Data collection: measurements were done with a Bruker Kappa APEX2 CCD diffractometer with a microfocus tube and Cu Kα radiation (λ = 1.54178 Å). Data collection, structure solving, refinement and the hydrogen atom positioning were all performed according to the recently reported procedure.17 The structural data have been deposited in the Cambridge Crystallographic Data Center (CCDC no 1994762†).
Molecular modeling studies
The proposed binding mode of isolated compounds with neutrophil elastase (NE) and SARS-CoV-2 main protease (Mpro) was studied using Autodock Vina and a method similar to what we reported earlier.12 Here, crystal structures of NE (PDB ID: 1H1B) and SARS-CoV-2 Mpro (PDB ID: 6LU7) were used. Prepared and co-crystalized ligands were docked in a grid box in the active site (25 × 25 × 25 Å3, centered on co-crystalized ligand) using exhaustiveness of 16. For each ligand, the top 9 binding poses were ranked according to their binding affinities and the predicted binding interactions were analyzed. The pose with the best binding affinity and binding mood similar to co-crystalized ligand was reported.Molecular dynamic (MD) simulation
MD simulations were done using GROMACS 2018.1 molecular dynamics package.18 CHARMM36 all-atom force field19 was used for protein simulations while ligands were parameterized using SwissParam.20 All simulations were performed in TIP3P explicit water21 in a dodecahedron box and neutralized by Na+ or Cl− ions. Steepest descent algorithm was used for minimization and maximum force was set to less than 1000 kJ mol−1 nm−1. Systems were initially equilibrated using NVT and NPT ensembles for 100 ps each. The temperature was maintained at 300 K using the V-rescale algorithm22 and pressure was controlled by the Parrinello–Rahman barostat.23 The LINear Constraint Solver (LINCS) algorithm24 was used for bond's length constraints and Particle mesh Ewald (PME) method25 was used for long-range electrostatics calculations. For all simulations 2 fs timestep was used. van der Waals cut-off distance (rvdw) was set to 1.2 nm. Initial coordinates was taken from docking poses or from crystal structure if available.Determination of elastase enzymatic activity
The compounds were further tested for direct inhibition of elastase enzymatic activity.26 The neutrophil suspension (6 × 105 cells per mL) was preheated for 5 min in the presence of CaCl2 (1 mM) at 37 °C. Priming agent CB (1.5 μg mL−1) was added for 2 min, followed by fMLF (0.1 μM) for 20 min to release elastase from the cells. After centrifugation at 1000g for 5 min at 4 °C, the supernatant containing elastase was preheated at 37 °C for 5 min, and the test compounds were added. Then, 0.1 mM of substrate methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide was added for 10 min. The effect of the compounds on elastase enzymatic activity was quantified by measuring the absorbance at 405 nm.Coronavirus 229E assay
The protective effects of the samples against human coronavirus 229E (HCoV-229) was determined similarly to the previously described method.27 Huh7 cells (human liver carcinoma cell line) were infected with 9TCID50 (Median Tissue Culture Infectious Dose) of each coronavirus 229E in the presence or absence of the compounds or vehicle. After incubation at 33 °C for 6 days, the surviving cells were then stained with MTT (3-[4.5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide). The percentage of surviving cells was then calculated.Results and discussion
Isolation and identification of major metabolites in the plant extract
A careful phytochemical investigation of the plant extract, implementing various chromatographic techniques, MS, 1D, 2D NMR spectroscopic analyses and by comparison with the reported literature, afforded four eudesmane sesquiterpene lactones (1–4). Two were unambiguously identified and reported previously as new natural products from the same extract namely, (6S,7S,10R)-3-oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid (1)28 and (6S,7S,10R)-3-oxo-6α-hydroxy-γ-costic acid (2)28 together with two known congeners, 3-oxo-γ-costic acid (3)12,29 and its methyl ester (4).12,30 In addition, (6S,9R)-roseoside (5)28 and several methoxylated flavonoids namely, jaceidin (6),12,31 kumatakenin (7),12,32 penduletin (8),12,33 pachypodol (9)12,34 and chrysosplenetin (10)12,35 were also isolated. Structure elucidation of the isolated compounds were unambiguously determined based on extensive 1D, 2D NMR analyses together with mass spectrometry (see ESI†).Compounds (1 and 2) were individually purified as white amorphous solids whose molecular formulas were determined to be C13H18O4 and C15H20O4 based on their HRESIMS spectra that revealed pseudomolecular ion peaks at m/z 237.1128 [M−H]− and m/z 263.1283 [M−H]− indicating the existence of five and six degrees of unsaturation, respectively. The larger molecular weight of 2 by 26 atomic units and its additional one degree of unsaturation compared to 1 suggested that the former might have an additional olefin moiety with one of its sp2 carbons turned into a quaternary one. The 1D NMR spectra of 1 including 1H and 13C NMR (see ESI, Table S1†) unravelled a close resemblance to the known natural metabolite 3-oxo-di-nor-eudesma-4-en-11-oic acid with an exocyclic carboxylic acid group (δC 176.8, C-11).36,37 By careful investigation of extensive 2D NMR spectra such as 1H–1H COSY, HMQC and HMBC, a clear difference was determined in 1 compared to the known metabolite that is the presence of an oxymethine group at δH 5.25 (d, 3.0, 1H, δC 67.6) instead of a methylene group at δH 2.29/δH 2.99 (d, 15.0, 2H, δC 29.2) ascribed for C-6.37 This notion along with other key 1H–1H COSY and HMBC correlations (see ESI, Fig. S3†) confirmed undoubtedly the oxygenation to be at the same position (C-6) in 1. In addition, 1H–1H COSY and HMQC spectra of 1 distinguished two clear spin systems, one exists between two adjacent methylene moieties at δH 1.72/δH 1.83 (m, 2H, δC 39.8) along with the two geminally coupled protons at δH 2.38 (ddd, 17.4, 15.2, 5.2) and at δH 2.66 (ddd, 17.4, 4.5, 2.5) bound to the same carbon peak at δC 34.6 that were assigned for C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2-1 and C
2-1 and C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2-2, respectively. A second spin system can be also featured extending over δH 5.25 (d, 3.0, 1H, δC 67.6), δH 2.46 (dt, 12.9, 3.0, 1H, δC 49.0) and two methylene groups at δH 2.21 (ddd, 13.4, 12.9, 3.2, 1H) and δH 1.74 (m, 1H) bound to a carbon peak at δC 18.5 in addition to δH 1.39 (dd, 13.3, 3.2) and δH 1.70 (dd, 6.7, 3.0, 1H) bound to a carbon peak at δC 41.4 that were ascribed to C
2-2, respectively. A second spin system can be also featured extending over δH 5.25 (d, 3.0, 1H, δC 67.6), δH 2.46 (dt, 12.9, 3.0, 1H, δC 49.0) and two methylene groups at δH 2.21 (ddd, 13.4, 12.9, 3.2, 1H) and δH 1.74 (m, 1H) bound to a carbon peak at δC 18.5 in addition to δH 1.39 (dd, 13.3, 3.2) and δH 1.70 (dd, 6.7, 3.0, 1H) bound to a carbon peak at δC 41.4 that were ascribed to C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) -6, C
-6, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) -7, C
-7, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2-8 and C
2-8 and C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2-9, respectively.
2-9, respectively.
A careful exploration of 1H and 13C NMR spectral data of 2 (see ESI, Table S1†) and by comparison with the reported literature, it was interpreted to be structurally resembling 3-oxo-γ-costic acid (3)29 with one additional oxygen atom in 2 as deduced from their molecular formulas. The presence of an additional oxygen atom was explained by comparing 1D and 2D NMR spectral data of 2 (see ESI, Table S1†) and 3 (ref. 19) that confirmed the presence of an oxymethine proton, similar to 1, at δH 5.52 (d, 6.5, 1H, δC 77.3) in 2 replacing a methylene group at δH 2.83 (br d, 13.5, 1H) and δH 2.02 (br t, 13.5, 1H) bound to a carbon peak at δC 33.3 assigned to C-6 in 3.29
To unambiguously determine the positions of the hydroxyl and carboxylic acid moieties in both 1 and 2, gHMBC experiments were performed (see ESI, Fig. S3, S9 and S17†). The key HMBC correlations of 1 and 2 (see ESI, Fig. S3†) undoubtedly confirmed the existence of a hydroxyl group at C-6 in both whereas a terminal carboxylic acid group was ascertained to be at C-11 and C-12, respectively.
The relative configuration of 1 was defined by interpreting the coupling constant (J) values and by key ROESY correlations (see ESI, Fig. S3†). The determined large J values for H-7 and H-8a (12.9 Hz) revealed a diaxial orientation, however, a small coupling constant value (3.0 Hz) for H-7 and H-6 suggested an axial-equatorial orientation, respectively.36–38 In addition, the ROESY spectrum of 1 disclosed clear correlations (see ESI, Fig. S3 and S11†) namely these from H-6 to H-7 and a singlet methyl group at δH 1.87 (br s, Me-13) together with other key correlations in particular those from a singlet methyl group at δH 1.42 (s, Me-12) to two proton peaks at δH 2.66 (H-2a) and at δH 2.21 (H-8a) indicated that 6-OH and Me-12 are in an axial orientation while 7-COOH adopts an equatorial one. Supported by the results of X-ray crystallography36 and modified Mosher method37 for exploring the absolute configuration of biosynthetically related eudesmane-type sesquiterpenes such as 3-oxo-di-nor-eudesma-4-en-11-oic acid and (1R,2S,6R,7S,10S)-1,2,6-trihydroxyeudesma-4-en-3-one, compound 1 was deduced to have similar absolute configuration to the related congeners36,37 apart from H-6 which was unambiguously distinguished to be in an equatorial position in 1 rather than an axial orientation in the related congeners. Accordingly, compound 1 was determined to be (6S,7S,10R)-3-oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid.
The relative and absolute configurations of 2 (see ESI, Fig. S3†) were defined as for 1 based on ROESY spectrum that revealed comparable key correlations to those found in 1 in addition to the J values analysis that revealed similar orientation pattern of H-6 and H-7 like that noted in 1. As a conclusion, compound 2 was unambiguously identified as (6S,7S,10R)-3-oxo-6α-hydroxy-γ-costic acid.
Superoxide anion generation and elastase release assays by human neutrophils
Assessment of the anti-inflammatory activity (Table 1) revealed that the isolated flavonols significantly suppressed the superoxide anion generation with IC50 values ranging between 4.32 ± 0.57 and 6.07 ± 0.65 μM. Flavonoids were previously reported to have potent inhibitory activity against pro-inflammatory mediators such as inducible nitric oxide synthase (iNOS), tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β) and interleukin-6 (IL-6).39 Among the tested compounds in this study, compound 10 revealed potent inhibitory effect on NE release and superoxide anion generation with IC50 values of 6.66 ± 1.03 and 4.32 ± 0.57 μM, respectively. The results indicated inhibitory effect on human neutrophils activation. Human neutrophil elastase plays a major role in neutrophils-associated diseases. According to a previous study, structurally similar flavonols including 8 (IC50 = 65.4 μM) showed inhibitory effect on human neutrophil elastase which correlates with 33.0% inhibitory effect of 8 on NE release in human neutrophils at 10 μM.40 The obtained results in this study strengthened the previous reports about the anti-inflammatory effects of flavonols combating the respiratory damage and NE release along with plausible antiviral activity as previously reported.41| Compound | Superoxide anion generation | Elastase release | ||
|---|---|---|---|---|
| % Inhibitiona | IC50b (μM) | % Inhibitiona | IC50b (μM) | |
| a Percentage of inhibition (Inh%) at 10 μM concentration.b Concentration necessary for 50% inhibition (IC50). Results are presented as mean ± S.E.M. (n = 3–6). *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control (0.1% DMSO).c Genistein inhibited 67.01 ± 2.38% of superoxide anion generation at 3 μM. | ||||
| (6S,7S,10R)-3-Oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid (1) | 12.05 ± 2.37 | >10 | 2.74 ± 1.90 | >10 |
| (6S,7S,10R)-3-Oxo-6α-hydroxy-γ-costic acid (2) | 6.23 ± 3.81 | >10 | 5.89 ± 1.07 | >10 |
| 3-Oxo-γ-costic acid (3) | 18.46 ± 5.06 | >10 | 3.89 ± 1.91 | >10 |
| 3-Oxo-γ-costic acid methyl ester (4) | 18.27 ± 4.97 | >10 | 0.70 ± 1.60 | >10 |
| (6S,9R)-Roseoside (5) | 9.43 ± 5.12 | >10 | 2.72 ± 1.78 | >10 |
| Jaceidin (6) | 80.26 ± 6.22 | 4.76 ± 0.83 | 48.22 ± 6.61 | >10 |
| Kumatakenin (7) | 24.06 ± 2.75 | >10 | 22.92 ± 4.29 | >10 |
| Penduletin (8) | 73.76 ± 5.24 | 5.66 ± 0.19 | 33.00 ± 7.94 | >10 |
| Pachypodol (9) | 67.46 ± 4.55 | 6.07 ± 0.65 | 31.56 ± 5.02 | >10 |
| Chrysosplenetin (10) | 92.03 ± 3.95 | 4.32 ± 0.57 | 65.82 ± 5.75 | 6.66 ± 1.03 |
| Genistein | c | 1.52 ± 0.17 | 31.57 ± 7.29 | 16.34 ± 4.26 |
Degranulation assay in mast cells
All isolated compounds (1–10) were assessed for their toxic effects on normal RBL-2H3 cells up to 50 μM. The results (Table 2) revealed that all tested compounds were non-toxic (viability more than 90%). Antiallergic activity of isolated compounds (1–10) was then tested via assessing their inhibitory activities against A23187- and antigen-induced β-hexosaminidase release in RBL-2H3 cells. Calcium ionophore A23187 represents an inducer facilitating calcium transport through mast cells membrane while antigen (IgE plus DNP-BSA) acts via FcεRI receptor similar to physiological environment.15 The obtained results (Table 2) revealed that among eudesmane sesquiterpene lactones (1–4), 3-oxo-γ-costic acid methyl ester (4) moderately suppressed A23187-induced degranulation (IC50 80.6 μM) indicating importance of carboxylic acid esterification for the activity. The obtained results (Table 2) revealed that compounds 8 and 10 displayed potent inhibitory activities against antigen-induced degranulation with IC50 values of 3.8 and 5.8 μM, respectively, exceeding that of azelastine as a standard antiallergic drug (IC50 = 15.4 μM). Also, compounds 4 and 7 exerted significant effects on antigen-induced degranulation of mast cells. In a previous report, a structurally-related flavonoid aglycone, kaempferol, was found to exert in vitro antiallergic actions in RBL-2H3 cells through inducing heme oxygenase-1 (ref. 42) which might be a putative mechanism for the antiallergic activity of 8 and 10 as well (Fig. 1).| Compound | % Viability, RBL-2H3a | % Inhibition of A23187-induced degranulationb | % Inhibition of antigen-induced degranulationc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 100 μM | 1 μM | 10 μM | 100 μM | IC50d (μM) | 1 μM | 10 μM | 100 μM | IC50d (μM) | |
| a The cytotoxicity of samples to RBL-2H3 was evaluated using MTT viability assay. Results are presented as mean (n = 3) compared to untreated control (DMSO). All samples were nontoxic towards RBL-2H3 cells.b Azelastine (20 μM) was used as a positive control and inhibited 78.4 ± 1.4% *** of A23187-induced degranulation. The inhibition of degranulation was assessed by A23187-induced β-hexosaminidase release in RBL-2H3 cells. Results are presented as mean ± S.E.M. (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001 (Prism, ANOVA, Dunnet's test) compared with the control value (A23187 only).c Azelastine (20 μM) was used as a positive control and inhibited 66.8 ± 9.8% *** of antigen-induced degranulation. The inhibition of degranulation was assessed by antigen-induced β-hexosaminidase release in RBL-2H3 cells. Results are presented as mean ± SEM (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001 (Prism, ANOVA, Dunnet's test) compared with the control value (antigen only).d Concentration necessary for 50% inhibition (IC50).e Compounds 8, 9 and 10 at concentration of 100 μM formed crystal-like precipitates upon the addition into medium, thus the effects could not be justified. | |||||||||
| (6S,7S,10R)-3-Oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid (1) | >90% | 2.1 ± 1.8 | 7.6 ± 6.6 | 7.7 ± 3.8 | 11.3 ± 3.6 | ||||
| (6S,7S,10R)-3-Oxo-6α-hydroxy-γ-costic acid (2) | >90% | 0.2 ± 0.2 | 0.3 ± 0.3 | 11.7 ± 4.2 | 19.8 ± 2.5 | ||||
| 3-Oxo-γ-costic acid (3) | >90% | 2.7 ± 2.3 | 1.8 ± 1.6 | 2.8 ± 1.0 | 0.3 ± 0.2 | ||||
| 3-Oxo-γ-costic acid methyl ester (4) | >90% | 7.2 ± 5.9 | 61.8 ± 7.4*** | 80.6 | 18.2 ± 4.4 | 36.2 ± 6.5*** | |||
| (6S,9R)-Roseoside (5) | >90% | 5.3 ± 4.3 | 16.8 ± 7.9 | 19.9 ± 4.3 | 21.6 ± 1.8 | ||||
| Jaceidin (6) | >90% | 5.3 ± 4.3 | 25.5 ± 5.0* | 3.9 ± 2.9 | 46.5 ± 10.2*** | ||||
| Kumatakenin (7) | >90% | 12.6 ± 9.1 | 36.8 ± 5.4 | 11.1 ± 7.6 | 60.9 ± 8.8*** | 80.4 | |||
| Penduletin (8) | >90% | 4.8 ± 3.9 | 25.1 ± 7.3 | e | 33.9 ± 8.6** | 85.8 ± 5.1*** | e | 3.8 | |
| Pachypodol (9) | >90% | 3.4 ± 2.8 | 22.6 ± 3.4 | e | 12.9 ± 6.5 | 47.2 ± 5.0*** | e | ||
| Chrysosplenetin (10) | >90% | 7.3 ± 5.9 | 41.6 ± 9.5*** | e | 12.4 ± 5.1 | 83.3 ± 7.5*** | e | 5.8 | |
| Azelastineb,c | >90% (20 μM) | 10.9 | 15.4 | ||||||
X-ray crystallographic analysis
Being unprecedentedly reported and based on the findings obtained from anti-inflammatory and antiallergic assays, we crystallized chrysosplenetin (10) using its pure methanol solution. Herein, we report for the first time its crystal structure (see ESI, Fig. S2 and Table S2†). From the single-crystal structure refinement, the structure for chrysosplenetin was obtained, containing two independent molecules in the asymmetric unit, which are connected by a hydrogen bond. Both molecules contain also intramolecular hydrogen bonds.Molecular modeling studies
In order to predict the binding mode of the isolated compounds in the active site of NE, compounds were docked using Autodock Vina. To validate docking method and parameters, co-crystalized ligand GW475151 was redocked in the active site. When the extracted crystal structure and the docked structure were aligned using DockRMSD server, a root mean square deviation (RMSD) value of 1.317 was obtained which indicates the validity of the method used.12 Results obtained from docking study of isolated compounds with NE showed that isolated methoxylated flavonols (6–10) possess better binding in the active site when compared to sesquiterpenes (1–4). Amongst flavonols, those with a methoxy group at C-7, compounds 7, 9 and 10, demonstrated better binding. This methoxy was found to be in close proximity to His57, one of the catalytic triad of the serine proteases like NE. Another important residue which is also one of the catalytic triad is Ser195 which was found to form hydrogen bond with the C-4 carbonyl group in all the three flavonols. The third residue among the catalytic triad, Asp102 was found to lie at the bottom of the binding site and is positioned beneath the docked flavonols. It is generally accepted that catalytic activity of NE is enhanced by these three residues (catalytic triads; His57, Asp102 and Ser195).43 The former two residues act as a base to increase nucleophilicity of Ser195 through accepting proton transferred from serine hydroxyl group.44 In addition to the catalytic triad, docked structures were found to form several hydrogen bonds, represented by dotted yellow line, with residues in the active site as illustrated in Fig. 2 and Table 3. These residues include Phe41, Cys191, Gly 193 and Val216. In addition, hydrophobic interactions between ring A and C from the flavonols with Phe192 and Val216 are observed, respectively. Hydrophobic interaction between ring C and Val216 could explain the differences in dihedral angles between rings B and C in the docked structure and the crystal structure of free flavonols. For example, when we crystalized 10 alone, the dihedral angle between ring B and C was either 0.5° (almost coplanar) or 40° with two independent molecules in the asymmetric unit. On the other hand, when 10 was docked in the active site of NE a dihedral angle of 85° (almost perpendicular) between rings B and C was obtained due interaction with Val216.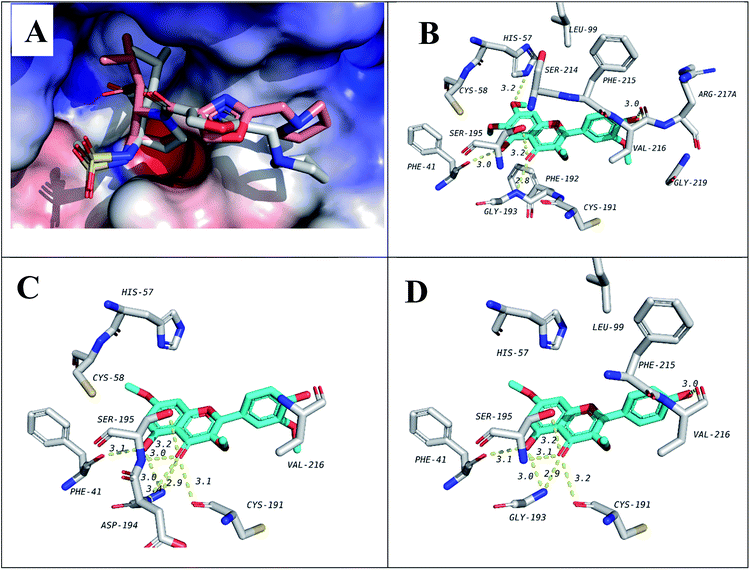 | ||
| Fig. 2 Docking results of isolated compounds with NE (1H1B). (A) Validation of docking through redocking of the co-crystalized ligand GW475151 (gray) overlaid with the docked structure (pink). (B, C and D) Docking poses of the best docked ligands 10, 9 and 7, respectively. | ||
| Compound/ligand | 1H1B (elastase) | 6LU7 (Mpro) | ||
|---|---|---|---|---|
| Binding affinity (kcal mol−1) | Interacting residues | Binding affinity (kcal mol−1) | Interacting residues | |
| (6S,7S,10R)-3-Oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid (1) | −5.5 | Ser195, Val216 | −5.9 | Gly143 |
| (6S,7S,10R)-3-Oxo-6α-hydroxy-γ-costic acid (2) | −6.2 | Ser195 | −6.1 | Gly143, Ser144, His163 |
| 3-Oxo-γ-costic acid (3) | −5.9 | Ser195 | −5.9 (4th pose) | Gly143, His163 |
| 3-Oxo-γ-costic acid methyl ester (4) | −5.9 | Ser195 | −6.0 (3rd pose) | Gly143, His163 |
| (6S,9R)-Roseoside (5) | −6.4 | Phe41, Asn61, Ser195, Val216 | −7.2 | Thr26, Leu141, Gly143, Ser144, Cys145, His163, Glu166 |
| Jaceidin (6) | −6.2 | Cys191, Gly193, Ser195 | −7.3 | Leu141, Gly143, Ser144, Cys145, Arg188 |
| Kumatakenin (7) | −6.7 | Phe41, Cys191, Gly193, Ser195, Val216 | −6.8 (3rd pose) | Leu141, Gly143, Ser144 |
| Penduletin (8) | −6.1 | Cys191, Gly193, Ser195 | −6.7 (4th pose) | Leu141, Gly143, Ser144, Cys145, His163 |
| Pachypodol (9) | −6.7 | Phe41, Cys191, Gly193, Asp194, Ser195 | −7.1 | Gly143, Ser144, Cys145 |
| Chrysosplenetin (10) | −6.5 | Phe41, His57, Gly193, Ser195, Val216 | −7.1 | Leu141, Gly143, Ser144, Cys145 |
| 1H1B-ligand | −6.9 | Ser195 | — | — |
| 6LU7-ligand (N3) | — | — | −7.1 (3rd pose) | Phe140, Gly143, His163, His164, Glu166, Asp187, Thr190 |
It is worth to mention that docking data has shown good agreement with the elastase release assay in human neutrophils indicating a possible direct effect on elastase enzymatic activity. To justify this notion, Pearson's correlation coefficient was determined whose value revealed a medium to strong negative association between the inhibition of human elastase release and the docking scores (r = −0.5). This indicates that the higher the inhibitory activity against human elastase release is, the lower the docking score (larger absolute value).
Considering NE as a protease, this encouraged us to assess the binding modes and affinity of the isolated compounds to SARS-CoV-2 main protease (Mpro).10 This enzyme was proposed to be a good target for treatment of COVID-19. Chen and co-workers reported the virtual screening of Mpro against a medicinal plant library containing 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 297 compounds including flavonoids.45
297 compounds including flavonoids.45
To test that virtually, the isolated compounds' docking into the active site of Mpro was studied. Before running the actual docking, validation was done by redocking of the co-crystalized ligand N3. The docked structure had a RMSD of 1.855 compared to the co-crystalized ligand structure (Fig. 3A).
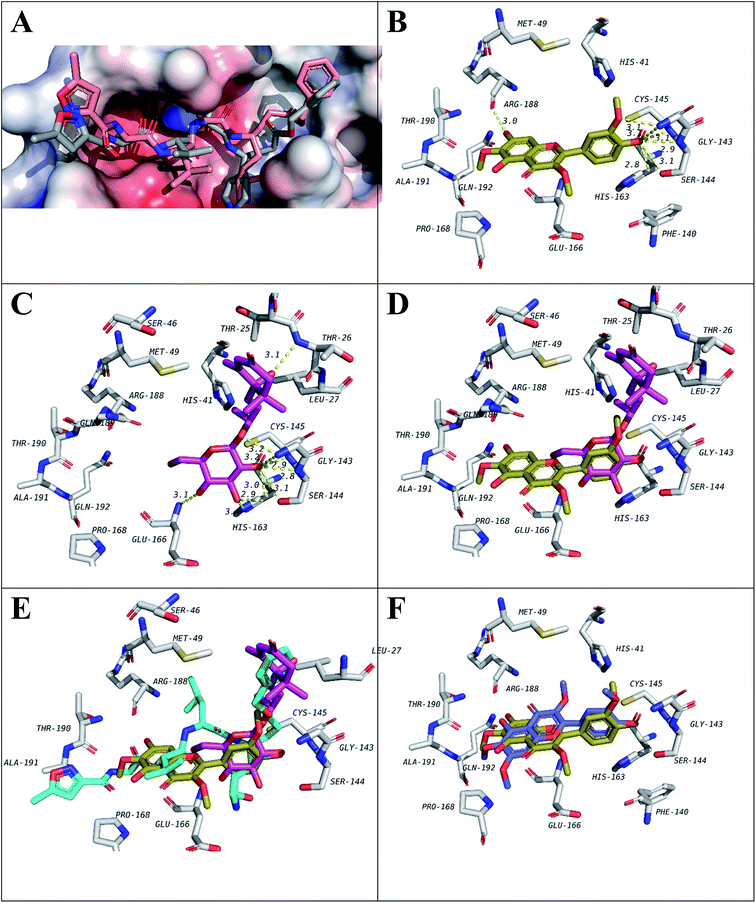 | ||
| Fig. 3 Docking results of isolated compounds with SARS-CoV-2 Mpro (6LU7). (A) Validation of docking through redocking of the co-crystalized ligand N3 (gray) overlaid with the docked structure (pink). (B and C) Docking poses of the best compounds 6 (golden) and 5 (pink) respectively and their overlapping in (D). (E) Docking poses of 5 (pink) and 6 (golden) overlapped with co-crystalized ligand N3 (blue). (F) Docking poses of 6 (golden) and 10 (violet) comparing their binding modes. | ||
Among the docked structures, tested flavonols, jaceidin (6), pachypodol (9) and chrysosplenetin (10) along with (6S,9R)-roseoside (5) revealed binding energy values equal or even higher than the co-crystalized ligand (6LU7, Table 3).
Among isolated flavonols, compounds 6, 9 and 10 feature a similar ring C structure with adjacent methoxy and hydroxyl groups that proved to form hydrogen bond network with residues Cys145, Ser144, Gly143, and Leu141 (Fig. 3B). A very recent report by Zhang and coworkers indicated the inevitable role of Cys145, Ser144 and Gly143 forming the canonical “oxyanion hole” of the cysteine protease, in the binding of SARS-CoV-2 Mpro inhibitors.10 In addition, only compound 6 possesses a free hydroxyl group at position 7 of ring A which was found to be involved in a hydrogen bond with Arg188 which may explain the slightly higher binding energy value compared to compounds 9 and 10 (Table 3).
For compound 5 (Fig. 3C), the binding pose is partially overlapped with that of flavonol (6) where the glycosidic sugar moiety in 5 is overlapping with ring C of 6. The hydroxy groups on the sugar unit in 5 are involved in hydrogen bond network with the same residues similar to 6, 9 and 10 binding (Cys145, Ser144, Gly143 and Leu141). In addition, hydrogen bonds with His163, Glu166 and Thr26 are also seen. Among those, His163 and Glu166 were reported in Zhang and his fellows' work mentioned above.10 It will be interesting to synthesize and test a compound that has the aglycon of (6S,9R)-roseoside linked to 3′-oxygen of 6 (Fig. 3D and E) as a proposed structure for a lead SARS-CoV-2 Mpro inhibitor.
Molecular dynamic simulation
Beside being the best compound in the elastase release assay with IC50 of 6.66 μM (Table 1), chrysosplenetin (10) was among the top compounds that showed the best docking in the active site of human elastase (1H1B) and has shown comparable binding affinity to the co-crystalized ligand in the active site of SARS-CoV-2 main protease (6LU7) as demonstrated in Table 3. These findings encouraged us to further investigate the bonding of 10 with both targets using molecular dynamic simulation studies. Four complexes were studied including 10 in both targets and the co-crystalized ligand corresponding to each target. Using production run trajectories, the average total interaction energy composed of coulombic interaction and Lennard-Jones (L-J) energy was calculated (Table 4).| Target | Complex | Energy (kJ mol−1) | ||
|---|---|---|---|---|
| Coulombic interaction | Lennard-Jones (L-J) energy | Total interaction energy | ||
| Human elastase | 1H1B-chrysosplenetin (10) | −31.12 ± 3.7 | −84.81 ± 10 | −115.93 ± 10 |
| 1H1B-control | −20.36 ± 3.8 | −99.14 ± 6.0 | −119.50 ± 7.1 | |
| SARS-CoV-2 main protease | 6LU7-chrysosplenetin (10) | −24.29 ± 6.9 | −81.58 ± 17 | −105.87 ± 18 |
| 6LU7-control | −173.04 ± 5.2 | −249.57 ± 2.8 | −422.61 ± 5.9 | |
| 6LU7-jaceidin (6) | −43.72 ± 2.0 | −113.80 ± 1.7 | −157.52 ± 2.6 | |
For elastase complexes, both compound 10 and the co-crystalized ligand have shown comparable average total interaction energy ranging from −115 to −120 kJ mol−1 supporting its potential activity as elastase inhibitor that was proved by the biological testing. In addition, plotting of RMSD of ligand heavy atoms during the MD production run (Fig. 4A), shows RMSD < 1 Å during more than 90% of production run time which suggests the stability of the proposed docking binding mode discussed earlier.
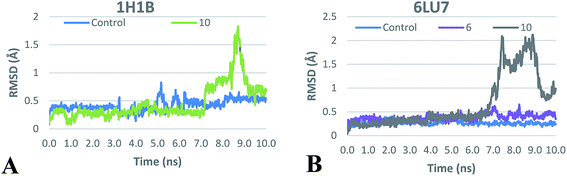 | ||
| Fig. 4 RMSD plot of ligands heavy atoms in the active sites of (A), human elastase 1H1B and (B), SARS-CoV-2 main proteae 6LU7 during the production MD run. | ||
On the other hand, the average total interaction energy between 10 and SARS-CoV-2 main protease was much less than the interaction energy seen in case of the co-crystalized ligand (Table 4). The RMSD plot (Fig. 4B) shows stability of the 6LU7–10 complex only during the initial MD production run. This suggested that compound 10 could be a lead for the design of compounds targeting main protease but need further improvement. To investigate the last hypothesis, we decided to run MD for 6LU7 complex with 6 whose choice was based on its close structural similarity to 10 and also on its better docking score. In elastase release inhibitory activity, compound 6 was also the second-most potent compound. We found that 6 has better total interaction energy (−157 kJ mol−1) which resulted from slight structural modification of compound 10. Moreover, jaceidin (6) has also shown less RMSD fluctuation during the MD production run (Fig. 4B). This clearly supports that further improvement can be done in this scaffold to design SARS-CoV-2 main protease inhibitors against COVID-19. This could be done through different medicinal chemistry techniques but it is beyond the scope of this work.
Elastase enzymatic activity
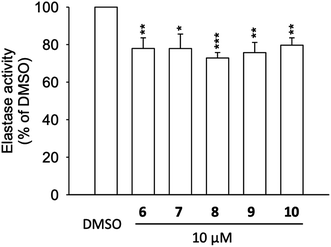
To address the results from molecular docking and molecular dynamic simulation studies, neutrophil elastase (NE) was extracted from activated neutrophils and cell-free assay on the elastase enzymatic activity was performed (Fig. 5). The results revealed, that compounds 6 to 10 significantly inhibited the activity of elastase enzyme by 20.3 to 27.1% at the concentration of 10 μM (Fig. 5). Thus, the effects of 6 to 10 on elastase release (22.9 to 65.8% inhibition at 10 μM), could be, at least partly, attributed to the direct interaction with elastase enzyme which is in agreement with the molecular docking of compounds with NE.Protective effects against human coronavirus 229E infection
We further tested the compounds that docked well with Mpro (compounds 5 to 10) in the human coronavirus 229E (HCoV-229E) assay in vitro (see ESI, Fig. S4†). The cells were infected with HCoV-229E strain (grey) and the protective effects of compounds (5–10) were evaluated. None of the tested compounds protected the cells from HCoV-229E infection. However, having no in vitro protective activity against viral host cell entry doesn't deny the Mpro inhibitory activity that converts viral polyprotein into functional units within host cells to be finally assembled into new viruses.Recently, natural products have been focused on as a source of inhibitors affecting SARS-CoV-2 Mpro, a functional protein considered as a potential target for developing new therapeutics to help combatting the current COVID-19 pandemic. Many natural compounds, belonging to various chemical classes such as alkaloids, coumarins, flavonoids, peptides, terpenoids and tannins, have been assessed for their SARS-CoV-2 Mpro inhibitory activity.46 Many potential Mpro inhibitors were chemically belonging to flavonoids such as quercetin, apigenin and luteolin (IC50 of 20–200 μM) in addition to the biflavonoid amentoflavone that was the most potent with IC50 of 8.3 μM.47 To further investigate and comprehend their Mpro inhibitory activity, in silico molecular docking and molecular dynamic simulation studies were also conducted.48,49 In our study, the obtained results enriched potential SARS-CoV-2 Mpro inhibitors with some more candidates supported by more details about their binding modes in addition to being proved in vitro as inhibitors of human elastase release. These features altogether may also escalate their efficiency in defeating the respiratory distress symptoms triggered by inflammatory responses to SARS-CoV-2 infection.
Conclusion
Phytochemical investigation of polyphenolic-rich extract of the Jordanian hawksbeard, prescribed by local herbalists for treatment of cough and flu, afforded several eudesmane sesquiterpenes and flavonols. They were assessed for their in vitro anti-inflammatory and antiallergic activities. Among the tested compounds, flavonols, in particularly chrysosplenetin (10), exhibited potent anti-inflammatory activity that was proved via in vitro experimental results through assessing its inhibitory activity against neutrophil elastase release along with further in silico molecular modeling that included docking and molecular dynamic simulation studies. By assessing the binding to SARS-CoV-2 (COVID-19) Mpro as an important target, chrysosplenetin (10) was also found to possess high binding energy comparable to that exhibited by its standard ligand.These findings are supporting the introduction of chrysosplenetin (10) as a potential lead candidate into the phytotherapeutic arena as an anti-inflammatory and antiallergic drug. Moreover, its inhibitory activity against SARS-CoV-2 (COVID-19) Mpro will support considering it as a potential lead structure paving the way toward finding out a natural remedy for treatment of the current COVID-19 pandemic.
Author contributions
SSE and PP: conceptualization, writing, reviewing and editing, SSE, NAA and FSY: Methodology, purification and isolation of pure metabolites; DHEK, TOK and CJ: NMR analysis, data curation and X-ray crystallography; AA: molecular modeling and MD simulation; MK and BHC: antiallergy assay; TLH: anti-inflammatory assay; GHL and CYL: anti-coronavirus 229E assay; SMA, AMD and PP; supervision and editing the manuscript.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
S. S. E. acknowledges the Junior Scientist and International Research Center (JUNO) at Heinrich-Heine University (Düsseldorf, Germany) for financial support. P. P. wants to thank Manchot Foundation for their support. The research was supported by the grants from the Ministry of Science and Technology (MOST 106-2320-B-255-003-MY3, MOST 108-2320-B-255-003-MY3, and MOST108-2320-B-037-004), Ministry of Education (EMRPD1G0231 and EMRPD1H0381), Kaohsiung Medical University (KMU-DK109002-3), Chang Gung University of Science and Technology (ZRRPF3H0101 and ZRRPF3H0111), and Chang Gung Memorial Hospital (CMRPF1F0011∼3, CMRPF1F0061∼3, CMRPF1G0241∼3, CMRPF1J0051∼3, and BMRP450), Taiwan. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.References
- P. A. Henriksen, The potential of neutrophil elastase inhibitors as anti-inflammatory therapies, Curr. Opin. Hematol., 2014, 21, 23–28 CrossRef CAS.
- K. Hoenderdos and A. Condliffe, The neutrophil in chronic obstructive pulmonary disease, Am. J. Respir. Cell Mol. Biol., 2013, 48, 531–539 CrossRef CAS.
- T. J. Moraes, C. W. Chow and G. P. Downey, Proteases and lung injury, Crit. Care Med., 2003, 31, S189–S194 CrossRef CAS.
- S. Hagiwara, H. , Iwasaka, K. , Togo and T. , Noguchi, A neutrophil elastase inhibitor, sivelestat, reduces lung injury following endotoxin-induced shock in rats by inhibiting HMGB1, Inflammation, 2008, 31, 227–234 CrossRef CAS.
- B. Ställberg, C. Janson, G. Johansson, K. Larsoon, G. Stratelis, G. Telg and K. H. Lisspers, Management, morbidity and mortality of COPD during an 11-year period: an observational retrospective epidemiological register study in Sweden (PATHOS), Prim. Care Respir. J., 2014, 23, 38–45 CrossRef.
- J. S. M. Peiris, Y. Guan and K. Y. Yuen, Severe acute respiratory syndrome, Nat. Med., 2004, 10, S88–S97 CrossRef CAS.
- A. M. Zaki, S. van Boheemen, T. M. Bestebroer, A. D. Osterhaus and R. A. Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia, N. Engl. J. Med., 2012, 367, 1814–1820 CrossRef CAS.
- H. A. Rothan and S. N. Byrareddy, The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak, J. Autoimmun., 2020, 108, 102433, DOI:10.1016/j.jaut.2020.102433.
- T. T.-Y. Lam, N. Jia, Y.-W. Zhang, M. H.-H. Shum, J.-F. Jiang, H.-C. Zhu, Y.-G. Tong, Y.-X. Shi, X.-B. Ni, Y.-S. Liao, W.-J. Li, B.-G. Jiang, W. Wei, T.-T. Yuan, K. Zheng, X.-M. Cui, J. Li, G.-Q. Pei, X. Qiang, W. Y.-M. Cheung, L.-F. Li, F.-F. Sun, S. Qin, J.-C. Huang, G. M. Leung, E. C. Holmes, Y.-L. Hu, Y. Guan and W.-C. Cao, Identifying SARA-CoV-2 related coronaviruses in Malayan pangloins, Nature, 2020, 583, 282–285 CrossRef CAS.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Crystal structure of SARS-Cov-2 main protease provides a basis for design of improved α-ketamide inhibitors, Science, 2020, 368, 409–412 CAS.
- S. Sansanelli and A. Tassoni, Wild food plants traditionally consumed in the area of Bologna (Emilia Romagna region, Italy), J. Ethnobiol. Ethnomed., 2014, 10(69), 1–11, DOI:10.1186/1746-4269-10-69.
- S. S. Ebada, N. A. Al-Jawabri, F. S. Youssef, A. Albohy, S. M. Aldalaein, A. M. Disi and P. Proksch, In vivo antiulcer activity, phytochemical exploration, and molecular modeling of the polyphenolic-rich fraction of Crepis sancta extract, Inflammopharmacology, 2020, 28, 321–331 CrossRef CAS.
- K. Michalska, E. Szneler and W. Kisiel, Structure elucidation of an unusual new sequiterpene lactone glucoside ester from Crepis pannonica, Magn. Reson. Chem., 2016, 54, 175–177 CrossRef CAS.
- M. Korinek, V. D. Wagh, I.-W. Lo, Y.-M. Hsu, H.-Y. Hsu, T.-L. Hwang, Y.-C. Wu, Y.-B. Cheng, B.-H. Chen and F.-R. Chang, Antiallergic phorbol ester from the seeds of Aquilaria malaccensis, Int. J. Mol. Med., 2016, 17, 398, DOI:10.3390/ijms17030398.
- M. Korinek, Y. H. Tsai, M. El-Shazly, K. H. Lai, A. Backlund, S. F. Wu, W. C. Lai, T. Y. Wu, S. L. Chen, Y. C. Wu, Y. B. Cheng, T. L. Hwang, B. H. Chen and F. R. Chang, Antiallergic hydroxy fatty acids from Typhonium blumei explored through ChemGPS-NP, Front. Pharmacol., 2017, 8, 356 CrossRef.
- S.-C. Yang, P.-J. Chung, C.-M. Ho, C.-Y. Kuo, M.-F. Hung, Y.-T. Huang, W.-Y. Chang, Y.-W. Chang, K.-H. Chan and T.-L. Hwang, Propofol inhibits superoxide production, elastase release, and chemotaxis in formyl peptide–activated human neutrophils by blocking formyl peptide receptor 1, J. Immunol., 2013, 190, 6511–6519 CrossRef CAS.
- D. H. El-Kashef, F. S. Youssef, R. Hartmann, T.-O. Knedel, C. Janiak, W. Lin, I. Reimche, N. Teusch, Z. Liu and P. Proksch, Azaphilones from the Red Sea fungus Aspergillus falconensis, Mar. Drugs, 2020, 18, 204, DOI:10.3390/md18040204.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1, 19–25 CrossRef.
- J. Huang and A. D. MacKerell, CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data, J. Comput. Chem., 2013, 34, 2135–2145, DOI:10.1002/jcc.23354.
- V. Zoete, M. A. Cuendet, A. Grosdidier and O. Michielin, SwissParam, a Fast Force Field Generation Tool for Small Organic Molecules, J. Comput. Chem., 2011, 32, 2359–2368, DOI:10.1002/jcc.21816 PMID: 21541964.
- P. Mark and L. Nilsson, Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 014101 CrossRef.
- M. Parrinello and A. Rahman, Polymorphic transitions in single crystals: a new molecular dynamics method, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, LINCS: a linear constraint solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: an N log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- Y. F. Tsai, H. P. Yu, W. Y. Chang, F. C. Liu, Z. C. Huang and T. L. Hwang, Sirtinol inhibits neutrophil elastase activity and attenuates lipopolysaccharide-mediated acute lung injury in mice, Sci. Rep., 2015, 5, 8347, DOI:10.1038/srep08347.
- C.-F. Hsieh, J.-R. Jheng, G.-H. Lin, Y.-L. Chen, J. Y. Ho, C.-J. Liu, K.-Y. Hsu, Y.-S. Chen, Y. F. Chan, H.-M. Yu, P.-W. Hsieh, J.-H. Chern and J.-T. Horng, Rosmarinic acid exhibits broad anti-enterovirus A71 activity by inhibiting the interaction between the five-fold axis of capsid VP1 and cognate sulfated receptors, Emerg. Microb. Infect., 2020, 9, 1194–1205 CrossRef CAS.
- S. S. Ebada, D. H. El-Kashef, W. E. G. Müller and P. Proksch, Cytotoxic eudesmane sesquiterpenes from Crepis sancta, Phytochem. Lett., 2019, 33, 46–48 CrossRef CAS.
- F. R. Garcez, W. S. Garcez, L. Hamerski and A. C. de M. Miranda, Eudesmane and rearranged eudesmane sesquiterpenes from Nectandra cissiflora, Quím. Nova, 2010, 33, 1739–1742 CrossRef CAS.
- M.-E. F. Hegazy, H. Matsuda, S. Nakamura, T. A. Hussein, M. Yoshikawa and P. W. Paré, Chemical constituents and their antibacterial and antifungal activity from the Egyptian herbal medicine Chiliadenus montanus, Phytochemistry, 2014, 103, 154–161 CrossRef CAS.
- G. Flamini, E. Antognoli and I. Morelli, Two flavonoids and other compounds from the aerial parts of Centaurea bracteata from Italy, Phytochemistry, 2001, 57, 559–564 CrossRef CAS.
- J.-H. Woo, J.-H. Ahn, D.-S. Jang, K.-T. Lee and J.-H. Choi, Effect of kumatakenin isolated from cloves on the apoptosis of cancer cells and the alternative activation of tumor-associated macrophages, J. Agric. Food Chem., 2017, 65, 7893–7899 CrossRef CAS.
- S. E. Sajjadi, S. M. Ghanadian, M. Rabbani and F. Tahmasbi, Isolation and identification of secondary metabolites from the aerial parts of Stachys lavandulifolia Vahl, Iran, J. Pharm. Res., 2017, 16, 58–63 Search PubMed.
- H.-A. Ali, A. K. A. Chowdhury, A. K. M. Rahman, T. Borkowski, L. Nahar and S. D. Sarker, Pachypodol, a flavonol from the leaves of Calycopteris floribunda, inhibits the growth of Caco 2 colon cancer cell line in vitro, Phytother Res., 2008, 22, 1684–1687 CrossRef CAS.
- X.-Y. Xie, R. Wang and Y.-P. Shi, Flavonoids from the flowers of Matricaria chamomilla, Chem. Nat. Compd., 2014, 50, 910–911 CrossRef CAS.
- E. Fattorusso, F. U. Santelia, G. Appendino, M. Ballero and O. Taglialatela-Scafati, Polyoxygenated eudesmanes and trans-chrysanthemanes from the aerial parts of Santolina insularis, J. Nat. Prod., 2004, 67, 37–41 CrossRef CAS.
- G.-C. Wang, G.-Q. Li, H.-W. Geng, T. Li, J.-J. Xu, F. Ma, X. Wu, W.-C. Ye and Y.-L. Li, Eudesmane-type sesquiterpene derivatives from Laggera alata, Phytochemistry, 2013, 96, 201–207 CrossRef CAS.
- M.-E. F. Hegazy, A. A. El-Beih, A. R. Hamed, A. A. Abd El Aty, N. S. Mohamed and P. W. Paré, 3-Oxo-γ-costic acid fungal-transformation generates eudesmane sesquiterpenes with in vitro tumor-inhibitory activity, Bioorg. Med. Chem. Lett., 2017, 27, 3825–3828 CrossRef CAS.
- R. Chen, Y. Yang, J. Xu, Y. Pan, W. Zhang, Y. Xing, H. Ni, Y. Sun, Y. Hou and N. Li, Tamarix hohenackeri Bung exerts anti-inflammatory effects on lipopolysaccharide activated microglia in vitro, Phytomedicine, 2018, 40, 10–19 CrossRef CAS.
- Z. Uddin, Z. Li, Y. H. Song, J. Y. Kim and K. H. Park, Visconata: a rare flavonol having long chain fatty acid from Dodonaea viscosa which inhibits human neutrophil elastase (HNE), Tetrahedron Lett., 2017, 58, 2507–2511 CrossRef CAS.
- V. S. Somerville, A. J. Braakhuis and W. G. Hopkins, Effect of flavonoids on upper respiratory tract infections and immune function: A systematic review and meta-analysis, Adv. Nutr., 2016, 7, 488–497, DOI:10.3945/an.115.010538.
- E. Hirose, M. Matsushima, K. Takagi, Y. Ota, K. Ishigami, T. Hirayama, Y. Hayashi, T. Nakamura, N. Hashimoto, K. Imaizumi, K. Baba, Y. Hasegawa and T. Kawabe, Involvement of heme oxygenase-1 in kaempferol-induced antiallergic actions in RBL-2H3 cells, Inflammation, 2009, 32, 99–108 CrossRef CAS.
- L. Crocetti, I. A. Schepetkin, A. Cilibrizzi, A. Graziano, C. Vergelli, D. Giomi, A. I. Khlebnikov, M. T. Quinn and M. P. Giovannoni, Optimization of N-benzoylindazole derivatives as inhibitors of human neutorphil elastase, J. Med. Chem., 2013, 56, 6259–6272 CrossRef CAS.
- I. M. Ayoub, M. Korinek, T.-L. Hwang, B.-H. Chen, F.-R. Chang, M. El-Shazly and A. N. B. Singab, Probing the antiallergic and anti-inflammatory activity of biflavonoids and dihydroflavonols from Dietes bicolour, J. Nat. Prod., 2018, 81, 243–253 CrossRef CAS.
- M. T. ul Qamar, S. M. Alqahtani, M. A. Alamri and L.-L. Chen, Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants, J. Pharm. Sci., 2020, 10, 313–319 Search PubMed.
- A. M. Sayed, A. R. Khattab, A. M. AboulMagd, H. M. Hassan, M. E. Rateb, H. Zaid and U. R. Abdelmohsen, Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale, RSC Adv., 2020, 10, 19790–19802 RSC.
- Y. B. Ryu, H. J. Jeong, J. H. Kim, Y. M. Kim, J. Y. Park, D. Kim, T. T. Nguyen, S. J. Park, J. S. Chang, K. H. Park, M. C. Rho and W. S. Lee, Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition, Bioorg. Med. Chem., 2010, 18, 7940–7947 CrossRef CAS.
- R. S. Joshi, S. S. Jagdale, S. B. Bansode, S. S. Shankar, M. B. Tellis, V. K. Pandya, A. Chugh, A. P. Giri and M. J. Kulkarni, Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease, J. Biomol. Struct. Dyn., 2020, 38 DOI:10.1080/07391102.2020.1760137.
- D. S. N. B. K. Prasanth, M. Murahari, V. Chandramohan, S. P. Panda, L. R. Atmakuri and C. Guntupalli, In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS-CoV-2, J. Biomol. Struct. Dyn., 2020, 38 DOI:10.1080/07391102.2020.1779129.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1994762. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0ra04876c |
| This journal is © The Royal Society of Chemistry 2020 |