Open Access Article
Open Access ArticleHomogeneous nanodiscs of native membranes formed by stilbene–maleic-acid copolymers†
Mansoore
Esmaili
 a,
Chanelle J.
Brown
b,
Rustem
Shaykhutdinov
a,
Claudia
Acevedo-Morantes
c,
Yong Liang
Wang
c,
Holger
Wille
a,
Chanelle J.
Brown
b,
Rustem
Shaykhutdinov
a,
Claudia
Acevedo-Morantes
c,
Yong Liang
Wang
c,
Holger
Wille
 c,
Richard D.
Gandour
c,
Richard D.
Gandour
 *b,
S. Richard
Turner
b and
Michael
Overduin
*b,
S. Richard
Turner
b and
Michael
Overduin
 *a
*a
aDepartment of Biochemistry, University of Alberta, Edmonton, AB T6G 2H7, Canada. E-mail: Overduin@ualberta.ca
bDepartment of Chemistry and Macromolecules Innovation Institute, Virginia Tech, Blacksburg, VA 24061, USA. E-mail: gandour@vt.edu
cCentre for Prions and Protein Folding Diseases, University of Alberta, Edmonton, AB T6G 2M8, Canada
First published on 11th August 2020
Abstract
Methylstilbene-alt-maleic acid copolymers spontaneously convert biological membranes into bilayer discs with ∼20 nm diameters. This readily functionalizable class of copolymers has the compositional homogeneity, hydrophobicity, dynamics, and charge that may help to achieve optimal structural resolution, membrane dissolution, stability, and broad utility.
Introduction
Developing native nanodiscs1 that solubilize intact membrane–protein assemblies (memteins) by using hydrolyzed styrene–maleic anhydride (SMA) copolymers has led researchers to synthesize various polymers and copolymers to enhance applications of this technology.2 Resolving structures of memteins will inform how integral membrane proteins function in native lipid bilayer systems. A big challenge remains! The highly disperse memteins limit resolution in any structural determination technique. Without uniformity of memteins, visualizing, at atomic resolution, the polymers, lipids, and proteins cannot occur.Conventional radical alternating copolymerization of styrene (S) and maleic anhydride (MA) has a long history.3 As in all radical copolymerizations, the resulting copolymers are polydisperse in molecular weight and vary in composition. SMA copolymers may be predominately alternating; however, as S can add to itself, some random distributions of S–S dyads and S–S–S triads exist in the copolymer chain. In contrast, MA does not homopolymerize; consequently, no MA–MA dyads exist in the chain. A continuous stirred tank reactor can control the composition of the copolymers, but not the polydispersity of chains.3
Controlled radical copolymerizations with RAFT to produce SMA copolymer show promise in improving compositional homogeniety.4,5 In one study, varying [total monomer]/[RAFT agent] at high ratios of 43–400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and [S]/[MA] ratios of 2–20
1 and [S]/[MA] ratios of 2–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 produces copolymers with controlled compositional gradients and low polydispersity.4 In extracting a protein from HEK293F cells, the hydrolyzed copolymer produced with [total monomer]/[RAFT agent] of 43
1 produces copolymers with controlled compositional gradients and low polydispersity.4 In extracting a protein from HEK293F cells, the hydrolyzed copolymer produced with [total monomer]/[RAFT agent] of 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and [S]/[MA] of 3
1 and [S]/[MA] of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 forms nanodiscs with lower polydispersity than those formed by SMA2000, a commercial SMA copolymer with [S]/[MA] of 2
1 forms nanodiscs with lower polydispersity than those formed by SMA2000, a commercial SMA copolymer with [S]/[MA] of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and molar mass range of 3–7.5 kDa. In the other study, at a [total monomer]/[RAFT agent] ratio of 250
1 and molar mass range of 3–7.5 kDa. In the other study, at a [total monomer]/[RAFT agent] ratio of 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and [S]/[MA] ratios of 1.5–4
1 and [S]/[MA] ratios of 1.5–4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, SMA copolymers with S-rich blocks form when [MA] reaches zero.5 When mixed with lipid vesicles, these hydrolyzed SMA-block-S copolymers produce nanodiscs of different sizes depending on the [S]/[MA] ratio.
1, SMA copolymers with S-rich blocks form when [MA] reaches zero.5 When mixed with lipid vesicles, these hydrolyzed SMA-block-S copolymers produce nanodiscs of different sizes depending on the [S]/[MA] ratio.
Biomedical applications usually require specific, chemically well-defined compositions to achieve a desired result. To date, hydrolyzed SMA copolymers, the most widely used reagents to form native nanodiscs, still have (a) compositional irregularities, (b) batch-to-batch variabilities, and (c) challenges with aggregation in aqueous solutions. A (co)polymer must overcome these deficiencies to solve the critical challenge of forming stable and homogenous nanodiscs. Such protein–lipid–polymer nanodiscs would enable downstream functional and structural characterization with methods including cryo-electron microscopy, which has successfully determined memtein structures, although lipid headgroups and ligands remain unresolved.6–8
Here we report initial studies with hydrolyzed methyl-substituted stilbene–maleic anhydride (STMA) copolymers (Scheme 1a) for extracting proteins from membranes by forming nanodiscs. Stilbene (ST), a 1,2-diphenylethylene monomer, differs from S, 1-phenylethylene monomer. In contrast to S, ST does NOT homopolymerize; neither does MA. As both monomers are not homopolymerizing, strictly alternating copolymers form. These ST-MA copolymers meet one of the challenges—a precisely defined composition—that SMA copolymers do not.
The strictly alternating copolymers of MA with ST and with many substituted STs have similar persistence lengths (∼3 nm).9,10 Both SMA and STMA copolymers have semi-rigid backbones; STMA copolymers have 30–50% greater persistence lengths than those (∼2.2 nm) of SMA analogues. This increase results from the chain stiffening caused by steric crowding in the backbone of STMA.10,11 The chemical composition of STMA copolymers has –(–CHPh–CHPh–MA–)– repeating units; SMA, –(–CH2–CHPh–MA–)–. Replacing –CH2– with –CHPh– increases the rigidity of the polymer.
Comparing SMA to STMA, a second phenyl group increases the hydrophobicity of the repeating unit. The most effective hydrolyzed SMA copolymers for forming nanodiscs have an [S]/[MA] ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, presumably containing (–CH2–CHPh–CH2–CHPh–MA–) repeating units. This repeating unit has considerably more hydrophobicity than that of a 1
1, presumably containing (–CH2–CHPh–CH2–CHPh–MA–) repeating units. This repeating unit has considerably more hydrophobicity than that of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 SMA copolymer and nearly identical to that of a methyl-substituted STMA copolymer. However, the additional S unit in 2
1 SMA copolymer and nearly identical to that of a methyl-substituted STMA copolymer. However, the additional S unit in 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 SMA has conformational freedom, which reduces the rigidity of the copolymer.
1 SMA has conformational freedom, which reduces the rigidity of the copolymer.
Both hydrolyzed SMA and STMA show a large difference in the pKa of the two carboxyl groups—∼4.4 and ∼9.0.12 This difference indicates strong stabilization of the monoanion. At low pH, SMA and STMA exist in compact form; upon reaching a pH of ∼6, they form loose coils with statistical segment lengths of 5.9 and 6.7 nm, respectively. The repeating unit for the monoanion resembles that of succinate, which forms a strong intramolecular hydrogen bond.13 This bond restricts rotation of the backbone and contributes to the rigidity of the copolymers in the loosely coiled state.
Increased backbone rigidity of hydrolyzed dicarboxylated STMA, (5) compared to hydrolyzed carboxylated SMA may enhance anti-infectivity against HIV.14 Further docking studies of HIV surface proteins to the repeating unit of carboxylated STMA suggest a strong affinity of (5) to these proteins.15 The water solubility of (5) obviates using it to form nanodiscs. The hydrophobicity of methylated STMA copolymers, (2)–(4), should be conducive to membrane insertion and formation of nanodiscs.
Results and discussion
Methylated STMA copolymers were synthesized by free-radical polymerization of the substituted stilbene monomers and maleic anhydride reactants,16 purified by reprecipitation and then hydrolyzed (Scheme 1b). Hydrolyzed SMA2000 copolymer, which is well known to form native nanodiscs,1 served as a control. FT-IR and NMR spectra (Fig. S1†) confirmed the conversion of maleic anhydride groups into maleic acids to form the water-soluble product. All hydrolyzed STMA copolymers were soluble at pH 8.0 in 10 mM Tris and 100 mM NaCl.The sizes of the copolymers range between 4–6 kDa, except (5) (40 kDa) (Table 1). The polydispersity index (PĐI) for the copolymers range from 1.19 to 1.54; the narrow PĐIs likely arise from precipitation–reprecipitation workups of the copolymers The PĐI of commercial SMA is usually 2.0 or more.
| Polymer number, name, abbreviation | M n (kDa) | PĐI |
|---|---|---|
| (1) Poly(stilbene-alt-maleic anhydride), STMA | Nd | Nd |
| (2) 2,2′-Dimethyl-STMA, 2,2′-STMA | 5.1 | 1.52 |
| (3) 4-Methyl-STMA, 4-STMA | 5.8 | 1.54 |
| (4) 2-Methyl-STMA, 2-STMA | 4.4 | 1.19 |
| (5) 4,4′-Dicarboxylato-STMA, 4,4′-STMA | 40 | 1.34 |
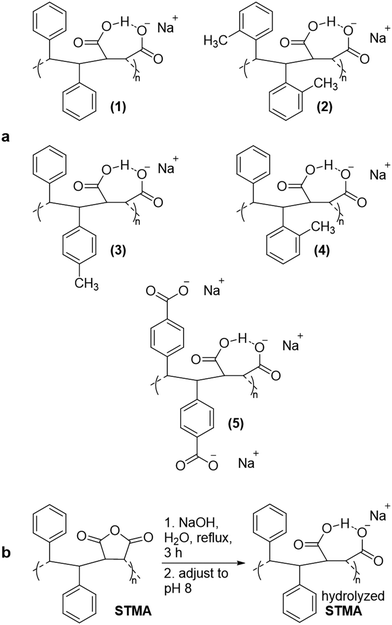
To test whether hydrolyzed STMA copolymers spontaneously form nanodiscs from lipid bilayers, we examined whether they solubilized vesicles composed of dimyristoylphosphatidylcholine (DMPC). The polymer concentrations used equaled that of SMA2000 (1–2% w/v). Only (3) and (4) efficiently dispersed multilayer vesicles of DMPC (Fig. 1a). In contrast, (1), (2) and (5) were relatively inactive, which was likely due to nonoptimal balances of hydrophobicity, negatively charge, high molecular weight17 and dynamics needed to insert into and disrupt membranes. Given the weak activity of these three, only (3) and (4) were assayed further.
31P NMR spectroscopy validated membrane-solubilizing activities of (3) and (4). Large vesicles composed of DMPC phospholipid exhibit 31P NMR signals that are broadened beyond detection; adding (3) or (4) clarified liposome suspensions and readily resolved 31P NMR signals (Fig. 1b). These results indicated formation of water-soluble nanodiscs and compared favorably to solubilization by the widely used SMA2000 copolymer.
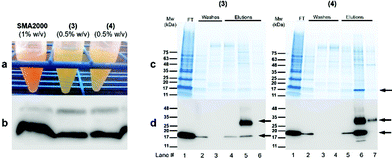
We expected STMA copolymers to behave in solution similarly to SMA because both have similar pKa values,12,13 similar charge distributions, and pH-dependent solubility profiles. SMA2000 was most water-soluble between pH 7.0 and 8.0. The STMA solubilities were tested. Surprisingly, (3) and (4) were soluble from pH 5 to 10 (Fig. 2).
 | ||
| Fig. 2 The pH sensitivity (left panel) and divalent cation (i.e., Ca2+, pH 8.0) tolerance (right panel) (a) (3) and (b) (4). | ||
These solubilities provide STMA copolymers a broader utility across the biological range of pH under which proteins operate in, for example, acidophiles, lysosomes and endosomes. The increased pH solubility of STMA likely results in part from the
• lack of polystyrene elements, which can seed aggregation,18
• strict alternation, which results in compact hydrophilic and hydrophobic groups, and
• more restricted backbone dynamics.
These advantages impart these hydrolyzed STMA copolymers with inherent practical advantages over hydrolyzed SMA copolymers.
Given the sensitivity of hydrolyzed SMA to divalent cations because of the maleic acid subunits, we expected both (3) and (4) to show similar sensitivities to levels of [Ca+2] that exceed 5 mM. Both (3) and (4) formed visible precipitates (Fig. S2†).19
This does limit using hydrolyzed STMA copolymers in functional assays that rely on high polycation levels over physiological levels. However, it does provide a method to coax membrane proteins from STMA-based nanodiscs and into liposomes or detergent phases.20
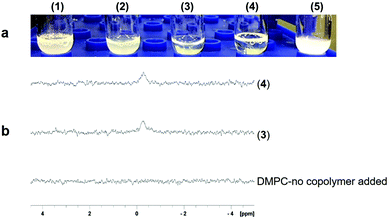
We investigated (3) and (4) for their abilities to solubilize E. coli outer membranes, which are tightly packed with proteins and glycolipids. The β-barrel protein PagP was expressed into the outer membrane to test whether (3) and (4) extract it in a physiologically relevant lipid bilayer complex. Copolymers (3) and (4) were added individually to raw outer membrane samples at a copolymer concentration of 0.5% w/v, which is two-fold lower than the level generally recommended for SMA2000 activity, as high polymer concentrations can induce large, undesirable aggregates. The yield of PagP monomers and dimers21,22 solubilized directly from the outer membrane into the supernatant was comparable to the levels of PagP extracted by SMA2000 at 1 w/v% concentration (Fig. 3b).
These data suggest that (3) and (4) function like SMA2000 by releasing protein–lipid complexes directly from biological membranes into native nanodiscs. Moreover, (3) and (4) appear to stabilize protein dimers, implying a gentler interaction with multimeric complexes. Low concentrations of (3) and (4) (0.5% w/v) can still solubilize PagP from native membrane with no effect on its SEC elution volume as compared with PagP solubilized in SMA2000 polymer (Fig. S3†). However, the considerable amount of PagP in flow-though (FT) fractions suggests that higher concentration of (3) and (4) is required to achieve optimal solubilization.
The nanodiscs containing His-tagged PagP were purified in a single step over a standard metal affinity resin, indicating compatibility with common affinity tags. The stain-free SDS-PAGE gel displays a prominent band for (4) after elution (lane 6) (Fig. 3c) (right) with apparent molecular weights of 18 and 36 kDa. The bands correspond to the monomeric unfolded and dimeric states, respectively, of PagP following heat denaturation.23 These states are less conspicuous in the purification when using (3) (Fig. 3c, lane 5) (left). The yield of PagP monomer and dimer appears to depend on which hydrolyzed STMA copolymer is used, with (4) generating a higher overall yield of purified protein. In both cases, PagP nanodiscs elute at the same retention volume (∼15 mL) by size exclusion chromatography (Fig. S3†), implying the similarity of sizes of nanodiscs.
Negative-stain TEM imaging of SEC fractions shows nanodiscs with diameters of ∼20 nm (Fig. 4b), with those formed by (4) being most homogeneous. These nanodiscs appear to be more regular and double the size of the PagP-containing nanodiscs formed by SMA2000 copolymer.1 Copolymer (4) appears to provide significantly more carrying capacity for larger memteins.
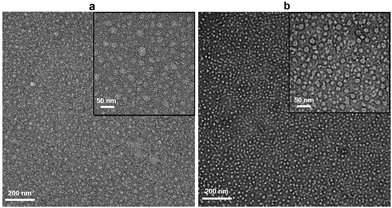 | ||
| Fig. 4 Negative stain EM images of native nanodiscs formed from outer membranes containing PagP by using (a) (3) and (b) (4). | ||
Conclusions
In summary, we report that, when activated, synthetic copolymers with strictly alternating ST and maleic acid subunits having minimal sequence and batch polydispersity can effectively solubilize biological membrane, including native bacterial membranes. Both (3) and (4) have less backbone flexibility than SMA copolymers. Yet both retain effective lipid-bilayer interactions, penetrate biological membranes to excise nanodiscs, and capture memteins. Hydrolyzed STMA copolymers can be utilized for detergent-free purification of native membrane proteins. As such, these copolymers add valuable diversity to the repertoire of amphipathic polymers available for native nanodisc production. Copolymers (3) and (4) offer notable advantages:• semi-rigidity of the polymer backbone,
• compositional regularity with strictly alternating periodicity of hydrophobic and hydrophilic groups,
• enhanced solubility over a wider range of pH values, and
• most significantly, generating large and homogeneous nanodiscs suitable for holding multimeric proteins in native bilayer sections.
Future refinements of the STMA copolymers can be envisaged. Various modifications of the maleic anhydride subunit24–27 could improve the polycation tolerance or enable incorporating thiols or other moieties (e.g. affinity tags and fluorescent labels). Such functional groups28–31 would further enhance the power of STMA copolymers and the utility of the resulting nanodiscs for large-scale production and high throughput assays of membrane protein targets. This unique potential of these STMA copolymers for detailed biophysical and structural analysis bodes well for future studies to increase the resolution of native protein–lipid–polymer complexes to the atomic level. The above results of (3) and (4) to enhance nanodisc solution behaviour, stability, and homogeneity represent a potential breakthrough for designing copolymers to maximize structural resolution of integral membrane proteins function in native lipid bilayers.
Conflicts of interest
M.E. and M.O. declare competing interests as they are co-inventors on a related patent filing.Acknowledgements
This work was supported by NSERC RGPIN-2018-04994, Campus Alberta Innovation Program (RCP-12-002C), Alberta Prion Research Institute/Alberta Innovates Bio Solutions (201600018) (M.O.) and National Science Foundation DMR-1609379 (S.R.T.) grants. NANUC was supported by TMIC, CFI and Genome Canada. We thank Russell Bishop for discussions and the PagP expression construct.References
- T. J. Knowles, R. Finka, C. Smith, Y.-P. Lin, T. Dafforn and M. Overduin, J. Am. Chem. Soc., 2009, 131(22), 7484–7485 CrossRef CAS PubMed.
- M. Overduin and M. Esmaili, Chem. Phys. Lipids, 2019, 218, 73–84 CrossRef CAS PubMed.
- B. Klumperman, Polym. Chem., 2010, 1, 558–562 RSC.
- A. A. A. Smith, H. E. Autzen, T. Laursen, V. Wu, M. Yen, A. Hall, S. D. Hansen, Y. Cheng and T. Xu, Biomacromolecules, 2017, 18, 3706–3713 CrossRef CAS PubMed.
- A. F. Craig, E. E. Clark, I. D. Sahu, R. Zhang, N. D. Frantz, M. S. Al-Abdul-Wahid, C. Dabney-Smith, D. Konkolewicz and G. A. Lorigan, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 2931–2939 CrossRef CAS PubMed.
- C. Sun, S. Benlekbir, P. Venkatakrishnan, Y. Wang, S. Hong, J. Hosler, E. Tajkhorshid, J. L. Rubinstein and R. B. Gennis, Nature, 2018, 557(7703), 123–126 CrossRef CAS PubMed.
- W. Qiu, Z. Fu, G. G. Xu, R. A. Grassucci, Y. Zhang, J. Frank, W. A. Hendrickson and Y. Guo, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(51), 12985–12990 CrossRef CAS PubMed.
- I. Tascon, J. S. Sousa, R. A. Corey, D. J. Mills, D. Griwatz, N. Aumuller, V. Mikusevic, P. J. Stansfeld, J. Vonck and I. Hanelt, Nat. Commun., 2020, 11(1), 626 CrossRef CAS PubMed.
- A. M. Savage, X. Zhou, J. Huang and S. R. Turner, Appl. Petrochem. Res., 2015, 5(1), 27–33 CrossRef CAS.
- J. Huang, X. Geng, C. Peng, T. Z. Grove and S. R. Turner, Macromol. Rapid Commun., 2018, 39, 1700530 CrossRef PubMed.
- Y. Li, M. Zhang, M. Mao, S. R. Turner, R. B. Moore, T. H. Mourey, L. A. Slater and J. R. Hauenstein, Macromolecules, 2012, 45, 1595–1601 CrossRef CAS.
- Y. Li, A. M. Savage, X. Zhou, S. R. Turner and R. M. Davis, J. Polym. Sci., Part B: Polym. Phys., 2013, 51(21), 1565–1570 CAS.
- M. S. Rudner, S. Jeremic, K. A. Petterson, D. R. Kent IV, K. A. Brown, M. D. Drake, W. A. Goddard and J. D. Roberts, J. Phys. Chem. A, 2005, 109(40), 9076–9082 CrossRef CAS PubMed.
- A. M. Savage, Y. Li, L. E. Matolyak, G. F. Doncel, S. R. Turner and R. D. Gandour, J. Med. Chem., 2014, 57(15), 6354–6363 CrossRef CAS PubMed.
- L. R. Hollingsworth, A. M. Brown, D. R. Bevan and R. D. Gandour, PLoS One, 2018, 13, e190658 CrossRef PubMed.
- Y. Li and S. R. Turner, Eur. Polym. J., 2010, 46(4), 821–828 CrossRef CAS.
- K. A. Morrison, A. Akram, A. Mathews, Z. A. Khan, J. H. Patel, C. Zhou, D. J. Hardy, C. Moore-Kelly, R. Patel, V. Odiba, T. J. Knowles, M. U. Javed, N. P. Chmel, T. R. Dafforn and A. J. Rothnie, Biochem. J., 2016, 473(23), 4349–4360 CrossRef CAS PubMed.
- S. Scheidelaar, M. C. Koorengevel, C. A. van Walree, J. J. Dominguez, J. M. Dörr and J. A. Killian, Biophys. J., 2016, 111, 1974–1986 CrossRef CAS PubMed.
- M. C. Fiori, Y. Jiang, G. A. Altenberg and H. Liang, Sci. Rep., 2017, 7(1), 1–10 CrossRef PubMed.
- S. J. Hesketh, D. P. Klebl, A. J. Higgins, M. Thomsen, I. B. Pickles, F. Sobott, A. Sivaprasadarao, V. L. G. Postis and S. P. Muench, Biochim. Biophys. Acta, Biomembr., 2020, 1862(5), 183192 CrossRef CAS PubMed.
- D. J. K. Swainsbury, S. Scheidelaar, N. Foster, R. van Grondelle, J. A. Killian and M. R. Jones, Biochim. Biophys. Acta, Biomembr., 2017, 1859(10), 2133–2143 CrossRef CAS PubMed.
- M. M. Xue, L. S. Cheng, I. Faustino, W. L. Guo and S. J. Marrink, Biophys. J., 2018, 115(3), 494–502 CrossRef CAS PubMed.
- R. E. Bishop, H. S. Gibbons, T. Guina, M. S. Trent, S. I. Miller and C. R. Raetz, EMBO J., 2000, 19(19), 5071–5080 CrossRef CAS PubMed.
- N. Z. Hardin, T. Ravula, G. D. Mauro and A. Ramamoorthy, Small, 2019, e1804813 CrossRef PubMed.
- S. Lindhoud, V. Carvalho, J. W. Pronk and M. E. Aubin-Tam, Biomacromolecules, 2016, 17(4), 1516–1522 CrossRef CAS PubMed.
- T. Ravula, N. Z. Hardin, S. K. Ramadugu and A. Ramamoorthy, Langmuir, 2017, 33(40), 10655–10662 CrossRef CAS PubMed.
- K. Yasuhara, J. Arakida, T. Ravula, S. K. Ramadugu, B. Sahoo, J. I. Kikuchi and A. Ramamoorthy, J. Am. Chem. Soc., 2017, 139(51), 18657–18663 CrossRef CAS PubMed.
- A. O. Oluwole, J. Klingler, B. Danielczak, J. O. Babalola, C. Vargas, G. Pabst and S. Keller, Langmuir, 2017, 33(50), 14378–14388 CrossRef CAS PubMed.
- S. Hall, C. Tognoloni, J. Charlton, É. Bragginton, A. Rothnie, P. Sridhar, M. Wheatley, T. Knowles, T. Arnold, K. Edler and D. TR, Nanoscale, 2018, 10(22), 10609–10619 RSC.
- T. Ravula, N. Z. Hardin, S. K. Ramadugu and A. Ramamoorthy, Langmuir, 2017, 33, 10655–10662 CrossRef CAS PubMed.
- T. Ravula, N. Z. Hardin, J. Bai, S. C. Im, L. Waskell and A. Ramamoorthy, Chem. Commun., 2018, 54, 9615–9618 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr03435e |
| This journal is © The Royal Society of Chemistry 2020 |