Immunoaffinity assisted LC-ICP-MS—a versatile tool in biomedical research
S.
Hann
,
K.
Boeck
and
G.
Koellensperger
*
Department of Chemistry, Division of Analytical Chemistry, University of Natural Resources and Applied Life Sciences-BOKU, Muthgasse 18, A-1190, Vienna. E-mail: gunda.koellensperger@boku.ac.at; Fax: +43-1-36006-6059; Tel: +43-1-36006-6086
First published on 21st October 2009
Abstract
A novel concept of combining immunochemistry, i.e., the specific recognition of antigens by antibodies, with elemental speciation analysis has potential use for targeted protein and peptide quantification in complex biological matrices. Accuracy, sensitivity and high selectivity is a major demand in this challenging task, both will be achieved when separating elemental labelled antigen-antibody complexes in solution via liquid chromatography in combination with inductively coupled plasma-mass spectroscopy (ICP-MS) detection. The on-line approach is rapid and automatable.
Recently, a methodological tool set available in elemental speciation analysis was proposed as a new opportunity for quantitative analysis of biomolecules such as proteins and peptides. The novelty relied on the combination of standard proteomic work flows with elemental detection by inductively coupled plasma-mass spectroscopy (ICP-MS).1,2 In this context, the term heteroatom-tagged proteomics was introduced, denoting the detection of a protein by use of an element being naturally present in a protein such as phosphorus (in phosphoproteins) or sulfur (in methionine and cysteine).3,4 New derivatization and staining techniques allowed extension of the application to biomolecules not containing heteroelements (elemental labeling).5–11 Several fundamental methodological studies addressed metalloproteins or metallodrug–protein adducts (which could be perceived as metal labelled proteins) showing the need for native multidimensional separation schemes.12–15 As a drawback most (analytical and preparative) native separations applied for ICP-MS based studies have significantly lower separation power compared to denaturing protocols. As a consequence the selectivity using the elemental speciation proteomic approach is often insufficient for the analysis of complex biological systems, especially when targeting binding partners of metals or metal containing compounds (e.g., cytostatic agents).
One key to significant methodological improvement of quantitative elemental proteomics is the use of antigen-antibody reactions. Immunochemistry offers a versatile tool, since the specific recognition of an antigen by an antibody can be exploited for the development of (1) selective elemental labels and (2) design of separation methods with unrivalled separation power. In their pioneering work Baranov et al.6 already combine conventional immunological methods with elemental analysis. The authors describe labelling strategies for ICP-MS using conventional fluorophores containing lanthanide elements or antibodies labelled by gold clusters. More applications of using commercially available elemental labels followed.7–9 Recently, a polymer-based elemental tagging kit was presented with the potential of even better sensitivity since the number of metal labels per binding site could be increased.16 Moreover, labelling techniques for proteins and antibodies based on iodination and DOTA-lanthanide chelates for application in ICP-MS were described.17,18 In these latter studies, the labelled antibodies were used for specific and simultaneous (multi-parameter) detection of different cytochrome P450 in animals after treatment with toxic or cancerogenic compounds.19 Thus, so far most elemental speciation studies integrated immunochemistry for the achievement of selective elemental labelling of the analytical targets. ICP-MS was used as a detector in immunoassays. Alternatively, laser ablation (LA) ICP-MS was applied after one-dimensional separation (western blotting).
The design of separation methods using immunochemistry in combination with ICP-MS is the next promising step to be taken. Immunoaffinity chromatography employing solid phase-immobilized antibodies is already known to be one of the most selective and rapid tools for the separation and purification of biomolecules. Recently, it could be shown that selective enrichment of proteins by immunoaffinity allowed quantification in the pg mL−1 range using liquid chromatography (LC)-MS.20 In this way detection limits comparable to immunoassays could be achieved. Another application of immunoaffinity in elemental speciation analysis addressed purification of samples, i.e., removal of high abundant proteins by immuno-depletion.21 However, the concept of targeting proteins by introduction of antibodies either as ligands in solution or as immobilized ligands on chromatographic stationary phases is new for elemental speciation analysis. The following fundamental experiments will prove the principle of such an approach and show the potential for future applications.
Liquid chromatographic separation of antigen-antibody complexes
For the first time solution based separations of antibody-antigen complexes in combination with on-line ICP-MS detection were addressed. Fundamental experiments were carried out using antigens amenable to ICP-MS, i.e., the elemental labeled peptide In-DOTA-Bß15–4210 and proteins labeled by metal containing anticancer drugs.Fig. 1 shows the size exclusion chromatography (SEC)-ICP-MS determination of the elemental labeled candidate drug In-DOTA-Bß15–42 which was incubated with monoclonal mouse anti Bß15–42 antibody. Rapid formation of a stable complex was achieved upon incubation in 100 mM NaCl, 20 mM TRIS-HCl (pH 7.4). The molar concentration ratio of peptide to antibody was 2:1. As can be readily observed the free antigen In-DOTA-Bß15–42, the antibody (a 55 kDa fragment as elucidated by PAGE) and the antibody-antigen complex AB-In-DOTA-Bß15–42 could be separated. The experiment clearly showed that the biospecificity of antibody recognition was retained for the elemental labelled In-DOTA-Bß15–42. The ratio of bound versus free peptide (0.127 ± 0.009) did not change significantly during an investigational period of 12 hours (see insert in Fig. 1). It is noteworthy that the complex formation was immediate. The absolute LOD for the detection of the complex AB-In-DOTA-Bß15–42 was 40 fmol. The detection limits given for biomarker detection by laser ablation (LA)-ICP-MS on Western blot are in the pmol range. Accordingly, we see the promising potential of solution based separations in the future for biomarker quantification using elemental labeled antibodies. The basic idea is to selectively label the analytical targets (e.g., biomarker) by incubation of the samples with elemental labeled antibodies and subsequent SEC-ICP-MS analysis. As a key advance, the problems arising in quantitative analysis with Western blots (e.g., losses during blotting procedure, time consuming separation and laser ablation processes, quantification problems of laser ablation technology) could be overcome. Moreover, the formation of an antibody-antigen complex is kinetically favored in solution compared to the formation in the heterogenic system “blot membrane–buffer containing antibodies”, which means that the analysis speed is significantly increased using SEC-ICP-MS allowing the design of fully automated high throughput systems.
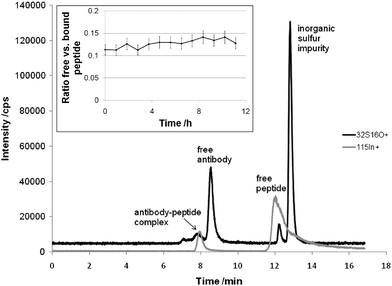 | ||
| Fig. 1 SEC-ICP-MS separation of an antigen-antibody complex formed by the elemental labeled candidate drug In-DOTA-Bß15–42 and the monoclonal mouse anti Bß15–42 antibody. The HPLC consisted of an AS 50 autosampler, a GP 40 gradient pump and the Chromeleon Chromatography Management System (Version 6.40), all from Dionex (Sunnyvale, California, USA). The SEC column was the BioSuite™ 125 (350 µL min−1 flow rate, 5 µL injection volume, 300 × 4.6 mm, 4 µm particle diameter; Waters, Milford, Massachusetts, USA). The SEC eluent was 150 mM NaCl and 20 mM Tris-HCl, pH 7. An ICP-QMS (ELAN DRC-II, PE SCIEX, Ontario, Canada) with a PFA-nebulizer and a cyclonic spray chamber was employed as the elemental detector. The figure inset shows the relative ratio of free versus bound peptide during an investigational period of 12 hours. The error bars correspond to a total combined uncertainty of 10%. | ||
Next, we used immunoaffinity assisted LC-ICP-MS experiments to address the quantification of metallodrug-protein adducts which is of the utmost importance in cancer research. SEC-ICP-MS offered in the past a simple and rapid tool for studying metallodrug-protein interactions with poor resolving power. A major disadvantage of the method is that the most important transporter proteins in serum samples, i.e., albumin and transferrin could not be separated. As a novelty, samples were incubated with specific antibodies prior to SEC-ICP-MS analysis. The retention time shift obtained upon antigen-antibody complex formation allowed the selectivity of the SEC-ICP-MS method to be increased. The approach was only valid provided the pure sample (without antibody addition) revealed no peak with the retention time of the metallodrug labelled antigen-antibody complex. The transferrin adducts of the experimental drug indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019, FFC14a) served as the ruthenium labelled antigen for method validation. The model antigen was prepared in vitro, purified by size exclusion, and incubated with a 5 fold molar excess of transferrin antibody. As can be readily observed in Fig. 2, despite the metallodrug bound to the protein the specific recognition between transferrin and the transferrin antibody was retained (hence, the adduct did not occlude the binding epitope). The Ru peak at 9 min was attributed to the antigen KP1019-transferrin, the peak at 7 min constituted the antigen-antibody complex (size exclusion limit of column, i.e., 150 kDa). Again, antigen-antibody complex formation was immediate and the yield of complex could not be increased upon increased incubation time. A ratio of 0.51 ± 0.03 (relative standard deviation 7%) for the KP1019–transferrin/KP1019–transferrin-antibody complex on average was assessed by repeated analysis of the in vitro standard over an analysis time of 700 min. As published earlier, in human plasma of cancer patients treated with KP1019 (clinical trial, phase 1) the drug was bound to the 60–80 kDa protein fraction. In order to determine the transferrin metallodrug interaction next to the high abundant albumin, a fully automated on-line two dimensional SEC-IC separation was combined with ICP-MS detection. The current novel method improves the limit of detection to 1 pmol (for the transferrin adduct). Hence, in future studies antibody reactions could be implemented for studying transferrin adducts in the presence of albumin by one dimensional SEC-ICP-MS.
![SEC-ICP-MS determination of Ru labeled antigen and antigen–antibody complex (experimental conditions as in Fig. 1): The transferrin (TF) adducts of indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019, FFC14a) served as the ruthenium labelled antigen. Purification by SEC resulted in a solution containing 4.5 nmol mL−1 adduct with a metallodrug/protein ratio of 1 which was incubated with 5 fold molar excess of antibody. The figure inset shows the relative ratio of TF-antibody complex versus free TF during an investigational period of 12 hours. The error bars correspond to a total combined uncertainty of 10%.](/image/article/2010/JA/b911462a/b911462a-f2.gif) | ||
| Fig. 2 SEC-ICP-MS determination of Ru labeled antigen and antigen–antibody complex (experimental conditions as in Fig. 1): The transferrin (TF) adducts of indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019, FFC14a) served as the ruthenium labelled antigen. Purification by SEC resulted in a solution containing 4.5 nmol mL−1 adduct with a metallodrug/protein ratio of 1 which was incubated with 5 fold molar excess of antibody. The figure inset shows the relative ratio of TF-antibody complex versus free TF during an investigational period of 12 hours. The error bars correspond to a total combined uncertainty of 10%. | ||
Finally, the potential of immunoaffinity chromatography for targeted protein quantification by elemental speciation analysis was addressed. In immunoaffinity, highly selective chromatographic systems are constructed by immobilization of antibodies on stationary phases. Our first methodological experiments concerned the quantification of KP1019–transferrin adduct proving the selectivity of the approach even in combination with elemental detectors. Columns pre-packed with Protein G Sepharose™ beads (HiTrap Protein G HP, 1 ml, bead size 34 µm, GE Healthcare Europe, Freiburg, Germany) were used for immobilization of transferrin antibody. Again a transferrin–KP1019 adduct containing 1 mol drug/mol protein was produced in vitro and purified by size exclusion. The corresponding albumin–KP1019 (without pre-fractionation by SEC) served for validation of selectivity and as the worst case scenario, blank. Loading of antibody was carried out off-line. Analysis of model compounds was performed on-line at flow rates of 0.5 mL min−1 using the PFA nebulizer and cyclonic spray chamber as the introduction system. Metallodrug protein adducts were applied to the affinity column at neutral pH (20 mM sodium phosphate, pH 7), and eluted at acidic pH (0.1 M glycine-HCl, pH 2.7). Since the antibody was not cross-linked on the Protein G Sepharose material, the columns were suitable only for a single analysis in these preliminary experiments. A linear calibration graph was obtained for KP1019–transferrin with the working range 50 pmol up to 700 pmol (R = 0.997, peak height calibration). Application of 700 pmol KP1019–albumin resulted in a blank signal corresponding to 30 pmol. These fundamental experiments clearly show the promising potential of immunoaffinity as the native separation method for LC-ICP-MS based applications. Future studies will aim at the implementation of such methods for selective enrichment of sensitive analysis of low abundant biomolecules amenable to ICP-MS analysis.
Conclusions
In fundamental experiments the principle of immunochemistry in LC-ICP-MS could be exploited to enhance sensitivity and selectivity of elemental proteomics. Both the separation of antibody-antigen complexes in solution and the design of stationary phases for targeted protein quantification proved to be promising analytical strategies. The potential of the LC separation of antibody-antigen complexes in solution relies in those applications where immunochemical reactions in combination with bulk analysis show limitations. This is the case where paired protein interactions are of interest, or simply, if it is not feasible to remove unbound antibodies by washing or spin filtration. The selectivity of the future LC methods will allow the same metal label for different antibodies to be used. Finally, only the application of LC opens the way to the separation of non-specific antibody complexes from desired ones.Acknowledgements
Financial support was granted by the Austria Science Fund (Projects L473-B11 and P21739-B11). Fibrex Medical Inc., and Marion Groeger and Peter Petzelbauer (Medical University of Vienna) are acknowledged for providing the monoclonal mouse anti Bß15–42 antibody, piCHEM R&D for synthesis of In-DOTA-Bβ15–42. We thank Bernhard Keppler (University of Vienna) for providing KP1019.References
- P. Marshall, O. Heudi, S. Bains, H. N. Freemann, F. Abou-Shakra and K. Reardon, Analyst, 2002, 127, 459 RSC.
- M. Wind, I. Feldmann, N. Jakubowski and W. D. Lehmann, Electrophoresis, 2003, 24, 1276 CrossRef CAS.
- J. Bettmer, N. Jakubowski and A. Prange, Anal. Bioanal. Chem., 2006, 386, 7 CrossRef CAS.
- A. Prange and D. Pröfrock, J. Anal. At. Spectrom., 2008, 23, 432 RSC.
- J. S. Becker, N. Jakubowski “Tutorial Review. The Synergy of Elemental and Biomolecular Mass Spectrometry: New Analytical Strategies in Life Sciences” to be published in Chem. Soc. Rev Search PubMed.
- V. I. Baranov, Z. Quinn, D. R. Bandura and S. D. Tanner, J. Anal. At. Spectrom., 2002, 17, 1148 RSC.
- S. D. Müller, R. A. Diaz-Bone, J. Felix and W. Goedecke, J. Anal. At. Spectrom., 2005, 20, 907 RSC.
- R. W. Hutchinson, R. Ma, C. W. McLeod, A. Milford-Ward and D. Lee, Can. J. Anal. Sci. Spectrosc., 2004, 49, 429 CAS.
- M. Careri, L. Elviri, A. Mangia and C. Mucchino, Anal. Bioanal. Chem., 2007, 387, 1851–1854 CrossRef CAS.
- G. Koellensperger, M. Groeger, D. Zinkl, P. Petzelbauer and S. Hann, J. Anal. At. Spectrom., 2009, 24, 97, 10.1039/b807397j.
- M. Krause, C. Scheler, U. Böttger, H. Weisshoff and M. Linscheid ( 2005) Verfahren und Reagenz zur spezifischen Identifizierung und Quantifizierung von einem oder mehreren Proteinen in einer Probe, Patentschrift DE 102 27 599 B4.
- S. Hann, G. Koellensperger, C. Obinger, P. Furtmueller and G. Stingeder, J. Anal. At. Spectrom., 2004, 19, 74 RSC.
- S. Hann, C. Obinger, G. Stingeder, M. Paumann, P. G. Furtmüller and G. Koellensperger, J. Anal. At. Spectrom., 2006, 21, 1224 RSC.
- G. Koellensperger, S. Daubert, R. Erdmann, S. Hann and H. P. Rottensteiner, Biol. Chem., 2007, 388, 1209 CrossRef CAS.
- M. Sulyok, S. Hann, C. Hartinger, B. K. Keppler, G. Stingeder and G. Koellensperger, J. Anal. At. Spectrom., 2005, 20, 856 RSC.
- X. Lou, G. Zhang, I. Herrera, R. Kinach, O. Ornatsky, V. Baranov, M. Nitz and M. A. Winnik, Angew. Chem., Int. Ed., 2007, 46, 1 CrossRef.
- N. Jakubowski, J. Messerschmidt, M. Garijo Anorbe, L. Waentig, H. Hayen and P. H. Roos, J. Anal. At. Spectrom., 2008, 23, 1487 RSC.
- N. Jakubowski, L. Waentig, H. Hayen, A. Venkatachalam, A. von Bohlen, P. H. Roos and A. Manz, J. Anal. At. Spectrom., 2008, 23, 1497 RSC.
- P. H. Roos, A. Venkatachalam, A. Manz, L. Waentig, C. U. Koehler and N. Jakubowski, Anal. Bioanal. Chem., 2008, 392, 1135 CrossRef CAS.
- B. L. Ackermann and M. J. Berna, Expert Rev. Proteomics, 2007, 4, 175 Search PubMed.
- J. Martosella, N. Zolotarjova, H. Liu, G. Nicol and B. E. Boyes, J. Proteome Res., 2005, 4, 1522 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2010 |
