Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Specialising and analysing instruction-tuned and byte-level language models for organic reaction prediction†
Jiayun
Pang
 *a and
Ivan
Vulić
*a and
Ivan
Vulić
 *b
*b
aSchool of Science, Faculty of Engineering and Science, University of Greenwich, Medway Campus, Central Avenue, Chatham Maritime, ME4 3RL, UK. E-mail: j.pang@gre.ac.uk
bLanguage Technology Lab, University of Cambridge, 9 West Road, Cambridge CB3 9DA, UK. E-mail: iv250@cam.ac.uk
First published on 19th August 2024
Abstract
Transformer-based encoder–decoder models have demonstrated impressive results in chemical reaction prediction tasks. However, these models typically rely on pretraining using tens of millions of unlabelled molecules, which can be time-consuming and GPU-intensive. One of the central questions we aim to answer in this work is: can FlanT5 and ByT5, the encoder–decoder models pretrained solely on language data, be effectively specialised for organic reaction prediction through task-specific fine-tuning? We conduct a systematic empirical study on several key issues of the process, including tokenisation, the impact of (SMILES-oriented) pretraining, fine-tuning sample efficiency, and decoding algorithms at inference. Our key findings indicate that although being pretrained only on language tasks, FlanT5 and ByT5 provide a solid foundation to fine-tune for reaction prediction, and thus become ‘chemistry domain compatible’ in the process. This suggests that GPU-intensive and expensive pretraining on a large dataset of unlabelled molecules may be useful yet not essential, to leverage the power of language models for chemistry. All our models achieve comparable Top-1 and Top-5 accuracy although some variation across different models does exist. Notably, tokenisation and vocabulary trimming slightly affect final performance but can speed up training and inference; the most efficient greedy decoding strategy is very competitive while only marginal gains can be achieved from more sophisticated decoding algorithms. In summary, we evaluate FlanT5 and ByT5 across several dimensions and benchmark their impact on organic reaction prediction, which may guide more effective use of these state-of-the-art language models for chemistry-related tasks in the future.
1 Introduction
Transformer-based deep learning techniques have revolutionised Natural Language Processing (NLP) in recent years. They are increasingly applied to chemical sciences where sequence representations of molecular structure, such as SMILES and SELFIES,1 bear similarity to language sequences. This makes it possible to adopt NLP algorithms to process and analyse molecules in a similar fashion as used to process and analyse text. This approach can be used for a wide range of tasks, such as molecular property prediction and data-driven molecular structure generation.The original Transformer architecture, introduced in the seminal paper of Vaswani et al.,2 contains two main components: (1) the encoder and (2) the decoder. All subsequent NLP models share some relationship with these components. For example, the widely used BERT (Bidirectional Encoder Representations from Transformers) only has the encoder component, while OpenAI’s GPT models and other very recent Large Language Models (LLMs) such as the Llama family3 or Gemini4 only have the decoder component. The encoder–decoder framework is ideally suited for sequence-to-sequence (seq2seq) learning, often referred to as text-to-text processing in NLP. In this framework, the encoder component captures the context of input sequences and sends it to the decoder which then generates output sequences.
Several studies have adapted seq2seq models for chemical reaction prediction tasks. Notably, the pioneering Molecular Transformer model by Schwaller et al.5 and the recent T5Chem model6 have achieved impressive performance. T5Chem adapted Google’s T5 (“Text-To-Text Transfer Transformer”) NLP model7 to chemical data represented in the SMILES format. T5 closely aligns with the encoder–decoder structure used in the original Transformer model but introduces the “Text-to-Text” framework which feeds text sequence (in a natural language such as English) as input and then generates text as output. This allows the same model to handle a variety of tasks simultaneously. To perform a specific task, a task-specific prefix is added to the input sequence, tailoring the model’s output. T5Chem pretrained the T5 encoder–decoder architecture with 97m SMILES from PubChem molecules and the USPTO_500_MT dataset, creating a multi-task reaction prediction model for five different types of reaction tasks. For example, it uses the task-specific prefix “Product:” for reaction product prediction, and a prefix “Reactants:” for single-step retrosynthesis. The advantage of multi-task learning is that it allows for simultaneous learning of multiple tasks by leveraging similarities between tasks and offers improved data efficiency, and fast learning without the need to predetermine a single specific prediction task.
T5Chem and a few other similar models8–10 have demonstrated the feasibility of applying a seq2seq framework to a variety of predictions in organic chemistry. However, several crucial issues have not been explored to enable more effective and accurate models. In the present study, we have trained multiple variants of two state-of-the-art seq2seq language models, namely instruction-tuned models Flan-T5 (ref. 11) and tokenisation-free byte-level models ByT5 (ref. 12), for standard organic reaction prediction tasks. With these two model architectures, our aim is to conduct a systematic empirical study on the following aspects:
(1) Adapted and adequate input preprocessing and tokenisation that reaches beyond natural language towards molecular structure. Tokenisation is usually the first step to train an NLP model. It is the process of breaking a sequence into discrete elements, called ‘tokens’ which are then converted to vectors/embeddings for machine learning models. In NLP, most pretrained language models over the past few years rely on tokenisation performed at the sub-word level, as it is effective with frequent tokens, capable of grouping sub-words while having some ability to deal with unknown words. However, sub-word tokenisation is still limited in its ability to deal with variants in spelling (e.g. typos) and unknown characters (e.g. from other languages). Recent approaches, such as ByT5, a variant of the multilingual T5 model which disposes of subword-level tokenisation, have shown the viability of token-free models which were trained on characters in the form of their UTF-8 byte encodings. ByT5 uses a standard Transformer architecture but is ‘tokenisation-free’ as it does not rely on a learned vocabulary to map words or sub-word units to tokens.
In addition, it has been demonstrated in NLP research that ByT5 is substantially more robust to noise in the data and performs better on tasks that are sensitive to spelling, grammar errors and ambiguous expressions, such as text on social media platforms. In terms of data noise, a chemical reaction dataset may have similarity with text on social media. For example, USPTO (United States Patent Office), the largest open-source chemical reaction dataset, contains noise: in this context, data noise is defined as incomplete reaction entry with missing or incorrect reactants, reagents and products; and could be quite common in all chemical reaction datasets due to the nature of how chemists record reactions, i.e. focusing on only the main product, and leaving unvaried reagents out when recording many similar reactions.13 Our work will thus implicitly assess whether ByT5’s advantage over dealing with noise in NLP data can be translated to better handling of noise in the chemical reaction data.
(2) Training data efficiency, i.e., how much annotated data is required in fine-tuning to generalise new sequences when working with chemical reaction datasets using seq2seq models.
(3) The use and impact of pretraining. T5-style language models are pretrained only on language data and/or language tasks, therefore are not “SMILES-aware”. T5Chem relies on self-supervised pretraining using 97m SMILES to learn the chemical space, which can be extremely GPU-intensive. Recently, language models pretrained on both language and chemical data have emerged. This cross-domain approach, adopted by models such as MolT5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 and nach0,15 creates a shared representation space. We thus also assess whether such hybrid pretraining offers better initialisation points for task-specific fine-tuning for reaction prediction tasks.
14 and nach0,15 creates a shared representation space. We thus also assess whether such hybrid pretraining offers better initialisation points for task-specific fine-tuning for reaction prediction tasks.
(4) Various other important modelling aspects that can impact the final task performance, e.g., model size, and decoding algorithm at inference.
2 Methodology
In this work, we aim to answer a range of questions related to the now established ability6,14–16 to apply encoder–decoder neural architectures, originally devised for language tasks in NLP research,7 to solve tasks related to organic reaction prediction. We vary models, modelling choices, as well as training and evaluation setups across several dimensions of comparison, which we outline next, and also summarise in Fig. 1.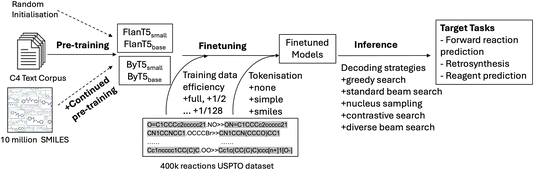 | ||
| Fig. 1 Illustration of the key areas explored along the flow of pretraining, fine-tuning and inference in our work. | ||
2.1 Model architectures
All the models follow the standard Transformer-based encoder–decoder structure where the input sequence is fed to the model (i.e., a sentence in a natural language in the case of language models or SMILES for organic reaction prediction), based on the original T5 architecture.7One of the central questions we aim to answer in this work is: can encoder–decoder models, originally pretrained only on language data and/or a variety of language tasks, be effectively specialised to organic reaction prediction tasks via task-specific fine-tuning? Will the model learn to encode and generate SMILES although it was originally pretrained for encoding and generating natural language? To this end, our starting points are different flavours of the T5-style, all pretrained on language data:
• The original T5 model,7 pretrained on the CommonCrawl-based C4 corpus covering ∼356b word tokens, via the span-mask denoising objective;
• The FlanT5 model11,17 is an instruction-tuned language model that starts from the pretrained T5 model of the same size, and then ‘instruction-tunes’ it on supervised data of 1800+ NLP tasks (Flan stands for Finetuning Language models).‡ It typically exhibits better performance than the underlying T5 model across a range of NLP tasks. It can also be used in a standard text-to-text fashion with task-specific fine-tuning if an ‘empty’ instruction is provided to the model (i.e., only the input sequence without an additional task description is provided). This is how we use the model for single task-specific fine-tuning.
• The ByT5 model12 obtains the same architecture, but disposes of standard subword-level tokenisation (see the next paragraph) and processes text as sequences of raw (UTF-8) bytes. Being originally designed to enhance multilingual NLP models, it was pretrained on the multilingual mC4 corpus spanning 101 diverse languages, again relying on the span-mask denoising objective.
All the models come in different sizes (in terms of model parameters), and due to high computational demands we mostly focus on benchmarking their Small and Base variants. The Small variant of T5 and FlanT5 comprises ∼60m parameters, while Base covers 220m parameters. ByT5 variants with the same label are not directly comparable to T5/FlanT5 as they contain a larger number of parameters: Small is 300m, and Base is 582m parameters.
Assuming the existence of task-specific training data for organic reaction prediction tasks, we also evaluate whether language-specific (and thus ‘chemical domain-incompatible’) pretraining is necessary at all, by also comparing to the same architectures of the same size which get randomly initialised and then fine-tuned for the task. We denote those variants of each model as random.
Furthermore, we also analyse whether continued pretraining in a self-supervised fashion on SMILES data offers any performance benefits before task-specific fine-tuning (denoted as cont). For the continued pretraining we rely on the standard masked language modelling objective adapted to SMILES. For each SMILES we sample 15% of its constituent tokens (where constituent tokens are based on the standard regular expression for SMILES from prior work,6 see also later) and then we do one of the following options: (a) replace the token with a special mask token (the ‘$’ character is used for the mask token) with the probability of 80%; or (b) replace the token with another random token from the same SMILES (10% probability); or (c) keep the token ‘as is’ (10% probability).
Finally, we test whether the ‘hybrid’ T5-style models of the same sizes pretrained in a multi-task fashion to handle both text and SMILES generation simultaneously, offer better initialisation points for single task-specific fine-tuning. For the latter, we select molT5 (ref. 14) and nach0 (ref. 15) (their Base variants spanning 220m parameters) as two representative recent models from this ‘SMILES-aware’ family of encoder–decoder models.
2.2 Input preprocessing, vocabulary, and tokenisation
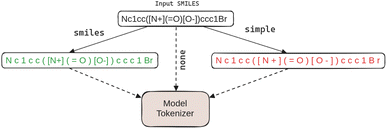
An often overlooked aspect of language models which may also have a profound impact on model behaviour and final performance, is the choice (and size) of vocabulary and corresponding tokenisation.18,19 We thus also delve deeper into their impact on final performance in organic reaction prediction tasks. With T5 and FlanT5 we experiment with different input preprocessing strategies, as also illustrated in Fig. 2. First, given a SMILES sequence as input, we can decide to directly feed the sequence into the model tokeniser: we refer to this variant as +none. We can also insert whitespaces into the input sequence in a trivial way, by simply dividing each character by a whitespace, ignoring any specific/chemistry domain knowledge; see Fig. 2. This means that the subsequent Br will be preprocessed into B r, and [Na+] will be turned into [ N a + ]. We refer to this input preprocessing strategy as +simple. It is possible to improve this via ‘SMILES-aware’ preprocessing which relies on a set of standard regular expressions as used in prior work6 to separate the input sequence into a whitespace-separated set of tokens. This strategy, referred to as +smiles, will not insert whitespaces into valid subsequences such as Br and [Na+]. The preprocessed input is then fed to the standard tokeniser of the pretrained model which has been created on and for natural language data. In this work, we thus test whether T5 and FlanT5 models can be used to process SMILES even with ‘ready-made’ tokenisers that have no adaptation or creation based on the (large collections of) raw SMILES data.Further, the original vocabularies of FlanT5 (and T5) typically span 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 subwords, but most of these subwords are associated with natural language subwords and can be safely discarded when processing SMILES with a much more restricted vocabulary. We thus trim the original vocabulary of FlanT5 by tokenising the large dataset of 116m SMILES sequences preprocessed via the +smiles strategy above, retaining only the subwords (and the corresponding embeddings) that occurred in those tokenised SMILES sequences.§ By doing this, we trimmed the vocabulary from the original 32k subwords to only 324 subwords, plus the three standard special tokens denoting the padding token, the start-of-sequence token and the ‘unknown’ token. This trimming of the vocabulary and the corresponding embeddings has a double effect: (1) it speeds up training and inference as it effectively constrains the search space, and (2) it reduces model size from 60m to 44.4m parameters (FlanT5Small) or from 220m to 198.7m (FlanT5Base) without losing any modelling capability and expressiveness in organic reaction prediction tasks. We denote variants that rely on the vocabulary and embed the trimming step as +trim, and variants with the original vocabulary as +orig. Different model variants are then possible when this step is combined with input preprocessing strategies (+none+orig as the simplest variant without any interventions versus, e.g., +smi+orig or +smi+trim).
000 subwords, but most of these subwords are associated with natural language subwords and can be safely discarded when processing SMILES with a much more restricted vocabulary. We thus trim the original vocabulary of FlanT5 by tokenising the large dataset of 116m SMILES sequences preprocessed via the +smiles strategy above, retaining only the subwords (and the corresponding embeddings) that occurred in those tokenised SMILES sequences.§ By doing this, we trimmed the vocabulary from the original 32k subwords to only 324 subwords, plus the three standard special tokens denoting the padding token, the start-of-sequence token and the ‘unknown’ token. This trimming of the vocabulary and the corresponding embeddings has a double effect: (1) it speeds up training and inference as it effectively constrains the search space, and (2) it reduces model size from 60m to 44.4m parameters (FlanT5Small) or from 220m to 198.7m (FlanT5Base) without losing any modelling capability and expressiveness in organic reaction prediction tasks. We denote variants that rely on the vocabulary and embed the trimming step as +trim, and variants with the original vocabulary as +orig. Different model variants are then possible when this step is combined with input preprocessing strategies (+none+orig as the simplest variant without any interventions versus, e.g., +smi+orig or +smi+trim).
Finally, given that SMILES comprise only special symbols, numbers, and alphabet letters where common atoms involved in organic reactions are represented as single or double letters, namely Br, Cl, N, O, S, P, F, I, b, c, n, o, s, p (note: upper case letters refer to non-aromatic atoms and lower case letters refer to aromatic atoms in SMILES), all the characters in SMILES sequences are UTF-8 compatible and can be easily fed into a tokenisation-free byte-level model such as ByT5. ByT5 is ‘tokenisation-free’ as it does not rely on a learned vocabulary to map words or sub-word units to vocabulary items and simply operates on the vocabulary of 256 UTF-8 characters. Therefore, in this work we also analyse the potential of such byte-level natural language-pretrained encoder–decoder models for organic reaction prediction tasks. We run ByT5 without any input preprocessing to keep the length of the sequences tractable, e.g., the only variant tested is +none+orig. In prior work in NLP, it was shown that ByT5 is significantly more robust to noise in the data and performs better on tasks that are sensitive to spelling, grammar errors and ambiguous expressions, such as text on social media platforms or speech transcribed to text.20 Our goal is to assess whether ByT5’s advantage related to dealing with noise in NLP data can be translated to better handling of noise in the chemical reaction data such as typically encountered in the USPTO datasets.
2.3 Decoding strategy
In NLP, the use of the decoding strategy (i.e., the method used to generate strings from the model) can have a profound impact on output text.21 However, the impact of the decoding strategy when working with SMILES as the input has not been properly investigated: prior work6 typically fixes the decoding strategy to the standard beam search (with beam width fixed, typically to 5), and does not provide any further evidence on how the chosen decoding strategy might impact the final output. In this paper, we thus compare the performance with much more efficient greedy search decoding to the standard beam search, and also analyse how varying beam width impacts the final results. Moreover, we also run additional experiments with more sophisticated text generation strategies adopted from NLP research such as nucleus sampling22 and contrastive search.23 For technical details on the respective generation strategies, we refer the reader to the original publications.3 Experimental setup
We examine a range of encoder–decoder models with distinct properties (e.g., tokenisation, model size, model architecture) in a variety of setups (e.g., full-model fine-tuning, parameter-efficient fine-tuning, training data size, inference strategies). After further combining with a range of possible reaction prediction tasks (e.g., forward reaction prediction, single-step retrosynthesis, reaction yield prediction, reaction type classification), this yields a huge space of possible experimental configurations. Therefore, due to the large computational demands associated with the full experimental space, we do not present the full spectrum of results across all possible tasks; we rather zoom in and provide a representative selection of models, tasks, and experimental configurations that offer useful insight into the main interactions and models’ inner workings without loss of generality.3.1 Target tasks and datasets
Following prior work,5,6 we train and evaluate the models on:(1) Forward reaction prediction (FWD-S) with reactant–reagent separation¶ on the USPTO_MIT dataset. The full training set consists of 409![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 035 input–output pairs.
035 input–output pairs.
(2) Single-step retrosynthesis (RETRO) on USPTO_50k, where the full training set comprises 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 029 input–output pairs.
029 input–output pairs.
(3) Reagent prediction (REAG) on the USPTO_500_MT dataset. While USPTO_500_MT has been created primarily as a multi-task dataset,6 unless stated otherwise we conduct task-specific tuning for the single task using only its corresponding data, which comprises 116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 input–output pairs.
360 input–output pairs.
We use the datasets and corresponding splits as provided by Lu et al.6 We evaluate all the models on the full test set of the RETRO task (5004 pairs), while we randomly sample 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 test instances from the full, larger test sets of FWD-S and REAG to speed up inference due to a large number of experiments.||
000 test instances from the full, larger test sets of FWD-S and REAG to speed up inference due to a large number of experiments.||
3.2 Input preprocessing
We experiment with different input preprocessing strategies primarily for FlanT5 (with some experiments on T5) as illustrated in Fig. 2. The maximum input sequence length is task-dependent and is defined based on the longest input sequence in each task-specific training set. Accordingly, the maximum output sequence length is also set according to the longest output sequence in the corresponding training sets.3.3 Evaluation metrics
We report the standard accuracy at rank K (Acc@K) scores, measuring if the gold sequence can be found in the top K output sequences generated by the model. We select K-s following prior work (i.e., it is {1, 3, 5} for RETRO and REAG, and {1, 2, 5} for FWD-S).3.4 Continued pretraining setup
For the cont variants that run self-supervised continued pretraining on SMILES data, we randomly sample a pretraining set of 10m SMILES from the full set of 116m SMILES, and run continued pretraining for 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps. It might be possible that longer pretraining and a larger pretraining set – as conducted e.g. by T5Chem6 – might yield models better adapted to SMILES input and output, but that setup exceeds our computational resources. We run continued pretraining for the FlanT5Base+trim+smi variant and ByT5Base; the batch size is set to 64 for both models, learning rate is set to 0.0001 with inverse square root decay, and the optimiser used is Adafactor.24
000 steps. It might be possible that longer pretraining and a larger pretraining set – as conducted e.g. by T5Chem6 – might yield models better adapted to SMILES input and output, but that setup exceeds our computational resources. We run continued pretraining for the FlanT5Base+trim+smi variant and ByT5Base; the batch size is set to 64 for both models, learning rate is set to 0.0001 with inverse square root decay, and the optimiser used is Adafactor.24
3.5 Training setup
Due to a large number of experiments, we run a very constrained hyperparameter search, based on the development sets of FWD-S and RETRO and FlanT5Small+none+orig as the underlying model variant. The found hyper-parameters are then ported to all the other models: we acknowledge that finer-grained hyper-parameter optimisation is warranted as part of future research. Unless stated otherwise, we fine-tune with the learning rate set to 0.003, batch size is 64, weight decay is set to 0.01, and warm-up is set to 5000 steps. For FWD-S in the standard setting we fine-tune for 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps (∼16 epochs), while we fine-tune for 100 epochs for RETRO (62.5k steps), and 50 epochs for REAG (∼90k steps).** The optimiser is Adafactor in all the experiments. For each model variant and task, we select the checkpoint based on performance on the corresponding development set: in most cases, it is the end checkpoint.
000 steps (∼16 epochs), while we fine-tune for 100 epochs for RETRO (62.5k steps), and 50 epochs for REAG (∼90k steps).** The optimiser is Adafactor in all the experiments. For each model variant and task, we select the checkpoint based on performance on the corresponding development set: in most cases, it is the end checkpoint.
3.6 Inference setup
The main decoding strategy is a standard beam search with beam width set to 5, again following prior work.5,6,15 In Section 4.6, we also analyse the impact of beam width on Acc@1 scores, and also run preliminary investigations with more sophisticated decoding strategies borrowed from NLP research (see Section 2).4 Results and discussion
4.1 Impact of the underlying encoder–decoder model
The results in the three tasks with different underlying encoder–decoder models are summarised in Table 1, where several findings emerge. First, results from all the models are ‘in the same ballpark’ although some variation across different models does exist. For instance, FlanT5 is a better initialisation point for task-specific fine-tuning than the corresponding original T5, yielding higher scores across all Acc@K metrics and all three tasks. Second, byte-level models offer the highest and most robust performance across the three tasks on average, indicating that a byte-level ‘tokenisation-free’ approach is a promising avenue for future work dealing with SMILES input and output. Next, the two models that were already fine-tuned on SMILES data and relevant tasks do not exhibit any advantage as starting points for task-specific fine-tuning over the ‘language-tuned’ models such as FlanT5 and ByT5. The exception is the REAG task where molT5Base and nach0Base show strong performance: we suspect that this is due to the fact that the exact REAG training data was already included in their multi-task tuning, while they did not see the data of the other two tasks beforehand. Finally, a general important finding is that ‘language-pretrained’ encoder–decoder models can be directly used for task-specific fine-tuning, even without any SMILES-oriented self-supervised pretraining, yielding competitive and robust performance.| Model ↓ | FWD-S | RETRO | REAG | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Acc@1 | Acc@2 | Acc@5 | Acc@1 | Acc@3 | Acc@5 | Acc@1 | Acc@3 | Acc@5 | |
| T5Small | 89.01 | 93.36 | 95.45 | 42.35 | 57.97 | 63.13 | 3.85 | 7.36 | 9.32 |
| T5Base | 89.28 | 93.36 | 95.47 | 42.59 | 58.31 | 62.89 | 20.33 | 29.72 | 33.90 |
| FlanT5Base | 89.83 | 93.73 | 95.73 | 44.86 | 61.45 | 66.55 | 23.27 | 31.86 | 35.82 |
| ByT5Small | 90.06 | 93.75 | 95.71 | 43.96 | 58.81 | 63.09 | 22.85 | 31.20 | 35.43 |
| ByT5Base | 90.10 | 93.90 | 96.07 | 44.74 | 60.25 | 64.89 | 24.18 | 32.27 | 36.18 |
| molT5Small14 | 88.98 | 93.23 | 95.60 | 42.63 | 59.09 | 63.53 | 20.89 | 27.81 | 31.39 |
| molT5Base14 | 89.90 | 93.68 | 95.75 | 42.71 | 58.45 | 63.77 | 25.0 | 32.87 | 36.82 |
| nach0Base15 | 87.33 | 92.12 | 94.72 | 41.33 | 57.35 | 62.59 | 25.0 | 33.54 | 37.26 |
Running the best-performing models on the full test set of FWD-S yields the peak Acc@1/Acc@5 scores of 90.2/96.2 which surpasses the performance of Molecular Transformer5 (88.8/92.6) and is close to the T5-Chem model (90.4/96.4) despite the fact that the models have not been pretrained on SMILES prior to task-specific fine-tuning. The advantage over Molecular Transformer also occurs on the full test set of RETRO (Acc@1 of 45.0 vs. 43.5), but the gap to T5-Chem is slightly larger (45.0/66.7 vs. 46.5/70.5). Additional results showing the minimal variation in relative trends in results between the subsampled 10k test set and full set are available later in Table 4.
4.2 Impact of input preprocessing, vocabulary size and tokenisation
The results of the analysis, based on the FlanT5 model family, are provided in Table 2. We observe that trimming the vocabulary and the corresponding embeddings (i.e., the +trim+smi variant) does not have any negative impact on the final performance: in contrast, due to the side-effect of constraining the search space, it even has a slight positive impact on the final scores. In terms of input preprocessing, +smi has a slight edge over +simple and +none (but the differences are sometimes minimal). This holds for FlanT5 of both tested sizes as well as for the original T5 model of those sizes (not shown). We speculate that this might be due to the fact that the language-pretrained model is better adapted to seeing input that contains whitespaces, but further investigations are warranted in future research. The random variant without any language-specific pretraining is unable to learn the task well, even with relatively large amounts of training data (e.g., ∼400k training instances for FWD-S), which again indicates the importance of non-random initialisation from the language-pretrained model.| Variant ↓ | FlanT5Small | FlanT5Base | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FWD-S | RETRO | FWD-S | RETRO | |||||||||
| Acc@1 | Acc@2 | Acc@5 | Acc@1 | Acc@2 | Acc@5 | Acc@1 | Acc@3 | Acc@5 | Acc@1 | Acc@3 | Acc@5 | |
| +orig+smi | 88.92 | 93.18 | 95.59 | 42.53 | 59.79 | 65.73 | 89.83 | 93.73 | 95.73 | 42.87 | 58.47 | 63.83 |
| +trim+smi | 89.32 | 93.29 | 95.66 | 44.68 | 60.71 | 66.13 | 89.91 | 93.72 | 95.70 | 44.86 | 61.45 | 66.55 |
| +orig+none | 88.70 | 93.22 | 95.56 | 43.07 | 58.05 | 63.21 | 89.84 | 93.51 | 95.58 | 42.03 | 57.71 | 62.05 |
| +trim+none | 0.03 | 0.04 | 0.06 | 0.16 | 0.22 | 0.22 | 0.04 | 0.05 | 0.07 | 0.16 | 0.2 | 0.24 |
| cont+trim+smi | — | — | — | — | — | — | 89.34 | 93.39 | 95.48 | 43.55 | 59.89 | 65.17 |
| +orig+simple | 88.95 | 93.31 | 95.52 | 43.86 | 60.71 | 65.67 | 89.25 | 93.31 | 95.52 | 42.61 | 58.83 | 64.39 |
| Random | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Finally, as expected, the +trim+none variant provides extremely low scores: we report it for didactic purposes to emphasise how the mismatch between input preprocessing, vocabulary and tokenisation can have extremely detrimental negative impact; this variant trims the vocabulary based on the +spaces preprocessing, while the actual input to the model does not undergo the same preprocessing step, which creates the mismatch and the model has to deal with (sub)sequences that result in the ‘unknown’ tokens.
4.3 Impact of model size
Zooming again into Table 1 and 2, we observe that, as expected, the Base variants of different models offer slightly higher performance than the Small variants. This holds for T5 and FlanT5 as well as for ByT5 and molT5. However, the gap between Small and Base is not large (the only exception is T5 on REAG, see Table 1): therefore, the choice of the model size also depends on the final need – if performance is paramount, Base is a stronger option, while the Small variants trade off some performance for improved training and inference efficiency.In order to verify if further performance benefits can be reaped from an even larger encoder–decoder model, we fine-tune FlanT5Large (780m parameters) in the RETRO task. However, the increase in model size does not result in any increase in performance, with obtained Acc@1/Acc@3/Acc@5 scores of 43.96/59.61/64.23. In fact, the scores even decrease a bit, which might be the result of overfitting to the RETRO training data.
4.4 Impact of continued SMILES-based pretraining
Continued pretraining (the +cont variant in Table 2) does not have a positive impact on final task performance in FWD-S and RETRO: in fact, it even yields slight performance drops. However, the +cont variant of ByT5Base does yield some small gains in both tasks when we decrease the learning rate for fine-tuning from the default 0.003 to 0.0003: it reaches 90.37/93.9 (Acc@2)/96.07 in FWD-S and 44.88/61.33 (Acc@3)/66.29. These mixed preliminary results call for further investigation and also indicate the importance of finer-grained hyper-parameter optimisation, which is required as part of future research.4.5 Visualisation of prediction with SHAP
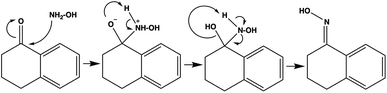
To check the chemistry validity of our models, we applied SHAP (SHapley Additive exPlanations)25 to explain the predictions made by ByT5 and FlanT5 (+orig+none variant of FlanT5 is used for simplicity). The SHAP explanations visualise the contributions of reactant and reagents to the structure of the predicted product at the token level. SHAP is a popular approach to explain the output of any machine learning model using game theory. The key idea behind SHAP is to calculate the Shapley values for each feature of the dataset and each Shapley value represents the impact of that feature on the model’s prediction. In the case of sequence input and output, such as the reactions we work with, multiple Shapley values are calculated for every token in the input sequence (i.e. reactant and reagents) for every token in the output sequence (i.e. the predict product). These SHAP values can be visualised in a matrix-like heatmap.We analysed a classic organic reaction: the generation of ketoxime from hydroxylamine and ketone in the presence of HCl and CH3OH. The SMILES representation of the reaction is as follows: NO.O=C1CCCc2ccccc21.CO.Cl>>ON=C1CCCc2ccccc21. This reaction involves first the nucleophilic attack of the nitrogen in hydroxylamine (NH2–OH) on the carbonyl carbon in the ketone, followed by two successive proton transfers from the nitrogen to the oxygen in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O to allow for elimination of water, resulting in the formation of the oxime functional group (C
O to allow for elimination of water, resulting in the formation of the oxime functional group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–OH) (Fig. 3).
N–OH) (Fig. 3).
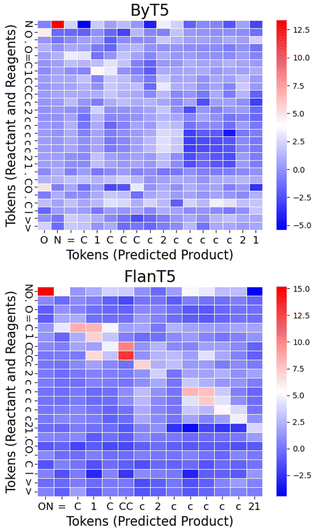
Fig. 4 displays the computed Shapley values for this reaction from the ByT5 and FlanT5 models. The figure is generated using Seaborn heatmap and coloured using the ‘bwr’ colormap from matplotlib. The y-axis represents the sequence of tokens from the reactants and reagents, while the x-axis represents the sequence of tokens from the predicted product. Additionally, we visualised the impact of tokens in the reactants and reagents on the first few tokens in the product by projecting the Shapley values onto their 2d structures (Fig. 5). Fig. 5 was generated using the GetSimilarityMapFromWeights function in RDkit26 and coloured using the ‘bwr’ colormap from matplotlib.
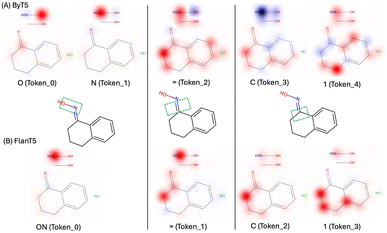 | ||
| Fig. 5 Visualisation of the impact of tokens in the reactants and reagents on the first few tokens in the predicted product. | ||
Both models highlight the hydroxylamine (N and O, token_0 and token_1 from ByT5; NO, token_0 in FlanT5) as having the most significant impact on the product, which aligns with the underlying reaction mechanism. Furthermore, both models correctly identified that ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–OH in the product originates from the hydroxylamine, while the oxygen in the reactant ketone has a much weaker impact. This suggests that the models may have learned the correct reaction pathway, i.e. the oxygen in the ketone as the leaving group. In addition, atoms present in both the reactant and product (such as the aromatic ring and the cyclic aliphatic ketone) also exhibit noticeable impact. For the double bond in C1
N–OH in the product originates from the hydroxylamine, while the oxygen in the reactant ketone has a much weaker impact. This suggests that the models may have learned the correct reaction pathway, i.e. the oxygen in the ketone as the leaving group. In addition, atoms present in both the reactant and product (such as the aromatic ring and the cyclic aliphatic ketone) also exhibit noticeable impact. For the double bond in C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–OH, neighbouring carbons in the cyclic aliphatic ring have a strong impact. Although the exact reason for this is not clear, it is well known that SN2 reactions, like this one, are sensitive to steric hindrance from neighbouring atoms. It is likely that our models have learned correlations between key substructures of the molecule, as has been observed in T5Chem.
N–OH, neighbouring carbons in the cyclic aliphatic ring have a strong impact. Although the exact reason for this is not clear, it is well known that SN2 reactions, like this one, are sensitive to steric hindrance from neighbouring atoms. It is likely that our models have learned correlations between key substructures of the molecule, as has been observed in T5Chem.
We provide two additional examples of SHAP analysis with more complex product structure and reaction conditions (e.g. reagents) in the SI.† FlanT5 seems slightly better at identifying key reagents involved in the reactions compared to ByT5. However, the impact of the reagents is not always associated with the reaction centre. Further work is needed to provide a more quantitative analysis of the SHAP method for chemical reaction prediction models.
4.6 Further analyses
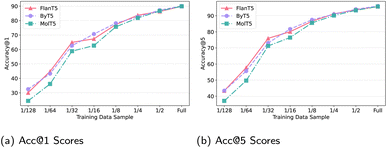
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 training instances), 1/4, …, 1/128 (3196 instances) of the full FWD-S training set. Each smaller set is a subset of all the larger (sub)sets. The models in comparison are Base versions of FlanT5 (+trim+smi), ByT5, and molT5: the summary of their performance across different FWD-S data sizes is provided in Fig. 6. The three models display very similar ‘performance trajectories’ where molT5 lags slightly behind the two other models for smaller data samples across both Acc@K metrics. Combined with the results already observed in Table 1 and 2, these plots point to a more general conclusion that the actual training data size and quality is more instrumental to the final performance than the chosen encoder–decoder architecture. While there is some variation that stems from the choice of input preprocessing, tokenisation, and model size, the data size is still a key factor determining the ‘performance magnitude’. This conjecture calls for future research across two axes: (1) creation of larger and higher-quality training and evaluation datasets15 for single-task and multi-task training; (2) work on more sample-efficient learning via transfer learning.
518 training instances), 1/4, …, 1/128 (3196 instances) of the full FWD-S training set. Each smaller set is a subset of all the larger (sub)sets. The models in comparison are Base versions of FlanT5 (+trim+smi), ByT5, and molT5: the summary of their performance across different FWD-S data sizes is provided in Fig. 6. The three models display very similar ‘performance trajectories’ where molT5 lags slightly behind the two other models for smaller data samples across both Acc@K metrics. Combined with the results already observed in Table 1 and 2, these plots point to a more general conclusion that the actual training data size and quality is more instrumental to the final performance than the chosen encoder–decoder architecture. While there is some variation that stems from the choice of input preprocessing, tokenisation, and model size, the data size is still a key factor determining the ‘performance magnitude’. This conjecture calls for future research across two axes: (1) creation of larger and higher-quality training and evaluation datasets15 for single-task and multi-task training; (2) work on more sample-efficient learning via transfer learning.
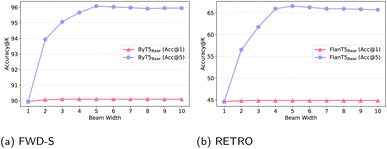 | ||
| Fig. 7 Beam analysis experiments: (a) FWD-S; (b) RETRO; we vary beam size from 1 (greedy) to 10; the FlanT5Base variant is +trim+smi. | ||
Additional experiments with top-K sampling, nucleus sampling and contrastive search with hyper-parameters suggested from NLP research (e.g., temperature for top-K sampling, top_p for nucleus sampling), did not yield any noteworthy benefits over the simple greedy search. Changing the hyper-parameters for the efficient nucleus sampling based on development set performance can yield slight benefits at inference over greedy search without damaging inference efficiency, but the gains are typically slight, ranging between 0.1 and 0.4 Acc@1 points. In summary, a more focused study on the impact of decoding strategy for SMILES generation is also warranted as part of future research.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000+ instances) in Table 3. The estimates were made in the following setup for all models: 100
000+ instances) in Table 3. The estimates were made in the following setup for all models: 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 training steps with the batch size of 16 and gradient accumulation of 4 (yielding the effective batch size of 64), input sequence length and output sequence length set to 144. All the estimates are based on single runs on a single 24GiB NVIDIA RTX 4090 GPU. The time estimates indicate that the main experiments with the chosen model architectures and their corresponding size can typically be run on consumer-level GPUs. Another finding is that some speed-ups can be achieved via trimming the vocabulary and the corresponding embeddings (moving from +orig to +trim) without any performance degradation (see Table 2 again).
000 training steps with the batch size of 16 and gradient accumulation of 4 (yielding the effective batch size of 64), input sequence length and output sequence length set to 144. All the estimates are based on single runs on a single 24GiB NVIDIA RTX 4090 GPU. The time estimates indicate that the main experiments with the chosen model architectures and their corresponding size can typically be run on consumer-level GPUs. Another finding is that some speed-ups can be achieved via trimming the vocabulary and the corresponding embeddings (moving from +orig to +trim) without any performance degradation (see Table 2 again).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 training steps) in the FWD-S task on a single 24GiB RTX 4090 GPU
000 training steps) in the FWD-S task on a single 24GiB RTX 4090 GPU
| Model variant | Time |
|---|---|
| FlanT5Small+orig+smi | 5 h 20 min |
| FlanT5Small+trim+smi | 4 h 25 min |
| FlanT5Base+orig+smi | 14 h 50 min |
| FlanT5Base+trim+smi | 13 h 35 min |
| ByT5Small | 16 h 20 min |
| ByT5Base | 30 h 55 min |
| Model ↓ | 10k | Full | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc@1 | 64 | 16 | 4 | Acc@1 | 64 | 16 | 4 | |
| Greedy search | ||||||||
| FlanT5Small | 89.10 | 1′15′′ | 3′57′′ | 11′56′′ | 89.20 | 4′53′′ | 15′53′′ | 46′25′′ |
| FlanT5Base | 89.85 | 2′27′′ | 5′49′′ | 17′59′′ | 89.73 | 9′59′′ | 22′53′′ | 1h12′14′′ |
| ByT5Small | 89.91 | 2′33′′ | — | — | 90.01 | 10′23′′ | 14′29′′ | 28′08′′ |
| ByT5Base | 89.95 | 3′23′′ | 4′21′′ | 9′6′′ | 90.01 | 13′36′′ | 17′25′′ | 36′38′′ |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Beam search (beam = 5) | ||||||||
| FlanT5Small | 89.32 | 6′48′′ | 8′36′′ | 18′22′′ | 89.36 | 27′23′′ | 35′55′′ | 1h12′48′′ |
| FlanT5Base | 89.91 | 12′59′′ | 13′41′′ | 27′4′′ | 89.86 | 52′32′′ | 54′1′′ | 1h44′19′′ |
| ByT5Small | 90.06 | 12′40′′ | 13′42′′ | 16′5′′ | 90.10 | 51′5′′ | 53′57′′ | 1h4′32′′ |
| ByT5Base | 90.10 | 14′20′′ | 15′23′′ | 18′39′′ | 90.20 | 58′18′′ | 1h4′4′′ | 1h14′9′′ |
Since we implemented customised inference batching, we acknowledge that further efficiency gains might be met through additional code optimisation and/or through the use of quantisation techniques;27 this goes beyond the scope of this work.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 training steps on multi-task data of the USPTO_500_MT dataset:6 it again covers three tasks (forward reaction prediction, retrosynthesis, and reagent prediction), where training data of each task constitutes 116
000 training steps on multi-task data of the USPTO_500_MT dataset:6 it again covers three tasks (forward reaction prediction, retrosynthesis, and reagent prediction), where training data of each task constitutes 116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 instances for the total of 349
360 instances for the total of 349![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 training instances for multi-task training. Each instance is marked by a specific prefix (Product:, Reactants:, Reagents:) which links it to the original task.
080 training instances for multi-task training. Each instance is marked by a specific prefix (Product:, Reactants:, Reagents:) which links it to the original task.
We fine-tune two models – FlanT5Base (+orig+none for simplicity) and ByT5Small – and run standard evaluation on the three tasks. Acc@1 scores of the multi-task model on the three tasks are 24.01 (REAG), 95.82 (FWD), 71.77 for FlanT5Base, and 24.15, 96.78, 72.24 for ByT5Small. This indicates that multi-task fine-tuning is also possible starting from language-pretrained model checkpoints. We noted that Acc@1 scores from multi-task models are higher on both FWD (95.82 and 96.78) and RETRO (71.77 and 72.24) compared to our single task models presented in Table 1 and 2 (∼90 for FWD and ∼44 for RETRO). The 70% RETRO accuracy from the multi-task models is above the typical 45–50% but is consistent with the findings from the recent paper of Lu and Zhang6 which used a similar multi-task approach. Our accuracy range is also broadly in line with several recent literature sources that explored various GNN models and data augmentation strategies (top-1 accuracy 60% to 70% for retrosynthesis).28–31 Possible reasons of our higher top-1 accuracy on RETRO task are:
(1) Different datasets: for single task-specific experiments, we used corresponding dedicated datasets (e.g., USPTO_MIT for FWD-S; USPTO_50k for RETRO). For multi-task fine-tuning we used the dataset USPTO_500_MT created by Lu and Zhang exactly for multi-task fine-tuning and evaluation. This dataset contains five objectives: forward reaction prediction, single-step retrosynthesis, reagent prediction, reaction type prediction, and reaction yield prediction. According to Lu and Zhang, “the training, validation and testing sets are well separated to ensure no reaction overlapping across all tasks” to avoid a possible data leakage problem.6 In terms of size, USPTO_50k includes 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 016 reactions which is much smaller than USPTO_500_MT which contains 143
016 reactions which is much smaller than USPTO_500_MT which contains 143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 535 reactions. However, the higher accuracy may not be fully accounted for just based on dataset size as it has been shown that a larger dataset such as USPTO_MIT (480k reactions) only brings small improvement in RETRO top 1 accuracy.28,29
535 reactions. However, the higher accuracy may not be fully accounted for just based on dataset size as it has been shown that a larger dataset such as USPTO_MIT (480k reactions) only brings small improvement in RETRO top 1 accuracy.28,29
(2) Advantage of the multi-task approach: multi-task reaction prediction has only emerged in the past few years, demonstrating that forward reaction, retrosynthesis and reagent predictions are different yet closely related tasks and can therefore be leveraged together to build a more robust model.6,32,33 Qiao et al. showed that transformer-based multi-task models can improve top-1 accuracy by 7–10% using just a few thousand reactions and the extent of improvement can depend on training data size.32 With our much larger multi-task training data (USPTO_500_MT, 143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 535 reactions), a higher improvement in accuracy is possible.
535 reactions), a higher improvement in accuracy is possible.
We then continue to fine-tune multi-task ByT5Small for the single REAG task. This yields marginal gains in the REAG task (from 24.15 to 24.38 with greedy search), but, as expected, it also yields catastrophic forgetting of the other two tasks (e.g., Acc@1 drops from 95.82 to 35.26) due to full-model specialisation. Motivated by these preliminary results, we plan to delve deeper into multi-task fine-tuning setups in future work, also coupled with recent advances in modular and parameter-efficient learning in NLP that by design avoid issues such as catastrophic forgetting and interference in multi-task setups.34–36
5 Reflection and conclusion
Our work operates at the intersection of several broad areas of NLP research and AI in chemistry, including multi-task learning, transfer learning and computer-assisted synthesis planning. Our preliminary results indicate that although FlanT5 and ByT5 are pretrained only on language tasks, they provide a solid foundation for fine-tuning in reaction prediction. This suggests that GPU-extensive pretraining on large unlabelled molecules may not be essential to leverage the power of these state-of-the-art language models for chemistry. While there is some variation that stems from the choice of input preprocessing, tokenisation, and model size, the training data size is instrumental to the final performance of the fine-tuned models. It is worth noting that the USPTO_500_MT dataset used in our work (a subset curated from the full USPTO (1976–2013) dataset37) covers only general organic reactions. It contains limited stereochemical information38 and data on catalysts is scarce. Further fine-tuning will be necessary to specialise our models to specific types of reactions or prediction tasks. In such cases, factors that are not significant to the performance of our models could be explored further. For example, when predicting catalysts, a larger beam size and perhaps more sophisticated decoding strategies will be needed to provide more exploratory predicted output. For future work, we plan to delve deeper into multi-task fine-tuning using recent advances in parameter-efficient and modular learning and improve our models for novel and more challenging reaction prediction tasks.5.1 Limitations
One limitation of this wide empirical study is that, due to a large number of experiments coupled with computational constraints, all the experimental configurations were run once (with a random seed set to 42), while averaging over multiple training runs is a typical good practice.39 However, in our preliminary experiments, we verified that score fluctuations were minimal, which should mitigate this concern. Furthermore, as mentioned in Section 3, a larger-scale per-model and per-variant hyper-parameter optimisation might also yield improved performance. Better performance may also be achieved via longer fine-tuning and checkpoint selection using the development sets. We also have not extensively explored the full range of possible decoding strategies, nor have we thoroughly optimised hyper-parameters associated with the tested decoding strategies (e.g., for nucleus sampling): this can also be further tuned on the task-specific development sets. Future work should also further analyse the interplay between longer and more GPU-intensive self-supervised pretraining on larger SMILES data and task-specific fine-tuning.Data availability
Code related to single-task fine-tuning and inference of T5-style models is available at https://www.github.com/cambridgeltl/chem-encdec/.Author contributions
JP and IV conceived and designed the study, carried out the research and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by the UK Engineering and Physical Sciences Research Council (EPSRC) (grant number EP/Y004167/1). Ivan Vulic would like to thank Benjamin Minixhofer for some fruitful discussions on tokenisation and acknowledges the support of a personal Royal Society University Research Fellowship (no. 221137).References
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Mach. Learn.: Sci. Technol., 2020, 1, 045024 Search PubMed.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. u. Kaiser and I. Polosukhin, Adv. Neural Inf. Process. Syst., 2017, 5998–6008 Search PubMed.
- H. Touvron, L. Martin, K. Stone, et al., CoRR, arXiv, 2023, preprint, arXiv:2307.09288, DOI:10.48550/arXiv.2307.09288.
- R. Anil, S. Borgeaud, Y. Wu, J. Alayrac, et al., CoRR, arXiv, 2023, preprint, arXiv:2312.11805, DOI:10.48550/arXiv.2312.11805.
- P. Schwaller, T. Laino, T. Gaudin, P. Bolgar, C. A. Hunter, C. Bekas and A. A. Lee, ACS Cent. Sci., 2019, 5, 1572–1583 CrossRef PubMed.
- J. Lu and Y. Zhang, J. Chem. Inf. Model., 2022, 62, 1376–1387 CrossRef PubMed.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, J. Mach. Learn. Res., 2020, 21, 1–67 Search PubMed.
- T. Sagawa and R. Kojima, ReactionT5: a Large-Scale Pre-trained Model towards Application of Limited Reaction Data, 2023 Search PubMed.
- D. S. Wigh, J. Arrowsmith, A. Pomberger, K. C. Felton and A. A. Lapkin, J. Chem. Inf. Model., 2024, 64, 3790–3798 CrossRef PubMed.
- A. Toniato, A. C. Vaucher, M. M. Lehmann, T. Luksch, P. Schwaller, M. Stenta and T. Laino, Chem. Mater., 2023, 35, 8806–8815 CrossRef PubMed.
- H. W. Chung, L. Hou, S. Longpre, B. Zoph, Y. Tay, W. Fedus, E. Li, X. Wang, M. Dehghani, S. Brahma, A. Webson, S. S. Gu, Z. Dai, M. Suzgun, X. Chen, A. Chowdhery, S. Narang, G. Mishra, A. Yu, V. Y. Zhao, Y. Huang, A. M. Dai, H. Yu, S. Petrov, E. H. Chi, J. Dean, J. Devlin, A. Roberts, D. Zhou, Q. V. Le and J. Wei, CoRR, arXiv, 2022, preprint, arXiv:2210.11416, DOI:10.48550/arXiv.2210.11416.
- L. Xue, A. Barua, N. Constant, R. Al-Rfou, S. Narang, M. Kale, A. Roberts and C. Raffel, Trans. Assoc. Comput. Linguist., 2022, 10, 291–306 CrossRef.
- A. Toniato, P. Schwaller, A. Cardinale, J. Geluykens and T. Laino, Unassisted Noise Reduction of Chemical Reaction Data Sets, 2021 Search PubMed.
- C. Edwards, T. Lai, K. Ros, G. Honke, K. Cho and H. Ji, Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, Abu Dhabi, United Arab Emirates, 2022, pp. 375–413 Search PubMed.
- M. Livne, Z. Miftahutdinov, E. Tutubalina, M. Kuznetsov, D. Polykovskiy, A. Brundyn, A. Jhunjhunwala, A. Costa, A. Aliper and A. Zhavoronkov, CoRR, arXiv, 2023, preprint, arXiv:2311.12410, DOI:10.48550/arXiv.2311.12410.
- G. Chilingaryan, H. Tamoyan, A. Tevosyan, N. Babayan, L. Khondkaryan, K. Hambardzumyan, Z. Navoyan, H. Khachatrian and A. Aghajanyan, CoRR, arXiv, 2022, preprint, arXiv:2211.16349, DOI:10.48550/arXiv.2211.16349.
- S. Longpre, L. Hou, T. Vu, A. Webson, H. W. Chung, Y. Tay, D. Zhou, Q. V. Le, B. Zoph, J. Wei and A. Roberts, International Conference on Machine Learning, ICML 2023, 23–29 July 2023, Honolulu, Hawaii, USA, 2023, pp. 22631–22648 Search PubMed.
- A. Petrov, E. La Malfa, P. Torr and A. Bibi, Adv. Neural Inf. Process. Syst., 2023, 36963–36990 Search PubMed.
- O. Ahia, S. Kumar, H. Gonen, J. Kasai, D. Mortensen, N. Smith and Y. Tsvetkov, Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing, Singapore, 2023, pp. 9904–9923 Search PubMed.
- S. Ruder, J. Clark, A. Gutkin, M. Kale, et al., Findings of the Association for Computational Linguistics: EMNLP 2023, Singapore, 2023, pp. 1856–1884 Search PubMed.
- G. Wiher, C. Meister and R. Cotterell, Trans. Assoc. Comput. Linguist., 2022, 10, 997–1012 CrossRef.
- A. Holtzman, J. Buys, L. Du, M. Forbes and Y. Choi, 8th International Conference on Learning Representations, ICLR 2020, Addis Ababa, Ethiopia, April 26–30, 2020, 2020 Search PubMed.
- Y. Su, T. Lan, Y. Wang, D. Yogatama, L. Kong and N. Collier, Advances in Neural Information Processing Systems 35: Annual Conference on Neural Information Processing Systems 2022, NeurIPS 2022, New Orleans, LA, USA, November 28 to December 9, 2022 Search PubMed.
- N. Shazeer and M. Stern, Proceedings of the 35th International Conference on Machine Learning, ICML 2018, Stockholmsmässan, Stockholm, Sweden, July 10–15, 2018, pp. 4603–4611 Search PubMed.
- S. M. Lundberg and S.-I. Lee, Advances in Neural Information Processing Systems 30, Curran Associates, Inc., 2017, pp. 4765–4774 Search PubMed.
- G. Landrum, RDKit: Open-Source Cheminformatics, http://www.rdkit.org, 2023, _03_1b1 (Q1 2023) Release, DOI:10.5281/zenodo.7828379.
- T. Dettmers, M. Lewis, Y. Belkada and L. Zettlemoyer, Advances in Neural Information Processing Systems 35: Annual Conference on Neural Information Processing Systems 2022, NeurIPS 2022, New Orleans, LA, USA, November 28 to December 9, 2022 Search PubMed.
- S. Xie, R. Yan, J. Guo, Y. Xia, L. Wu and T. Qin, Proc. AAAI Conf. Artif. Intell., 2023, 37, 5330–5338 Search PubMed.
- X. Zhang, Y. Mo, W. Wang and Y. Yang, Retrosynthesis prediction enhanced by in-silico reaction data augmentation, arXiv, 2024, preprint, arXiv:2402.00086, DOI:10.48550/arXiv.2402.00086.
- H. Lee, S. Ahn, S.-W. Seo, Y. Y. Song, E. Yang, S.-J. Hwang and J. Shin: A Selection-based Approach for Retrosynthesis via Contrastive Learning, arXiv, 2021, preprint, arXiv:2105.00795, DOI:10.48550/arXiv.2105.00795.
- Z. Zhong, J. Song, Z. Feng, T. Liu, L. Jia, S. Yao, T. Hou and M. Song, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2024, 14, e1694 Search PubMed.
- H. Qiao, Y. Wu, Y. Zhang, C. Zhang, X. Wu, Z. Wu, Q. Zhao, X. Wang, H. Li and H. Duan, RSC Adv., 2022, 12, 32020–32026 RSC.
- C. J. Taylor, K. C. Felton, D. Wigh, M. I. Jeraal, R. Grainger, G. Chessari, C. N. Johnson and A. A. Lapkin, ACS Cent. Sci., 2023, 9, 957–968 CrossRef PubMed.
- Z. Han, C. Gao, J. Liu, J. Zhang and S. Q. Zhang, CoRR, arXiv, 2024, preprint, arXiv:abs/2403.14608, DOI:10.48550/arXiv.2403.14608.
- N. Ding, Y. Qin, G. Yang, F. Wei, Z. Yang, Y. Su, S. Hu, Y. Chen, C. Chan, W. Chen, J. Yi, W. Zhao, X. Wang, Z. Liu, H. Zheng, J. Chen, Y. Liu, J. Tang, J. Li and M. Sun, CoRR, arXiv, 2022, preprint, arXiv:2203.06904, DOI:10.48550/arXiv.2203.06904.
- J. Pfeiffer, S. Ruder, I. Vulic and E. M. Ponti, CoRR, arXiv, 2023, preprint, arXIv:2302.11529, DOI:10.48550/arXiv.2302.11529.
- D. M. Lowe, Extraction of Chemical Structures and Reactions From the Literature, PhD thesis, University of Cambridge, 2012.
- G. Pesciullesi, P. Schwaller, T. Laino and J.-L. Reymond, Nat. Commun., 2020, 11, 4874 CrossRef PubMed.
- J. Dodge, S. Gururangan, D. Card, R. Schwartz and N. A. Smith, Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, 2019, pp. 2185–2194 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fd00104d |
| ‡ Instruction-tuning is a specialised form of fine-tuning in which a model is fine-tuned using pairs of input–output instructions, enabling it to learn specific tasks guided by these instructions. |
| § In practice, we did not tokenise the entire set of 116m SMILES sequences but stopped tokenisation when we encountered that the set of ‘seen subwords’ has not been extended after seeing 500k new SMILES sequences. |
| ¶ Preliminary experiments in the task version with mixed reactants and reagents,6 yields very similar relative trends in results and comparisons; we thus omit it for brevity and to save computation. |
| || We have empirically validated that the relative trends in results do not change due to the test set sampling. We also run a smaller selection of models on full test sets to enable direct comparison to prior methods that operated on the same datasets. |
| ** We capped the maximum number of steps to 100k to keep training times manageable and the work computationally feasible given the large number of trained models. We note that FWD-S has by far the largest training set out of the three tasks and there is a possibility that the models on FWD-S are ‘under-trained’ and would benefit from additional fine-tuning. |
| This journal is © The Royal Society of Chemistry 2025 |