Open Access Article
Open Access ArticleA toolbox approach for multivalent presentation of ligand–receptor recognition on a supramolecular scaffold†
Svenja
Ehrmann‡
 ab,
Chih-Wei
Chu‡
ab,
Chih-Wei
Chu‡
 c,
Shalini
Kumari‡
a,
Kim
Silberreis
d,
Christoph
Böttcher
b,
Jens
Dernedde
c,
Shalini
Kumari‡
a,
Kim
Silberreis
d,
Christoph
Böttcher
b,
Jens
Dernedde
 d,
Bart Jan
Ravoo
d,
Bart Jan
Ravoo
 *c and
Rainer
Haag
*c and
Rainer
Haag
 *a
*a
aInstitute for Chemistry and Biochemistry, Freie Universität Berlin, Takustr. 3, 14195 Berlin, Germany. E-mail: haag@chemie.fu-berlin.de
bResearch Center of Electron Microscopy, Freie Universität Berlin, Fabeckstr. 36a, 14195 Berlin, Germany
cOrganic Chemistry Institute and Center for Soft Nanoscience, Westfälische Wilhelms-Universität Münster, Corrensstr. 40, 48149 Münster, Germany. E-mail: b.j.ravoo@uni-muenster.de
dCharité-Universitätsmedizin Berlin, Corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, Berlin Institute of Health, Institute of Laboratory Medicine, Clinical Chemistry and Pathobiochemistry, CVK Augustenburger Platz 1, 13353 Berlin, Germany
First published on 21st May 2018
Abstract
A supramolecular toolbox approach for multivalent ligand–receptor recognition was established based on β-cyclodextrin vesicles (CDVs). A series of bifunctional ligands for CDVs was synthesised. These ligands comprise on one side adamantane, enabling the functionalisation of CDVs with these ligands, and either mannose or sulphate group moieties on the other side for biological receptor recognition. The physicochemical properties of the host–guest complexes formed by β-cyclodextrin (β-CD) and adamantane were determined by isothermal titration calorimetry (ITC). Ligand–lectin interactions were investigated by surface plasmon resonance experiments (SPR) for the mannose ligands and the lectin Concanavalin A (ConA). Microscale thermophoresis (MST) measurements were applied for sulphate-dependent binding to L-selectin. In both cases, a multivalent affinity enhancement became apparent when the ligands were presented on the CDV scaffold. Furthermore, not only the clustering between our supramolecular mannosylated complex and Escherichia coli (E. coli), expressing the lectin FimH, was visualised by cryo-TEM, but also the competitive character to detach bound E. coli from a cell line, representing the uroepithelial cell surface, was demonstrated. In summary, a facile and effective supramolecular toolbox was established for various ligand–receptor recognition applications.
Introduction
In biological systems, cell-linked carbohydrates generate the glycocalyx, which is involved in diverse fundamental processes, such as cell–cell recognition and downstream signalling in living organisms.1,2 The interactions, e.g., carbohydrate–lectin binding, are usually weak and non-covalent. A multivalent display of ligands and receptors on complementary surfaces, however, is a well-known concept in nature to realise higher specificity and stronger relative binding affinities.3 For instance, the adhesion of bacteria to cell surfaces requires multivalent protein–carbohydrate interactions.4,5 In particular, the adhesion of uropathogenic E. coli (UPEC) to uroepithelial cells in the urinary tract is mediated by the binding of α-D-mannosides, presented on cell surface proteins, to the bacterial lectin FimH of type I pili of UPECs.6 In this specific case, urinary tract infections (UTI) are mostly caused by antibiotic resistant E. coli strains7–9 and therefore alternative treatment options are badly needed.To achieve an effective inhibition of bacterial colonisation of tissues, synthetic multivalent scaffolds as binding competitors are a promising option. Different variables including valency, topology, density and cluster effects of carbohydrates at synthetic scaffolds are currently under investigation.10–14 Biomimetic materials have obtained increasing interest in this regard in recent years. Glycoconjugates and supramolecular self-assembled glycoclusters have been used to generate multivalent systems,15 including liposomes,16 glycopeptides,17 dendrimersomes,18 and fluorescent glycoprobes.19 The development of a toolbox system, mimicking the glycocalyx and being easily variable by adapting different suitable ligands is therefore of high interest.
Bilayer vesicles are dynamic supramolecular structures that can be used as mimic for the glycocalyx by functionalising the surface of the vesicles with carbohydrates.20 Cyclodextrins are cyclic oligosaccharides, containing D-glucopyranose as repeating units. The special orientation of glucopyranose units in cyclodextrins shapes a conical structure with a hydrophobic cavity. In addition, the hydroxyl groups on both sides at the open ring allows for further functionalization.21 By amphiphilic functionalisation with alkyl chains and oligo(ethylene glycol) chains, the formation of bilayer vesicles was reported.22 Depending on the number of glucopyranose units (6 for α-, 7 for β- and 8 for γ-cyclodextrins), the size of the hydrophobic cavity ranges from 5.7 to 9.5 Å, thus enabling the formation of inclusion complexes with different hydrophobic guest molecules in water.23 For β-CD, adamantane is known to be a well-fitting inclusion guest (Ka ∼ 104 M−1). By choosing different guest conjugates, the self-assembled cyclodextrin vesicles (CDV) are versatile scaffolds for various applications.24–26 A similar host–guest system based on amphiphilic cucurbit[6]uril and spermidine derivatives was reported for Concanavalin A (ConA) recognition27 and photoresponsive capture and release of lectins could be achieved by introducing azobenzene motifs28 into the ligand structure.
The enhanced development of a toolbox scaffold system that enables diverse variation of its ligands and the investigation of subsequent ligand recognition were the aims of this work. The versatility concept was demonstrated by a few examples. Therefore, CDVs were non-covalently functionalised with two different kinds of lectin-recognising ligands. Mannose-presenting ligands were chosen for the binding to ConA and FimH expressing E. coli, whereas sulphate-presenting ligands were used to show L-selectin binding. ConA is a plant lectin, homotetrameric at neutral pH and has four carbohydrate recognition domains for mannosides. L-selectin is an adhesion protein that supports the attachment of leukocytes to endothelial cells during the inflammation process.29–32 PSGL-1 is one of the physiological ligands for L-selectin, bearing an anionic sulphotyrosine residue, which binds to the positively charged region of L-selectin.33,34 Due to this electrostatic interaction, sulphated ligands with a high local anionic charge density have been reported previously for the effective binding to L-selectin35 including derivatives of heparin36 and dendritic polyglycerol sulphates37 (dPGS), which were blocking the interaction between leukocytes and endothelial cells.
To enhance the binding affinity of ligands towards the CDV surface, also divalent adamantane anchoring moieties were considered.38,39 In addition, we synthesised ligands with different amounts of mannosides and sulphate groups, respectively, to investigate the influence of ligand density on CDVs in terms of target recognition. Therefore, interactions with the desired lectins were investigated by surface plasmon resonance (SPR) and microscale thermophoresis experiments (MST) as well as turbidimetric analysis of rising aggregates (OD400). Furthermore, all host–guest complexes were analysed regarding their physicochemical properties by ITC measurements. In a second step, binding of mannose-decorated CDVs to FimH expressing E. coli was investigated by cryo-TEM experiments and studied in a competitive binding assay to the uroepithelial cell line RT4.
Results and discussion
Synthesis
In order to explore the multivalent supramolecular binding of cyclodextrin vesicles (CDVs), seven different ligands were synthesised, four of them bearing mannosides and three with sulphate group functionalisation. The valency of the ligands included mono-, tri- and octavalent mannosides as well as tetra- and octavalent sulphates in order to study the effect of multivalent ligand–receptor binding. The ligands were designed so that on one side of the structure a motif of one or two adamantyl residues allowed for the complexation to β-cyclodextrins (β-CD) at the surface of CDVs. Adamantyl groups are known to form inclusion complexes with β-CD with an affinity constant of around Ka ∼ 104 M−1 for monovalent 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding. Divalent binding with two adamantane residues per ligand, however, can lead to an apparent affinity constant of up to Ka ∼ 107 M−1 plus additional kinetic stabilization of the complex.40 On the opposite side of the ligands, either mannose or sulphate groups were introduced. The synthetic ligands are shown in Scheme 1 and their detailed synthesis procedures are described in the ESI.†
1 binding. Divalent binding with two adamantane residues per ligand, however, can lead to an apparent affinity constant of up to Ka ∼ 107 M−1 plus additional kinetic stabilization of the complex.40 On the opposite side of the ligands, either mannose or sulphate groups were introduced. The synthetic ligands are shown in Scheme 1 and their detailed synthesis procedures are described in the ESI.†
 | ||
| Scheme 1 Schematic representation of the synthesised adamantyl ligands with mannose and sulphate motifs and amphiphilic cyclodextrin vesicles. | ||
The octavalent mannoside ligand Ad2Man8 was synthesised as shown schematically in Fig. S4 (ESI†). A polyglycerol-based dendron was synthesised according to published procedures41 and coupled to a two-adamantane precursor by amide bond formation. Afterwards, the acetal-protection groups of the dendron were cleaved off using Dowex® resin. The free hydroxyl groups were then converted into terminal alkyne groups by the addition of 4-pentynoic acid via esterification. Subsequently, azido-mannosides were clicked onto the dendron's terminal alkyne groups via copper-catalysed azide–alkyne cycloaddition (CuAAC) under optimised conditions. The reaction was stirred for four days at elevated temperature (50 °C). The addition of a piece of copper wire42 along with CuSO4 and sodium ascorbate turned out to facilitate the reaction drastically and led to full octavalent functionalisation. Sulphate-adamantyl conjugates were synthesised in a similar procedure. Here, the terminal hydroxyl groups were sulphated using a sulphur trioxide pyridine complex.
Amphiphilic β-cyclodextrins functionalised with alkyl and polyethylene glycol chains were synthesised according to previous reports.24 β-Cyclodextrin vesicles (CDV) were obtained by extrusion, yielding vesicles with diameters between 100 and 200 nm.
Physicochemical characterisation
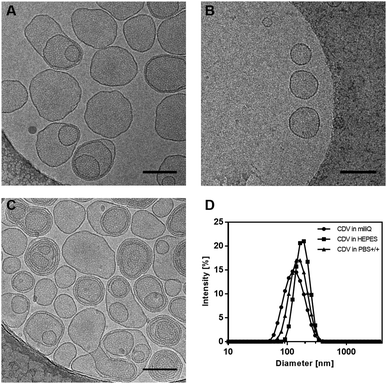
In order to investigate the vesicle's size and morphology, dynamic light scattering (DLS) and cryogenic transmission electron microscopy (cryo-TEM) experiments were carried out.Cryo-TEM images show mono- and multilayered vesicles with diameters of (177 ± 40) nm (Fig. 1A). These values closely correlate with the range of 140 to 180 nm obtained by DLS measurements (Fig. 1D), regardless of the solvent used (Milli-Q, HEPES, PBS+/+). Even after addition of mannoside ligands, the shape and size of the vesicles remained similar as can be seen in Fig. 1B and C.
The host–guest interaction was investigated between adamantane mannoside ligands (Ad1Man1, Ad2Man1, Ad2Man3, and Ad2Man8) and β-CD as non-aggregated β-CD allows unhindered inclusion complex formation with adamantane. Measurements were carried out by isothermal titration calorimetry (ITC) using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes of adamantane and β-CD (Table 1). Each titration was performed with a 10-fold higher host concentration in relation to the guest molecule. All ligands showed association constants in the range of 104 M−1, which are in the typical range of adamantane to β-CD binding and are in line with previous results.26 The thermodynamic parameters of Ad1Man1, Ad2Man1, and Ad2Man3 are consistent (negative ΔH and positive ΔS), while for Ad2Man8 the change in entropy (ΔS) is slightly negative and the change in enthalpy (ΔH) is slightly increased. We attribute this behaviour to the bulky and strongly hydrated octavalent mannose dendron, which will significantly reduce the entropic bonus of releasing hydration water from the adamantane. Instead, it will be first solvated by the dendron and then included by the β-CD, so that the net gain in entropy due to the desolvation is minimal or even negative. In addition, ΔH is higher assumingly because of multiple weak but favourable hydrogen bonds between the dendron and the β-CD. However, the resulting association constant Ka lies within the same range of the other ligands, which results in a stable inclusion complex formation for all ligands. Furthermore, due to the divalent binding of the ligands Ad2Man1, Ad2Man3 and Ad2Man8 to the membrane of CDVs, increased binding affinities (Ka ∼ 107 M−1) and additional stabilization effects for the complexes can be assumed as shown by Huskens et al.39 Additional titration plots including fitting curves are given in ESI,† (Fig. S5–S7).
1 complexes of adamantane and β-CD (Table 1). Each titration was performed with a 10-fold higher host concentration in relation to the guest molecule. All ligands showed association constants in the range of 104 M−1, which are in the typical range of adamantane to β-CD binding and are in line with previous results.26 The thermodynamic parameters of Ad1Man1, Ad2Man1, and Ad2Man3 are consistent (negative ΔH and positive ΔS), while for Ad2Man8 the change in entropy (ΔS) is slightly negative and the change in enthalpy (ΔH) is slightly increased. We attribute this behaviour to the bulky and strongly hydrated octavalent mannose dendron, which will significantly reduce the entropic bonus of releasing hydration water from the adamantane. Instead, it will be first solvated by the dendron and then included by the β-CD, so that the net gain in entropy due to the desolvation is minimal or even negative. In addition, ΔH is higher assumingly because of multiple weak but favourable hydrogen bonds between the dendron and the β-CD. However, the resulting association constant Ka lies within the same range of the other ligands, which results in a stable inclusion complex formation for all ligands. Furthermore, due to the divalent binding of the ligands Ad2Man1, Ad2Man3 and Ad2Man8 to the membrane of CDVs, increased binding affinities (Ka ∼ 107 M−1) and additional stabilization effects for the complexes can be assumed as shown by Huskens et al.39 Additional titration plots including fitting curves are given in ESI,† (Fig. S5–S7).
| Ligand at CDV | ΔH (kJ mol−1) | ΔG (kJ mol−1) | ΔS (J K−1 mol−1) | K a (M−1) |
|---|---|---|---|---|
| Ad1Man1 | −16.7 | −26.0 | 31.0 | 3.55 × 104 |
| Ad2Man1 | −18.1 | −26.3 | 17.6 | 1.24 × 104 |
| Ad2Man3 | −18.4 | −23.6 | 17.5 | 1.38 × 104 |
| Ad2Man8 | −25.2 | −23.3 | −6.1 | 1.23 × 104 |
In a second set of experiments, the non-covalent interactions between β-CD and the sulphated adamantyl ligands were investigated by ITC. Additionally, the non-sulphated analogues (OH-group terminated) were included in this assay to examine the effect of sulphation on the thermodynamic parameters. Again, reasonable binding affinities (Ka ∼ 104 M−1) for adamantane to β-CD were observed for all ligands (Table 2). In the case of Ad2OH8, a binding affinity comparable to the mannose-functionalised ligands (cf.Table 1) was obtained. A similar effect as before could be observed, i.e., a negative change in entropy and an increased change in enthalpy, which we again attribute to the strongly hydrated and bulky octavalent dendron in combination with two adamantane moieties. By comparing the titrations of Ad2OH8 and Ad2Su8, an unexpected behaviour was observed, i.e., the sulphated ligand showed not only a lower heat rate change by a factor of five than Ad2OH8, but also reached the thermal equilibrium already after three titrations (Fig. S7, ESI†). This observation suggests that a self-assembled structure of Ad2Su8 was present as the amphiphilicity of Ad2Su8 increased due to the sulphation of the highly branched headgroup in comparison to the uncharged Ad2OH8. Thus, the adamantane groups were hindered from binding to β-CD. For all other ligands, this behaviour was not observed.
| Ligand at CDV | ΔH (kJ mol−1) | ΔG (kJ mol−1) | ΔS (J K−1 mol−1) | K a (M−1) | ζ (mV) |
|---|---|---|---|---|---|
| β-CDV | — | — | — | — | −10.0 ± 1.7 |
| Ad1OH4 | −17.4 | −25.9 | 28.5 | 3.45 × 104 | −9.5 ± 0.4 |
| Ad1Su4 | −16.8 | −25.2 | 28.1 | 2.59 × 104 | −14.6 ± 1.1 |
| Ad1OH8 | −18.0 | −25.1 | 23.6 | 2.46 × 104 | −8.5 ± 0.9 |
| Ad1Su8 | −17.3 | −24.1 | 22.6 | 1.66 × 104 | −16.3 ± 2.4 |
| Ad2OH8 | −25.9 | −23.6 | −7.6 | 1.38 × 104 | −5.3 ± 0.2 |
Zeta-potential measurements were carried out to investigate the charge of CDVs after the complexation with neutral and sulphated ligands at the CDV surface (Table 2). For the dendritic neutral ligands, Ad1OH4 and Ad1OH8, the zeta-potential remained unchanged after addition of the ligands in comparison to the bare CDVs (ζ = −8–10 mV), which also contain ethylene glycol side chains at the vesicle surface.
In contrast, the decoration of CDVs with the sulphated ligands Ad1Su4 and Ad1Su8 led to an increased negative zeta-potential (ζ = −14–16 mV), as the negatively charged ligands changed the surface environment after their complexation to CDVs.
In the following, supramolecular complexes, consisting of CDVs and mannose-functionalised or sulphated adamantyl ligands, respectively, were investigated regarding their binding affinities towards lectins.
Lectin binding
In this assay, the aim was to compare binding affinities between the synthetic mannose-functionalised ligands as well as the respective ligand-functionalised vesicles to ConA. Therefore, ConA–biotin was immobilised on a streptavidin-functionalised SPR sensor chip, whereas mannose ligands and ligand-functionalised vesicles, dissolved in HEPES buffer, were run in a continuous flow over the sensor chip (Fig. 2A). In a kinetic titration series, binding measurements were conducted with increasing concentrations of the respective sample. Subsequently, KD values for each sample were determined by single cycle kinetics and the resulting binding isotherms (Fig. 2B, C and Fig. S10, S11, ESI†). The results are summarised in Table 3.
| Ligand | K D (μM) | |
|---|---|---|
| Bare ligand | Ligand displayed on CDVs | |
| Ad1Man1 | 234 | 89 |
| Ad2Man1 | 50 | 7 |
| Ad2Man3 | 69 | 13 |
| Ad2Man8 | 50 | 9 |
Due to non-specific binding of dissociated adamantane to the dextran layer on the SPR chip, additional cyclodextrin was added to the SPR running buffer. In order to efficiently shield the adamantyl moiety, β-CD and γ-CD were added as surfactants to the buffer during the measurements with mannoside ligands and ligand-decorated vesicles, respectively. Further information about the experiments, showing that the system was not affected by the additives, is given in the ESI.†
At first, binding affinities of mannose-functionalised conjugates (Ad1Man1, Ad2Man1, Ad2Man3 and Ad2Man8) to ConA were measured. The determined KD values ranged from 50–234 μM (Table 3).
Subsequently, CDVs were decorated with the ligands through the formation of adamantane-β-CD inclusion complexes. The concentrations of ligands were chosen such that full decoration of the vesicles by complexation of every cyclodextrin was theoretically enabled (referred to as 100% surface coverage). Here, determined KD values ranged from 7–89 μM. By comparison with the corresponding values for the conjugates itself, the binding was strengthened by a factor of approx. three to seven through the multivalent display of ligands at CDVs, highlighting the effectivity of multivalent organisation of ligands. However, differences in the binding affinities resulting from different carbohydrate valences (one to eight) could not be discriminated at that point.
The obtained values are comparable to recent studies also using SPR.43–46 However, in contrast to the present approach, these works used covalently synthesised glycoclusters and glycopolymers. By using a supramolecular multivalent approach, a facile adaption to different targets in terms of e.g. valency and type of ligand is enabled. Additional aggregation experiments with turbidimetric measurements (OD400) investigated the optimisation of the surface coverage of vesicles with mannoside ligands by altering the ratio of ligand to host moiety. Here, a threshold of 20% surface coverage was identified for effective aggregation for all ligands (Fig. S8 and S9, ESI†). Further details of the experiments can be found in the ESI.†
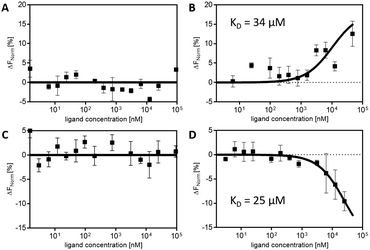
The sulphated ligands Ad1Su4 and Ad1Su8 without CDVs showed randomly fluctuating values for the MST response throughout the ligand titration range. However, likewise to the mannose-functionalised ligands, an optimised binding behaviour due to multivalent display of the ligands on CDVs was observed. In the presence of vesicles, characteristic s-shaped and mirrored-s-shaped sigmoidal binding curves could be obtained (Fig. 3). The different appearances of the binding curves can be attributed to different thermophoretic movements of the respective ligands.49 From these experimental data, KD values of 34 μM for Ad1Su4 and 25 μM for Ad1Su8 were determined. These values are in the same range of those observed from mannoside–ConA binding experiments (cf. SPR experiments) and underline the broad and easy applicability of non-covalently functionalised CDVs in biological binding approaches by simply changing the functionalised ligands.
The potential of our system to detach UPECs from uroepithelial cells was investigated using an in vitro assay. Uroepithelial RT-4 cells were incubated with E. coli strain ORN178 for 1 h. Afterwards, mannose-functionalised vesicles were added and incubated for 15 min together with cells and bacteria. In case of successful competitive binding, the vesicles would detach the bacteria from cell surfaces. The assay principle is depicted in Fig. 4A. Subsequently, the supernatant with detached bacteria was harvested and plated in serial dilutions on agar plates. Colonies were counted after overnight incubation at 37 °C and the detaching ability was calculated as relative activity in relation to PBS buffer solution as negative control. All bi-adamantane ligands, carrying one, three and eight mannose residues were tested (Ad2Man1, Ad2Man3, Ad2Man8). Furthermore, ligand concentrations were chosen such that both half and full coverage of the vesicles was achieved. For comparison, also the monovalent methyl-α-D-mannopyranoside (Me-Man) was included. The non-carbohydrate ligand Ad-TEG-OH served as a control to exclude unspecific binding. As can be seen in Fig. 4B, tri- and octavalent mannoside ligands were able to release significantly more bacteria from RT-4 cell layers than the monovalent and control ligands. Furthermore, Me-Man and Ad2Man1 did show the same or even less detaching activity compared to the inactive control ligand Ad-TEG-OH.
In the following, different coverage densities of the vesicles with ligands were tested. As can be seen in Fig. 4B, the coverage density, i.e., half and full coverage with ligands on the CDV's surface, had no influence on the detaching efficiency. These findings seem to be non-intuitive at the first glance, i.e., one would expect a direct dependence between the detaching efficiency and ligand density. However, the above-described results can be interpreted by considering the effect of local density of binding sites. The pili of the UPECs exhibit only a single mannose-binding site, which is located at the outermost protein, i.e., FimH. At the beginning of our cell experiments, this binding site was bound to the mannosides at the uroepithelial RT4 cell membrane. By adding the functionalised vesicles, alternative mannosides were offered for binding. Since binding is a dynamic process, binding and debinding occurs occasionally. If an alternative binding site other than the natural mannosides at the cell membrane were present during debinding, FimH may bind to these alternatives. Binding to these alternatives becomes even more favourable, if they are arranged in multivalent clusters. If this is the case, they exhibit a high local concentration of binding sites. An increase of the high local concentration of binding sites leads to a decreased dissociation rate53,54 and statistically favoured rebinding. Hence, FimH stayed bound to the dendritic mannoside clusters. These mannoside clusters are present in the tri- and octavalent mannose ligands, explaining the increased detaching efficiency compared to the monovalent mannosides and the inactive ligand. Thus, multivalency is an important aspect for enhancing binding efficiencies in biological systems.
In addition, cryo-TEM images of UPECs and CDVs were taken (Fig. 4C–E). Here, CDVs were functionalised with either the inactive ligand Ad-TEG-OH or the octavalent mannoside ligand Ad2Man8. For UPECs incubated with CDVs plus the inactive Ad-TEG-OH ligand, no binding of vesicles to the pili of the bacteria could be observed (Fig. 4C and D). However, vesicles, functionalised with Ad2Man8 ligands, show clear co-localization to the bacteria and their pili (Fig. 4E), supporting the above results from the in vitro assays. Furthermore, the cryo-TEM images prove that binding occurs to the functionalised vesicles and not to free and non-complexed ligands in solution.
Conclusion
In conclusion, we performed a thorough study on multivalently functionalised CDVs for lectin binding. Functionalisation of the vesicles was realised by novel carbohydrate and sulphate ligands, namely mono-, tri- and octavalent mannosides as well as tetra- and octavalent sulphates. These different ligands were used in a toolbox approach to demonstrate enhanced lectin binding through multivalent display of the ligands at the vesicle's surface. Our toolbox approach easily allowed addressing different biologically relevant receptors by modifying the supramolecular system with corresponding ligands, i.e., mannosides for ConA binding as well as sulphated ligands for L-selectin binding. Finally, we demonstrated in an in vitro assay that our mannose-functionalised vesicles are an efficient system for detaching E. coli from uroepithelial cells. FimH antagonists as anti-adhesive compounds reduce the bacterial load and therefore could be further developed for wash out strategies as a treatment option with respect to antibiotic resistant UPEC strains. In the future, new strategies to target different pathogens using the toolbox approach presented in this work will be anticipated.Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
The authors would like to thank the German Research Foundation (DFG, SFB 765) (SE, KS, CB, JD, and RH) and the – EC H2020 – Marie Skłodowska-Curie Actions – Innovative Training Network, Multi-App (project number: 642793) (CWC, SK, RH, and BJR) for financial support. The authors thank Katharina Goltsche for support in the synthesis of dendrons and Dr Christian Kühne for assisting with SPR measurements. Dr Pamela Winchester is gratefully acknowledged for language polishing the manuscript.References
- H. Lis and N. Sharon, Chem. Rev., 1998, 98, 637–674 CrossRef PubMed.
- H. J. Gabius, S. André, J. Jiménez-Barbero, A. Romero and D. Solís, Trends Biochem. Sci., 2011, 36, 298–313 CrossRef PubMed.
- M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754–2794 CrossRef PubMed.
- J. Pizarro-Cerdá and P. Cossart, Cell, 2006, 124, 715–727 CrossRef PubMed.
- A. M. Krachler, H. Ham and K. Orth, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11614–11619 CrossRef PubMed.
- M. Hartmann and T. K. Lindhorst, Eur. J. Org. Chem., 2011, 3583–3609 CrossRef.
- E. C. Svanborg and P. de Man, Infect. Dis. Clin. N. Am., 1987, 1, 731–750 Search PubMed.
- J. R. Johnson, Clin. Microbiol. Rev., 1991, 4, 80–128 CrossRef PubMed.
- L. Hagberg, U. Jodal, T. K. Korhonen and C. E. Svanborg, Infect. Immun., 1981, 31, 564–574 Search PubMed.
- J. W. Wehner, M. Hartmann and T. K. Lindhorst, Carbohydr. Res., 2013, 371, 22–31 CrossRef PubMed.
- C. Müller, G. Despras and T. K. Lindhorst, Chem. Soc. Rev., 2016, 45, 3275–3302 RSC.
- D. Deniaud, K. Julienne and S. G. Gouin, Org. Biomol. Chem., 2011, 9, 966–979 Search PubMed.
- E. M. Munoz, J. Correa, E. Fernandez-Megia and R. Riguera, J. Am. Chem. Soc., 2009, 131, 17765–17767 CrossRef PubMed.
- M. L. Talaga, N. Fan, A. L. Fueri, R. K. Brown, Y. M. Chabre, P. Bandyopadhyay, R. Roy and T. K. Dam, Biochemistry, 2014, 53, 4445–4454 CrossRef PubMed.
- M. Delbianco, P. Bharate, S. Varela-Aramburu and P. H. Seeberger, Chem. Rev., 2016, 116, 1693–1752 CrossRef PubMed.
- W. Curatolo, A. O. Yau, D. M. Small and B. Sears, Biochemistry, 1978, 17, 5740–5744 CrossRef PubMed.
- J. R. Kramer, A. R. Rodriguez, U.-J. Choe, D. T. Kamei and T. J. Deming, Soft Matter, 2013, 9, 3389–3395 RSC.
- Q. Xiao, S. S. Yadavalli, S. Zhang, S. E. Sherman, E. Fiorin, L. da Silva, D. A. Wilson, D. A. Hammer, S. André, H.-J. Gabius, M. L. Klein, M. Goulian and V. Percec, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 1134–1141 CrossRef PubMed.
- K.-B. Li, N. Li, Y. Zang, G.-R. Chen, J. Li, T. D. James, X.-P. He and H. Tian, Chem. Sci., 2016, 7, 6325–6329 RSC.
- R. V. Vico, J. Voskuhl and B. J. Ravoo, Langmuir, 2011, 27, 1391–1397 CrossRef PubMed.
- B. V. K. J. Schmidt and C. Barner-Kowollik, Angew. Chem., Int. Ed., 2017, 56, 8350–8369 CrossRef PubMed.
- B. J. Ravoo and R. Darcy, Angew. Chem., Int. Ed., 2000, 39, 4324–4326 CrossRef PubMed.
- J. Szejtli, Chem. Rev., 1998, 98, 1743–1754 CrossRef PubMed.
- P. Falvey, W. Lim, R. Darcy, T. Revermann, U. Karst, M. Giesbers, A. T. M. Marcelis, A. Lazar, A. W. Coleman, D. N. Reinhoudt and B. J. Ravoo, Chem. – Eur. J., 2005, 11, 1171–1180 CrossRef PubMed.
- J. Voskuhl, M. C. A. Stuart and B. J. Ravoo, Chem. – Eur. J., 2010, 16, 2790–2796 CrossRef PubMed.
- U. Kauscher and B. J. Ravoo, Beilstein J. Org. Chem., 2012, 8, 1543–1551 CrossRef PubMed.
- H. K. Lee, K. M. Park, Y. J. Jeon, D. Kim, D. H. Oh, H. S. Kim, C. K. Park and K. Kim, J. Am. Chem. Soc., 2005, 127, 5006–5007 CrossRef PubMed.
- A. Samanta, M. C. A. Stuart and B. J. Ravoo, J. Am. Chem. Soc., 2012, 134, 19909–19914 CrossRef PubMed.
- L. M. Coussens and Z. Werb, Nature, 2002, 420, 860–867 CrossRef PubMed.
- R. P. McEver, K. L. Moore and R. D. Cummings, J. Biol. Chem., 1995, 270, 11025–11028 CrossRef PubMed.
- L. A. Lasky, Annu. Rev. Biochem., 1995, 64, 113–139 CrossRef PubMed.
- S. D. Rosen and C. R. Bertozzi, Curr. Biol., 1996, 6, 261–264 CrossRef PubMed.
- W. S. Somers, J. Tang, G. D. Shaw and R. T. Camphausen, Cell, 2000, 103, 467–479 CrossRef PubMed.
- B. J. Graves, R. L. Crowther, C. Chandran, J. M. Rumberger, S. Li, K.-S. Huang, D. H. Presky, P. C. Familletti, B. A. Wolitzky and D. K. Burns, Nature, 1994, 367, 532–538 CrossRef PubMed.
- E. E. Simanek, G. J. McGarvey, J. A. Jablonowski and C.-H. Wong, Chem. Rev., 1998, 98, 833–862 CrossRef PubMed.
- J. Fritzsche, S. Alban, R. J. Ludwig, S. Rubant, W.-H. Boehncke, G. Schumacher and G. Bendas, Biochem. Pharmacol., 2006, 72, 474–485 CrossRef PubMed.
- J. Dernedde, A. Rausch, M. Weinhart, S. Enders, R. Tauber, K. Licha, M. Schirner, U. Zügel, A. von Bonin and R. Haag, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 19679–19684 CrossRef PubMed.
- A. Mulder, T. Auletta, A. Sartori, S. Del Ciotto, A. Casnati, R. Ungaro, J. Huskens and D. N. Reinhoudt, J. Am. Chem. Soc., 2004, 126, 6627–6636 CrossRef PubMed.
- A. Perl, A. Gomez-Casado, D. Thompson, H. H. Dam, P. Jonkheijm, D. N. Reinhoudt and J. Huskens, Nat. Chem., 2011, 3, 317–322 CrossRef PubMed.
- J. Huskens, A. Mulder, T. Auletta, C. A. Nijhuis, M. J. W. Ludden and D. N. Reinhoudt, J. Am. Chem. Soc., 2004, 126, 6784–6797 CrossRef PubMed.
- M. Wyszogrodzka and R. Haag, Chem. – Eur. J., 2008, 14, 9202–9214 CrossRef PubMed.
- K. Petkau-Milroy and L. Brunsveld, Eur. J. Org. Chem., 2013, 3470–3476 CrossRef.
- B. Bertolotti, I. Sutkeviciute, M. Ambrosini, R. Ribeiro-Viana, J. Rojo, F. Fieschi, H. Dvorakova, M. Kasakova, K. Parkan, M. Hlavackova, K. Novakova and J. Moravcova, Org. Biomol. Chem., 2017, 15, 3995–4004 Search PubMed.
- G. Goti, A. Palmioli, M. Stravalaci, S. Sattin, M.-G. De Simoni, M. Gobbi and A. Bernardi, Chem. – Eur. J., 2016, 22, 3686 CrossRef PubMed.
- I. Morbioli, V. Porkolab, A. Magini, A. Casnati, F. Fieschi and F. Sansone, Carbohydr. Res., 2017, 453-454, 36–43 CrossRef PubMed.
- D. Diwan, K. Shinkai, T. Tetsuka, B. Cao, H. Arai, T. Koyama, K. Hatano and K. Matsuoka, Molecules, 2017, 22, 157 CrossRef PubMed.
- M. Jerabek-Willemsen, C. J. Wienken, D. Braun, P. Baaske and S. Duhr, Assay Drug Dev. Technol., 2011, 9, 342–353 CrossRef PubMed.
- S. Duhr and D. Braun, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19678–19682 CrossRef PubMed.
- S. A. I. Seidel, C. J. Wienken, S. Geissler, M. Jerabek-Willemsen, S. Duhr, A. Reiter, D. Trauner, D. Braun and P. Baaske, Angew. Chem., Int. Ed., 2012, 51, 10656–10659 CrossRef PubMed.
- S. D. Fihn, N. Engl. J. Med., 2003, 349, 259–266 CrossRef PubMed.
- R. H. Mak and H. J. Kuo, Curr. Opin. Pediatr., 2006, 18, 148–152 CrossRef PubMed.
- D. F. Sahm, C. Thornsberry, D. C. Mayfield, M. E. Jones and J. A. Karlowsky, Antimicrob. Agents Chemother., 2001, 45, 1402–1406 CrossRef PubMed.
- M. Kanai, K. H. Mortell and L. L. Kiessling, J. Am. Chem. Soc., 1997, 119, 9931–9932 CrossRef.
- D. M. Crothers and H. Metzger, Immunochemistry, 1972, 9, 341–357 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8tb00922h |
| ‡ The authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |