Open Access Article
Open Access ArticleMultienzyme cascade synthesis of ω-amino fatty acids from vegetable-derived precursors for use as nylon monomers
Yueyue
Zhou
a,
Ran
Lu
a,
Xiaoxia
Gao
a,
Lu
Lin
a,
Yongjun
Wei
 *b and
Xiao-Jun
Ji
*b and
Xiao-Jun
Ji
 *a
*a
aState Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, China. E-mail: xiaojunji@njtech.edu.cn
bSchool of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou 450001, China. E-mail: yongjunwei@zzu.edu.cn
First published on 28th March 2025
Abstract
Omega-amino fatty acids (ω-AmFAs) are non-natural organic molecules with amino and carboxyl groups located at the ends of unbranched carbon chains. They are widely used in the synthesis of polymers such as polyesters and polyamides, as well as in the production of chemical products such as biofuels and pharmaceutical intermediates. In recent years, the production of such materials and other chemicals via the fermentation of renewable resources using engineered microorganisms has become a hot spot of research, as examples of emerging green and low-carbon technologies. Traditional petrochemical synthesis methods of nylon monomers often face problems such as environmental pollution, increased energy consumption and high cost. By contrast, the catalytic production of ω-AmFAs from fatty acids such as oleic acid, ricinoleic acid and lauric acid found in vegetable oils using a multienzyme cascade has the unique advantages of being environmentally friendly and having high process economics. This paper reviews multienzyme synthesis strategies of ω-AmFAs used as nylon monomers.
Sustainability spotlightAs the international community becomes increasingly concerned about climate, environmental and resource issues, the traditional chemical industry is shifting towards a renewable and sustainable approach. Using engineered microorganisms as catalysts and renewable resources as raw materials, it has been possible to synthesis high value-added chemicals under mild reaction conditions. Compared with chemical processes, the multienzyme cascade methods for Bio-based nylon synthesis are more sustainable and environmentally friendly, meeting the requirements of green, low-carbon and sustainable development. In this paper, ω-amino fatty acids used as nylon monomers are synthesized from vegetable-derived precursors using multienzyme cascade methods. This review aligns with the following UN sustainable development goals: responsible consumption and production (SDG12) and climate action (SDG13). |
Introduction
Polymers and their monomers constitute the largest portion of all chemical products produced today. The global polymer market was valued at $522.7 billion in 2024 and is expected to grow at a rate of 4.0% per annum from 2024–2030. The fastest growing market segment includes polyamides and polyesters, with a compound annual growth rate (CAGR) of 8%.1 Polyamide, also known as nylon, is a generic term for thermoplastic resins that contain repeating amide groups (–[NHCO]–) in the main chain of the molecule. Nylon monomers can be biosynthesized or produced chemically.2 According to their molecular structure, the most common nylons can be divided into two categories. The first type of nylon structure is obtained by the copolymerization of two types of monomers, one of which is an α, ω-diamine, and the other is an α, ω-dicarboxylic acid. For example, nylon 66 is made by copolymerization of 1,6-hexanediamine and adipic acid. The other type of nylon is a homopolymer of a single bifunctional monomer. For example, nylon 6 is derived from the ring-opening polymerization of caprolactam.3 Nylons are widely used in the manufacture of parts for machinery, chemicals, electrical appliances and other plastic products due to their excellent resistance to heat, wear, corrosion, and chemicals, combined with a low friction coefficient and other comprehensive properties. The synthesis of nylons was a major breakthrough in the synthetic fiber industry and an important milestone of polymer chemistry3 (Table 1).| Polyamide monomer | Polyamide product | Nylon functions | Commercial producers | References |
|---|---|---|---|---|
| 8-Aminooctanoic acid | Nylon 8 | Used in industry to make small machine parts, gears, etc.; used in textiles to make high-quality fiber products | Geeyes | 7 |
| 9-Aminononanoic acid | Nylon 9 | Fiber monofilaments, extrusions, molded products, cable wraps, medical biomimetic materials for osteogenesis | KURARAY | 8 |
| 11-Aminoundecanoic acid | Nylon 11 | Automotive industry, electronic and biomedical devices, sensors, energy storage systems | Arkema, UBE, Mitsubishi, Toray | 9 and 10 |
| 12-Aminododecanoic acid | Nylon 12 | 3D printing, automotive manufacturing, oil and gas extraction, medical devices, electrical and electronic appliances | Evonik, UBE, Arkema, EMS | 11 |
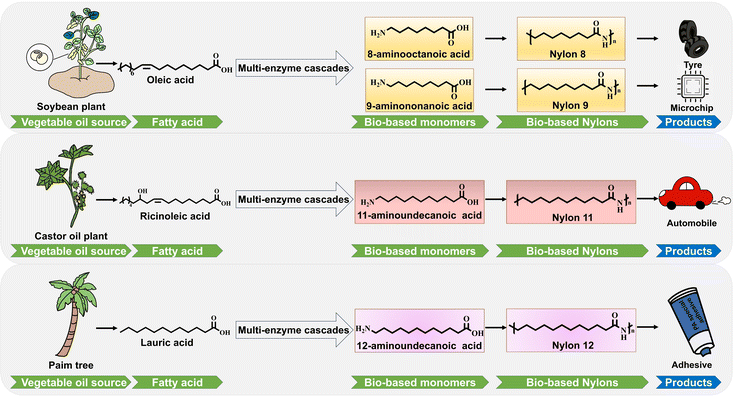
Omega-amino fatty acids (ω-AmFAs) are an important class of bifunctional fatty acid derivatives with α-carboxy and ω-amino groups, which can be further condensed to form polyamides. They are important monomers for the synthesis of polyamides, with examples including 8-aminooctanoic acid, 9-aminononanoic acid, 11-aminoundecanoic acid and 12-aminododecanoic acid, which are widely used at present. The traditional chemical routes for the industrial synthesis of ω-AmFAs are based on ozone-as well as nickel-catalyzed redox reactions or cross–complexation reactions of enoic acids with acrylonitrile.4 For example, the reaction of fatty acids containing carbonyl groups with amines catalyzed by nickel metal can produce the corresponding amino fatty acids. Alkenoates and acrylonitrile generate unsaturated fatty acid intermediates containing nitrile groups under the action of catalysts, after which the treated unsaturated fatty acid intermediates are subjected to a hydrogenation reaction, in which the nitrile group is reduced to an amino group while the unsaturated bond is reduced to a saturated bond, resulting in an amino fatty acid. These synthesis processes mainly rely on highly polluting and energy-consuming chemical oxidation, which is harmful to the environment and is not in line with the modern goals of “green and sustainable development”.5 In addition, chemical synthesis can also lead to the formation of reaction by-products, which is not conducive to product separation and process control. The use of precious and transition metals as catalysts has limitations such as poor stability, high cost, and difficulty in recovering precious metals. For these reasons, scientists have been exploring new green methods and process technologies for the efficient synthesis of ω-AmFAs. Enzymatic synthesis offers advantages of mild conditions, environmental friendliness, and unmatched regioselectivity, making it an attractive alternative.6 Multienzyme cascade catalysis has become a research hotspot in recent years as a “one-pot” synthesis modality avoiding additional unit operation, thus saving manpower, material inputs, and waste.
At a time when bio-based materials are booming, there are various sources of bio-based feedstocks for the synthesis of ω-AmFAs. Bio-based nylon monomers can be produced by two pathways, the sugar pathway and the vegetable oil pathway. The sugar pathway has the disadvantages of long process and low yield. For example, for the production of 12-aminododecanoic acid, Gao et al.12 developed a fermentation process for the synthesis of 12-aminododecanoic acid from glucose and cellobiose[6], but the yield was only 6%. Similarly, Ge et al.13 achieved de novo biosynthesis from glucose to 12-aminododecanoic acid with a yield of only 0.01 g per gram. In contrast, the vegetable oil pathway shows significant advantages over the sugar pathway. Lauric acid extracted from palm oil was used as raw material to synthesize 12-aminododecanoic acid, and the conversion rate was as high as 96%.14 In addition, there are differences in the length of carbon chains, the position and number of unsaturated bonds in the fatty acids contained in different types of vegetable oils. These provide a variety of options for the synthesis of ω-AmFAs with different structures. For example, 11-aminoundecanoic acid can be synthesized from ricinoleic acid in castor oil. Ricinoleic acid has a hydroxyl group at the 12-position of its carbon chain. Through transamination reactions, this hydroxyl group can be converted into an amino group, ultimately leading to the formation of 11-aminoundecanoic acid. Plants are widely distributed globally and their important metabolites, vegetable oils, can be obtained from the seeds of various lipid-rich crops. These crops are easily expanded, providing a reliable supply of vegetable oils.15 The annual global production of vegetable oils exceeded 200 million tons in 2015, with palm and soybean oil accounting for more than 50% of the total production.16,17 This is also an important application of lipid resources in biocatalysis. Taking all these factors into consideration, vegetable oils, as a representative of plant sources, have unique application value and broad prospects in the biosynthesis of ω-AmFAs. This paper reviews recent research advances in the biosynthesis of ω-AmFAs and demonstrates a synthetic pathway for the sustainable production of bio-based nylon monomers through the bioconversion of renewable fatty acids by means of multienzyme cascade reactions (Fig. 1).
Multienzyme cascade reactions
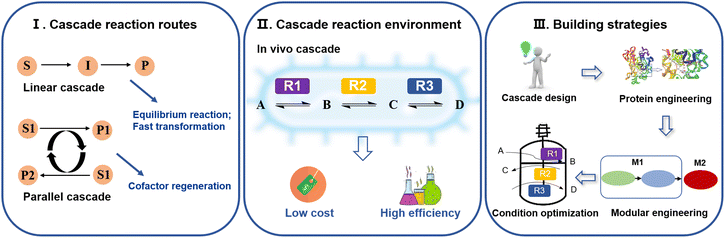
A multienzyme cascade is a process in which multiple enzymatic reactions are combined in an orderly manner to achieve efficient conversion of simple substrates into complex target products.18,19 In contrast to traditional chemical synthesis methods, the enzymes used in cascade reactions are renewable and biodegradable, offering an environmentally friendly biosynthesis method that meets the requirements of sustainable development.20 The synthesis of ω-AmFAs, as a class of polyamide precursors with important biological functions and wide application prospects, has become a research hotspot in the field of biotechnology. The traditional synthesis method of ω-AmFAs has disadvantages of harsh reaction conditions and environmental pollution, whereas the multienzyme cascade catalyzed synthesis of ω-AmFAs provides a new pathway for synthesis under environmentally friendly and mild conditions.21Types of multienzyme cascade reactions
Based on the different catalytic reaction routes of multienzyme cascades, they can be classified into linear, parallel, orthogonal and cyclic cascades.22,23 Among them, linear cascades are the main way to produce bio-based nylon monomers. For example, the production of 12-aminododecanoic acid is catalyzed sequentially by monooxygenase, alcohol dehydrogenase (ADH), and ω-transaminase (ω-TA). Linear cascades enable rapid conversion of unstable intermediates,24 thus avoiding isolation steps.25 Parallel cascades are mostly used for cofactor regeneration.26 Examples include systems for the synthesis of ω-AmFAs using glucose dehydrogenase (GDH)27 and formate dehydrogenase (FDH).28 In addition, according to the different environments of multienzyme cascade reactions, they can be classified into in vivo, in vitro and mixed cascade reactions. The in vivo multienzyme cascade reaction for biocatalysis is called “whole-cell biocatalysis”. By constructing a cell factory and using intact cells as catalysts, there is no need for cell disruption and purification, resulting in low production costs.29,30In vivo multienzyme cascade catalysis is usually chosen for the production of bio-based nylon monomers. This is due to the fact that in vivo multienzyme cascades have access of a variety of cofactors inside the cell that can be easily recycled, which reduces the economic cost and increases the reaction rate while avoiding the accumulation of harmful intermediates31 (Fig. 2). For example, in the process of multienzyme cascade stages to form 11-aminoundecanoic acid,32 continuous recycling of NADP+/NADPH is achieved through alcohol dehydrogenase (ChnD) and NAD(P)H flavin oxidoreductase (DrNFO). This regenerative mechanism not only reduces production costs, but also significantly accelerates the reaction rate.Strategies for the construction of multienzyme cascade reactions
The construction of multienzyme cascade reactions requires thermodynamic and kinetic analyses of the reaction processes and synthetic pathways33 (Fig. 2). The first step is to design the cascade route by performing retrosynthetic analysis of the target product and screening the cascade enzymes for selectivity and compatibility.34 For example, in the synthesis of 12-aminododecanoic acid, the monooxygenase CYP153A, which is highly selective for the ω-hydroxylation of lauric acid, was chosen to catalyze the process. Secondly, the catalytic performance and stability of cascade enzymes generally need to be further improved using methods such as rational design or directed evolution.35 For example, Baeye–Villiger monooxygenase from Pseudomonas putida KT2440 (PpBVMO) was engineered for the synthesis of 11-aminoundecanoic acid, and the fusion of maltose-binding protein (MBP) to BVMO with an E6-tag and C302L mutation resulted in a significant increase in the soluble expression level of the engineered enzyme (MBP-E6BVMOC302L).32 After determining the component enzymes and cascade routes, a self-sufficiency-based modularization strategy has also been shown to be an effective solution to improve the overall pathway efficiency.36 Through modular engineering, complex multienzyme cascade reactions can be disassembled into multiple relatively independent modules according to their functions or reaction steps, requiring the consideration of the equilibrium of protein expression and coenzyme cycling.37 For example, the three enzymes required for the catalytic production of 12-aminododecanoic acid in a multienzyme cascade are CYP153A-NCP (cytochrome P450 153A with the NADPH: cytochrome P450 oxidoreductase from Bacillus megaterium), ADH from Geobacillus stearothermophilus (BsADHC257L), and ω-transaminase from Chromobacterium violaceum DSM30191 (CV2025). They were assigned to two catalytic modules, with module 1 expressed CYP153A-NCP individually, while module 2 co-expressed BsADHC257L and CV2025, which was decided taking into account both protein expression stress and cofactor balance.14 The exogenous synthetic pathway was connected to central metabolism via enzymes and metabolites involved in sugar metabolism, while regenerating NAD(H), NADPH, L-alanine, and PLP, to further increase the synthetic efficiency of the cascade reaction. Finally, systematic tests were conducted to analyze the conditions of the reaction system, such as temperature, pH, as well as co-substrate and enzyme ratios and concentrations, etc., after which the reaction was optimized by determining the changes in the concentrations of the substrates and products during the catalytic process of the cascade using chromatography and other analytical methods. For example, the performance of P450 in the amination cascade reaction is influenced by the amino donor.38 Therefore, the productivity of 12-hydroxydodecanoic acid may be hampered by an excess of benzylamine (BA) as an amino donor for ω-transaminase (ω-TA). Ahsan et al.39 tested different concentrations of BA in order to obtain optimal conditions. When 20 mM BA was added as the amino donor, 12-aminodododecanoic acid production was increased 5-fold, reaching a product titer of 1.48 mM from 10 mM lauric acid.Synthesis of ω-amino fatty acids for use as nylon monomers
Currently, most nylon monomers are derived from fossil resources, e.g. the nylon 12 monomer ω-dodecalactam is produced from butadiene. However, the process involves toxic and corrosive raw materials such as benzene and fuming sulfuric acid, as well as generating large amounts of waste, which puts pressure on the environment. By contrast, nylon monomers obtained from biomass-derived resources, such as vegetable oils, not only have significant low-carbon properties and environmental advantages, but also can produce unique structural features that can be used to meet different performance requirements.40 For example, in the synthesis of nylon 11, the molecular chains polymerized from castor oil-derived 11-aminoundecanoic acid castor oil have a better flexibility than ordinary nylons due to their long carbon-chain structure.10There are two routes for the production of bio-based nylon monomers: the sugar route and the vegetable oil route.41 The sugar route refers to the fermentation of sugars by microbiological techniques and is used to produce short-chain nylon monomers (C3–C6). The vegetable oil pathway involves the use of fatty acids extracted from vegetable oils as substrates, which involves a series of biological or chemical transformations, the latter usually requiring harsh reaction conditions and often involving organic solvents and toxic substances. By contrast, the bioconversion of fatty acids into nylon monomers using E. coli expressing multiple enzymes as a cascade catalyst is a promising and more environmentally friendly process that usually requires no organic solvents or toxic substances, and can be conducted under milder reaction conditions.42 The bioconversion of fatty acids into nylon monomers can be divided into two categories of multienzyme cascade reactions depending on the fatty acid substrate used: (1) oxidation of fatty acid terminal C–H bonds, e.g. synthesis of nylon 12 monomer; (2) oxidative cleavage of fatty acids as in the synthesis of the nylon 8, 9 and 11 monomers. Both strategies are described here.
Multienzyme cascade synthesis via terminal C–H bond oxygenation
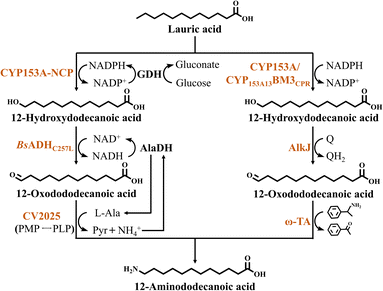
The biosynthesis of nylon monomers by oxidative oxidation of terminal C–H bonds usually uses medium-chain fatty acids such as lauric acid (DDA) as substrates. The synthesis of 12-aminododecanoic acid (ω-AmDDA, nylon 12 monomer) from DDA extracted from palm oil usually involves three reaction steps (Fig. 3). The first step is the hydroxylation of the ω-terminus of DDA using a monooxygenase to generate 12-hydroxydodecanoic acid (ω-OHDDA), the second step is the generation of 12-oxodododecanoic acid (ω-ODDA) from ω-OHDDA by ADH, and finally amination to give ω-AmDDA. As an example of this strategy, Ge et al.14 screen enzymes that catalyze the hydroxylation, oxidation and amination of the DDA terminus, and found that CYP153A-NCP, BsADHC257L and CV2025 were the best candidates. These three enzymes were sequentially assembled into two catalytic modules. CYP153A-NCP was expressed individually in module 1, while BsADHC257L and CV2025 were co-expressed in module 2. Then these two catalytic modules were transformed into the same E. coli BL21 (DE3) for the production of ω-AmDDA in a whole-cell catalytic cascade system. In addition, to overcome problems such as by-product accumulation due to cofactor imbalance and overoxidation during biotransformation, the author applied a number of methods. (1) NADH-dependent alanine dehydrogenase (AlaDH) from Bacillus subtilis was introduced to construct a cycle of NAD(H) and L-Ala to solve the amino donor supply problem. AlaDH promotes the formation of L-Ala from intracellular pyruvate, which is formed during glucose catabolism or released during transamination, thus regenerating the intracellular L-Ala pool. Since NAD+-dependent BsADHC257L reduces NAD+ to NADH during catalysis, NADH-dependent AlaDH was introduced to maintain intracellular redox balance. (2) Introducing GDH from Bacillus cereus for intracellular in situ regeneration of NADPH. (3) Introducing the heterologous ribulose 5-phosphate-dependent PLP synthesis pathway to increase PLP supply. (4) Blocking the β-oxidation pathway and introducing the outer membrane protein AlkL. This study culminated in the construction of a cofactor self-sufficient multienzyme cascade system, which achieved the efficient synthesis of ω-AmDDA from 5 mM DDA with 96.5% yield (Table 2).| Substrate | Multienzyme cascade catalysta | Products | Time (h) | Substrate conversion | Product titer (mmol·L−1) | References |
|---|---|---|---|---|---|---|
| a ADH: alcohol dehydrogenase; BVMO: Baeyer–Villiger monooxygenase; ALDH: aldehyde dehydrogenase; NOX: NADPH oxidase; CYP153A: cytochrome P450 153A; FDH: formate dehydrogenase; TA: transferase; OhyA: oleic acid hydratase; NFO: NAD(P)H flavin oxidoreductase; CamAB: redox companion; CYP153A13BM3CPR: cytochrome P450 153A fused with a naturally self-contained cytochrome P450 reductase (CPR). ChnD: alcohol dehydrogenase from Acinetobacter sp; AlkJ: alcohol dehydrogenase from Pseudomonas putida GPo1; TLL: lipase from Thermomyces lanuginosus; PMTA: ω-transaminase from Silicibacter pomeroyi; CV2025: ω-transaminase from Chromobacterium violaceum DSM30191; CYP153A-NCP: cytochrome P450 153A with the NADPH: cytochrome P450 oxidoreductase from Bacillus megaterium. Pa: Paracoccus aminophilus; Ml: Micrococcus luteus; Pa: Pseudomonas aeruginosa; Ar: Acinetobacter radioresistens; Sm: Streptococcus mutans; Cb: Candida boidinii; Sm: Stenotrophomonas maltophilia; Pp: Pseudomonas putida; Af: Agrobacterium fabrum; Gs: Gordonia sihwensis; Dr: Deinococcus radiodurans; Rp: Ruegeria pomeroyi; Mpr: Mycobacterium parascrofulaceum; Bs: Geobacillus stearothermophilus; Ml: Mesorhizobium loti; Ab: Alcanivorax borkumensis; Sp: Silicibacter pomeroyi. | ||||||
| Plant oil | PaOHM3, MlADHM4, PaBVMO, TLL, ArADH, ArALDH, SmNOXM2, CYP153A7, CbFDH, ChnD, PMTA | 8-Aminooctanoic acid | n.m. | n.m. | 8.60 | 7 |
| Olive oil | SmOhyA, MlADH, PpBVMO, FadL, TLL, ChnD, AfTA | 9-Aminononanoic acid | 6 | 54.0% | 10.30 | 44 |
| Olive oil | PaOHM3, MlADHM4, GsBVMOC308L, TLL, CYP153A7, CbFDH, ChnD, SmNOXM2, PMTA | 9-Aminononanoic acid | n.m. | n.m. | 4.10 | 45 |
| Ricinoleic acid | MlADH, PpBVMO, DrNFO, ChnD, RpTA | 11-Aminoundecanoic acid | 36 | 83.0% | 232.00 | 32 |
| Lauric acid | MprCYP153A-NCP, BsADHC257L, CV2025 | 12-Aminododecanoic acid | 8 | 96.5% | 4.83 | 14 |
| Lauric acid | MprCYP153A, CamAB, AlkJ, MlTA | 12-Aminododecanoic acid | 6 | n.m. | 0.60 | 43 |
| Lauric acid | AbCYP153A13BM3CPR, AlkJ, SpTA | 12-Aminododecanoic acid | 24 | n.m. | 1.48 | 39 |
Ahsan et al.43 used another monooxygenase, CYP153A from Mycobacterium parascrofulaceum (MprCYP153A), to catalyze terminal C–H bond oxidation (Fig. 3). CamAB was used as the oxidative chaperone. Initially, the authors utilized MprCYP153A and CamAB to convert DDA into ω-OHDDA. Subsequently, ω-ODDA was generated from ω-OHDDA through alcohol dehydrogenase from Pseudomonas putida GPo1 (AlkJ). Ultimately, ω-AmDDA was obtained via the transamination reaction catalyzed by ω-TA from Mesorhizobium loti. In this study, MprCYP153A/CamAB and AlkJ/ω-TA were constructed in different strains. However, the two-strain catalytic system constructed by separating the cascade into two whole-cell catalytic modules suffered from mass transfer barrier problems, as well as a failure to optimize the supply of cofactors required for the reaction. The conversion of 2 mM DDA by a high concentration of whole-cell catalysts at 300 gCDW·L−1 for 5 h yielded only 0.6 mM ω-AmDDA (Table 2). To simplify the hydroxylation cycle of CYP153A, the group subsequently constructed a self-contained P450 enzyme (CYP153A13 BM3CPR) by fusing CYP153A13 with a naturally self-contained cytochrome P450 reductase (CPR). Co-expression of this enzyme with AlkJ and ω-TA from Silicibacter pomeroyi enabled the whole-cell conversion of DDA into ω-AmDDA. By optimizing the reaction system and BA concentration, the final system catalyzed the production of 1.48 mM ω-AmDDA from 10 mM DDA in 24 h, representing a 5-fold increase in the product concentration compared to the initial system39 (Table 2).
Multienzyme cascade synthesis via oxidative cleavage of fatty acids
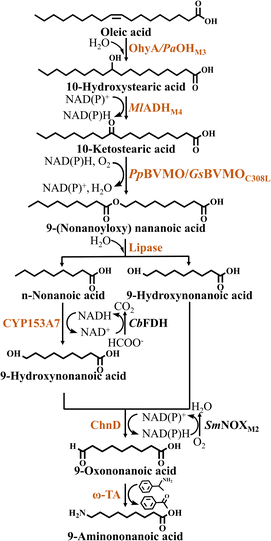
The substrates used in the biosynthesis of nylon monomers by oxidative cleavage are usually long-chain fatty acids such as oleic acid and ricinoleic acid. In contrast to lauric acid, oleic acid and ricinoleic acid are unsaturated, long-chain C18 fatty acids that must undergo oxidative cleavage before they can form a medium-chain structure. For this purpose, C18 fatty acids are converted into different types of ester intermediates by functionalization of the double bond using a variety of enzymes including hydratase, ADH and BVMO, followed by hydrolysis of the corresponding esters by esterases or lipases, to produce ω-AmFAs. In these enzymatic reactions, the regioselectivity of BVMO is critical in determining the type of ester produced, which should be considered in the development of downstream processing.46In multienzyme cascade studies using oleic acid as a substrate, the first step is the generation of 10-hydroxystearic acid. An example of this is the conversion of oleic acid-rich olive oil into 10-hydroxystearic acid by a strain co-expressing lipase from Thermomyces lanuginosus (TLL) and oleic acid hydratase (OhyA).47 However, the activity of OhyA was relatively poor in this reaction system. To solve this problem, researchers screened the reported OhyA homologs based on their sequence similarity.48 A highly efficient OhyA homolog (PaOH) was identified in Paracoccus aminophilus DSM 8538, the most active oleic acid hydratase reported to date. After optimizing the reaction conditions, 90 g·L−1 of oleic acid was successfully converted into 10-hydroxystearic acid with a volumetric productivity of 522 g·L−1 d−1. In a subsequent study, PaOH was subjected to whole-gene random mutagenesis by error-prone PCR.49 The thermal stability and combined catalytic performance of the mutants for oleic acid conversion were also evaluated. The final triple mutant PaOHF233L/F122L/T15N exhibited higher activity and better thermal stability, and its catalytic efficiency constant (Kcat/Km) increased significantly from 33 to 119 s−1 mM−1.
Next, two types of ester intermediates were produced from 10-hydroxystearic acid due to the selection of different BVMOs for the esterification reaction. When using BVMO from Pseudomonas putida KT2440, 10-hydroxystearic acid is esterified using ADH and PpBVMO to produce 9-(nonanoyloxy) nonanoic acid, which is subsequently hydrolyzed by esterase to produce n-nonanoic acid and 9-hydroxynonanoic acid50,51(Fig. 4). To further enhance the esterification efficiency, on the basis of an engineered BVMO enzyme (E6BVMO),52 Jeon et al.53 overexpressed FadL, a long-chain fatty acid transporter protein located in the extracellular membrane, which increased the esterification efficiency by 4-fold, with the rate of ester production reaching 1.2 mmol·L−1·h−1. Finally, 9-hydroxynonanoic acid was further converted into the nylon 9 monomer 9-aminononanoic acid using the alcohol dehydrogenase ChnD and ω-TA. However, ω-TA exhibited relatively low activity in this reaction system. In order to overcome the bottleneck of the ω-TA reaction, Wang et al.44 conducted screened transaminases that catalyze the reductive amination of ω-oxoalkanoic acids. Among them, ω-TA from Agrobacterium fabrum (AfTA) was selected for its ability to effectively catalyze the conversion of 9-oxononanoic into 9-aminononanoic acid. Eventually 15 mM oleic acid was converted into 10.3 mM 9-aminononanoic acid in 54% yield (Table 2). Moreover, Chong et al.45 also introduced innovative operations in the synthesis of 9-aminononanoic acid. They rationalized the cascade reaction into different modules to manage the protein expression burden by distribution of the complete pathway into four separate recombinant strains. Strain A co-expressed TLL and oleic acid hydratase from Paracoccus aminophilus (PaOHM3), to hydrolyze olive oil to oleic acid and further produce 10-hydroxystearic acid. Strain B co-expressed BVMO from Gordonia sihwensis (GsBVMOC308L) and ADH from Micrococcus luteus (MlADHM4) to esterify 10-hydroxystearic acid into produce 9-(nonanoyloxy) nonanoic acid. Strain C co-expressed CYP153A7, the iron-oxidizing protein Fdx and its reducing protein Fdr, in combination with FDH from Candida boidinii (CbFDH), which converted n-nonanoic acid into 9-hydroxynonanoic acid. Strain D co-expressed ChnD, aldehyde dehydrogenase from Geobacillus thermodenitrificans (GtALDH) and the NAD(P)H oxidase from Streptococcus mutans (SmNOXM2), which converted 9-hydroxynonanoic acid to 1, 9-nonanedioic acid. Strain E co-expressed ChnD, ω-transaminase from Silicibacter pomeroyi (PMTA) and SmNOXM2 to convert 9-hydroxynonanoic acid into 9-aminononanoic acid. Strain E, acting independently, was able of converting 10 mM 9-hydroxynonanoic acid to 2.5 mM 9-aminononanoic acid within 6 h. In addition, the authors optimized the cell ratio and cell density of strains A and B in this reaction system. It was found that a 12 gcdw·L−1 of cell loading and a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of cell catalysts showed the highest productivity. The authors speculated that the ultra-high density of cells might have affected the mass transfer of oxygen and hydrophobic substrates. Combining strains A, B, C and E under optimal conditions resulted in the catalytic production of 4.8 mM 9-aminononanoic acid from olive oil (Table 2).
1 ratio of cell catalysts showed the highest productivity. The authors speculated that the ultra-high density of cells might have affected the mass transfer of oxygen and hydrophobic substrates. Combining strains A, B, C and E under optimal conditions resulted in the catalytic production of 4.8 mM 9-aminononanoic acid from olive oil (Table 2).
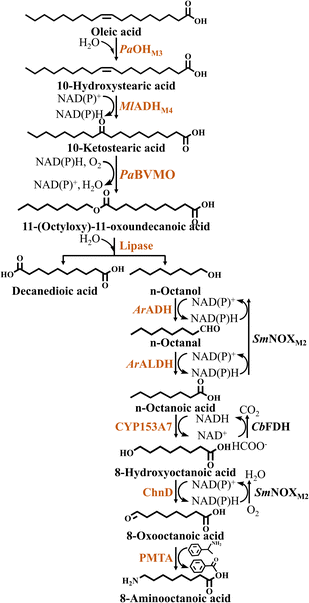
When using BVMO from Pseudomonas aeruginosa, 10-hydroxystearic acid is esterified using ADH and PaBVMO to produce 11-(octyloxy)-11-oxoundecanoic acid. Similarly, 11-(octyloxy)-11-oxoundecanoic acid is hydrolyzed by esterase to produce n-octanol and sebacic acid. Subsequently, n-octanoic acid is further converted into n-octanoic acid by ADH, aldehyde dehydrogenase (ALDH) and NOX. N-octanoic acid is hydroxylated by CYP153A7 to produce 8-hydroxynonanoic acid. Finally, the nylon 8 monomer 8-aminooctanoic acid is generated by ChnD and PMTA7 (Fig. 5, Table 2).
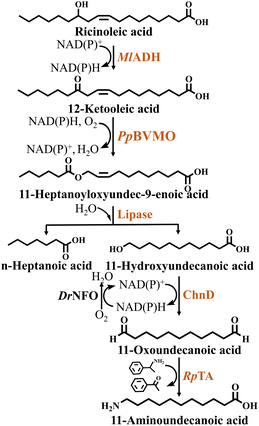
In a study on the synthesis of the nylon 11 monomer 11-aminoundecanoic acid from ricinoleic acid, Kim et al.32 designed a multienzyme cascade catalytic system based on an E. coli whole-cell biocatalyst and lipase. The enzymes involved in this reaction system included MlADH, PpBVMO, DrNFO (NAD(P)H flavin oxidoreductase from Deinococcus radiodurans), ChnD and RpTa (ω-Ta from Ruegeria pomeroyi). In order to improve the expression and structural stability of the cascade enzymes in the heterologous host, the authors engineered PpBVMO. Firstly, the Cys residue, which is susceptible to oxidative stress, was replaced with Leu, i.e. the C302L mutation. The aim was to strengthen the resistance to oxidants of BVMO to improve its stability. Then fusion of maltose-binding protein (MBP) to BVMO via an E6-tag and mutated to C302L variant, resulting in a significant increase in the soluble expression level of the engineered enzyme (MBP-E6BVMOC302L). However, when the substrate concentration exceeded 10 mM, it resulted in obvious inhibition of the cascade reaction. In order to overcome this bottleneck, the authors added in situ adsorption resin. After screening, it was found that the resin SP825 binds more tightly to the substrate and has the highest adsorption effect. Ultimately, 232 mM 11-aminoundecanoic acid was obtained from 300 mM ricinoleic acid using a one-pot multienzyme cascade system. This was a remarkable 640-fold increase compared to the initial system (Fig. 6, Table 2).
Emerging strategies for increasing the ω-amino fatty acid synthesis efficiency
Compared to traditional chemo-catalytic processes, multienzyme cascade reactions still suffer from lower productivity. To improve the efficiency, economy and sustainability of multienzyme cascades, recent studies applied multiple strategies. The production of ω-AmFAs from vegetable oils requires cofactor-dependent redox reactions. Several cofactor regeneration strategies have been developed to make it economically feasible to use expensive cofactors, without requiring large quantities. In addition, protein engineering strategies were employed to improve enzyme stability and catalytic performance.Cofactor regeneration
Oxidoreductases are the largest class of enzymes,54 accounting for a quarter of all entries in the Expert Protein Analysis System (ExPASy). Reactions catalyzed by oxidoreductases involve the transfer of electrons between donors and acceptors, which usually requires the participation and facilitation of cofactors.55 However, biological cofactors are expensive and constantly consumed during the reaction, resulting in the cofactor limitation of enzymatic redox reactions. To address this problem, the construction of cofactor regeneration systems has been extensively studied in recent years, as the constant addition of expensive exogenous cofactors would not be economically feasible. NAD(P)+-dependent ADH and ALDH can achieve cofactor regeneration by coupling with NOX. For example, to achieve the synthesis of 11-aminoundecanoic acid, DrNFO was introduced into E. coli expressing ChnD and RpTA, whereby NAD(P)+-dependent ChnD could regenerate NAD(P)H to realize cofactor regeneration.32 Compared to ChnD and RpTA-expressing E. coli without a cofactor regeneration system, the specific activity of the engineered strain with a cofactor regeneration system was increased 2-fold. For the synthesis of 12-aminododecanoic acid, Ye et al.14 adopted two cofactor regeneration strategies. First, they introduced GDH for intracellular in situ regeneration of NADPH. Second, NADH-dependent AlaDH was introduced to construct an alanine-NADH cycle. Pyruvate is formed during glucose catabolism, and the introduction of AlaDH promotes the formation of L-Ala from intracellular pyruvate, leading its intracellular regeneration. NAD+-dependent BsADHC257L reduces NAD+ to NADH during catalysis, so that adding an NADH-dependent AlaDH is conducive to maintaining intracellular redox balance. By combining these two strategies in E. coli, a self-sufficient cofactor and co-substrate regeneration system was constructed.The multi-enzyme cascade reaction with self-sufficient cofactors is achieved through the coupling between cofactor-regenerating enzymes and cofactor-consuming enzymes. However, the introduction of cofactor-regenerating enzymes increases the expression pressure and requires the addition of co-substrates, increasing the complexity and cost of the catalytic process.20,56 To address this issue, one study modified the cofactor specificity of NAD+-dependent MlADH to convert oleic acid into 9-aminononanoic acid intermediates.57 The biochemical properties of MlADH were first characterized to identify engineering targets to alter its cofactor specificity. The corresponding mutants were then constructed based on the substrate binding mode and structural features to successfully shift the cofactor preference from NAD+ to NADP+. The modified NADP+-dependent engineered enzyme MlADHD37S/A38R/V39S/T15I successfully was coupled with NADP+-dependent PpBVMO to convert C18 fatty acids into C9 compounds, resulting in a remarkable 1800-fold increase of NADP+ economy.
Protein engineering
As one of the fundamental tools in modern biotechnology, protein engineering is commonly used to construct engineered enzymes with high stability, activity, specificity, as well as chemo-, regio- and stereoselectivity.58,59 Protein engineering has been successfully applied to the development of high-efficiency whole-cell biocatalysts for ω-AmFA production by increasing the solubility and expression levels of pathway enzymes. An example of such a system is the engineering of BVMO-based E. coli biocatalysts for the oxidative cleavage of long-chain fatty acids for the production of ω-AmFAs. The low stability of BVMO is the main reason for its limited use.60 BVMO has been reported to contain 12 Met and 8 Cys residues,61 which are highly sensitive to reactive oxygen species (ROS). One of them, Cys302, is located at approximately 8.7 A inside the active site and may be oxidized by ROS. Therefore, Woo et al.62 selected Cys302 as an engineered target for protein modification.63–65 They replaced the Cys residues susceptible to oxidative stress with Leu to alleviate the oxidation of the active site and improve the stability of the engineered enzyme (E6BVMOC302L) to oxidation and thermal stress. In another study, Zhang et al.66 identified a novel GsBVMO with high regioselectivity. The structure of GsBVMO was predicted using homology modelling. The variant GsBVMOC308L was tested and found to maintain good thermal stability while significantly improving the catalytic activity and regioselectivity. This was attributed to the replacement of Cys, which is susceptible to oxidative stress, by the inert amino acid Leu. In addition, the C308L mutation created additional hydrophobic interactions with Ala200 and Thr224, which promoted enzyme–substrate interactions to accelerate the reaction. Using GsBVMOC308L as a biocatalyst, 10-ketostearic acid was efficiently converted into 9-(nonanoyloxy) nonanoic acid with a space-time yield of 60.5 g·L−1 d−1. A study was conducted to predict the enzymatic pathway of PpBVMO using CAVER and/or protein energy landscape exploration (PELE) software.67 Amino acid residues susceptible to oxidation by H2O2 (e.g. methionine and tyrosine) as well as amino acid residues located near the predicted H2O2 migration pathway were replaced with less active or inert amino acids (e.g. leucine and isoleucine). The enzyme variants were ultimately designed to be resistant to H2O2. PpBVMOC302L/M340L/M380L produced a 2-fold higher concentration of 9-(nonanoyloxy) nonanoic acid in cascade biotransformation. In addition to modification of key residues, enzyme fusion with a soluble polyionic peptide tag (hexaglutamate, E6) or a highly soluble enzyme (MlADH) significantly improved the functional expression and structural stability of BVMO in E. coli. The use of the engineered enzyme E6BVMO as a biocatalyst yielded 85 mM 11-heptanoyloxyundec-9-enoic acid, an ester that could be subsequently hydrolyzed to yield 11-aminoundecanoic acid.32 Maltose-binding protein (MBP) acts as a solubility tag that contributes not only to the stability of BVMO but also to its catalytic efficiency.68 When MBP was fused with E6BVMOC302L (MBP-E6BVMOC302L), its catalytic efficiency (Kcat/Km) was increased 1.8 and 2.2 times compared to E6BVMOC302L and E6BVMO, respectively. The whole-cell biocatalyst constructed by co-expressing MBP-E6BVMOC302L with MlADH was able to convert 200 mM ricinoleic acid into 152 mM 11-heptanoyloxyundec-9-enoic acid in 12 h, representing a 30% improvement over the controls train expressing only E6BVMOC302L.The thermostable NAD+-dependent alcohol dehydrogenase from Geobacillus stearothermophilus (BsADH) was used in the biocatalytic synthesis of 12-aminodododecanoic acid. Considering the poor stability of BsADH under oxidative stress, sequence homology and computational simulations revealed that among the three Cys residues not involved in zinc coordination, the Cys257 site is the most unstable and susceptible to oxidation. Consequently, this residue was mutated to Leu, and the resulting C257L variant exhibited greater oxidative stability combined with a higher inactivation temperature.69 In contrast to wild-type BsADH, biocatalytic conversion using the engineered enzyme BsADHC257L reached up to 23% without overoxidation. The engineered enzyme BsADHC257L therefore represents a promising biocatalyst for the synthesis of 12-aminododecanoic acid.
Modular engineering
Module-based modular engineering is mainly applied at the pathway level to adjust the expression of enzyme modules in order to increase the yield of target products.70 Although module-based modular engineering makes the catalytic system more complex, it also offers greater opportunities.71 The modular design aims to create a biocatalyst by assembling a reusable or replaceable modular unit in a plug-and-play fashion.72 For example, in the multienzyme cascade pathway for 8-aminooctanoic acid production, 9 reactions catalyzed by 12 enzymes were involved in the construction of the cascade scheme.7 Considering that co-expression of all genes encoding the required enzymes in a single cell will lead to a high protein expression load and redox imbalance problems, the researchers divided it into five separate modules based on the three principles of rapid substrate conversion, reducing the accumulation of intermediates or by-products, and self-sufficient cofactor recycling. Based on the modular systems, the authors designed a two-stages-in-one-pot process, obtaining 8.6 mM 8-aminooctanoic acid. In another study, Chong et al.45 divided the 9-aminononanoic acid synthesis pathway into four modules. Strain A co-expressing TLL and PaOHM3 hydrolyzed olive oil to yield oleic acid and further oxidized it to produce 10-hydroxystearic acid. Strain B co-expressed GsBVMOC308L and MlADHM4 to esterify 10-hydroxystearic acid, thus producing 9-(nonanoyloxy) nonanoic acid. Strain C co-expressed CYP153A7, Fdx and Fdr, in combination with the introduced coenzyme CbFDH, in order to convert n-nonanoic acid into 9-hydroxynonanoic acid. Finally, strain E co-expressed ChnD, PMTA and SmNOXM2 to convert 9-hydroxynonanoic acid into 9-aminononanoic acid. Ultimately, 10 mM 9-hydroxynonanoic acid was converted into 2.5 mM 9-aminononanoic acid in 6 h. Ye et al.14 reported a modular engineering strategy for the production of 12-aminododecanoic acid. The design of these modules was based on the two principles of stable inputs and outputs, combined with fast conversion of unstable intermediates. The authors expressed CYP153A-NCP alone in module 1 to produce ω-OHDDA from DDA. Due to the readily reducible nature of ω-ODDA, BsADHC257L and CV2025 were expressed in module 2 to convert ω-ODDA into ω-AmDDA immediately after its production, thereby pulling the reaction equilibrium towards ω-AmDDA formation rather than ω-OHDDA accumulation. As a results of this modularization strategy, 5 mM ω-OHDDA was converted into 4.83 mM ω-AmDDA, corresponding to a 96% conversion rate. Kim et al.32 designed two consecutive modules to construct a multienzyme cascade pathway for the production of 11-aminoundecanoic acid from ricinoleic acid. Module 1 consisted of MlADH and PpBVMO, while module 2 consisted of ChnD and RpTA. After the ratio between module 1 and 2 was further optimized to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.25 (w/w), the multienzyme cascade system converted 300 mM ricinoleic acid into 232 mM 11-aminoundecanoic acid.
2.25 (w/w), the multienzyme cascade system converted 300 mM ricinoleic acid into 232 mM 11-aminoundecanoic acid.
Conclusions
High-value-added nylon monomers can be obtained from vegetable oil-derived fatty acids through multienzyme cascades producing ω-AmFAs. This process has opened up a new green path for the production of nylon industry by eliminating the dependence on fossil resources. However, most of the nylon monomer biosynthesis processes are still at the laboratory stage and have not yet reached industrial scale production. This is mainly due to the insufficient catalytic activity and stability of the utilized enzymes compared to chemical catalysts. At the same time, in vivo multi-enzyme cascades can lead to excessive metabolic burden and poor enzyme expression due to co-expression of multiple genes. In addition, the low tolerance of E. coli to industrial conditions and the toxicity of substrates and products to cells are factors that limit large-scale production. In the future, several bottlenecks need to be overcome to enhance the industrial potential of multienzyme cascade synthesis for bio-based nylon monomers. Firstly, in the dynamic regulation of cofactor balance, the metabolic network of intracellular cofactors needs to be studied in depth, and sensors capable of real-time monitoring of cofactor concentration need to be developed. And through the feedback control system, the expression level or activity of relevant enzymes should be dynamically adjusted to ensure that cofactors are always maintained in the appropriate concentration range during the multienzyme cascade reaction to meet the reaction demand and enhance the overall reaction efficiency. Secondly, the development of thermally stable enzymes adapted to factory reaction conditions is also a key direction. Through the thermal stability modification of existing enzymes, they can still maintain good catalytic activity and stability under the more severe reaction conditions such as higher temperature and high substrate concentration, which are common in industrial production. It reduces the cooling cost during the reaction process, improves the production efficiency, and promotes the efficient multi-enzyme synthesis of nylon monomers from vegetable oils from the laboratory to the industrial scale. By addressing these problems, it is expected to break through the dilemma faced by the current nylon monomer biosynthesis process, and realize the efficient and stable production of high value-added nylon monomers by multienzyme cascade catalysis using vegetable-derived fatty acids as substrates. This will help the nylon fibre industry to develop in a more sustainable direction and open up broad prospects for industrial applications.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 22178173), and the State Key Laboratory of Materials-Oriented Chemical Engineering (No. SKL-MCE-23B04 and SKL-MCE-24A10).Notes and references
- S. Schaffer and T. Haas, Org. Process Res. Dev., 2014, 18, 752–766 CAS.
- V. Hirschberg and D. Rodrigue, J. Polym. Sci., 2023, 61, 1937–1958 CAS.
- L. Lin, R. Ledesma-Amaro, X.-J. Ji and H. Huang, Trends Biotechnol., 2023, 41, 150–153 CAS.
- B. M. Stadler, C. Wulf, T. Werner, S. Tin and J. G. de Vries, ACS Catal., 2019, 9, 8012–8067 CrossRef CAS.
- J.-W. Song, J.-H. Lee, U. T. Bornscheuer and J.-B. Park, Adv. Synth. Catal., 2014, 356, 1782–1788 CrossRef CAS.
- S. Lim, H.-w. Yoo, S. Sarak, B.-g. Kim and H. Yun, J. Ind. Eng. Chem., 2021, 98, 358–365 CrossRef CAS.
- G.-G. Chong, L.-Y. Ding, Y.-Y. Qiu, X.-L. Qian, Y.-L. Dong, C.-X. Li, A. Li, J. Pan and J.-H. Xu, J. Agric. Food Chem., 2022, 70, 10543–10551 CrossRef CAS.
- K. Nagao, A. Murakami and M. Umeda, Chem. Pharm. Bull., 2019, 67, 327–332 Search PubMed.
- L. Huang, D. Yuan, Y. Yang and X. Cai, RSC Adv., 2020, 10, 39654–39661 RSC.
- S. Anwar, M. Hassanpour Amiri, S. Jiang, M. M. Abolhasani, P. R. F. Rocha and K. Asadi, Adv. Funct. Mater., 2021, 31, 2004326 CrossRef CAS.
- B. de Jager, T. Moxham, C. Besnard, E. Salvati, J. Chen, I. P. Dolbnya and A. M. Korsunsky, Polymers, 2020, 12, 1169 CrossRef CAS.
- H. Gao, Q. Fang, Y. Bai, C. Hu and H. H. Chou, Metab. Eng., 2025, 89, 87–96 CrossRef CAS.
- J. Ge, T. Wang, H. Yu and L. Ye, Nat. Commun., 2025, 16, 175 CrossRef PubMed.
- J. Ge, X. Yang, H. Yu and L. Ye, Metab. Eng., 2020, 62, 172–185 CrossRef CAS.
- J. Y. Kim, Y.-J. Ahn, J. A. Lee and S. Y. Lee, Curr. Opin. Green Sustainable Chem., 2023, 40, 100777 Search PubMed.
- S. M. Danov, O. A. Kazantsev, A. L. Esipovich, A. S. Belousov, A. E. Rogozhin and E. A. Kanakov, Catal. Sci. Technol., 2017, 7, 3659–3675 RSC.
- J.-H. Seo, S.-M. Lee, J. Lee and J.-B. Park, J. Biotechnol., 2015, 216, 158–166 Search PubMed.
- Z. Wang, B. Sundara Sekar and Z. Li, Bioresour. Technol., 2021, 323, 124551 Search PubMed.
- D. M. Barber, A. Ďuriš, A. L. Thompson, H. J. Sanganee and D. J. Dixon, ACS Catal., 2014, 4, 634–638 Search PubMed.
- J. Muschiol, C. Peters, N. Oberleitner, M. D. Mihovilovic, U. T. Bornscheuer and F. Rudroff, Chem. Commun., 2015, 51, 5798–5811 Search PubMed.
- E. Ricca, B. Brucher and J. H. Schrittwieser, Adv. Synth. Catal., 2011, 353, 2239–2262 Search PubMed.
- R. Cutlan, S. De Rose, M. N. Isupov, J. A. Littlechild and N. J. Harmer, Biochim. Biophys. Acta, Proteins Proteomics, 2020, 1868, 140322 Search PubMed.
- J. H. Schrittwieser, S. Velikogne, M. Hall and W. Kroutil, Chem. Rev., 2018, 118, 270–348 Search PubMed.
- K. A. Baritugo, H. T. Kim, Y. David, T. U. Khang, S. M. Hyun, K. H. Kang, J. H. Yu, J. H. Choi, J. J. Song, J. C. Joo and S. J. Park, Microb. Cell Fact., 2018, 17, 129 Search PubMed.
- A. Bruggink, R. Schoevaart and T. Kieboom, Org. Process Res. Dev., 2003, 7, 622–640 Search PubMed.
- S. Sung, H. Jeon, S. Sarak, M. M. Ahsan, M. D. Patil, W. Kroutil, B.-G. Kim and H. Yun, Green Chem., 2018, 20, 4591–4595 Search PubMed.
- C. Rodríguez, I. Lavandera and V. Gotor, Curr. Org. Chem., 2012, 16, 2525–2541 Search PubMed.
- X. Yu, X.-Y. Chen, H.-L. Yu, J.-H. Xu and Z.-J. Zhang, Green Chem., 2023, 25, 3469–3474 Search PubMed.
- A. B. Ramzi, in Microbial Cell Factories Engineering For Production Of Biomolecules, ed. V. Singh, Academic Press, 2021, pp. 393–406 Search PubMed.
- Y. Mo, X. Li, Q. Li, Y. Han, T. Su, P. Zhao, L. Qiao, M. Xiang, F. Li, X. Guo, M. Liu and Q. Qi, J. Agric. Food Chem., 2025, 73, 5320–5327 Search PubMed.
- K. C. Shin and D. K. Oh, World J. Microbiol. Biotechnol., 2021, 37, 105 CrossRef CAS PubMed.
- T.-H. Kim, S.-H. Kang, J.-E. Han, E.-J. Seo, E.-Y. Jeon, G.-E. Choi, J.-B. Park and D.-K. Oh, ACS Catal., 2020, 10, 4871–4878 CAS.
- B. O. Bachmann, Nat. Chem. Biol., 2010, 6, 390–393 Search PubMed.
- B. Delépine, T. Duigou, P. Carbonell and J. L. Faulon, Metab. Eng., 2018, 45, 158–170 Search PubMed.
- A. Kumar, L. Wang, C. Y. Ng and C. D. Maranas, Nat. Commun., 2018, 9, 184 Search PubMed.
- J. Shi, Y. Wu, S. Zhang, Y. Tian, D. Yang and Z. Jiang, Chem. Soc. Rev., 2018, 47, 4295–4313 Search PubMed.
- Y. Gao, F. Li, Z. Luo, Z. Deng, Y. Zhang, Z. Yuan, C. Liu and Y. Rao, Nat. Commun., 2024, 15, 30 Search PubMed.
- M. Tavanti, J. Mangas-Sanchez, S. L. Montgomery, M. P. Thompson and N. J. Turner, Org. Biomol. Chem., 2017, 15, 9790–9793 Search PubMed.
- M. M. Ahsan, M. D. Patil, H. Jeon, S. Sung, T. Chung and H. Yun, Catalysts, 2018, 8, 400 CrossRef.
- J. Rydz, W. Sikorska, M. Kyulavska and D. Christova, Int. J. Mol. Sci., 2014, 16, 564–596 CrossRef.
- M. Winnacker and B. Rieger, Macromol. Rapid Commun., 2016, 37, 1391–1413 CrossRef CAS PubMed.
- J.-W. Song, J.-H. Seo, D.-K. Oh, U. T. Bornscheuer and J.-B. Park, Catal. Sci. Technol., 2020, 10, 46–64 RSC.
- M. M. Ahsan, H. Jeon, P. N. S, T. Chung, H. W. Yoo, B. G. Kim, M. D. Patil and H. Yun, Biotechnol. J., 2018, 13, e1700562 CrossRef PubMed.
- S.-Y. Hwang, J.-M. Woo, G. E. Choi, D.-K. Oh, J.-H. Seo and J.-B. Park, ACS Catal., 2024, 14, 4130–4138 CAS.
- G.-G. Chong, L.-Y. Ding, Y.-Y. Qiu, X.-L. Qian, C.-X. Li, J. Pan and J.-H. Xu, ACS Sustain. Chem. Eng., 2022, 10, 13125–13132 CAS.
- J. M. Yu, Y. Y. Liu, Y. C. Zheng, H. Li, X. Y. Zhang, G. W. Zheng, C. X. Li, Y. P. Bai and J. H. Xu, Chembiochem, 2018, 19, 2049–2054 CAS.
- E.-J. Seo, Y. J. Yeon, J.-H. Seo, J.-H. Lee, J. P. Boñgol, Y. Oh, J. M. Park, S.-M. Lim, C.-G. Lee and J.-B. Park, Bioresour. Technol., 2018, 251, 288–294 CAS.
- Y.-X. Wu, J. Pan, H.-L. Yu and J.-H. Xu, J. Biotechnol., 2019, 306, 100008 Search PubMed.
- Q.-F. Sun, Y.-C. Zheng, Q. Chen, J.-H. Xu and J. Pan, Biochem. Biophys. Res. Commun., 2021, 537, 64–70 CAS.
- J.-W. Song, E. Jeon, D.-H. Song, H. Y. Jang, U. T. Bornscheuer, D. K. Oh and J. B. Park, Angew. Chem., Int. Ed., 2013, 52(9), 2534–2537 CAS.
- A. Kirschner, J. Altenbuchner and U. T. Bornscheuer, Appl. Microbiol. Biotechnol., 2007, 73, 1065–1072 CrossRef CAS PubMed.
- J.-H. Seo, H.-H. Kim, E.-Y. Jeon, Y.-H. Song, C.-S. Shin and J.-B. Park, Sci. Rep., 2016, 6, 28223 CAS.
- E.-Y. Jeon, J.-H. Seo, W.-R. Kang, M.-J. Kim, J.-H. Lee, D.-K. Oh and J.-B. Park, ACS Catal., 2016, 6, 7547–7553 CrossRef CAS.
- A. Guarneri, W. J. van Berkel and C. E. Paul, Curr. Opin. Biotechnol., 2019, 60, 63–71 CrossRef CAS.
- Y. Cárdenas-Moreno, J. González-Bacerio, H. García Arellano and A. del Monte-Martínez, Appl. Biochem. Biotechnol., 2023, 70, 2108–2135 CrossRef.
- A. Kohl, V. Srinivasamurthy, D. Böttcher, J. Kabisch and U. T. Bornscheuer, Enzyme Microb. Technol., 2018, 108, 53–58 CrossRef CAS PubMed.
- E.-J. Seo, H.-J. Kim, M.-J. Kim, J.-S. Kim and J.-B. Park, Chem. Commun., 2019, 55, 14462–14465 RSC.
- Z.-G. Zhang, L. P. Parra and M. T. Reetz, Chemistry, 2012, 18, 10160–10172 CrossRef CAS PubMed.
- K. Balke, A. Beier and U. T. Bornscheuer, Biotechnol. Adv., 2018, 36, 247–263 CrossRef CAS.
- S. Schmidt and U. T. Bornscheuer, in The Enzymes, ed. P. Chaiyen and F. Tamanoi, Academic Press, 2020, vol. 47, pp. 231–281 Search PubMed.
- D. J. Opperman and M. T. Reetz, ChemBioChem., 2010, 11, 2589–2596 CrossRef CAS PubMed.
- J.-M. Woo, E.-Y. Jeon, E.-J. Seo, J.-H. Seo, D.-Y. Lee, Y. J. Yeon and J.-B. Park, Sci. Rep., 2018, 8, 10280 CrossRef PubMed.
- W. P. Dijkman, G. de Gonzalo, A. Mattevi and M. W. Fraaije, Appl. Microbiol. Biotechnol., 2013, 97, 5177–5188 CAS.
- G. Gadda, Biochemistry, 2012, 51, 2662–2669 CAS.
- J. A. Imlay, Nat. Rev. Microbiol., 2013, 11, 443–454 CAS.
- G.-X. Zhang, Z.-N. You, J.-M. Yu, Y.-Y. Liu, J. Pan, J.-H. Xu and C.-X. Li, ChemBioChem., 2021, 22, 1190–1195 CAS.
- E.-J. Seo, M.-J. Kim, S.-Y. Park, S. Park, D.-K. Oh, U. Bornscheuer and J.-B. Park, Adv. Synth. Catal., 2022, 364, 555–564 CrossRef CAS.
- D. S. Waugh, Postepy Biochem., 2016, 62, 377–382 CrossRef CAS.
- L. Kirmair, D. L. Seiler and A. Skerra, Appl. Microbiol. Biotechnol., 2015, 99, 10501–10513 CrossRef CAS PubMed.
- J. M. Sperl and V. Sieber, ACS Catal., 2018, 8, 2385–2396 CrossRef CAS.
- M. Jeschek, D. Gerngross and S. Panke, Curr. Opin. Biotechnol., 2017, 47, 142–151 CrossRef CAS PubMed.
- S. Garcia and C. T. Trinh, Biotechnol. Adv., 2019, 37, 107403 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |