Open Access Article
Open Access ArticlePicoplatin binding to proteins: X-ray structures and mass spectrometry data on the adducts with lysozyme and ribonuclease A†
Giarita
Ferraro
a,
Tereza
Lyčková
b,
Lara
Massai
c,
Pavel
Štarha
 b,
Luigi
Messori
b,
Luigi
Messori
 c and
Antonello
Merlino
c and
Antonello
Merlino
 *a
*a
aDepartment of Chemical Sciences, University of Naples Federico II, Complesso universitario di Monte Sant'Angelo, via Cinthia, 21, 80126, Naples, Italy. E-mail: antonello.merlino@unina.it
bDepartment of Inorganic Chemistry, Faculty of Science, Palacký University Olomouc, 17. listopadu 12, 771 46 Olomouc, Czech Republic
cDepartment of Chemistry “Ugo Schiff”, University of Florence, via della Lastruccia, 3-13, 50019, Sesto Fiorentino, Florence, Italy
First published on 6th May 2024
Abstract
The reactivity of the anticancer drug picoplatin (cis-amminedichlorido(2-methylpyridine)platinum(II) complex) with the model proteins hen egg white lysozyme (HEWL) and bovine pancreatic ribonuclease (RNase A) was investigated by electrospray ionisation mass spectrometry (ESI MS) and X-ray crystallography. The data were compared with those previously obtained for the adducts of these proteins with cisplatin, carboplatin and oxaliplatin under the same experimental conditions. ESI-MS data show binding of Pt to both proteins, with fragments retaining the 2-methylpyridine ligand and, possibly, a chloride ion. X-ray crystallography identifies different binding sites on the two proteins, highlighting a different behaviour of picoplatin in the absence or presence of dimethyl sulfoxide (DMSO). Metal-containing fragments bind to HEWL close to the side chains of His15, Asp18, Asp119 and both Lys1 and Glu7, whereas they bind to RNase A on the side chain of His12, Met29, His48, Asp53, Met79, His105 and His119. The data suggest that the presence of DMSO favours the loss of 2-methylpyridine and alters the ability of the Pt compound to bind to the two proteins. With both proteins, picoplatin appears to behave similarly to cisplatin and carboplatin when dissolved in DMSO, whereas it behaves more like oxaliplatin in the absence of the coordinating solvent. This study provides important insights into the pharmacological profile of picoplatin and supports the conclusion that coordinating solvents should not be used to evaluate the biological activities of Pt-based drugs.
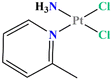
Picoplatin (cis-amminedichlorido (2-methylpyridine) platinum(II), Fig. 1)1–3 is a cisplatin derivative designed primarily to overcome thiol-mediated drug resistance phenomena.4 Indeed, the presence of a pyridine ligand should provide a steric hindrance to a nucleophilic attack. Over the past decades, picoplatin has been evaluated as a single anticancer agent5–7 or in combination with other cytotoxic compounds in a wide variety of tumour types.8–10In vitro studies have shown its ability to overcome platinum drug resistance in cisplatin, carboplatin and oxaliplatin resistant cell lines.11 For these reasons, it was evaluated in the clinical trials for the treatment of small-cell lung and colorectal cancers reaching the Phase III.12
As with the other Pt-based anticancer compounds, once administered, picoplatin can bind to plasma proteins in the blood stream, in particular to albumin, the most abundant plasma protein, which could critically determine its bioavailability and toxicology.13,14 The effect of the protein platination by these platinum drugs is still an open question. In fact, on the one hand, binding of the metal compound to the protein can induce the deactivation/degradation of the drug, making it unavailable for the final targets and causing severe side effects, or it can favour its excretion. On the other hand, the protein can act as a carrier for the bound Pt drug, delivering it to the cells, or as a reservoir for the drug.14 To gain more insight into the role of proteins in the pharmacokinetics of Pt drugs, understanding of their mutual interaction is fundamental. There are several papers and reviews reporting on the effects of the reaction of Pt compounds with proteins;15,16 however, only a few studies have been published on the adducts formed between picoplatin and proteins17 and no structural data are available. For these reasons, picoplatin reactivity with the model systems hen egg white lysozyme (HEWL) and bovine pancreatic ribonuclease (RNase A) was investigated in detail. The two proteins have been chosen, since they have been frequently used to study protein metalation processes.6,18 Moreover, they are cheap, stable, well folded and easy to crystallize and represent a simple model system for studies using electrospray ionization mass spectrometry (ESI MS).6 In the present study, the binding of picoplatin to the proteins was studied in solution by ESI MS and in the solid state by X-ray crystallography using a previously described combined approach.19 Prior to the ESI MS experiments, the metal complex was dissolved in DMSO to guarantee correct solubilisation of the sample. A freshly prepared picoplatin solution was then incubated with the selected proteins in ammonium acetate containing 1% DMSO. The in-solution behaviour of picoplatin in DMSO, 10% DMSO in D2O, 1% DMSO in D2O, 10% DMSO in PBS/D2O and 1% DMSO in PBS/D2O was studied by 1H NMR and ESI-MS (see ESI, Fig. S1–S6 and Table S1†). In all cases, picoplatin degradation was observed and its extent depended on the DMSO concentration. In the case of DMSO and PBS-buffered solutions (10% DMSO in PBS/D2O and 1% DMSO in PBS/D2O), the release of the carrier ligand α-picoline was clearly proved by 1H NMR (see ESI† for more details). The results of studies of picoplatin stability in the presence of DMSO indicate that picoplatin is similar to cisplatin in terms of instability in pure DMSO.20 On the other hand, in contrast to cisplatin and similarly to oxaliplatin, the stability of picoplatin depends on the DMSO concentration and the presence of PBS.
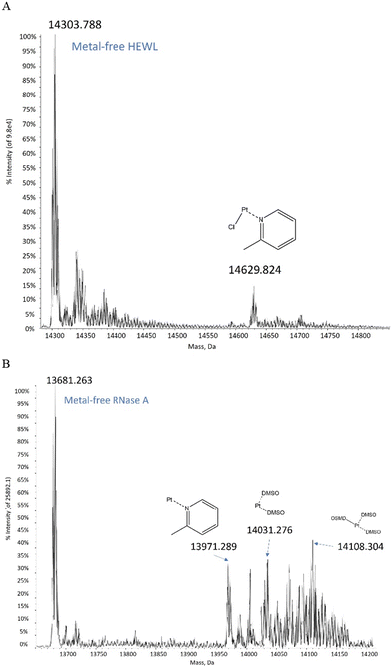
ESI MS data collected on the picoplatin/HEWL system (details in ESI†) revealed that a [Pt(Cl)(2-methylpyridine)]+ fragment bound to the protein after 24 h (Fig. 2A). Similar results were observed after 48 h. The data collected on the picoplatin/RNase A system showed that a [Pt(2-methylpyridine)]2+ fragment bound to RNase A after 24 h or 48 h (Fig. 2B). The spectra also suggested that platinum-containing fragments containing DMSO could bind to the latter protein. Altogether, these data confirmed that at least one Pt-containing fragment could bind to the analysed proteins after 24 h of incubation under the investigated experimental conditions. Notably, these Pt-containing fragments appear to retain the 2-methylpyridine ligand and potentially a chloride ligand as well. To obtain further information on the metal binding site and on the effect of the picoplatin binding on the overall structure of the two proteins, crystallographic studies were carried out.
The structures of platinated HEWL (Fig. S7†) were obtained by reacting the protein with picoplatin under two different experimental conditions. In one case, the Pt complex was pre-dissolved in pure DMSO, diluted with the mother liquor, and then used to soak native protein crystals. The obtained protein-to-Pt molar ratio was about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (structure A, Fig. S7A†). In the other case, picoplatin was added as a powder to a drop containing native HEWL crystals (structure B, Fig. S7A†). In this way, it was in a large molar excess. Both metalated HEWL structures were solved by molecular replacement using the coordinates from PDB code 193L21 as a starting model.
3 (structure A, Fig. S7A†). In the other case, picoplatin was added as a powder to a drop containing native HEWL crystals (structure B, Fig. S7A†). In this way, it was in a large molar excess. Both metalated HEWL structures were solved by molecular replacement using the coordinates from PDB code 193L21 as a starting model.
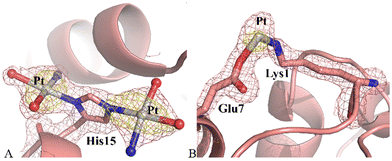
Structure A contains 1165 atoms and refines to an R-factor of 0.185 (Rfree 0.230) at 1.60 Å resolution (see ESI† for further details). The final model revealed that the protein remained unaltered upon Pt binding. Indeed, Cα r.m.s. deviation (rmsd) of HEWL was as low as 0.25 Å. Fourier difference and anomalous difference electron density maps suggested the presence of two peaks due to the binding of Pt-containing fragments close to the side chain of His15 (Fig. 3A) and of a minor peak due to a Pt atom bridging the side chains of Lys1 and Glu7 (Fig. 3B). Close to His15 the two picoplatin fragments were found bound to the NE2 and ND1 atoms (0.50 occupation for both Pt centres), as previously observed in the case of cisplatin22–24 and carboplatin.23,25 In this site, the electron density map was clear and allowed establishing the presence of three ligands around the Pt centre: in both sites these ligands were interpreted as one NH3 and two water molecules. In this way the Pt centre completes its coordination sphere keeping the square planar geometry (Fig. 3A). The fragment bound to ND1 atom of His15 interacts via NH3 with the OD1 atom of Asn93.
Close to the side chains of Lys1 and Glu7 the electron density map was not well-defined, maybe also due to the low occupation of the metal centre (0.30). Thus, only the Pt atom was modelled (Fig. 3B).
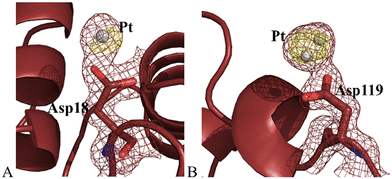
Structure B contains 1230 atoms and refines to an R-factor of 0.174 (Rfree 0.208) at 1.36 Å resolution (see ESI† for further details). The overall structure of the protein in the adduct was substantially unaltered when compared to that of the metal-free protein (Fig. S7A†). Cα r.m.s. deviation (rmsd) of HEWL in the present structure from the model of the metal-free protein (PDB code 193L) is as low as 0.29 Å. Pt atoms were found close to the side chains of Asp18 and Asp119 (Fig. 4). The electron density around the metal centre did not allow completing the metal coordination sphere in these two binding sites, where naked Pt atoms, with partial occupation (0.20 for both metal atoms), were modelled. Asp119 has been previously identified as oxaliplatin binding site.26
Thus, the crystallographic analysis of HEWL adducts with picoplatin in the absence and in the presence of DMSO gave different results suggesting that the Pt compound has a different reactivity towards the protein under the two different experimental conditions studied. The comparison with the structures of the adducts that HEWL forms with cisplatin,23,27 carboplatin23,25 and oxaliplatin26 suggests that, when dissolved in DMSO, picoplatin acts similarly to cisplatin and carboplatin. Indeed, as evidenced by Tanley and co-workers,28 upon addition of DMSO, cisplatin and carboplatin fragments coordinate to the solvent accessible His15 side chain, as observed for picoplatin in structure A. Conversely, in the absence of DMSO, picoplatin mimics oxaliplatin reactivity: it binds the Asp side chains. Indeed, Asp119 has been identified as Pt binding site in structure B and in the adduct formed upon reaction of HEWL with oxaliplatin.26,29
RNase A adducts with picoplatin were then obtained by using the same approach employed to produce platinated HEWL. Structure C was obtained using data collected on a crystal of Pt/RNase A adduct obtained when metal-free RNase A crystals were exposed to picoplatin in the presence of DMSO. In another experiment picoplatin powder was added to a drop containing metal-free RNase A crystals without addition of DMSO (structure D). Phase problem was solved by molecular replacement using coordinates from PDB file 1JVT30 as a starting model. The final models of platinated RNase A contain two protein molecules (chain 1 and chain 2) in the asymmetric unit (Fig. S8A and B†).
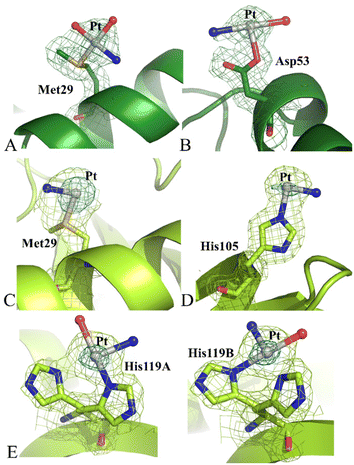
Structure C contains 2164 atoms and refines at 1.99 Å resolution up to R-factor of 0.193 (Rfree 0.243) (see ESI† for further details). The overall structure of RNase A in the adduct presents some differences when compared to the metal-free protein deposited in the PDB under the accession code 1JVT.30 Cα r.m.s.d. values of chains 1 and 2 from those of the metal-free protein are within the range 0.40–0.52 Å. Peaks were observed in the anomalous difference and 2Fo–Fc electron density maps close to the side chains of Met29 and Asp53 of chain 1 (Fig. 5A and B) and close to the side chain of Met29, His105 and His119 of chain 2 (Fig. 5C–E). In this structure, the coordination sphere of the Pt-containing fragments bound to RNase A indicates that picoplatin loses the 2-methylpyridine ligand and undergoes extensive degradation upon protein binding. Indeed, close to side chains of Met29 and Asp53 of chain 1, Pt coordination sphere is completed by three ligands interpreted as ammonia and water molecules, which have replaced the original Pt ligands (Fig. 5A and B). In chain 2, close to the side chains of Met29 and His105 only one ligand (ammine ligand) was modelled. Finally, His119 adopts a double conformation, and the Pt atom can bind both the His side chain conformations, alternatively (Fig. 5E).
Structure D contains 2144 atoms, refines at 1.76 Å resolution up to R-factor of 0.199 (Rfree 0.235) (see ESI† for further details).
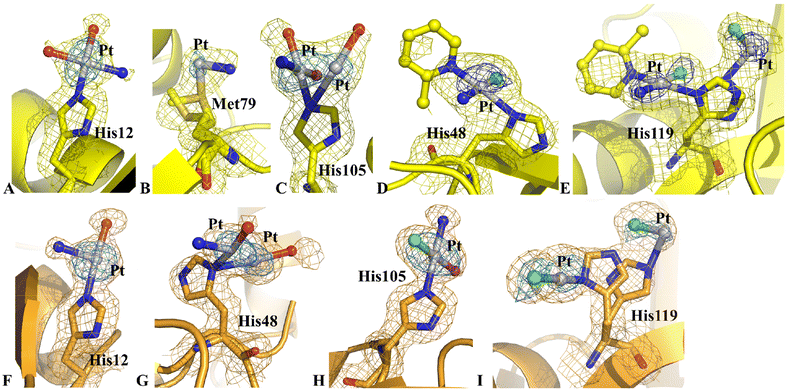
The inspection of the electron density map revealed significant differences in this structure when compared to structure C. In particular, diffraction data of structure D indicated that the side chains of His12, His48 and Met79 could also be platinated, while the side chains of Met29 and Asp53 were metal-free (Fig. 6). The Pt atoms did not retain the 2-methylpyridine ligand and a partial hydrolysis of picoplatin could be observed, as found for HEWL, close to the side chains of His12 (Fig. 6A), Met79 (Fig. 6B) and His105 (Fig. 6C) of chain 1 and close to the side chains of His12 (Fig. 6F), His48 (Fig. 6G), His105 (Fig. 6H) and His119 (Fig. 6I) of chain 2. On the contrary, an almost complete picoplatin molecule is observed close to the side chains of His48 and His119 of chain 1 (Fig. 6D and E). Here, picoplatin bound to the ND1 atom of the His side chains upon losing one of the two chloride ligands. The NE2 atom of His119 was also platinated, even though in this case only a chloride could be interpreted as a Pt ligand (Fig. 6E). Interestingly, chloride ligands were found also close to the Pt centres bound to the side chains of His105 and His119 of chain 2 (Fig. 6H and I).
A detailed comparison between the structure of metal-free RNase A and structure D revealed that the Pt binding to His48 induced significant variations at the level of the His environment. In the metal-free protein, the side chain of His48 is hydrogen bonded to OG1 atom of Thr82 and to the carbonyl oxygen of Asp14 and in contact with the side chain of Thr17.30 Upon platination, the hydrogen bond with Asp14 and the contact with the side chain of Thr17 are lost. The loss of these interactions induced complete disordering of the high flexible loop constituted by residues 16–22, so that residues 17–21 in chain 1 and residues 17–20 in chain 2 could not be modelled in the electron density map.
In agreement with the observations in the case of HEWL, picoplatin showed a different reactivity with RNase A in the presence (structure C) and absence of DMSO (structure D). The presence of DMSO in structure C favoured the loss of Pt ligands in picoplatin, thus changing the protein binding properties of the metal compound.
Notably, cisplatin31,32 and carboplatin33 bind to RNase A close to Met29 in isomorphous RNase A crystals, while oxaliplatin33 binds to the side chains of Asp14 and Met29, His119 and, to a lesser extent, of His105.
In summary, we studied the formation of the adducts resulting from the reaction of picoplatin with HEWL or RNase A. There are no other structural studies of the interaction between this compound and proteins. Our data show that picoplatin fragments bind to HEWL without altering the overall protein structure; the interaction of the metallodrug with RNase A is associated with significant variations in the Pt binding site environment. Crystallographic and mass spectrometry data indicate that different Pt-containing fragments derived from the Pt-based drug can bind to the proteins close to Met, Asp and/or His side chains. The 2-methylpyridine ligand and possibly a chloride ion can be retained.
Comparison with the structural data collected on the same proteins in the presence of cisplatin, carboplatin and oxaliplatin under the same experimental conditions reveals that picoplatin reactivity with proteins seems more similar to that of oxaliplatin in the absence of DMSO, and more similar to cisplatin and carboplatin upon extensive degradation, i.e. in the presence of DMSO. These findings, once again,34 suggest caution in the use of coordinating solvents for establishing the biological activities of potential metal-based drugs and underline that different experimental conditions have to be tested to properly evaluate the reactivity of metallodrugs with proteins.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Elettra staffs for technical assistance during data collections. P. Š. thanks Mr Lukáš Masaryk for his help with the preparation of picoplatin. A. Merlino and L. Messori thank MIUR PRIN 2022-Cod. 2022JMFC3X, “Protein Metalation by Anticancer Metal-based Drugs” for financial support.References
- F. I. Raynaud, F. E. Boxall, P. M. Goddard, M. Valenti, M. Jones, B. A. Murrer, M. Abrams and L. R. Kelland, Cis-Amminedichloro(2-Methylpyridine) Platinum(II) (AMD473), a Novel Sterically Hindered Platinum Complex: In Vivo Activity, Toxicology, and Pharmacokinetics in Mice, Clin. Cancer Res., 1997, 3(11), 2063–2074 CAS.
- P. Beale, I. Judson, A. O'Donnell, J. Trigo, C. Rees, F. Raynaud, A. Turner, L. Simmons and L. A. Etterley, Phase I Clinical and Pharmacological Study of Cis-Diamminedichloro(2-Methylpyridine) Platinum II (AMD473), Br. J. Cancer, 2003, 88(7), 1128–1134, DOI:10.1038/sj.bjc.6600854.
- L. Kelland, Broadening the Clinical Use of Platinum Drug–Based Chemotherapy with New Analogues: Satraplatin and Picoplatin, Expert Opin. Invest. Drugs, 2007, 16(7), 1009–1021, DOI:10.1517/13543784.16.7.1009.
- J. Reedijk, Why Does Cisplatin Reach Guanine-N7 with Competing S-Donor Ligands Available in the Cell?, Chem. Rev., 1999, 99(9), 2499–2510, DOI:10.1021/cr980422f.
- L. Kelland, I. Judson, M. Koehler, T. Stevens and D. Roberts, Preclinical and Clinical Overview of the Novel Platinum Complex, ZD0473 (Cis-Amminedichloro[2-Methylpyridine] Platinum[II]), Lung Cancer, 2000, 29(1), 70, DOI:10.1016/S0169-5002(00)80227-5.
- L. Messori and A. Merlino, Protein Metalation by Metal-Based Drugs: X-Ray Crystallography and Mass Spectrometry Studies, Chem. Commun., 2017, 53(85), 11622–11633, 10.1039/C7CC06442J.
- C.-H. Tang, C. Parham, E. Shocron, G. McMahon and N. Patel, Picoplatin Overcomes Resistance to Cell Toxicity in Small-Cell Lung Cancer Cells Previously Treated with Cisplatin and Carboplatin, Cancer Chemother. Pharmacol., 2011, 67(6), 1389–1400, DOI:10.1007/s00280-010-1435-5.
- R. Earhart, S. Cheporov, O. Gladkov, M. Biakhov, H. Breitz and R. De Jager, Randomized, Phase II Study of Picoplatin in Combination with 5-Fluorouracil and Leucovorin (FOLPI) as a Neuropathy-Sparing Alternative to Modified FOLFOX-6 as First-Line Therapy for Colorectal Cancer (CRC), J. Clin. Oncol., 2009, 27(15_suppl), 4026–4026, DOI:10.1200/jco.2009.27.15_suppl.4026.
- R. L. De Jager, L. Roman, N. Lopatkin, P. Karlov, H. Breitz and R. Earhart, Results of a Phase II Study of Picoplatin with Docetaxel and Prednisone in First-Line Treatment of Castration-Resistant Prostate Cancer (CRPC), J. Clin. Oncol., 2009, 27(15_suppl), 5140–5140, DOI:10.1200/jco.2009.27.15_suppl.5140.
- O. Gladkov, G. Manikhas, M. Biakhov, S. Tjulandin and D. Karlin, Phase 1 Study of Picoplatin (Pico) in Combination with 5-Fluorouracil (FU) and Leucovorin (LV) as Initial Therapy in Subjects with Metastatic Colorectal Cancer (CRC), J. Clin. Oncol., 2007, 25(18_suppl), 14510–14510, DOI:10.1200/jco.2007.25.18_suppl.14510.
- P. Tang, J. Wang and P. Bourne, Molecular Classifications of Breast Carcinoma with Similar Terminology and Different Definitions: Are They the Same?, Hum. Pathol., 2008, 39(4), 506–513, DOI:10.1016/j.humpath.2007.09.005.
- T. Ciuleanu, M. Samarzjia, Y. Demidchik, V. Beliakouski, M. Rancic, D. L. Bentsion, S. V. Orlov, B. A. Schaeffler, R. L. De Jager and H. B. Breitz, Randomized Phase III Study (SPEAR) of Picoplatin plus Best Supportive Care (BSC) or BSC Alone in Patients (Pts) with SCLC Refractory or Progressive within 6 Months after First-Line Platinum-Based Chemotherapy, J. Clin. Oncol., 2010, 28(15_suppl), 7002–7002, DOI:10.1200/jco.2010.28.15_suppl.7002.
- Y. Wang, P. Wu, X. Zhou, H. Zhang, L. Qiu and J. Cao, Exploring the Interaction between Picoplatin and Human Serum Albumin: The Effects on Protein Structure and Activity, J. Photochem. Photobiol., B, 2016, 162, 611–618, DOI:10.1016/j.jphotobiol.2016.07.031.
- B. P. Espósito and R. Najjar, Interactions of Antitumoral Platinum-Group Metallodrugs with Albumin, Coord. Chem. Rev., 2002, 232(1–2), 137–149, DOI:10.1016/S0010-8545(02)00049-8.
- G. Ferraro, D. Loreto and A. Merlino, Interaction of Platinum-Based Drugs with Proteins: An Overview of Representative Crystallographic Studies, Curr. Top. Med. Chem., 2021, 21(1), 6–27, DOI:10.2174/1568026620666200624162213.
- L. Messori and A. Merlino, Cisplatin Binding to Proteins: A Structural Perspective, Coord. Chem. Rev., 2016, 315, 67–89, DOI:10.1016/j.ccr.2016.01.010.
- Y. Wang, P. Wu, X. Zhou, H. Zhang, L. Qiu and J. Cao, Exploring the Interaction between Picoplatin and Human Serum Albumin: The Effects on Protein Structure and Activity, J. Photochem. Photobiol., B, 2016, 162, 611–618, DOI:10.1016/j.jphotobiol.2016.07.031.
- A. Merlino, Recent Advances in Protein Metalation: Structural Studies, Chem. Commun., 2021, 57(11), 1295–1307, 10.1039/D0CC08053E.
- A. Merlino, T. Marzo and L. Messori, Protein Metalation by Anticancer Metallodrugs: A Joint ESI MS and XRD Investigative Strategy, Chem. – Eur. J., 2017, 23(29), 6942–6947, DOI:10.1002/chem.201605801.
- H. P. Varbanov, D. Ortiz, D. Höfer, L. Menin, B. K. Keppler and P. J. Dyson, Oxaliplatin Reacts with DMSO only in the Presence of water, Dalton Trans., 2017, 46, 8929–8932, 10.1039/C7DT01628J.
- M. C. Vaney, S. Maignan, M. Riès-Kautt and A. Ducruix, High-Resolution Structure (1.33 Å) of a HEW Lysozyme Tetragonal Crystal Grown in the APCF Apparatus. Data and Structural Comparison with a Crystal Grown under Microgravity from SpaceHab-01 Mission, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1996, 52(3), 505–517, DOI:10.1107/S090744499501674X.
- A. Casini, G. Mastrobuoni, C. Temperini, C. Gabbiani, S. Francese, G. Moneti, C. T. Supuran, A. Scozzafava and L. Messori, ESI Mass Spectrometry and X-Ray Diffraction Studies of Adducts between Anticancer Platinum Drugs and Hen Egg White Lysozyme, Chem. Commun., 2007,(2), 156–158, 10.1039/B611122J.
- S. W. M. Tanley, A. M. M. Schreurs, L. M. J. Kroon-Batenburg and J. R. Helliwell, Room-Temperature X-Ray Diffraction Studies of Cisplatin and Carboplatin Binding to His15 of HEWL after Prolonged Chemical Exposure, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2012, 68(11), 1300–1306, DOI:10.1107/S1744309112042005.
- G. Ferraro, A. Pica, I. Russo Krauss, F. Pane, A. Amoresano and A. Merlino, Effect of Temperature on the Interaction of Cisplatin with the Model Protein Hen Egg White Lysozyme, J. Biol. Inorg. Chem., 2016, 21(4), 433–442, DOI:10.1007/s00775-016-1352-0.
- S. W. M. Tanley, K. Diederichs, L. M. J. Kroon-Batenburg, C. Levy, A. M. M. Schreurs and J. R. Helliwell, Carboplatin Binding to Histidine, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2014, 70(9), 1135–1142, DOI:10.1107/S2053230X14016161.
- L. Messori, T. Marzo and A. Merlino, The X-Ray Structure of the Complex Formed in the Reaction between Oxaliplatin and Lysozyme, Chem. Commun., 2014, 50(61), 8360, 10.1039/c4cc02254h.
- A. Casini, G. Mastrobuoni, C. Temperini, C. Gabbiani, S. Francese, G. Moneti, C. T. Supuran, A. Scozzafava and L. Messori, ESI Mass Spectrometry and X-Ray Diffraction Studies of Adducts between Anticancer Platinum Drugs and Hen Egg White Lysozyme, Chem. Commun., 2007,(2), 156–158, 10.1039/B611122J.
- S. W. M. Tanley, A. M. M. Schreurs, L. M. J. Kroon-Batenburg, J. Meredith, R. Prendergast, D. Walsh, P. Bryant, C. Levy and J. R. Helliwell, Structural Studies of the Effect That Dimethyl Sulfoxide (DMSO) Has on Cisplatin and Carboplatin Binding to Histidine in a Protein, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68(5), 601–612, DOI:10.1107/S0907444912006907.
- D. Marasco, L. Messori, T. Marzo and A. Merlino, Oxaliplatin vs. Cisplatin: Competition Experiments on Their Binding to Lysozyme, Dalton Trans., 2015, 44(22), 10392–10398, 10.1039/C5DT01279A.
- L. Vitagliano, A. Merlino, A. Zagari and L. Mazzarella, Reversible Substrate–induced Domain Motions in Ribonuclease A, Proteins, 2002, 46(1), 97–104, DOI:10.1002/prot.10033.
- D. Picone, F. Donnarumma, G. Ferraro, I. Russo Krauss, A. Fagagnini, G. Gotte and A. Merlino, Platinated Oligomers of Bovine Pancreatic Ribonuclease: Structure and Stability, J. Inorg. Biochem., 2015, 146, 37–43, DOI:10.1016/j.jinorgbio.2015.02.011.
- L. Messori and A. Merlino, Cisplatin Binding to Proteins: Molecular Structure of the Ribonuclease A Adduct, Inorg. Chem., 2014, 53(8), 3929–3931, DOI:10.1021/ic500360f.
- L. Messori, T. Marzo and A. Merlino, Interactions of Carboplatin and Oxaliplatin with Proteins: Insights from X-Ray Structures and Mass Spectrometry Studies of Their Ribonuclease A Adducts, J. Inorg. Biochem., 2015, 153, 136–142, DOI:10.1016/j.jinorgbio.2015.07.011.
- M. D. Hall, K. A. Telma, K.-E. Chang, T. D. Lee, J. P. Madigan, J. R. Lloyd, I. S. Goldlust, J. D. Hoeschele and M. M. Gottesman, Say No to DMSO: Dimethylsulfoxide Inactivates Cisplatin, Carboplatin, and Other Platinum Complexes, Cancer Res., 2014, 74(14), 3913–3922, DOI:10.1158/0008-5472.CAN-14-0247.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt00773e |
| This journal is © The Royal Society of Chemistry 2024 |