Open Access Article
Open Access Article3D self-assembly of cyclic peptides into multilayered nanosheets
Sandra Díaza,
Adrian Sanchez-Fernandez b,
Juan R. Granja
b,
Juan R. Granja *a,
Ignacio Insua
*a,
Ignacio Insua *cd and
Javier Montenegro
*cd and
Javier Montenegro *ad
*ad
aCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, 15705 Santiago de Compostela, Spain. E-mail: juanr.granja@usc.es; javier.montenegro@usc.es
bCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Enxeñaría Química, Universidade de Santiago de Compostela, 15705 Santiago de Compostela, Spain
cCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Farmacoloxía, Farmacia e Tecnoloxía Farmacéutica, Universidade de Santiago de Compostela, 15705 Santiago de Compostela, Spain. E-mail: ignacio.insua.lopez@usc.es
dOportunius, Galician Innovation Agency (GAIN), 15702 Santiago de Compostela, Spain
First published on 2nd February 2026
Abstract
The hierarchical self-assembly of three-dimensional (3D) supramolecular materials presents a significant challenge in molecular design. This process requires monomers with specific packing geometries and orthogonal contacts to enable multidimensional elongation. Here, we describe a new cyclic peptide design that allows hierarchical self-assembly in all three spatial dimensions to produce nanosheets composed of ordered nanotube layers. The uniformity of the nanosheet aspect ratio and thickness is consistent with the high packing order of the monomers observed in this 3D assembly.
Introduction
Supramolecular self-assembly allows the bottom–up fabrication of nanomaterials with high molecular order, both in packing geometry and surface topology.1 By rational design of self-assembling monomers, it is possible to control their spontaneous organisation into different nanostructures such as nanospheres,2–4 one-dimensional (1D) nanotubes5–7 and nanofibers,8–12 two-dimensional (2D) nanosheets,13–19 to complex three-dimensional (3D) lattices.20 The greater the structural complexity of these nanomaterials, the bigger the challenge of incorporating the required supramolecular motifs in the correct orientations. The addition of more dimensions of supramolecular propagation (1D < 2D < 3D) increases the complexity of monomer design, demanding additional non-covalent forces (e.g. electrostatics, hydrophobic effects and hydrogen bonds) that can bind complementarily without interference. These supramolecular contacts must be precisely positioned around the monomer's structure to control the direction of propagation.21 For these reasons, the rational design of monomers capable of undergoing sequential self-assembly across multiple dimensions is evidently challenging. Furthermore, the development of supramolecular systems that can precisely control the final size and 3D aspect ratio of the resulting nanostructure – such as biological components – remains elusive, particularly without the use of templates to limit their size.In the pursuit of biocompatible supramolecular systems, peptides provide the necessary backbone rigidity and chemical versatility in their side chains to create self-assembling monomers with geometric control.22–24 The adoption of secondary structures by peptides, like α-helices or β-sheets,25 can be exploited to arrange amino acid side chains on their surface with specific orientations, thereby controlling self-assembly.26–28 For instance, α-helical motifs can be assembled into spheres,4,29 tetrahedra,30 fibres31 and nanosheets,32,33 while β-sheet domains can also direct 1D34–36 and 2D37,38 self-assembly.39 Additionally, reactive peptides can produce dynamic assemblies where structure and function can be controlled by physical and chemical stimuli through monomer conversion.40,41 However, the rational design of peptides that undergo ordered elongation in the three dimensions of space remains challenging, with reported examples mostly focused on peptide foldamer assemblies42 and peptide crystals.20,43
Cyclic sequences of alternating D/L amino acids can adopt flat macrocyclic conformations to enable backbone stacking via H-bonds to form 1D nanotubes (Fig. 1).44,45 During controlled stacking, cyclic peptides (CPs) will preferentially segregate amino acid side chains with favourable interactions (e.g. hydrophobic effects, electrostatic forces, etc.), creating different self-sorted domains along the resulting nanotube. We have exploited the particular geometry of this monomer to control the sequential self-assembly of CPs into nanotubes and nanosheets (Fig. 1).46–48 In these strategies, axial inter-backbone hydrogen bonding primarily drives nanotube elongation, while 2D nanotube packing is induced by hydrophobic nanotube surfaces in aqueous medium. The disposition of hydrophobic domains around the nanotube can be used to control the 2D ordering of nanotubes. For example, a single hydrophobic domain occupying one third of the cyclic peptide's surface can generate bilayers (Fig. 1a),46,47 while two hydrophobic domains on opposite sides of the macrocycle can lead to a monolayer (Fig. 1b).48 The substitution of a tetraphenylethene hydrophobic core with four cyclic peptide rings, strongly induces the assembly of large 2D self-assembled structures.49
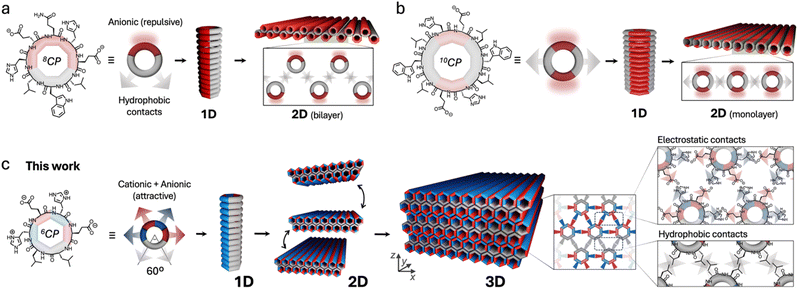 | ||
| Fig. 1 Hierarchical self-assembly of D/L-alternating cyclic peptides into 1D nanotubes and 2D/3D nanosheets. (a) and (b) Previous designs using octapeptides (8CP)46,47 and decapeptides (10CP)48 do not allow elongation along the Z axis, only producing nanotube bilayers and monolayers, respectively. (c) This work presents a smaller cyclic hexapeptide (6CP), capable of establishing six supramolecular contacts at 60° from each other around the nanotube's circumference, thus allowing the complementary hexagonal packing of nanotubes propagating in the third dimension as nanosheet multilayers. | ||
To engineer sequential 1D–2D–3D self-assembly, we here report the use of a smaller monomer, a hexapeptide (6CP). The restricted backbone flexibility and well-defined chemical topology of 6CP make it a suitable building block for multidimensional self-assembly (Fig. 1c).50 Unlike previous studies, where charged domains were used to generate repulsive surfaces, this new design strategy arranges supramolecular forces; we here exploit charge complementarity to create an internal network that matches all non-covalent interactions around the monomer. Therefore, our aim was to balance electrostatic forces between CP monomers to induce growth in the third dimension. This simple yet critical modification opened access to a new dimension of supramolecular growth, from 2D to 3D. Thus, 6CP was rationally designed to form nanotubes for subsequent assembly into flat nanosheets, which can further stack complementarily to form regular multilayered architectures. Following this rationale, the resulting 3D nanosheets were assembled in aqueous buffer and characterised by microscopy and X-ray scattering techniques, exhibiting remarkably homogeneous heights and aspect ratios.
Results and discussion
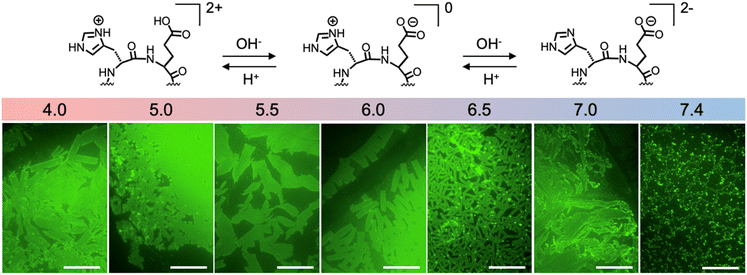
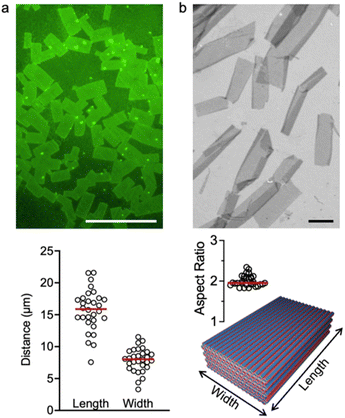
The molecular design of 6CP [cyclo-(D-Leu-L-Leu-D-His-L-Glu-D-His-L-Glu-)] consists of six amino acids with alternating chirality and distributed into two domains: a hydrophobic Leu dyad and a polar Glu–His repeat (Fig. 1c). As such, the self-assembly of 6CP expands the amphiphilic nature of this monomer along the surface of 1D nanotubes, creating hydrophilic and hydrophobic domains by polarity-based self-organisation. Since D/L-alternating cyclic peptides can only stack homochiral residues within nanotubes,51 6CP was designed to restrict peptide stacking into a parallel β-sheet by including two consecutive leucines with opposite chirality. Thus, antiparallel self-assembly would be disfavoured by the chiral mismatch of hydrophobic domains. Consequently, the designed nanotubes would align hydrophilic residues with identical properties – either acidic (Glu) or basic (His) – forming alternating charged sections around the nanotubes at angles of 60° (Fig. 1c). These rational monomer design considerations enable the aqueous self-assembly of 6CP into a 3D nanotube lattice with hexagonal packing, which was previously inaccessible from larger octapeptide (8CP) and decapeptide (10CP) monomers with narrower angles between adjacent side chains.46–48To probe this molecular design, 6CP was synthesised by standard Fmoc solid-phase peptide synthesis as previously reported.52 The purified peptide was dissolved at a concentration of 50 µM in a phosphate buffer solution (20 mM). Self-assembly was induced by temperature annealing: the peptide solution was heated to 80 °C for 1.5 h and then cooled to room temperature on the bench. Due to the presence of acidic glutamic and basic histidine residues, the self-assembly of 6CP was tested across pH (4.0–7.4) to assess the effect of monomer ionisation on supramolecular association (Fig. 2). All samples were analysed by epifluorescence microscopy using thioflavin-T (ThT) to stain any assembled nanostructures, as this dye shows enhanced emission when intercalated between β-sheets,53 like those generated by stacked CPs. As shown in Fig. 2, the morphology of the drop-cast 6CP samples varied with pH. Discrete rectangular nanostructures were produced at pH 6.0–6.5, where zwitterionic monomers could elongate in 3D due to complementary electrostatic interactions between the surface of nanotubes with segregated domains (Fig. 1c). At pH values where 6CP acquires a self-repulsive net charge other than zero, like pH 7.4, this peptide forms amorphous disordered aggregates. Although side chain (de)protonation occurs at pH values defined by their pKa (Glu ≈ 4.2 and His ≈ 6.0),54 cooperative effects (e.g. neighbouring monomer interactions or hydrophobic pockets within the assembly) can result in deviations from these intrinsic pKa values, as has been observed in other supramolecular systems and protein folds.55–58 Interestingly, nanosheets assembled between pH 6.0 and 6.5 displayed remarkable homogeneity in size and shape. In particular, at pH 6.5, the consistent 2D aspect ratio and low polydispersity observed (0.04–0.05)59 suggest a highly ordered self-assembling process (Fig. 3), which is consistent with the cooperative 2D self-assembly previously reported for the cyclic decapeptide 10CP.48 The anisotropic (i.e. rectangular) growth of 2D nanosheets can be explained by the different supramolecular forces that drive propagation in each direction. While inter-backbone H-bonding is the primary driving force along one axis, perpendicular electrostatic and hydrophobic contacts direct lateral nanotube association (Fig. 1c). The number of H-bonds involved in axial peptide stacking remains constant during the polymerisation of nanotubes, while lateral nanotube contacts gain multivalency cooperatively with increasing nanotube length.48
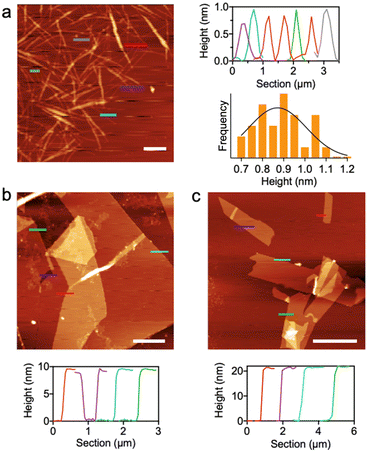
Atomic force microscopy (AFM) was employed to measure the heights of these nanosheets, revealing discrete heights of ca. 10 and 20 nm (Fig. 4). Individual nanotubes were obtained after sonication of 6CP nanosheets for 30 min, confirming our hierarchical growth model based on nanotubes, which showed an average height of 0.90 ± 0.12 nm. This size is consistent with the expected diameter of a nanotube assembled from a cyclic hexapeptide.45,60 Importantly, nanotubes isolated by sonication can be converted into nanosheets by thermal annealing, a strategy that allows interconversion between supramolecular stages of this class of cyclic peptides.46–48 By comparing the heights of individual nanotubes (1 nm) and nanosheets (10 nm) as determined by AFM, propagation along the Z axis was confirmed, hence validating the design of 6CP for hierarchical self-assembly from 1D to 2D and 3D (Fig. 1). The observed nanosheet heights (10 and 20 nm) were consistent with a step-growth elongation along the Z axis, most likely through the association of thinner pre-formed nanosheets that join their multivalent 2D surfaces in order to grow in 3D thickness. Overall, the regular aspect ratio and height profile of 6CP nanosheets suggest homogeneous growth kinetics in all three spatial dimensions, which may arise from the cooperative self-assembly and packing order imposed by the geometry of this particular monomer.
Complementary scattering techniques were then used to elucidate the internal structure of 6CP nanosheets. In solution, small-angle X-ray scattering (SAXS) confirmed the presence of two coexisting nanostructures: elongated objects at high q values (>0.3 nm−1) with a characteristic −1 Porod slope, and 2D films at low q values (<0.3 nm−1) with a slope of −2.2 (Fig. 5a). Shape-independent analysis confirms the presence of these two structures, where the 1D assemblies are present at a very low volume fraction (Fig. S6).61 These results confirm the hierarchical assembly of the system in solution, where the 2D structures are formed by nanotubes. Applying a model-based analysis (Fig. 5a), average diameters of 1.2 ± 0.5 nm and thicknesses of 19.7 ± 0.5 nm were extracted for the nanotubes and nanosheets, respectively. These dimensions are in good agreement with those found by AFM (ca. 1 and 20 nm, Fig. 4). However, it should be noted that the exponential scaling of SAXS signal with size may mask the presence of thinner nanosheets, as those observed by AFM (10 nm in height, Fig. 4b), which are required for the proposed hierarchical self-assembly mechanism. Importantly, SAXS results confirmed that the hierarchical 3D self-assembly of 6CP occurs in solution and it is not dependent on surface effects. Additionally, wide-angle X-ray scattering (WAXS) was performed to analyse the internal network of the supramolecular 3D architecture. WAXS analysis revealed two predominant spacings in 6CP packing (Fig. 5b): 9.7 Å, corresponding to the spacing between nanotube interfaces and matching the 1 nm thickness of nanotubes determined by AFM and SAXS, and 4.3 Å, which matches the distance between stacked cyclic peptides in β-sheets, as previously reported in the literature.44,46,62 Combined, these results confirmed the hierarchical assembly and proposed packing model for 6CP nanosheets, made up of nanotube layers (Fig. 1c). The internal order of the nanosheets also explains their uniformity in thickness and aspect ratio, as they may impose geometrical constraints on this cooperative self-assembly, which could control elongation rates in all dimensions of space.63 Nevertheless, while local packing defects may be present, these imperfections are likely to undergo spontaneous reorganisation into the favoured 3D nanosheet architecture by the dynamic reordering of non-covalent interactions.
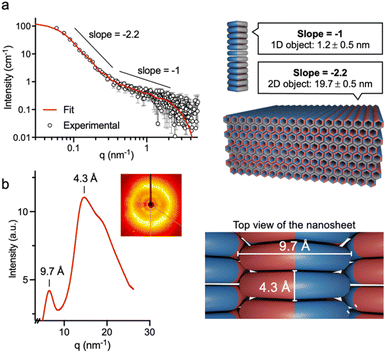 | ||
| Fig. 5 X-ray scattering analyses of 6CP samples. (a) SAXS profile fitted to a combination of a cylindrical and a lamellar model,64 consistent with coexisting 1D nanotubes and 2D nanosheets. (b) WAXS profile, showing spacings at 9.7 and 4.3 Å, in agreement with the expected packing of cyclic peptide monomers as laterally-interfacing nanotubes. All samples were prepared at pH 6.5. | ||
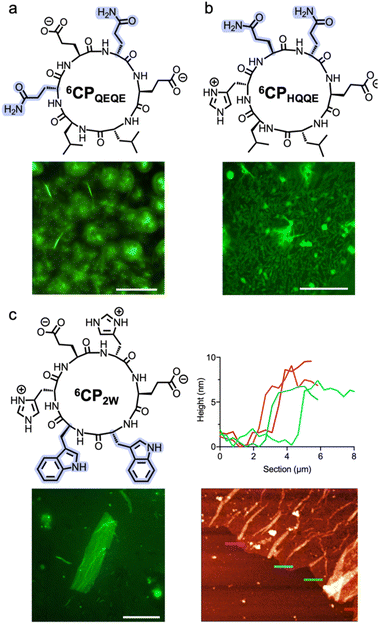
Sequence modifications of 6CP were studied to further validate the structural design requirements for 3D self-assembly. Control peptides 6CPQEQE [cyclo-(D-Leu-L-Leu-D-Gln-L-Glu-D-Gln-L-Glu-)] and 6CPHQQE [cyclo-(D-Leu-L-Leu-D-His-L-Gln-D-Gln-L-Glu-)] were unable to assemble any sort of nanosheets due to their truncated electrostatic domains, now blocked by neutral glutamines (Fig. 6a and b). These control peptides confirmed that matching ion pairing is critical to establishing the designed packing order and complementarity in 3D assembly. In contrast, replacement of hydrophobic contacts, from leucines to larger tryptophan residues in peptide 6CP2W [cyclo-(D-Trp-L-Trp-D-His-L-Glu-D-His-L-Glu-)], did produce 3D multilayered nanosheets of 6–8 nm in thickness (Fig. 6c). Nevertheless, assemblies of 6CP2W showed more irregular surfaces and rugged edges than those made from 6CP, suggesting that leucine is better suited to the hydrophobic contacts required for this supramolecular lattice than the bulkier tryptophan. Overall, these control peptides demonstrate the tolerance of this monomer design to accommodate other non-polar groups, and the unavoidable requirement of complementary electrostatic contacts to achieve sequential 1D-to-3D self-assembly.
Conclusions
We describe here the first cyclic peptide, 6CP, able to undergo hierarchical self-assembly into ordered, three-dimensional (3D) multilayered nanosheets. For this, 6CP was designed to form supramolecular contacts at 60° around its circumference, creating, for the first time, perfect complementarity between the hydrophobic and hydrophilic domains around the nanotubes formed by the parallel stacking of CP monomers. Consequently, the nanotubes that comprise these 3D multilayers are packed in a hexagonal lattice due to the complementarity and orthogonality of their hydrophobic and electrostatic interfaces. Remarkably, 6CP nanosheets exhibited uniform 2D aspect ratios and thicknesses, likely due to the high internal order and cooperativity observed in the 3D packing of peptide monomers. This supramolecular design constitutes a key milestone in the development of self-assembled nanomaterials with precise dimensional control without the use of external templates. Here, we demonstrate how the same fundamental monomer – a cyclic peptide of alternating D/L chirality – can be modified in terms of backbone length and amphiphilic topology to precisely engineer self-assembly in 1D, 2D, and 3D (Fig. 1c). The supramolecular fate of these monomers, from nanotubes to multilayered nanosheet structures, can be rationally controlled by adjusting the non-covalent interactions and angles between the side chains on their surface. The design parameters presented here offer a new supramolecular strategy for the simple preparation of organic self-assembling systems with defined cavities, having potential applications in nanotechnology and a potential impact in biomolecular chemistry and soft materials engineering.Author contributions
S. D. synthesised and characterised all peptides. A. S. F. acquired and analysed all SAXS data. S. D. performed all other experiments. All authors contributed to the design of the experiments and to the writing of this manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: materials, methods and chemical characterisation. See DOI: https://doi.org/10.1039/d5sc09489e.Acknowledgements
This research was partially funded by Agencia Estatal de Investigación (PDC2021-121192-I00, PID2023-152181OB-I0, RYC2021-031367-I, PID2022-142440NB-I00, and PID2023-147885OA-I00), Xunta de Galicia (ED431G 2023/03, ED431C 2024/03, ED431C 2025/15 IN855A 2021/06, ED431F 2023/24, ED431C 2024/09, and 2025-CP047), Fundación la Caixa (TROPIC and HR23-00221), and the European Research Council (StG-DYNAP-677786 and StG-REPLICATE-101163359). We acknowledge the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities under proposal number MX-2662, and we would like to thank Dr Petra Pernot for assistance and support in using beamline BM29.Notes and references
- H. K. Lin, T.-H. Yan, S. Bashir and J. L. Liu, in Advanced Nanomaterials and Their Applications in Renewable Energy, 2022, pp. 61–110 Search PubMed.
- M. Coste and S. Ulrich, Chem. Sci., 2025, 16, 22438–22446 RSC.
- H.-R. Jia, Y.-X. Zhu, X. Liu, G.-Y. Pan, G. Gao, W. Sun, X. Zhang, Y.-W. Jiang and F.-G. Wu, ACS Nano, 2019, 13, 11781–11792 CrossRef CAS PubMed.
- J. M. Fletcher, R. L. Harniman, F. R. H. Barnes, A. L. Boyle, A. Collins, J. Mantell, T. H. Sharp, M. Antognozzi, P. J. Booth, N. Linden, M. J. Miles, R. B. Sessions, P. Verkade and D. N. Woolfson, Science, 2013, 340, 595–599 CrossRef CAS PubMed.
- N. C. Burgess, T. H. Sharp, F. Thomas, C. W. Wood, A. R. Thomson, N. R. Zaccai, R. L. Brady, L. C. Serpell and D. N. Woolfson, J. Am. Chem. Soc., 2015, 137, 10554–10562 CrossRef CAS PubMed.
- A. Mendez-Ardoy, A. Bayón-Fernández, Z. Yu, C. Abell, J. R. Granja and J. Montenegro, Angew. Chem., Int. Ed., 2020, 59, 6902–6908 CrossRef CAS PubMed.
- J. W. Fredy, A. Méndez-Ardoy, S. Kwangmettatam, D. Bochicchio, B. Matt, M. C. A. Stuart, J. Huskens, N. Katsonis, G. M. Pavan and T. Kudernac, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11850–11855 CrossRef CAS PubMed.
- Z. Chen, Y. Suzuki, A. Imayoshi, X. Ji, K. V. Rao, Y. Omata, D. Miyajima, E. Sato, A. Nihonyanagi and T. Aida, Nat. Mater., 2022, 21, 253–261 CrossRef CAS PubMed.
- A. Singh, J. P. Joseph, D. Gupta, I. Sarkar and A. Pal, Chem. Commun., 2018, 54, 10730–10733 RSC.
- M. P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50, 2440–2448 CrossRef CAS PubMed.
- P. Besenius, G. Portale, P. H. H. Bomans, H. M. Janssen, A. R. A. Palmans and E. W. Meijer, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17888–17893 CrossRef CAS PubMed.
- M. Yolamanova, C. Meier, A. K. Shaytan, V. Vas, C. W. Bertoncini, F. Arnold, O. Zirafi, S. M. Usmani, J. A. Müller, D. Sauter, C. Goffinet, D. Palesch, P. Walther, N. R. Roan, H. Geiger, O. Lunov, T. Simmet, J. Bohne, H. Schrezenmeier, K. Schwarz, L. Ständker, W.-G. Forssmann, X. Salvatella, P. G. Khalatur, A. R. Khokhlov, T. P. J. Knowles, T. Weil, F. Kirchhoff and J. Münch, Nat. Nanotechnol., 2013, 8, 130–136 Search PubMed.
- Z. Tong, Y. Xie, M. C. Arno, Y. Zhang, I. Manners, R. K. O'Reilly and A. P. Dove, Nat. Chem., 2023, 15, 824–831 Search PubMed.
- L. Gallego, J. F. Woods and M. Rickhaus, Org. Mater., 2022, 4, 137–145 CrossRef CAS.
- T. Kim, J. Y. Park, J. Hwang, G. Seo and Y. Kim, Adv. Mater., 2020, 32, e2002405 CrossRef PubMed.
- B. Shen, Y. Kim and M. Lee, Adv. Mater., 2020, 32, 1905669 Search PubMed.
- J. del Barrio, J. Liu, R. A. Brady, C. S. Y. Tan, S. Chiodini, M. Ricci, R. Fernández-Leiro, C.-J. Tsai, P. Vasileiadi, L. D. Michele, D. Lairez, C. Toprakcioglu and O. A. Scherman, J. Am. Chem. Soc., 2019, 141, 14021–14025 Search PubMed.
- K. T. Nam, S. A. Shelby, P. H. Choi, A. B. Marciel, R. Chen, L. Tan, T. K. Chu, R. A. Mesch, B.-C. Lee, M. D. Connolly, C. Kisielowski and R. N. Zuckermann, Nat. Mater., 2010, 9, 454–460 Search PubMed.
- N. Sasaki, J. Kikkawa, Y. Ishii, T. Uchihashi, H. Imamura, M. Takeuchi and K. Sugiyasu, Nat. Chem., 2023, 15, 922–929 CrossRef CAS PubMed.
- C. Yuan, W. Ji, R. Xing, J. Li, E. Gazit and X. Yan, Nat. Rev. Chem., 2019, 3, 567–588 CrossRef CAS PubMed.
- I. Insua, J. Bergueiro, A. Méndez-Ardoy, I. Lostalé-Seijo and J. Montenegro, Chem. Sci., 2022, 13, 3057–3068 Search PubMed.
- N. J. Sinha, M. G. Langenstein, D. J. Pochan, C. J. Kloxin and J. G. Saven, Chem. Rev., 2021, 121, 13915–13935 Search PubMed.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Nat. Rev. Chem., 2020, 4, 615–634 CrossRef CAS PubMed.
- E. Krieg, M. M. C. Bastings, P. Besenius and B. Rybtchinski, Chem. Rev., 2016, 116, 2414–2477 Search PubMed.
- S. Lou, X. Wang, Z. Yu and L. Shi, Adv. Sci., 2019, 6, 1802043 Search PubMed.
- Y. Wang, S. Rencus-Lazar, H. Zhou, Y. Yin, X. Jiang, K. Cai, E. Gazit and W. Ji, ACS Nano, 2024, 18, 1257–1288 CrossRef CAS PubMed.
- S. Cavalli, F. Albericio and A. Kros, Chem. Soc. Rev., 2009, 39, 241–263 Search PubMed.
- F. Sheehan, D. Sementa, A. Jain, M. Kumar, M. Tayarani-Najjaran, D. Kroiss and R. V. Ulijn, Chem. Rev., 2021, 121, 13869–13914 CrossRef CAS PubMed.
- X. Chen, Y. He, Y. Kim and M. Lee, J. Am. Chem. Soc., 2016, 138, 5773–5776 Search PubMed.
- H. Gradišar, S. Božič, T. Doles, D. Vengust, I. Hafner-Bratkovič, A. Mertelj, B. Webb, A. Šali, S. Klavžar and R. Jerala, Nat. Chem. Biol., 2013, 9, 362–366 Search PubMed.
- E. F. Banwell, E. S. Abelardo, D. J. Adams, M. A. Birchall, A. Corrigan, A. M. Donald, M. Kirkland, L. C. Serpell, M. F. Butler and D. N. Woolfson, Nat. Mater., 2009, 8, 596–600 Search PubMed.
- A. D. Merg, G. Touponse, E. van Genderen, X. Zuo, A. Bazrafshan, T. Blum, S. Hughes, K. Salaita, J. P. Abrahams and V. P. Conticello, Angew. Chem., Int. Ed., 2019, 58, 13507–13512 Search PubMed.
- C. Wang, D. Lu, J. Li, L. Chen, X. Zuo, C. Wu, X.-K. Wei, B. Wang, Y.-B. Jiang and T. Jiang, J. Am. Chem. Soc., 2025, 147, 24510–24518 CrossRef CAS PubMed.
- A. Chatterjee, C. Mahato and D. Das, Angew. Chem., Int. Ed., 2021, 60, 202–207 Search PubMed.
- S. Bal, C. Ghosh, T. Ghosh, R. K. Vijayaraghavan and D. Das, Angew. Chem., Int. Ed., 2020, 59, 13506–13510 Search PubMed.
- N. Singh, K. Zhang, C. A. Angulo-Pachón, E. Mendes, J. H. van Esch and B. Escuder, Chem. Sci., 2016, 7, 5568–5572 RSC.
- Y. Lin, M. R. Thomas, A. Gelmi, V. Leonardo, E. T. Pashuck, S. A. Maynard, Y. Wang and M. M. Stevens, J. Am. Chem. Soc., 2017, 139, 13592–13595 CrossRef CAS PubMed.
- B. Dai, D. Li, W. Xi, F. Luo, X. Zhang, M. Zou, M. Cao, J. Hu, W. Wang, G. Wei, Y. Zhang and C. Liu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2996–3001 Search PubMed.
- A. Solomonov, A. Kozell and U. Shimanovich, Angew. Chem., Int. Ed., 2024, 63, e202318365 Search PubMed.
- M. Wenisch, Y. Li, M. G. Braun, L. Eylert, F. Späth, S. M. Poprawa, B. Rieger, C. V. Synatschke, H. Niederholtmeyer and J. Boekhoven, Chem, 2025, 11, 102578 CAS.
- N. Singh, A. Lopez-Acosta, G. J. M. Formon and T. M. Hermans, J. Am. Chem. Soc., 2022, 144, 410–415 CrossRef CAS PubMed.
- S. H. Yoo and H.-S. Lee, Acc. Chem. Res., 2017, 50, 832–841 CrossRef CAS PubMed.
- S. Kralj, O. Bellotto, E. Parisi, A. M. Garcia, D. Iglesias, S. Semeraro, C. Deganutti, P. D'Andrea, A. V. Vargiu, S. Geremia, R. D. Zorzi and S. Marchesan, ACS Nano, 2020, 14, 16951–16961 Search PubMed.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 CrossRef CAS PubMed.
- Q. Song, Z. Cheng, M. Kariuki, S. C. L. Hall, S. K. Hill, J. Y. Rho and S. Perrier, Chem. Rev., 2021, 121, 13936–13995 CrossRef CAS PubMed.
- I. Insua and J. Montenegro, J. Am. Chem. Soc., 2020, 142, 300–307 Search PubMed.
- S. Díaz, I. Insua, G. Bhak and J. Montenegro, Chem.–Eur. J., 2020, 26, 14765–14770 CrossRef PubMed.
- I. Insua, A. Cardellini, S. Díaz, J. Bergueiro, R. Cappeli, G. M. Pavan and J. Monetenegro, Chem. Sci., 2023, 14, 14074–14081 Search PubMed.
- A. Bayón-Fernández, A. Torrón-Celada, A. Méndez-Ardoy, M. Coste, D. Delgado-Gestoso, S. Ulrich, J. Montenegro and J. R. Granja, Angew. Chem., Int. Ed., 2025, e14543 Search PubMed.
- T. Kurita and K. Numata, Phys. Chem. Chem. Phys., 2024, 26, 28776–28792 Search PubMed.
- J. D. Hartgerink, J. R. Granja, R. A. Milligan and M. R. Ghadiri, J. Am. Chem. Soc., 1996, 118, 43–50 Search PubMed.
- A. Méndez-Ardoy, I. Insua, J. R. Granja and J. Montenegro, Cyclization and Self-Assembly of Cyclic Peptides in Peptide Macrocycles, Methods Mol. Biol., 2022, 2371, 449–466 CrossRef PubMed.
- L. S. Wolfe, M. F. Calabrese, A. Nath, D. V. Blaho, A. D. Miranker and Y. Xiong, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16863–16868 CrossRef CAS PubMed.
- D. L. Nelson, M. M. Cox and M. M. Cox, Lehninger principles of biochemistry, W H Freeman & Company, 5th edn, 2008 Search PubMed.
- E. R. Cross and D. J. Adams, Soft Matter, 2019, 15, 1522–1528 RSC.
- N. B. Hamilton, S. Arns, M. Shelley, I. Bechis and J. C. Shelley, Mol. Pharm., 2025, 22, 588–593 CrossRef CAS PubMed.
- D. G. Isom, C. A. Castañeda, B. R. Cannon, P. D. Velu and B. E. Garcia-Moreno, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16096–16100 Search PubMed.
- D. G. Isom, C. A. Castañeda, B. R. Cannon and B. E. Garcia-Moreno, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5260–5265 Search PubMed.
- Polydispersity indices were calculated as the square of the ratio of the standard deviation to the mean value of each nanosheet dimension measured (i.e. length and width, n = 30).
- M. Saviano, L. Zaccaro, A. Lombardi, C. Pedone, B. D. Blasio, X. Sun and G. P. Lorenzi, J. Incl. Phenom. Mol. Recognit. Chem., 1994, 18, 27–36 Search PubMed.
- J. B. Hopkins, J. Appl. Crystallogr., 2024, 57, 194–208 Search PubMed.
- M. R. Silk, J. Newman, J. C. Ratcliffe, J. F. White, T. Caradoc-Davies, J. R. Price, S. Perrier, P. E. Thompson and D. K. Chalmers, Chem. Commun., 2017, 53, 6613–6616 Search PubMed.
- T. Jiang, E. L. Magnotti and V. P. Conticello, Interface Focus, 2017, 7, 20160141 Search PubMed.
- J. S. Pedersen, Adv. Colloid Interface Sci., 1997, 70, 171–210 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |