Tunable, bacterio-instructive scaffolds made from functional graphenic materials†
Karoline E.
Eckhart
 a,
Anne M.
Arnold
a,
Anne M.
Arnold
 b,
Francesca A.
Starvaggi
b,
Francesca A.
Starvaggi
 c and
Stefanie A.
Sydlik
c and
Stefanie A.
Sydlik
 *ad
*ad
aDepartment of Chemistry, Carnegie Mellon University, Pittsburgh, PA 15213, USA. E-mail: ssydlik@andrew.cmu.edu
bChemical and Biological Signature Sciences Group, National Security Directorate, Pacific Northwest National Laboratory, Richland, WA 99354, USA
cDepartment of Chemistry, Stanford University, Stanford, CA 94305, USA
dDepartment of Biomedical Engineering, Carnegie Mellon University, Pittsburgh, PA 15213, USA
First published on 11th December 2020
Abstract
The balance of bacterial populations in the human body is critical for human health. Researchers have aimed to control bacterial populations using antibiotic substrates. However, antibiotic materials that non-selectively kill bacteria can compromise health by eliminating beneficial bacteria, which leaves the body vulnerable to colonization by harmful pathogens. Due to their chemical tunablity and unique surface properties, graphene oxide (GO)-based materials – termed “functional graphenic materials” (FGMs) – have been previously designed to be antibacterial but have the capacity to actively adhere and instruct probiotics to maintain human health. Numerous studies have demonstrated that negatively and positively charged surfaces influence bacterial adhesion through electrostatic interactions with the negatively charged bacterial surface. We found that tuning the surface charge of FGMs provides an avenue to control bacterial attachment without compromising vitality. Using E. coli as a model organism for Gram-negative bacteria, we demonstrate that negatively charged Claisen graphene (CG), a reduced and carboxylated FGM, is bacterio-repellent through electrostatic repulsion with the bacterial surface. Though positively charged poly-L-lysine (PLL) is antibacterial when free in solution by inserting into the bacterial cell wall, here, we found that covalent conjugation of PLL to CG (giving PLLn-G) masks the antimicrobial activity of PLL by restricting polypeptide mobility. This allows the immobilized positive charge of the PLLn-Gs to be leveraged for E. coli adhesion through electrostatic attraction. We identified the magnitude of positive charge of the PLLn-G conjugates, which is modulated by the length of the PLL peptide, as an important parameter to tune the balance between the opposing forces of bacterial adhesion and proliferation. We also tested adhesion of Gram-positive B. subtilis to these FGMs and found that the effect of FGM charge is less pronounced. B. subtilis adheres nondiscriminatory to all FGMs, regardless of charge, but adhesion is scarce and localized. Overall, this work demonstrates that FGMs can be tuned to selectively control bacterial response, paving the way for future development of FGM-based biomaterials as bacterio-instructive scaffolds through careful design of FGM surface chemistry.
Introduction
Over the course of human history, we have co-evolved with the bacteria that inhabit our bodies.1 The collection of “commensal” bacterial species that inhabit our bodies is termed our microbiome. These commensals are involved in a highly mutualistic relationship with humans:2 we (the host) rely on the colonization of commensals for nutrient uptake and storage3 as well as immune development and function.1,4–6 Consequently, the commensals that make up the microbiome play a crucial role in our health and survival, highlighting a need to protect, support, and control these bacteria.Researchers have begun to consider the roles of commensal bacteria and their promotion through probiotic therapies. Numerous reports correlate microbiome composition with various diseases such as cancer, rheumatoid arthritis, obesity, Parkinson's disease, and more.7 The ability to restore a healthy microbiome composition by supplementing commensal strains could be a revolutionary treatment for these ailments.8 Delivery and retention of commensal bacteria could be facilitated by bacterio-adhesive surfaces. However, examples of nontoxic, bacterio-adhesive biomaterials remain limited despite the increasing demand to support and capitalize on the mutualistic relationship between humans and bacteria to promote better health.
To address this, we aimed to design a class of materials that instruct bacterial adhesion while maintaining the vitality of both adherent and nonadherent bacteria. Prior research has resulted in the development of nontoxic, bacterio-repulsive biomaterials9 for use as biomedical implants that accelerate wound healing and lessen the risk of implant failure10,11 by preventing the formation of bacterial biofilms.12 These technologies are meant to provide an alternative to systemic antibiotics,13 which non-selectively destroy harmful pathogens along with our commensal “good” bacteria, leaving our intestinal microbiome vulnerable to colonization by resistant pathogens14 long after the antibiotic loses efficacy.15 Bacterio-repellent surfaces are designed to chemically or physically block bacterial adhesion, for example, by possessing a low surface energy, hydrophilic moieties, or negatively charged groups.9,16,17 These biomaterials leverage the innate properties of the bacterial cell wall to prevent bacterial adhesion. Inspired by this concept, we hypothesized that tuning the surface charge of an inherently cytocompatible substrate would provide an avenue to control bacterial cell attachment without compromising bacterial vitality.
Bacterial cell walls are decorated with anionic polymers that create a net negative surface charge. Specifically, the outermost layer of a Gram-positive bacterial cell wall is densely functionalized with anionic lipoteichoic acid polymers.18 Likewise, Gram-negative bacterial cell walls contain negatively charged lipopolysaccharides embedded in the outer membrane.19 Numerous studies have demonstrated that negatively and positively charged surfaces influence bacterial repulsion and adhesion, respectively, through electrostatic interactions with the bacterial cell wall.20–23 While anionic, bacterio-repellent materials that are nontoxic to bacteria have been developed, cationic, bacterio-adhesive materials often kill bacteria on contact.24,25 This property is beneficial in the context of surgical implant applications, where antimicrobial or bacteriostatic activity is desirable to prevent biofilm formation. However, the bacterio-toxicity of cationic surfaces hinders their use in commensal delivery systems where supporting bacterial vitality is crucial.
A nontoxic bacterio-instructive substrate could be designed using graphene oxide (GO) as a platform, due to the high surface area and remarkable adsorption capacity of this nanocarbon material. These properties have made GO capable of unique cellular interactions that have prompted its use in a range of cell scaffold applications.26,27 The high surface area of GO provides space for bacterial attachment as well as proliferation. Meanwhile, the high adsorption capacity of the GO backbone, which stems from its aromatic nature, facilitates cell adhesiveness.27 Recent reports have studied the factors that influence bacterial adherence to GO surfaces, including the work function, hydrophobicity, noncovalent surface functionalization, spatial orientation of the graphenic sheets (sheets parallel versus perpendicular to the surface), electrostatic charge, and electron-accepting capability of the GO surface.28–34 We aimed to build on this burgeoning area of research by investigating how covalent installation of an electrostatic charge of the GO surface can instruct bacterial adhesion while maintaining bacterial vitality.
The facile chemical tunability of GO makes it an appropriate platform for the design of a bacterio-instructive substrate. Structurally, GO is a two-dimensional material comprised of a conjugated carbon sheet that is decorated with oxygen-containing functional groups, where the quantity of functional groups (also known as the “degree of oxidation” of the GO) can be easily tuned in the synthesis of the material.35 These functional groups can serve as chemical handles to covalently modify GO, creating functional graphenic materials (FGMs) with specialized and sustained surface properties.
Here, we studied a series of FGMs with covalently installed negative and positive surface charges to evaluate their ability to selectively repel or adhere bacteria by interacting with the inherent negative charge of the bacterial cell wall. Negatively charged FGMs included GO and Claisen graphene (CG), which is a reduced and carboxylated GO. Positively charged FGMs were fabricated by covalently conjugating poly-L-lysine (PLL) to CG to give PLLn-G with a PLL degree of polymerization (n) of either 6 or 50. We found that all of the tested graphenic materials (GO, CG, PLL6-G, and PLL50-G) were nontoxic to Gram-negative Escherichia coli and Gram-positive Bacillus subtilis. While B. subtilis was more sensitive to the FGMs, compared to E. coli, the FGMs are not considered antimicrobial due to the persistence of viable bacteria following FGM exposure. Positively charged polymers, such as PLL, are typically used to kill bacteria by creating pores in the outer membrane;36–40 but we found that the graphenic component of the PLLn-G conjugate masks the toxicity of the polycationic PLL by restricting polypeptide mobility.
Further, we demonstrate a strong correlation between FGM surface charge and bacterio-instruction of E. coli. CG, which possesses a more negative surface charge than GO (CG = −54.30 mV; GO = −44.73 mV zeta potential), exhibits bacterio-repellent behavior with E. coli. Meanwhile, both PLLn-G constructs, which possess increasing magnitudes of positive charge (PLL6-G = +3.11 mV; PLL50-G = +19.90 mV zeta potential), promote E. coli adhesion with higher proliferation on the PLL6-G scaffold, compared to the PLL50-G scaffold. B. subtilis adheres indiscriminately to all FGM scaffolds regardless of charge, but cell growth is limited and localized on these surfaces. This difference in Gram-negative and Gram-positive bacterial affinity to the charged FGMs is likely due to the difference in structural arrangement of negative charge in the cell walls of these bacteria. Notably, the vitality of both E. coli and B. subtilis is retained in the presence of all FGM scaffolds. The FGMs presented herein represent a promising class of materials that can influence bacterial cell fate by acting as an instructive surface that encourages a different response from Gram-negative and Gram-positive bacteria.
Design of FGM bacterio-instructive properties through surface functionalization
The net negative surface charge of bacterial cell walls19 creates an opportunity to leverage the charge of a substrate to promote bacterial adhesion or repulsion through electrostatic interactions with the bacterial cell wall.20–23 While both Gram-negative and Gram-positive bacteria possess a net negative surface charge, the cell walls of these bacteria possess distinguishing features that influence the way they interact with surfaces, antibiotics, and the environment (Fig. 1A).18 The affinity of bacterial cell wall components to a surface influences both the strength of bacterial adhesion (which also impacts proliferation) and the effectiveness of the bacterial cell wall to protect the plasma membrane from penetration.18 In Gram-positive bacteria, the plasma membrane is surrounded by a thick peptidoglycan (PGN) layer, which contributes to cell wall rigidity and maintains the shape of the bacteria.41 Polyanionic lipoteichoic acid (LTA) polymers are anchored into the plasma membrane and thread through the PGN layer to extend beyond the cell wall, imparting a negative charge to the surface of Gram-positive bacteria.42 Compared to Gram-positive bacteria, Gram-negative bacteria possess a significantly thinner PGN layer surrounding the plasma membrane.41 This difference in PGN thickness allows Gram-positive and Gram-negative bacteria to be distinguished by gram staining.43 Another distinguishing cell wall feature, is that Gram-negative bacteria possess an outer membrane while Gram-positive bacteria do not. The outer membrane of a Gram-negative bacteria is a lipid bilayer with an inner leaflet comprised of phospholipids and an outer leaflet comprised of lipopolysaccharides (LPS).18 The phosphate constituents of LPS impart a negative charge to the surface of a Gram-negative bacteria.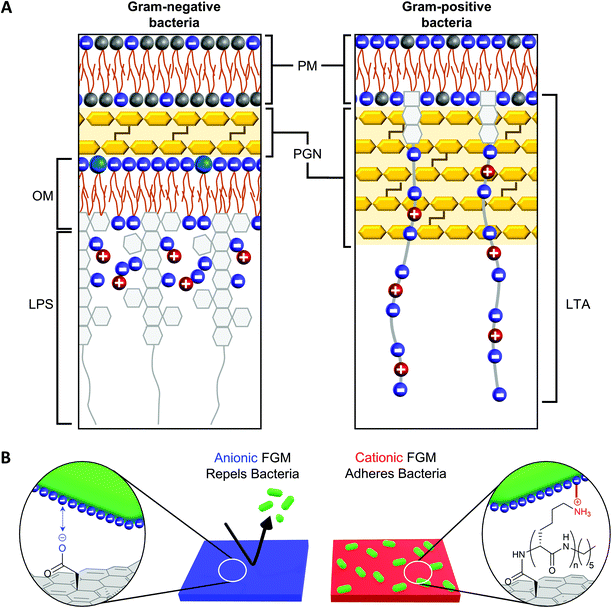 | ||
| Fig. 1 (A) Gram-negative and Gram-positive bacteria have different cell wall architectures (based on information from Malanovic and Lohner18). PM = plasma membrane; PGN = peptidoglycan; OM = outer membrane; LPS = lipopolysaccharide; LTA = lipoteichoic acid. (B) The charge of a functional graphenic material (FGM) surface, which can be tailored in the synthesis of the material, can influence the interaction of the FGM with the net negative charge of the bacterial cell wall. | ||
Considering this, we hypothesized GO would serve as a superior substrate to promote bacterial adhesion or repulsion given its bacterial compatibility,44–47 excellent adsorption capacity, and tunable surface chemistry that would allow us to create FGMs with a controlled surface charge.23,24 Here, covalent functionalization of FGMs is important so that the surface chemistry remains intact over timescales long enough to measure substrate-directed, irreversible bacterial adhesion rather than reversible attachment events. Specifically, we hypothesized that a negatively charged FGM would inhibit bacterial attachment through electrostatic repulsion, while a positively charged FGM would promote bacterial adhesion through electrostatic attraction (Fig. 1B).
Materials and methods
Materials and methods are explained in the ESI.†Characterization of FGM functionalization
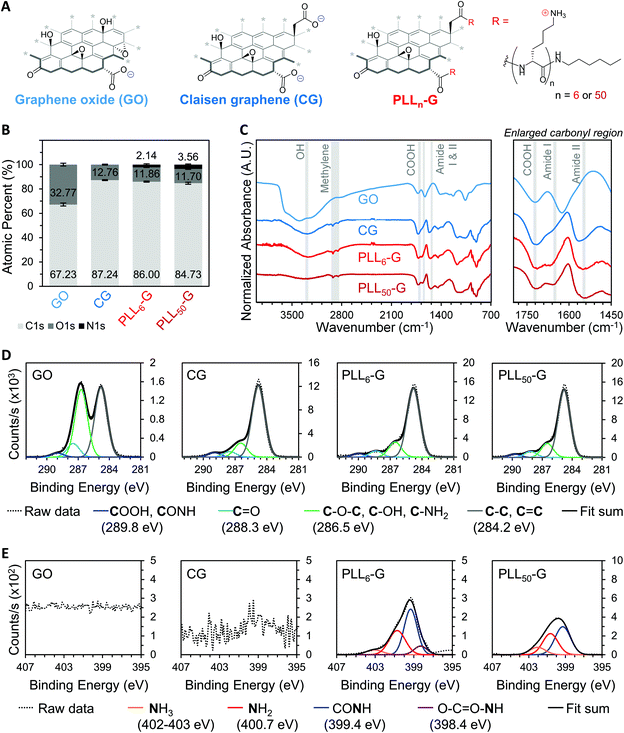
We synthesized two negatively charged FGMs and two positively charged FGMs (Fig. 2A). The negatively charged FGMs were GO and Claisen graphene (CG), which both possess anionic carboxylic acids. To produce CG, the tertiary alcohols on the basal plane of GO are converted to carboxylic acids through a [3, 3] sigmatropic rearrangement.48 This transformation makes CG more reduced and carboxylated, compared to the GO starting material. The positively charged FGMs were produced by covalently conjugating poly-L-lysine (PLL) to CG to give PLLn-G as previously described (Scheme S1†),49 where larger polypeptide length (n) results in a higher magnitude of positive charge due to the cationic primary amines on the lysine side chains. We synthesized PLLn-G conjugates with n = 6 and 50 to evaluate the effect of peptide length, and thus magnitude of positive charge of the FGM, on bacterial response. To confirm the difference in polypeptide length of the PLLn-G conjugates, PLL polypeptides were characterized by gel permeation chromatography (GPC) and proton nuclear magnetic resonance (1H-NMR) spectroscopy prior to grafting to CG (Fig. S2†). GPC and 1H-NMR spectroscopy end-group analysis confirmed that the PLL polypeptides had a degree of polymerization of 6 and 50, generating PLL6-G and PLL50-G conjugates, respectively.Fourier transform infrared (FTIR) spectroscopy and X-ray photoelectron spectroscopy (XPS) were used to verify the above chemical modifications to the FGMs. The carboxylation of GO to give CG is confirmed by FTIR, which shows a peak at 1720 cm−1 indicating the presence of anionic carboxylic acids in both materials (Fig. 2C). Deconvolution of high-resolution carbon (C1s) spectra obtained via XPS enables quantification of the carboxylic acid functionality. Quantification of the 289.8 eV C1s peak reveals that CG has more carboxylic acids (3.99% of all atoms are carboxylic acid carbons) than GO (1.17% of all atoms are carboxylic acid carbons) (Fig. 2D and S3†). XPS survey scans demonstrate that both PLLn-Gs contain nitrogen (N1s), which is a unique element to PLL polypeptides that is not present in the CG graphenic starting material (Fig. 2B and S4†). PLL50-G, which contains longer PLL peptides, has a higher nitrogen content than PLL6-G, suggesting that PLL50-G possesses more PLL functionality than PLL6-G. The FTIR spectra of PLLn-Gs contains unique amide stretches (Amide I at 1670 and Amide II at 1540 cm−1), indicating the presence of PLL polypeptides in the conjugate materials (Fig. 2C). Deconvolution of high-resolution nitrogen (N1s) XPS reveals, quantitatively, that PLL50-G has more cationic amine and ammonium functional groups (1.79% of all atoms are amine or ammonium nitrogen atoms) compared to PLL6-G (0.87% of all atoms are amine or ammonium nitrogen atoms) (Fig. 2E and S5†). Energy-dispersive X-ray spectroscopy (EDS) mapping of the FGMs demonstrates a uniform distribution of elements across the surface of the FGMs, indicating uniform chemical functionalization (Fig. S6†).
The identity and quantity of functional groups present on the FGMs dictates the charge of the materials. The chemical transformation of GO to produce CG results in additional negatively charged carboxylate groups on CG, as discussed above. This corresponds with a higher magnitude of negative charge in CG compared to GO, as evidenced by zeta potential measurements where CG has a −54.30 mV charge and GO has a −44.73 mV charge (Fig. 3A and S7†). Covalent PLLn-G conjugates possess positively charged primary amine groups from the PLL side chains. We predicted that a larger PLL length in the PLLn-G conjugate would result in a larger net positive charge due to the greater amount of amine and ammonium groups. This hypothesis was confirmed via zeta potential measurements (Fig. 3A and S7†), where PLL50-G has a +19.90 mV charge and PLL6-G has a +3.11 mV charge. With a zeta potential of +22.80 mV, unconjugated PLL50 serves as a control to demonstrate that the positive zeta potential of the PLLn-G conjugates stems from their PLL functionality (Fig. 3A). Graphenic materials are capable of adsorbing proteins and ions onto their surface through van der Waals, π–π, electrostatic, and hydrogen bonding interactions.50–52 Zeta potential of our FGMs in bacterial culture media (LB Miller and Trypticase Soy broth) demonstrates this phenomenon: the zeta potential of all FGMs becomes indiscriminately negative in media, which is likely due to adsorption of media components (charged ions and proteins) (Fig. S8†). The affinity of media components to the FGMs is expected to enhance the FGMs ability to serve as cell scaffolds by concentrating nutrients at the scaffold surface.53 Bacteria that bind to the FGM surface by displacing adsorbed media components will have an essentially limitless source of nutrients that are localized near the scaffold surface.
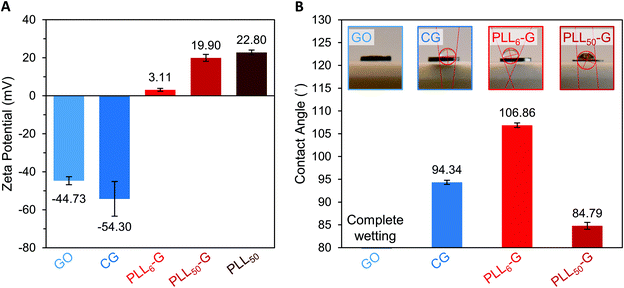 | ||
| Fig. 3 (A) Zeta potential of FGM dispersions in buffered water. (B) Water contact angle of FGM surfaces. | ||
Water contact angle measurements were performed to evaluate how FGM surface functionalization impacts the hydrophobicity of the materials (Fig. 3B). These measurements were performed on FGM surfaces made by pressing the FGM powders into cylindrical pellets (Fig. S1†). For CG and the PLLn-Gs, we found that the magnitude of zeta potential of the FGM, which results from the charged functional groups on the graphenic surface, impacts its hydrophobicity. Of all the FGMs reported herein, PLL6-G has the lowest magnitude of charge (+3.11 mV) and is the most hydrophobic with a water contact angle of 106.86°. CG and PLL50-G have larger magnitudes of charge (−54.30 and +19.90 mV, respectively), are less hydrophobic than PLL6-G with water contact angles of 94.34° and 84.79°, respectively. With numerous oxygen-containing functional groups, GO is the most hydrophilic FGM: the GO pellet fully absorbed the water droplet before a contact angle could be determined. Bacterial cells are known to adhere strongly to hydrophobic surfaces.31,33,54–56 This positions CG and the PLLn-Gs as promising bacterial scaffolds due to their relative hydrophobicity compared to GO. The hydrophobicity of CG and the PLLn-Gs, despite their charged surface functionalization, is because they are reduced FGMs.
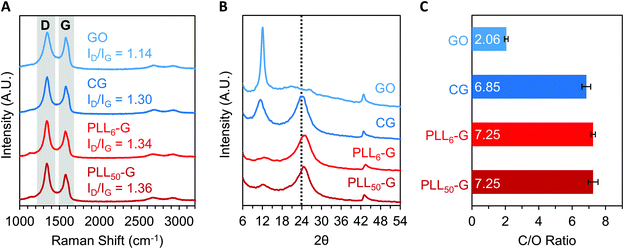
The level of reduction of the FGMs was evaluated using Raman spectroscopy, X-Ray diffraction (XRD), XPS, and FTIR spectroscopy. Raman spectroscopy is a widely used technique to characterize graphenic materials. Graphenic materials possess two major peaks in their Raman spectra: the G band at ∼1580 cm−1 is produced by scattering of E2g mode in sp2 carbon domains, while the D band at ∼1345 cm−1 represents sp3 hybridization resulting from functional groups or structural defects on the graphenic surface.57 When GO is reduced, new sp2 domains are created; these numerous yet small sp2 domains translate to large quantities of structural defects and a more intense D band compared to the original GO material.57 The ratio of D-to-G band intensities (ID/IG) is a measure of the level of reduction in graphenic materials: reduced FGMs have a higher ID/IG ratio than oxidized FGMs.58,59 By comparing the ID/IG ratios of the FGMs studied here, it can be determined that GO is the most oxidized (ID/IG = 1.14) while CG, PLL6-G, and PLL50-G are more reduced (ID/IG = 1.30, 1.34, and 1.36, respectively) (Fig. 4A). XRD spectra of the FGMs supports this conclusion: CG, PLL6-G, and PLL50-G possess a peak at 24 2θ, which is commonly observed in reduced FGMs (Fig. 4B).60,61 These results are further validated by comparing the carbon-to-oxygen (C/O) ratio of the FGMs, which was obtained from XPS survey scans: GO is the most oxidized FGM, as evidenced by its low C/O ratio, while CG, PLL6-G, and PLL50-G are significantly more reduced as indicated by their larger C/O ratio (Fig. 4C). FTIR spectroscopy agrees with this analysis of FGM reduction level. Specifically, GO has more intense hydroxyl (3290 cm−1) and epoxide (1000 cm−1) stretches and lacks methylene stretches (2920–2860 and 900 cm−1), indicating a higher level of oxidation. CG, PLL6-G, and PLL50-G have less intense hydroxyl (3290 cm−1) and epoxide (1000 cm−1) stretches and feature methylene peaks (2920–2860 and 900 cm−1) (Fig. 2C).
Reduced graphenic materials, or those with higher sp2 hybridized character, tend to be more conductive than oxidized graphenic materials.62 We have previously established the electrical conductivity of CG and the PLLn-Gs.49 The conductivity of these FGMs, which stems from the fact that they are reduced, has implications in terms of their ability to influence bacterial fate by conducting electrical current. Previous research has found that bacteria respond to electrical stimulation and use electrical pulses to sense their environment;63,64 bacterial and mammalian cell adhesion and proliferation can be mediated by electrical stimulation of a piezoelectric scaffold.49,65,66 The conductive nature of CG and the PLLn-Gs, positions them as promising bacterial scaffolds that conduct electrical signals to influence bacterial response. Future work in our lab seeks to investigate this possibility.
FGMs are nontoxic to bacterial and mammalian cells
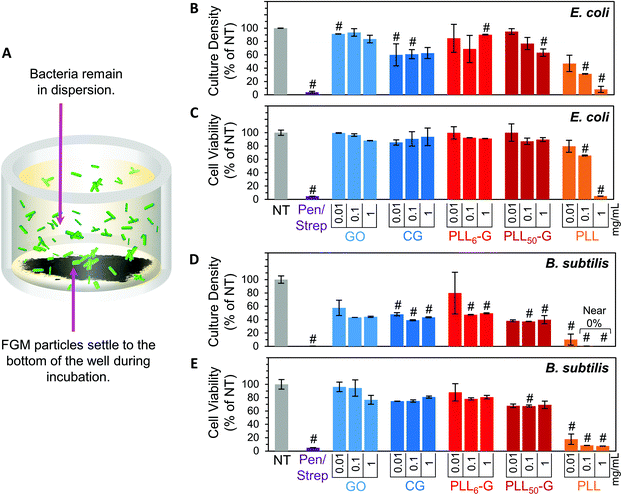
To evaluate the bacterial compatibility of the FGMs, we co-cultured E. coli K12 or B. subtilis Marburg strain bacteria with FGM dispersions (GO, CG, PLL6-G, and PLL50-G). We focused our studies on these bacterial strains because they are nonpathogenic, noninvasive, and widely accepted model organisms for Gram-negative or Gram-positive bacteria, respectively.67–70 As such, these bacteria serve as appropriate model organisms to begin investigating the interactions of our FGMs with commensal bacteria. Bacterial compatibility was evaluated using two metrics: proliferation and viability. Bacterial proliferation was assessed using turbidimetry, specifically by measuring the absorbance of bacterial solutions at 670 nm. Since absorbed light is proportional to the number of bacterial cells in a solution, absorbance can be used to easily and rapidly quantify the relative cell density of a culture.71 However, turbidimetric methods do not distinguish between viable and nonviable bacteria, which both cause turbidity in solution. To account for this discrepancy, bacterial viability was evaluated using a dual-dye kit,72 which distinguishes living and dying bacteria with fluorescent dyes and allows quantitative determination of bacterial cell viability using fluorescence. Both the absorbance (proliferation) and fluorescence (viability) data sets were normalized to the “no treatment” (NT) condition. A positive control was also evaluated using a mixture of 100 μg mL−1 streptomycin and 100 U mL−1 (∼60 μg mL−1) penicillin (Pen/Strep). The Pen/Strep mixture was selected as the positive control because this combination is widely used in cell culture to conclusively and reliably eliminate bacterial contamination.73,74 Further, a solution of free PLL polypeptide (PLL50) was analyzed to compare the cytocompatibility of an unconjugated polypeptide to the FGM-conjugated polypeptides (PLLn-G). PLL is an established antimicrobial polypeptide,75–77 so this material serves as an additional positive control.Our experimental setup was designed to allow the FGMs to settle to the bottom of the culture well over the course of the experiment (Fig. 5A), enabling facile isolation of the bacteria from the FGMs following incubation. Since nanocarbon materials are known to cause interference in many in vitro assays,78,79 this experimental design allows for bacterial sampling from the top of the cell culture well without FGM interference.
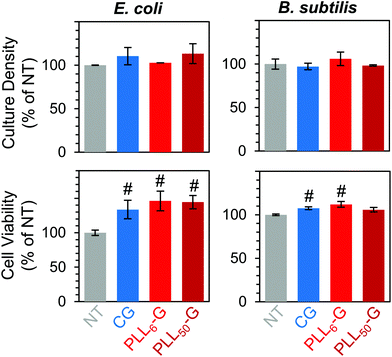
Overall, the FGMs evaluated here (GO, CG, PLL6-G, or PLL50-G) were found to be cytocompatible with both E. coli and B. subtilis model organisms. Interestingly, some reports assert that GO is antibacterial;80 but our results, like many others, indicate that GO is nontoxic to bacteria.44–46 Expectedly, bacterial toxicity was observed in the free (unconjugated) PLL. The antimicrobial effects of PLL are supported by a large body of literature.75–77 PLL ultimately causes bacterial cell death by infiltrating the cell wall, displacing divalent cations that stabilize the outer membrane, and inhibiting primary metabolic pathways. Unsurprisingly, our results corroborate the known dose-dependent toxicity of PLL towards both E. coli and B. subtilis: at 1 mg mL−1, culture density was reduced to 8.2% for E. coli and approaches 0% for B. subtilis, and cell viability fell to 5.1% for E. coli and 7.5% for B. subtilis (Fig. 5B–E).
Importantly, when PLL is covalently conjugated to a graphenic material, such as in PLL6-G and PLL50-G, the resulting FGM is not antimicrobial. This contrasting result can be explained by the orientation of PLL on the graphenic sheet in PLLn-G. Unlike in the free polymer, in the conjugate material, covalently bound PLL is intercalated between graphenic sheets as well as on the graphenic surface. PLL lays flat on the graphenic sheet through hydrogen bonding and electrostatic interactions between cationic amines on the PLL and residual anionic functional groups on the graphenic material. The reduced nature of the PLLn-Gs (Fig. 4) results in π–π stacking between graphenic sheets, which further locks the PLL chains in a flat orientation on the graphenic basal plane. This adhered orientation of the PLL chains on the PLLn-G conjugate restricts the mobility of PLL, preventing it from infiltrating the bacterial cell wall and that inciting toxic effects on the bacteria. Neither PLL6-G nor PLL50-G hindered the viability of E. coli. On the other hand, B. subtilis experienced statistically significant reductions in culture density at PLL6-G and PLL50-G concentrations of 0.1 and 1 mg mL−1 (p < 0.05), yet viability remained above 67.5% in these conditions. This result suggests that, compared to Gram-negative bacteria, Gram-positive bacteria may be more susceptible to the positive charge or slight PLL mobility contributed by the PLLn-G conjugates. Researchers have theorized that the lipoteichoic acid (LTA) polyanions that extend through Gram-positive cell wall may act as a ladder to usher antimicrobial polycations through the cell wall to the plasma membrane, allowing them to disrupt the plasma membrane.18 These LTA structures do not exist in Gram-negative bacterial cell walls, which could explain the greater compatibility of E. coli with the PLLn-G conjugates. To this end, the tunability of the PLL length on the PLLn-G conjugate could be a parameter to control microbial populations by selecting for specific bacterial strains that are tolerant to the FGM.
In sum, our data suggests that covalent conjugation of the PLL to a graphenic backbone masks its toxicity by preventing cell wall infiltration. The relatively low magnitude of positive charge may also contribute to the reduced toxicity of the covalent PLLn-G conjugates in comparison with free PLL polypeptide. This phenomenon has been previously observed, where noncovalent PLL-graphenic composites (with higher positive charge) exhibited high antimicrobial efficacy while a covalent PLL-conjugate (with a lower positive charge) was far less antimicrobial.45 Since the mechanism for PLL toxicity has been negated in these conjugates, the net positive charge of our PLLn-G conjugates can be leveraged for bacterial adhesion without sacrificing bacterial proliferation and vitality.
Mammalian biocompatibility of an FGM is critical for its implementation in a medical application. The FGMs presented herein (GO, CG, and both PLLn-Gs) have been previously evaluated for their biocompatibility with three different mammalian cell lines that play a role in immune function (murine RAW 264.7 macrophages), wound healing (murine NIH-3T3 fibroblasts), and tissue regeneration (hMSCs), respectively.49 We determined that, while cellular viability is dose-dependent, physiologically relevant concentrations of the FGMs do not negatively affect the viability of these mammalian cell types.49 These results support our assertion that the FGMs described here could be applied in biomedical applications without inciting deleterious effects on mammalian cells.
FGMs as bacterio-instructive scaffolds
We fabricated three-dimensional pellets of the FGMs to evaluate how bacteria interact with the FGM surfaces (Fig. S1†). We focused this experiment on pellets made from the more reduced FGMs (CG, PLL6-G, and PLL50-G) because GO, which is more oxidized, was too dispersible in water to form a stable pellet. After co-culturing the FGM pellets with either E. coli or B. subtilis, the bacteria that were adhered to the FGM pellet were analyzed separately from the non-adherent bacteria that remained in suspension (Fig. 6).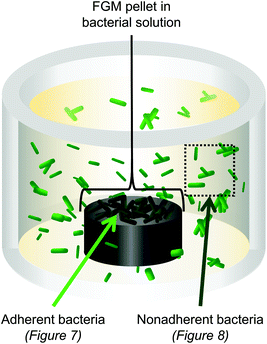 | ||
| Fig. 6 3D pellets of the FGMs (CG, PLL6-G, and PLL50-G) were co-cultured with E. coli or B. subtilis. Bacteria adhered to the FGM pellet were analyzed separately from nonadherent bacteria. | ||
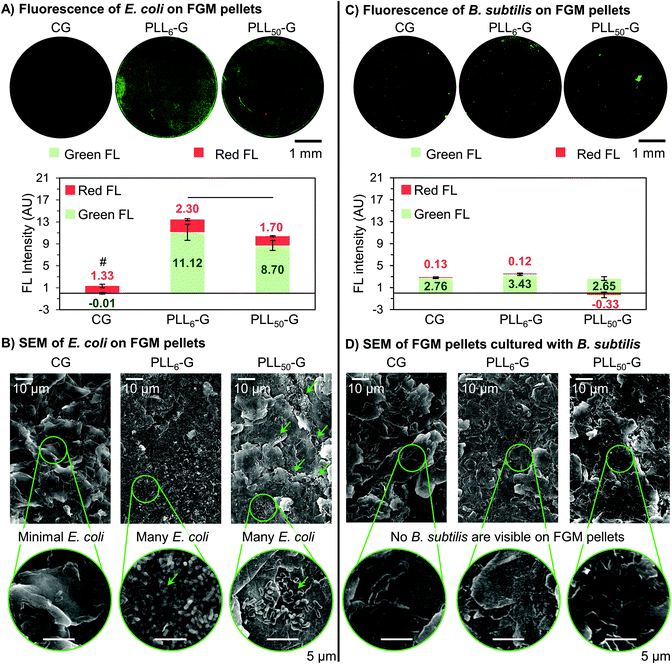
Following incubation of the FGM pellets with bacteria, the pellets were stained using a dual-dye kit to distinguish live bacteria from membrane compromised bacteria, and each pellet was analyzed via fluorescence microscopy, which could be evaluated qualitatively and quantitatively. Quantitative analysis of the fluorescence images allowed for the determination of the absolute fluorescence intensity of the red and green channels, which is proportional to the quantity of dying and living bacteria, respectively. E. coli and B. subtilis exhibited different behaviors with respect to adhesion to the FGM scaffolds, which may be explained by differences in the affinity of the FGMs to the different cell wall components (LPS in Gram-negative and LTA in Gram-positive).
Adherent E. coli cells were present on the surface of the PLLn-Gs but scarce on the surface of CG, demonstrating the ability of the FGM functionalization and charge to mediate bacterial adhesion. This is seen qualitatively in the fluorescence images, where both PLLn-G pellets show visible fluorescence, while very little fluorescence can be seen on CG (Fig. 7A). Quantitatively, the total fluorescence intensity (Green FL + Red FL) of the FGM pellets supports the conclusion that CG is bacterio-repellant and the PLLn-Gs are bacterio-adhesive: there are significantly fewer cells adhered to the CG pellet when compared to the PLLn-G pellets (p < 0.05) (Fig. 7A). These results are expected, due to electrostatic interactions between negatively charged bacterial cell walls and the FGM surfaces: negatively charged CG repels bacteria, while positively charged PLLn-Gs adhere bacteria (Fig. 1).
Importantly, of the bacteria adhered to either PLLn-G pellet, there are significantly more live bacteria than dying bacteria (p < 0.05). This supports our finding that the antibiotic nature of PLL polypeptides is masked when covalently conjugated to a graphenic material. Interestingly, more cells were observed on the PLL6-G substrate compared to the PLL50-G substrate (p < 0.05) (Fig. 7A), which can be explained by the larger positive zeta potential of PLL50-G (+19.90 mV) compared to PLL6-G (+3.11 mV). This result is consistent with previous research, which has demonstrated that while positively charged surfaces promote initial bacterial adhesion,20 they can also limit bacterial proliferation and viability depending on the magnitude of charge.45,81 Strong bacterial adhesion can prevent elongation, which is an essential step in cell division.21,82 Further, greater mobility of the longer PLL chains in PLL50-G may deter proliferation. For Gram-negative bacteria, such as E. coli, PLLn-G provides an appropriate platform to support bacterial adhesion and proliferation, where the proliferation can be regulated by the magnitude of positive charge on the FGM surface.
Scanning electron microscopy (SEM) was used to evaluate the morphology of the bacterial cells on the FGM pellets. The rod-shaped E. coli on the surface of each FGM appear to have intact outer membranes, supporting our position that none of the FGMs evaluated here are antibacterial (Fig. 7B and S9†). SEM also supports the bacterial localization trends seen in the fluorescence microscopy. Very few E. coli are adhered to the CG pellet, supporting the bacterio-repulsive characteristic of this material. E. coli are adhered to both PLL50-G and PLL6-G, illustrating their bacterio-adhesiveness; however, the localization of E. coli on these scaffolds is different. Fewer E. coli can be seen on the PLL50-G pellet, compared to the PLL6-G pellet, and the bacteria are localized; meanwhile, copious amounts of bacteria are adhered to the entire surface of the PLL6-G pellet (Fig. 7B).
The FGM scaffolds possess a facet of bacterial selectivity due to the differences in surface properties of Gram-positive B. subtilis and Gram-negative E. coli. Because Gram-positive bacteria are also negatively charged,18 we expected to see the same bacterio-repulsive and bacterio-adhesive behavior that we observed when E. coli was cultured with the FGM scaffolds. Instead, we found no significant difference in the amount of B. subtilis cells adhered to the FGM pellets, regardless of FGM charge. Further, fluorescence on the B. subtilis pellet images was considerably less intense and more localized, compared to the E. coli pellet images (Fig. 7C). No bacterial cells could be found in the field of view of SEM images of the FGM pellets following overnight culture with B. subtilis (Fig. 7D). The scarcity of B. subtilis growth on the FGM substrates is consistent with the culture density results from the FGM dispersion experiment (Fig. 5D), where B. subtilis proliferation was limited in the presence of CG and the PLLn-G conjugates. Strong initial adhesion of B. subtilis to the PLLn-G conjugates, due to a larger net negative charge than E. coli,83 would prevent subsequent cell division and proliferation to a greater extent; this would result in fewer overall cells and localization of the cells, which is observed in the fluorescence microscopy images (Fig. 7C). Although, this theory does not explain the presence of negatively charged B. subtilis adhered to the negatively charged CG surface. This indicates that forces beyond electrostatics, such as van der Waals, work function, surface free energy, and hydrophobic forces,28,84 may be dominating the interaction of B. subtilis with the FGMs. A deeper investigation into the interactions of the FGMs with the LTA polymers decorating the Gram-positive B. subtilis cell wall could shed light on the mechanism behind this result.
Finally, we confirmed that non-adherent E. coli and B. subtilis retained their ability to survive and proliferate in the presence of the FGM pellets. When cultured with CG and PLLn-G pellets, these bacteria proliferated (between 102.8–113.3% for E. coli and between 97.2–106.0% for B. subtilis, relative to the no treatment condition) and maintained high viability (between 133.6–146.1% for E. coli and between 105.8–112.0% for B. subtilis, relative to the no treatment condition) (Fig. 8). These results support our goal of creating a nontoxic scaffold for bacteria.
Conclusions and perspective
In sum, we investigated a class of functional graphenic materials (FGMs) that can selectively instruct bacterial adhesion and proliferation through electrostatic interactions with the bacterial cell wall. Specifically, the FGMs studied include negatively charged graphene oxide (GO) and Claisen graphene (CG) as well as two positively charged poly-L-lysine-graphenic conjugates (PLLn-G), where the magnitude of the positive charge in the PLLn-G conjugates is tuned by the degree of polymerization (n) of the PLL polypeptide. Notably, the FGMs in dispersion were found to support both Gram-negative and Gram-positive vitality regardless of their charge.The CG, PLL6-G, and PLL50-G FGMs were fabricated into water-stable 3D constructs to evaluate their bacterio-instructive surface properties. Negatively charged CG is bacterio-repellant to Gram-negative E. coli. On the other hand, both PLLn-G conjugates with varying magnitudes of positive zeta potential promote adhesion and proliferation of E. coli. We found that the PLL6-G, with a shorter polypeptide and lower zeta potential of +3.11 mV, enables more E. coli proliferation on the surface of the construct than PLL50-G, which has a longer polypeptide and higher zeta potential of +19.90 mV. Meanwhile, Gram-positive B. subtilis adheres to both CG and the PLLn-Gs, yet proliferation is limited and localized. These results highlight differences in the mechanism of Gram-negative and Gram-positive adhesion to a surface, likely due to structural differences between the cell walls of these bacterial types. Given the inherently tunable surface chemistry of FGMs, these materials can be utilized to selectively adhere or repel Gram-negative or Gram-positive bacteria, while maintaining bacterial vitality.
It is known that the microbiome, and its composition of commensal bacteria, play a crucial role in human health by preventing pathogenic takeover and modulating the immune system. Further development of bacterio-instructive surfaces, specifically those that are intrinsically bacterio-selective, can lead to the establishment of vehicles that deliver specific commensal bacteria while supporting their growth and proliferation. This would allow us to begin to manipulate the composition of the microbiome and study how this composition influences human health.
The research presented herein establishes FGMs as promising candidates for generating compatible, bacterio-instructive materials that can be utilized as scaffolds and coatings. The tunable surface chemistry and innate cytocompatibility of FGMs creates an opportunity to expand upon this work to develop bacterio-instructive surfaces for specialized applications. For example, FGM platforms could be utilized to create tailored materials that control bacterial growth rates and target additional bacterial strains. Further research in bacterio-instructive FGMs could progress the field of delivery vehicles for commensal bacteria and revolutionize the way bacteria are leveraged in the context of human health.
Funding
This work was funded by start-up grants from Carnegie Mellon University and the NSF DMR (S. A. S.) award #1905665.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Joel Gillespie for providing training and use of the XPS in the Materials Characterization Laboratory at the University of Pittsburgh. We also thank Alexandra (Sasha) Kennedy and Anna Watson for assistance with materials characterization. The authors acknowledge use of the XRD and SEM in the Materials Characterization Facility at Carnegie Mellon University under grant # MCF-677785. Further, we thank Subha Das (microcentrifuge), Marcel Bruchez (microplate reader), Krzysztof Matyjaszewski (GPC, Zetasizer), and Roberto Gil (NMR, funded in part by NSF grant # CHE-9808188, CHE-1039870, and CHE1726525) for the use of their facilities. We thank Anthony Varni (Raman spectroscopy), Rongguan Yin (zeta potential), Mingkang Sun (GPC), Zhao Lu and Yue Zhai (SEM and EDS mapping) for acquisition of data.References
- J. Chow, S. M. Lee, Y. Shen, A. Khosravi and S. K. Mazmanian, Adv. Immunol., 2010, 107, 243–274 CrossRef CAS.
- S. Z. Haque and M. Haque, Clin. Exp. Gastroenterol., 2017, 10, 91–103 CrossRef CAS.
- F. Bäckhed, H. Ding, T. Wang, L. V. Hooper, G. Y. Koh, A. Nagy, C. F. Semenkovich and J. I. Gordon, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15718–15723 CrossRef.
- D. Kelly, S. Conway and R. Aminov, Trends Immunol., 2005, 26, 326–333 CrossRef CAS.
- R. Khan, F. C. Petersen and S. Shekhar, Front. Immunol., 2019, 10, 1203, DOI:10.3389/fimmu.2019.01203.
- L. V. Hooper, T. S. Stappenbeck, C. V. Hong and J. I. Gordon, Nat. Immunol., 2003, 4, 269–273 CrossRef CAS.
- J. W. Keith and E. G. Pamer, J. Exp. Med., 2019, 216, 10–19 CrossRef CAS.
- E. G. Pamer, Science, 2016, 352, 535–538 CrossRef CAS.
- I. Francolini, G. Donelli, F. Crisante, V. Taresco and A. Piozzi, in Biofilm-based Healthcare-associated Infections: Volume II, ed. G. Donelli, Springer International Publishing, Cham, 2015, pp. 93–117 Search PubMed.
- C. M. González-Henríquez, M. A. Sarabia-Vallejos and J. Rodriguez-Hernandez, Materials, 2017, 10, 232 CrossRef.
- A. S. Veiga and J. P. Schneider, Pept. Sci., 2013, 100, 637–644 CrossRef CAS.
- C. R. Arciola, D. Campoccia and L. Montanaro, Nat. Rev. Microbiol., 2018, 16, 397–409 CrossRef CAS.
- M. S. Butler and D. L. Paterson, J. Antibiot., 2020, 73, 329–364 CrossRef CAS.
- R. Lima, F. S. Del Fiol and V. M. Balcão, Front. Pharmacol., 2019, 10, 692, DOI:10.3389/fphar.2019.00692.
- M. Y. Yoon and S. S. Yoon, Yonsei Med. J., 2018, 59, 4–12 CrossRef CAS.
- I. Francolini, G. Donelli, C. Vuotto, F. A. Baroncini, P. Stoodley, V. Taresco, A. Martinelli, L. D'Ilario and A. Piozzi, Pathog. Dis., 2014, 70, 401–407 CrossRef CAS.
- G. Cheng, Z. Zhang, S. Chen, J. D. Bryers and S. Jiang, Biomaterials, 2007, 28, 4192–4199 CrossRef CAS.
- N. Malanovic and K. Lohner, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 936–946 CrossRef CAS.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2, a000414 Search PubMed.
- G. Harkes, J. Feijen and J. Dankert, Biomaterials, 1991, 12, 853–860 CrossRef CAS.
- B. Gottenbos, D. W. Grijpma, H. C. van der Mei, J. Feijen and H. J. Busscher, J. Antimicrob. Chemother., 2001, 48, 7–13 CrossRef CAS.
- G. Harkes, J. Dankert and J. Feijen, J. Biomater. Sci., Polym. Ed., 1992, 3, 403–418 CrossRef CAS.
- E. R. Kenawy, F. I. Abdel-Hay, A. E. R. R. El-Shanshoury and M. H. El-Newehy, J. Polym. Sci., Part A: Polym. Chem., 2002, 40, 2384–2393 CrossRef CAS.
- K. Colville, N. Tompkins, A. D. Rutenberg and M. H. Jericho, Langmuir, 2010, 26, 2639–2644 CrossRef CAS.
- A. E. Madkour, J. M. Dabkowski, K. Nüsslein and G. N. Tew, Langmuir, 2009, 25, 1060–1067 CrossRef CAS.
- Z. M. Wright, A. M. Arnold, B. D. Holt, K. E. Eckhart and S. A. Sydlik, Regener. Eng. Transl. Med., 2019, 5, 190–209 CrossRef CAS.
- Kenry, W. C. Lee, K. P. Loh and C. T. Lim, Biomaterials, 2018, 155, 236–250 CrossRef CAS.
- W. Pajerski, J. Duch, D. Ochonska, M. Golda-Cepa, M. Brzychczy-Wloch and A. Kotarba, Mater. Sci. Eng., C, 2020, 113, 110972 CrossRef CAS.
- J. Duch, M. Golda-Cepa and A. Kotarba, Appl. Surf. Sci., 2019, 463, 1134–1140 CrossRef CAS.
- F. Perreault, H. Jaramillo, M. Xie, M. Ude, L. D. Nghiem and M. Elimelech, Environ. Sci. Technol., 2016, 50, 5840–5848 CrossRef CAS.
- J. Xue, S. BinAhmed, Z. Wang, N. G. Karp, B. L. Stottrup and S. R. V. Castrillón, Environ. Sci. Technol. Lett., 2018, 5, 14–19 CrossRef CAS.
- J. Ming, D. Sun, J. Wei, X. Chen and N. Zheng, ACS Appl. Bio Mater., 2020, 3, 704–712 CrossRef CAS.
- E. Tegou, M. Magana, A. E. Katsogridaki, A. Ioannidis, V. Raptis, S. Jordan, S. Chatzipanagiotou, S. Chatzandroulis, C. Ornelas and G. P. Tegos, Biomaterials, 2016, 89, 38–55 CrossRef CAS.
- P. Subramanian, F. Barka-Bouaifel, J. Bouckaert, N. Yamakawa, R. Boukherroub and S. Szunerits, ACS Appl. Mater. Interfaces, 2014, 6, 5422–5431 CrossRef CAS.
- A. M. Arnold, K. R. Crytzer, B. D. Holt and S. A. Sydlik, ACS Appl. Mater. Interfaces, 2019, 11, 20881–20887 CrossRef CAS.
- K. S. Huang, C. H. Yang, S. L. Huang, C. Y. Chen, Y.-Y. Lu and Y.-S. Lin, Int. J. Mol. Sci., 2016, 17, 1578 CrossRef.
- N. F. Kamaruzzaman, L. P. Tan, R. H. Hamdan, S. S. Choong, W. K. Wong, A. J. Gibson, A. Chivu and M. de F. Pina, Int. J. Mol. Sci., 2019, 20, 2747 CrossRef CAS.
- F. A. G. da Silva Jr., J. C. Queiroz, E. R. Macedo, A. W. C. Fernandes, N. B. Freire, M. M. da Costa and H. P. de Oliveira, Mater. Sci. Eng., C, 2016, 62, 317–322 CrossRef.
- R. M. A. P. Lima, J. J. Alcaraz-Espinoza, F. A. G. da Silva Jr. and H. P. de Oliveira, ACS Appl. Mater. Interfaces, 2018, 10, 13783–13795 CrossRef CAS.
- F. A. G. da Silva Jr., C. M. S. de Araújo, J. J. Alcaraz-Espinoza and H. P. de Oliveira, J. Polym. Sci., Part B: Polym. Phys., 2018, 56, 1063–1072 CrossRef.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2(5), a000414 Search PubMed.
- G. K. Auer and D. B. Weibel, Biochemistry, 2017, 56, 3710–3724 CrossRef CAS.
- R. Coico, Curr. Protoc. Microbiol., 2006, A.3C.1–A.3C.2 Search PubMed.
- I. Barbolina, C. R. Woods, N. Lozano, K. Kostarelos, K. S. Novoselov and I. S. Roberts, 2D Mater., 2016, 3, 025025 CrossRef.
- S. Some, S.-M. Ho, P. Dua, E. Hwang, Y. H. Shin, H. Yoo, J.-S. Kang, D. Lee and H. Lee, ACS Nano, 2012, 6, 7151–7161 CrossRef CAS.
- O. N. Ruiz, K. A. S. Fernando, B. Wang, N. A. Brown, P. G. Luo, N. D. McNamara, M. Vangsness, Y.-P. Sun and C. E. Bunker, ACS Nano, 2011, 5, 8100–8107 CrossRef CAS.
- P.-C. Wu, H.-H. Chen, S.-Y. Chen, W.-L. Wang, K.-L. Yang, C.-H. Huang, H.-F. Kao, J.-C. Chang, C.-L. L. Hsu, J.-Y. Wang, T.-M. Chou and W.-S. Kuo, J. Nanobiotechnol., 2018, 16, 1 CrossRef.
- S. A. Sydlik and T. M. Swager, Adv. Funct. Mater., 2013, 23, 1873–1882 CrossRef CAS.
- K. E. Eckhart, B. D. Holt, M. G. Laurencin and S. A. Sydlik, Biomater. Sci., 2019, 7, 3876–3885 RSC.
- P. C. Henriques, A. T. Pereira, A. L. Pires, A. M. Pereira, F. D. Magalhães and I. C. Gonçalves, ACS Appl. Mater. Interfaces, 2020, 12, 21020–21035 CrossRef CAS.
- Y. Huang, A. Hara, C. Terashima, A. Fujishima and M. Takai, Carbon, 2019, 152, 354–362 CrossRef CAS.
- K. E. Eckhart, S. J. Schmidt, F. A. Starvaggi, M. E. Wolf, W. M. Vickery and S. A. Sydlik, Regener. Eng. Transl. Med., 2020 DOI:10.1007/s40883-020-00182-y.
- W. C. Lee, C. H. Y. X. Lim, H. Shi, L. A. L. Tang, Y. Wang, C. T. Lim and K. P. Loh, ACS Nano, 2011, 5, 7334–7341 CrossRef CAS.
- R. S. Friedlander, N. Vogel and J. Aizenberg, Langmuir, 2015, 31, 6137–6144 CrossRef CAS.
- Y. F. Dufrêne, Trends Microbiol., 2015, 23, 376–382 CrossRef.
- Y. L. Ong, A. Razatos, G. Georgiou and M. M. Sharma, Langmuir, 1999, 15, 2719–2725 CrossRef CAS.
- Z. Bo, X. Shuai, S. Mao, H. Yang, J. Qian, J. Chen, J. Yan and K. Cen, Sci. Rep., 2014, 4, 4684 CrossRef.
- A. Malas, A. Bharati, O. Verkinderen, B. Goderis, P. Moldenaers and R. Cardinaels, Polymers, 2017, 9(11), 613 CrossRef.
- C. Chen, M. Long, M. Xia, C. Zhang and W. Cai, Nanoscale Res. Lett., 2012, 7, 101 CrossRef.
- X. Qiao, S. Liao, C. You and R. Chen, Catalysts, 2015, 5, 981–991 CrossRef CAS.
- L. Stobinski, B. Lesiak, A. Malolepszy, M. Mazurkiewicz, B. Mierzwa, J. Zemek, P. Jiricek and I. Bieloshapka, J. Electron Spectrosc. Relat. Phenom., 2014, 195, 145–154 CrossRef CAS.
- W. Zhang, Y. Zhang, Y. Tian, Z. Yang, Q. Xiao, X. Guo, L. Jing, Y. Zhao, Y. Yan, J. Feng and K. Sun, ACS Appl. Mater. Interfaces, 2014, 6, 2248–2254 CrossRef CAS.
- J. P. Stratford, C. L. A. Edwards, M. J. Ghanshyam, D. Malyshev, M. A. Delise, Y. Hayashi and M. Asally, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 9552–9557 CrossRef CAS.
- G. N. Bruni, R. A. Weekley, B. J. T. Dodd and J. M. Kralj, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9445–9450 CrossRef CAS.
- E. O. Carvalho, M. M. Fernandes, J. Padrao, A. Nicolau, J. Marqués-Marchán, A. Asenjo, F. M. Gama, C. Ribeiro and S. Lanceros-Mendez, ACS Appl. Mater. Interfaces, 2019, 11, 27297–27305 CrossRef CAS.
- C. Shuai, G. Liu, Y. Yang, F. Qi, S. Peng, W. Yang, C. He, G. Wang and G. Qian, Nano Energy, 2020, 74, 104825 CrossRef CAS.
- H. Vlamakis, Y. Chai, P. Beauregard, R. Losick and R. Kolter, Nat. Rev. Microbiol., 2013, 11, 157–168 CrossRef CAS.
- V. M. N. Idalia and F. Bernardo, in Escherichia coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications, ed. A. Samie, IntechOpen, 2017, DOI:10.5772/67306.
- Z. D. Blount, eLife, 2015, 4, e05826 CrossRef.
- U. Sahu and S. Kar, Bioeng. Bugs, 2012, 3, 131–135 Search PubMed.
- A. Turano and F. Pirali, in Laboratory Diagnosis of Infectious Diseases, ed. A. Balows, W. J. Hausler, M. Ohashi, A. Turano and E. H. Lennete, Springer New York, New York, NY, 1988, pp. 8–13 Search PubMed.
- LIVE/DEAD BacLight bacterial viability kit protocol, https://assets.thermofisher.com/TFS-Assets/LSG/manuals/mp07007.pdf, (accessed 10 June 2019).
- H. Jannadi, W. Correa, Z. Zhang, K. Brandenburg, R. Oueslati and M. Rouabhia, Microb. Pathog., 2019, 133, 103546 CrossRef CAS.
- A. H. Ryu, W. L. Eckalbar, A. Kreimer, N. Yosef and N. Ahituv, Sci. Rep., 2017, 7, 1–9 CrossRef CAS.
- Z. Tan, Y. Shi, B. Xing, Y. Hou, J. Cui and S. Jia, Bioresour. Bioprocess., 2019, 6, 11 CrossRef.
- S. Shima, H. Matsuoka, T. Iwamoto and H. Sakai, J. Antibiot., 1984, 37, 1449–1455 CrossRef CAS.
- M. Hyldgaard, T. Mygind, B. S. Vad, M. Stenvang, D. E. Otzen and R. L. Meyer, Appl. Environ. Microbiol., 2014, 80, 7758–7770 CrossRef.
- M. A. Creighton, J. R. Rangel-Mendez, J. Huang, A. B. Kane and R. H. Hurt, Small, 2013, 9, 1921–1927 CrossRef CAS.
- K. H. Liao, Y. S. Lin, C. W. Macosko and C. L. Haynes, ACS Appl. Mater. Interfaces, 2011, 3, 2607–2615 CrossRef CAS.
- P. Kumar, P. Huo, R. Zhang and B. Liu, Nanomaterials, 2019, 9, 737 CrossRef CAS.
- A. Terada, A. Yuasa, T. Kushimoto, S. Tsuneda, A. Katakai and M. Tamada, Microbiology, 2006, 152, 3575–3583 CrossRef CAS.
- B. Gottenbos, H. C. van der Mei and H. J. Busscher, J. Biomed. Mater. Res., 2000, 50, 208–214 CrossRef CAS.
- J. S. Dickson and M. Koohmaraie, Appl. Environ. Microbiol., 1989, 55, 832–836 CrossRef CAS.
- H. H. Tuson and D. B. Weibel, Soft Matter, 2013, 9, 4368–4380 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01471k |
| This journal is © The Royal Society of Chemistry 2021 |