Advancing the fundamental understanding and practical applications of photo-bioelectrocatalysis
Matteo
Grattieri
 ,
Kevin
Beaver
,
Kevin
Beaver
 ,
Erin M.
Gaffney
,
Erin M.
Gaffney
 ,
Fangyuan
Dong
,
Fangyuan
Dong
 and
Shelley D.
Minteer
and
Shelley D.
Minteer
 *
*
Department of Chemistry, University of Utah, 315 S 1400 E Rm 2020, Salt Lake City, UT 84112, USA. E-mail: minteer@chem.utah.edu
First published on 17th June 2020
Abstract
Photo-bioelectrocatalysis combines the natural and highly sophisticated process of photosynthesis in biological entities with an abiotic electrode surface, to perform semi-artificial photosynthesis. However, challenges must be overcome, from the establishment and understanding of the photoexcited electron harvesting process at the electrode to the electrochemical characterization of these biotic/abiotic systems, and their subsequent tuning for enhancing energy generation (chemical and/or electrical). This Feature Article discusses the various approaches utilized to tackle these challenges, particularly focusing on powerful multi-disciplinary approaches for understanding and improving photo-bioelectrocatalysis. Among them is the combination of experimental evidence and quantum mechanical calculations, the use of bioinformatics to understand photo-bioelectrocatalysis at a metabolic level, or bioengineering to improve and facilitate photo-bioelectrocatalysis. Key aspects for the future development of photo-bioelectrocatalysis are presented alongside future research needs and promising applications of semi-artificial photosynthesis.
1. Introduction
Over billions of years of evolution, nature developed an elegant approach to convert solar energy into chemical energy, while generating oxygen, by means of photosynthesis.1,2 Inspired by nature, scientists developed synthetic strategies to artificially perform the photosynthetic process, utilizing sunlight to drive the water-splitting reaction and to reduce an abundant electron acceptor (i.e., CO2) into valuable compounds (i.e., CO, formaldehyde, methanol). This process is defined as “artificial photosynthesis”,3–6 and provides the opportunity to fabricate robust catalysts,7 as well as the use of ligand design to tune them for selectivity. Furthermore, artificial light absorbers can be synthesized to cover a broad portion of the solar spectrum, increasing their light-to-product conversion efficiency. However, the outstanding performance of artificial photosynthesis often comes at the cost of high-purity catalysts. Additionally, several molecular catalysts performing artificial photosynthesis under aqueous conditions require noble metals (i.e., ruthenium), sacrificial reagents, and present challenges for their heterogenization that limits the performance of molecular-catalyst-based electrodes. We refer the reader to a recent review discussing advantages and challenges of artificial photosynthesis.8 Focusing on the natural photosynthesis, chlorophyll-based light harvesting at the basis of photosynthesis in plants, algae, and bacteria, covers only a specific part of the solar spectrum that varies depending on the photosynthetic entity utilized, thus significantly decreasing the final efficiency of the process. However, the natural photosynthetic apparatuses of biological entities offer the unique advantage of having sophisticated self-repair mechanisms,9 as well as mechanisms to protect against excessive illumination.10 Another critical advantage of biological entities is the ability to utilize sunlight to drive complex reactions performed by highly sophisticated “enzymatic machinery”, which is not yet feasible with synthetic approaches, particularly under mild aqueous conditions. In order to take advantage of the benefits of both natural and artificial photosynthesis, while minimizing their drawbacks, there has been increasing interest in combining abiotic systems with photosynthetic biological entities to perform semi-artificial photosynthesis.11 Accordingly, the goal of semi-artificial photosynthesis is to merge the strengths of natural photosynthesis (i.e., selectivity towards complex products and regeneration features), and artificial photosynthesis (i.e., improved light absorption, efficiency towards a desired pathway, facilitated charge transfer). Furthermore, it should be considered that plants, algae, purple bacteria, and cyanobacteria constitute an abundant, diverse, and broadly available and sustainable source of bio-photocatalysts, paving the way for industrialization of photo-bioelectrochemical systems. The recent progressions in synthetic biology could further expand the synthetic diversity enabled by biohybrid systems, making semi-artificial photosynthesis even more appealing with exciting new research possibilities. As discussed in detail below, a variety of biological entities have been coupled to an electrode surface, having various advantages and strengths.12–14 In this Feature Article, the basics of photo-bioelectrocatalysis are first introduced, exploring intriguing possibilities provided by biohybrid systems for the renewable energy field. Thereafter, the specific challenges for the application of biological entities in photo-bioelectrochemical systems are presented. The other sections of the article focus on discussing recent works performed to address the presented challenges, posing particular focus on the importance of multidisciplinary approaches, which combine the knowledge of chemists, biologists, computational scientists, material scientists and engineers. We strongly believe that only by gaining a better understanding of the fundamental aspects of photo-bioelectrocatalysis, will it be possible to pave the way for the employment of semi-artificial photosynthesis in the field of renewable energy.Photo-bioelectrocatalysis and the future of the renewable energy field
Perhaps, when analyzing the relationship between photo-bioelectrocatalysis and renewable energy, the first aspect that should be considered is that Earth receives solar energy in the range of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–178
000–178![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 TW (where 1 TW corresponds to 1012 W).15–17 For comparison, the worldwide primary energy consumption in 2018 was in the range of 635 × 1018 J, for an average energy consumption rate of approximately 20 TW. This value is projected to rise to about 30 TW by 2050.18,19 Thus, it is clear that the world energy requirement would be largely exceeded if sustainable approaches to efficiently harvest solar energy became available. Silicon-based photovoltaics have reached impressive efficiencies for solar to electrical energy conversion and widespread worldwide application.20 However, in the effort to increase the amount of usable solar energy, multiple approaches should be employed, expanding beyond the sunlight to electricity conversion. Semi-artificial photosynthesis provides several pathways for the conversion of solar energy and sustainable alternatives to common energy-intensive processes, including the sunlight-driven synthesis of valuable compounds from broadly available substrates and monitoring water decontamination with sunlight-powered biosensors.
000 TW (where 1 TW corresponds to 1012 W).15–17 For comparison, the worldwide primary energy consumption in 2018 was in the range of 635 × 1018 J, for an average energy consumption rate of approximately 20 TW. This value is projected to rise to about 30 TW by 2050.18,19 Thus, it is clear that the world energy requirement would be largely exceeded if sustainable approaches to efficiently harvest solar energy became available. Silicon-based photovoltaics have reached impressive efficiencies for solar to electrical energy conversion and widespread worldwide application.20 However, in the effort to increase the amount of usable solar energy, multiple approaches should be employed, expanding beyond the sunlight to electricity conversion. Semi-artificial photosynthesis provides several pathways for the conversion of solar energy and sustainable alternatives to common energy-intensive processes, including the sunlight-driven synthesis of valuable compounds from broadly available substrates and monitoring water decontamination with sunlight-powered biosensors.
Basics of photo-bioelectrocatalysis
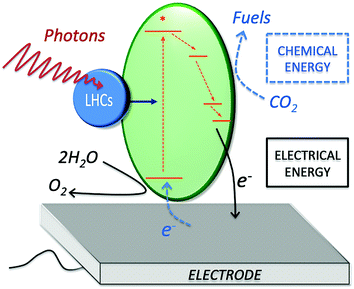
Photo-bioelectrocatalysis covers a broad research field, as it refers to the employment of biological entities capable of bridging sunlight energy and electrical energy, while catalyzing a chemical reaction at an electrode surface. Three main components are at play during photo-bioelectrocatalysis (Fig. 1): (i) light-harvesting complexes (blue circle), which harvest light energy (photons); (ii) protein complexes (green ellipse) that convert light into a charge-separated state after receiving the excitation energy (generation of an electron–hole pair at high energy); and (iii) an electrode acting as the terminal electron acceptor (or primary electron donor for a photocathode) for the redox reactions enabled by the charge-separated state. The high-energy electron obtained after the charge-separation event undergoes a series of electron transfer processes in the metabolisms of photosynthetic microorganisms and plants. At the same time, the high-energy hole allows the oxidation of various substrates, which span from water to organic carbon depending on the photosynthetic entity, as discussed in Section 3. By diverting the photosynthetically generated electrons to the electrode (or from the electrode to the photosynthetic electron transfer chain), the process of photo-bioelectrocatalysis can be selectively tuned toward facilitating specific reactions, opening opportunities for several energy-related applications.Practical applications of photo-bioelectrocatalysis
Below, some reports of practical applications of photo-bioelectrocatalysis are presented to provide a broad overview of the recent trends and opportunities offered by this research field. Both isolated photosynthetic apparatuses and intact organisms have been employed for the sunlight-driven online sensing of various toxic compounds, avoiding the requirement of a power source, and providing an easily measured signal.21 In some previous work from our group, the photo-bioelectrocatalytic features of both intact cyanobacteria or isolated thylakoids membranes were employed for the online detection of herbicides.22 A self-powered system obtained by coupling a thylakoid-based photoanode with an air-breathing cathode enabled detection of three commercial herbicides at concentrations below 1 µg L−1.23 On-going experiments in our group are focused on expanding the range of photosynthetic entities that can be utilized to develop self-powered system for the detection of herbicides, focusing on the sustainability of the entire photo-bioelectrochemical system. Recent reports from other groups showed the possibility to employ both isolated reaction centers or intact organisms for the detection of herbicides and other toxic compounds,24 as well as a self-powered system with microalgae.25Moving to the possibility to utilize photobioelectrocatalysis for sustainable power generation, considerable improvements have been reported employing cyanobacterial mutants in biophotovoltaic systems, which allowed the achievement of a power density of 0.5 W m−2.26,27
Another extremely interesting application of photo-bioelectrocatalysis is to accomplish the bioelectrosynthesis of valuable products with high energy efficiency compared to the synthetic approaches commonly utilized. Leddy and co-workers initially reported the possibility to electrochemically stimulate the cyanobacterium Anabaena variabilis SA-1 (A. variabilis) immobilized on an electrode to enhance ammonia production.28 As discussed in detail below, we recently studied this photo-bioelectrocatalytic process to unveil the redox intermediates allowing sunlight driven ammonia production.29 Other photosynthetic entities were also utilized by other groups for product synthesis, such as purple bacteria in a bioelectrochemical system allowing sustainable H2 production and CO2 reduction utilizing wastewater as substrate.30 Self-assembly of photosynthetic entities in protocells (artificial cell models) enabled obtaining highly oriented photosynthetic reaction centers from Rhodobacter sphaeroides (R. sphaeroides) in giant unilamellar vesicles. The orientation of the reaction centers allowed the generation of a proton gradient that could be used for ATP synthesis in a biohybrid photoelectrochemical system.31,32
2. Challenges of semi-artificial photosynthesis
The opportunities offered by photo-bioelectrocatalysis for converting sunlight energy come with several critical challenges to be overcome. Fig. 2 presents some of the challenges, which will be specifically discussed in this Feature Article. The first and, perhaps, most important challenge of photo-bioelectrocatalysis is gaining a deeper understanding of the photoexcited electron harvesting process at an electrode surface. While this is, relatively, easier when isolated photosystems are employed, extremely little information is available for the case of more complex apparatuses or intact organisms. Gaining a comprehensive understanding of the process would allow specifically tackling the various bottlenecks, thus optimizing the process. Another fundamental challenge is to understand how environmental parameters (i.e., temperature, pH, pollutants, salinity, etc.) influence photo-bioelectrocatalysis. This is critical for the future applications of semi-artificial photosynthesis in industrial scenarios. Microbial photo-bioelectrocatalysis could be employed for energy recovery, decontamination, and monitoring of industrial wastewater that encounter fluctuations in salinity, temperature, pH, and contaminant concentrations. Additionally, for enzymatic photo-bioelectrocatalysis systems, changes in solvent polarity, ionic strength, and temperature can challenge the applications.Expanding to the more applicative field of photo-bioelectrocatalysis, several other challenges can be identified. First, the photosynthetic apparatus of biological entities has been optimized by nature to utilize a specific wavelength of the incoming photons. However, from an efficiency point of view, maximizing the range of usable light is critical.
As we consider maximizing the efficiency of the process, another critical aspect is to artificially facilitate the photoexcited electron transfer (PEET) to the electrode surface. In fact, biological entities are not designed to exchange electrons with an external solid electron acceptor (i.e., the electrode), and for the case of intact microorganisms, to funnel charge transfer to the desired pathway rather than to biomass accumulation. When focusing on product synthesis, an important challenge is defined by the need to tune the semi-artificial photosynthesis process in order to selectively obtain a desired product, which would greatly facilitate its purification and isolation. Finally, to make photo-bioelectrochemical systems appealing to the industrial community, it will be critical to enhance their stability to enable long-term application, and to develop cost-effective systems (considering both their assembly and operation).
As discussed below, there is no single approach that would allow solving all of the presented challenges. However, the implementation of multi-disciplinary approaches has been recently reported in literature, providing critical insights toward exciting future research directions.
3. Understanding the photo-excited electron harvesting process and the effect of environmental parameters
Isolating photosynthetic apparatuses constitutes a model system for the study of the PEET with an electrode surface, thanks to their simpler structure and the possibility to directly deposit them on the electrode surface. Various photosynthetic entities are initially presented, starting from the most up to date understanding of their electrical communication with an electrode surface. The case of intact photosynthetic microorganisms is then presented in a detailed section, which initially describes anoxygenic (purple bacteria) and oxygenic (cyanobacteria) microorganisms, and concludes with the case of eukaryotic algae. It is important to note that when utilizing intact bacterial cells or algae, the challenge of accomplishing the PEET with an electrode surface is magnified by the presence of various membranes separating the photosynthetic apparatus and the electrode, as well as by the metabolism of the microorganisms requiring electrons for their own sustainment. As a result, the fundamental aspects of the PEET process for this category of photosynthetic entities remain largely unexplored.The approaches utilized for the detailed study of photo-bioelectrocatalysis discussed below include: (i) bioengineering for selective inhibition (or removal) of redox intermediates of the photosynthetic electron transfer chain to study their involvement in the PEET process; (ii) theoretical models to predict the concentrations of redox species in the biohybrid photoelectrochemical systems; (iii) localized electrochemical techniques (i.e., scanning (photo)electrochemical microscopy); (iv) quantum mechanical calculations that enable studying structures and states of the various components involved in photo-bioelectrocatalysis (i.e., redox mediators, electrodes, substrates, and products) to probe reaction mechanisms; (v) bioinformatic tools to unveil gene and transcript expression in organisms exposed to various stimuli/conditions to study their influence on the PEET process.
3.1 Thylakoids
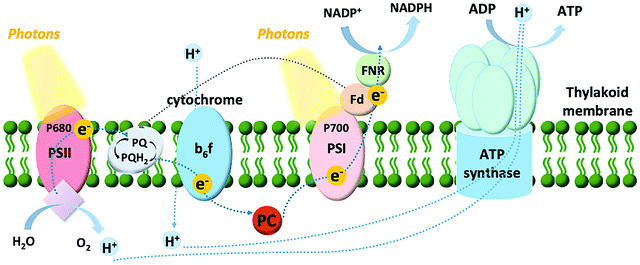
Among the model systems for investigating photo-bioelectrocatalysts are thylakoids, pigmented membranous compartments localized within the chloroplasts of plant and algal cells, and situated free in the cytosol of cyanobacteria. As shown in Fig. 3, the thylakoid membrane consists of a series of two photosystems functioning in tandem: photosystem II and I (PSII and PSI, respectively). The oxidation of water to oxygen in oxygenic photosynthesis can be accomplished at PSII thanks to three main components: (i) a chlorophyll dimer referred to as P680, which has an extremely high midpoint redox potential of 1.1–1.4 V vs. SHE; (ii) a tyrosine residue; and (iii) the tetranuclear Mn cluster that stores four-oxidative equivalents and reduces the tyrosine residue.33 PSII has both light-harvesting and charge-separation functions, utilizing ambient light to catalyze the oxidation of water. After the charge-separation event, the photoexcited electrons are shuttled down an electron transport chain to PSI. From there, a second electron transport chain shuttles the electrons from PSI to ferredoxin, a small protein containing a single 2Fe–2S center, to catalyze the reduction of nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH, which powers the metabolisms of plants, algae, and cyanobacteria. This photosynthetic electron transfer is commonly referred to as the “Z scheme”.Thylakoids can be isolated from a diversity of organisms. Accordingly, our group investigated the photo-bioelectrocatalytic properties of thylakoids obtained from different sources (including chard and beets),34 showing that spinach thylakoids yielded the highest current density. Following, the efforts from our group were focused on unveiling the detailed electron transfer process of thylakoid membranes with an abiotic carbon paper electrode.35 A multidisciplinary approach was utilized, combining electrochemical studies with the bioengineering of the thylakoids membranes by removal, activation, and inhibition of several components involved in light-dependent reactions (plastoquinone, ferredoxin, or ferredoxin–NADP+ reductase). Interestingly, it was possible to show that direct electron transfer (DET) can be obtained, where both PSII and PSI contribute to the biophotocurrent, along with a significant contribution from plastoquinone, the cytochrome b6f complex, and plastocyanin. For the sake of clarity, we define DET between a biological entity and an electrode as a condition where no exogenous redox mediator is added to the investigated system, and the redox molecule responsible for the establishment of the electron transfer is not an endogenous redox-active compound secreted outside the cellular membrane of a living organism (i.e., Shewanella oneidensis that secretes riboflavin and uses it as a redox shuttle between the membrane-bound cytochromes and an electrode surface36). Conversely, mediated electron transfer (MET) is obtained in the presence of exogenous redox mediators artificially added to the system (i.e., diffusible redox mediators such as methyl viologen, or redox polymers), or endogenous redox-active compounds secreted outside the cellular membrane of a living organism.
3.2 Isolated reaction centers and photosystems
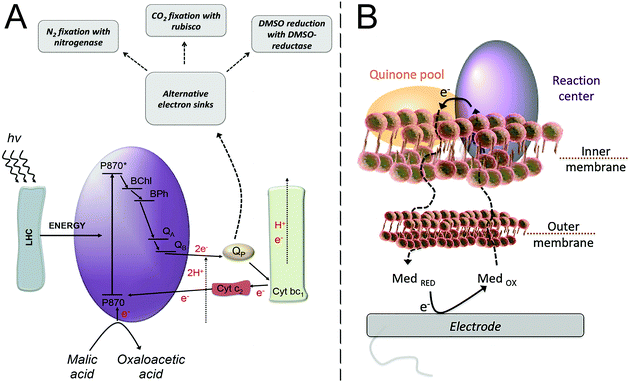 | ||
| Fig. 4 (A) Purple bacteria photosynthetic electron transfer chain. Light harvesting complexes (LHC); the reaction center (purple ellipse, RC) where a “special pair”, consisting of two bacteriochlorophyll molecules, absorbs light at 870 nm (P870); an excited state of the “special pair” (P870*); monomeric bacteriochlorophyll (BChl); bacteriopheophytin (BPh); a primary quinone acceptor (QA); a secondary quinone acceptor (QB); the quinone pool (QP); cytochrome bc1 (Cyt bc1); cytochrome c2 (Cyt c2). From the LHC, the excitation energy is transferred to the “special pair” of bacteriochlorophyll in the RC, leading to the charge separation event that is then followed by electron transfer steps, ultimately resulting in the reduction of quinone molecules in the quinone pool. A reduced quinone molecule transfers an e− and a proton to the Cyt bc1 complex that generates a proton gradient for ATP synthesis and a reduction of Cyt c2, thereby regenerating the cycle. Purple bacteria have alternative electron sinks to divert excess of electrons, through N2 fixation, CO2 fixation or dimethyl sulfoxide (DMSO) reduction. Adapted with permission from E. M. Gaffney, M. Grattieri, K. Beaver, J. Pham, C. McCartney and S. D. Minteer, Unveiling salinity effects on photo-bioelectrocatalysis through combination of bioinformatics and electrochemistry, Electrochim. Acta, 2020, 337, 135731. Copyright 2020 Elsevier.63 (B) The photosynthetic apparatus of purple bacteria is embedded in the inner membrane of the bacterial cell, requiring a mediator to cross both the outer and inner membranes in order to harvest the photoexcited electrons at an electrode surface. Adapted from M. Grattieri, Purple bacteria photo-bioelectrochemistry: enthralling challenges and opportunities, Photochem. Photobiol. Sci., 2020, 19, 424–43512 with permission from the European Society for Photobiology, the European Photochemistry Association, and The Royal Society of Chemistry. | ||
The redox processes taking place between the RC and diffusible quinone redox mediators were investigated in detail by scanning electrochemical microscopy (SECM).47 The study allowed measuring kinetic constants for the interaction between RC and mediators, which highlighted the importance of the chemical environment in which the RC is embedded. In a recent study from the same group, the photo-bioelectrocatalytic process for solubilized reaction centers was described by means of a theoretical model, unveiling kinetic parameters and physico-chemical factors affecting the light conversion process.48 The importance of achieving preferential orientation of the RC on indium tin oxide electrodes modified with cytochrome c for the generation of a photovoltage has also been discussed in a recent work.49
An outstanding 3-D architecture was reported by Ciornii et al. where PSI is combined with a redox enzyme and cytochrome c resulting in a well-defined interprotein electron transfer allowing different pathways to switch on and off based on experimental conditions.56 Recently, Robinson et al. reported an electrochemical reaction–diffusion model for PSI multilayer film atop a metal electrode that allowed shedding light on the electrochemical, physico-chemical, and transport processes taking place at the photoelectrode.57 The reader is also referred to a recent review discussing the recent trends in PSI-based biophotovoltaics.58
3.3 Intact photosynthetic microorganisms
Advantageous to bioelectrochemical systems is the use of whole-cells on an electrode surface due to the enhanced long term stability, owing to the ability of microorganisms to self-replicate and repair. Whole-bacterial cells can easily be deposited on an electrode surface and form bacterial communities known as biofilms. The biofilm is composed of the whole-cells and self-secreted extracellular polymeric substances that function as structural support and protection from the external environment, providing enhanced resistance to inhibiting agents.59 Electroactive biofilms arise from having electroactive microorganisms present in the biofilm, which establish the extracellular electron transfer with the electrode surface.60,61Accordingly, particular interest has been posed on unveiling the capability of diffusible redox mediators to cross the membranes and reach the photosynthetic apparatus. A pioneering study was performed by Cai et al. in 2002, where SECM was used to clarify the properties of redox molecules capable of crossing both the outer and inner membrane.64 A class of reduced hydrophobic redox mediators crossing the inner cellular membrane to react with redox components of the photosynthetic electron transfer chain was defined. Furthermore, the study unveiled the potential of the redox cofactor interacting with the diffusible redox mediators being in a range of −110 ± 90 mV vs. Ag|AgCl. Another interesting aspect was that, utilizing the formal redox potential of the selected redox mediators, a correlation between the heterogeneous rate constant and the redox potential could not be obtained, pointing to the presence of additional limitations in the PEET process. Recently, our group focused on combining electrochemical experimental evidence and quantum mechanical calculations, specifically density functional theory (DFT), to clarify the rate-determining step in the PEET process.65 First, the electronic and structural parameters of various quinone forms were obtained by DFT calculations to investigate their redox processes. Then, the computationally-derived electrochemical properties of the mediators were utilized to fit the experimentally measured current density obtained from purple bacteria employing the various redox mediators during malic acid oxidation. It was possible to identify a model correlating to the PEET rates, unveiling that the rate determining step is the one-electron, proton-decoupled electron transfer taking place in the lipophilic membrane of purple bacteria. The result also suggests that the mediators interact, accepting one electron, from the active site QA in the photosynthetic electron transfer chain, rather than the quinone pool, a finding that is supported also from the SECM results of Cai et al.64 In view of future studies, modeling the binding between biological entities and electrodes by means of quantum mechanical calculations would provide the possibility to gain an atomic-level understanding of the biotic/abiotic interface, which has been reported for redox proteins on carbon-based electrodes.66,67
The extreme versatile metabolism of purple bacteria allows for their employment also in contaminated environments,68 paving the way for the use of photo-bioelectrocatalysis for the light-driven degradation of various wastewater pollutants. However, determining the effects of environmental parameters—such as the presence of contaminants, or variations in pH, salinity, and temperature—on photo-bioelectrocatalysis from a fundamental point of view is critical to elucidate the PEET taking place under environmentally relevant conditions. Extreme variations in pH, temperature, and salinity are harmful to non-tolerant bacteria; however, small variations can have drastic effects on the metabolisms of bacteria. Additionally, tolerant bacteria such as extremophilic organisms, provide a useful platform to study and overcome these conditions.69 Although gene expression and metabolic changes vary widely among species, temperature and pH changes generally result in a variation of the lipid membrane composition.70 Few studies elucidated these effects in photosynthetic microorganisms, with the effects of salinity studied at the genomic level. Salinity is a critical parameter affecting the metabolism of biological entities. As a result, studies have been focused on quantifying gene expression in purple bacteria exposed to saline conditions to unveil their adaptation mechanisms. R. sphaeroides was shown to increase the expression of glycine–betaine uptake systems, due to glycine–betaine acting as an osmotic protectant.71 Although the electrochemical performance of R. sphaeroides has not been evaluated in high salinity, closely related purple bacterium, Rhodobacter capsulatus (R. capsulatus), has been shown to have interesting electrochemical performance resulting from saline adaptation. In a study from our group, it was shown that R. capsulatus adaptation to salinity, and the resulting increased photo-bioelectrocatalysis, occurred only with gradual increases in salinity achieved during several growth cycles.72 Following, bioinformatic analysis was performed and coupled to electrochemical evidence to analyze differential gene expression for understanding the adaptation process from a metabolic point of view.63 Several key metabolic groups showed differences in their expression values between the adapted and non-adapted strains. The genes important to photo-bioelectrocatalysis included those involved in the photosynthetic electron transfer chain and the nitrogen metabolisms of this bacterium. With saline adaptation, there was a decrease in expression of key genes in the photosynthetic electron transport chain, including cytochrome c2, cytochrome c550, and phytoene synthase. Additionally, decreased expression of several genes involved in nitrogen metabolism regulations and many genes in the actual nitrogen metabolism revealed a key link between these systems. The nitrogen metabolism is a known electron sink of the photosynthetic electron transfer chain, as shown in Fig. 4A. This justifies its over-expression during the hindered photosynthetic electron transfer inferred from the decreased photocurrent produced by the saline-adapted strain. Interestingly, the overexpression of an osmoprotectant system offered the ability to artificially tune for rapid saline tolerance of this bacterium and an instant increase in photo-bioelectrocatalysis when compared to the non-adapted strain.
Transcriptomic approaches were utilized also by Volpicella et al. to clarify the mechanism of adaptation for R. sphaeroides to high concentrations of cobalt ions at a metabolic level.73 It was possible to reveal the involvement of energetic metabolisms on the capability of the bacterial cells to cope with high cobalt concentrations. Such a detailed understanding of the effects of contaminants on the metabolisms of photosynthetic organisms could pave the way for the development of biohybrid systems where the interaction between bacterial cells and the electrode surface could be used to tune the metabolism in a fashioned way.
Expanding the study of the effects of environmental parameters on the PEET process, the simultaneous presence of contaminants and redox mediators in a solution containing R. capsulatus cells was recently investigated by Borghese et al.74 Interestingly, the use of a diffusible naphthoquinone mediator allowed R. capsulatus cells to extracellularly reduce tellurite, which also enabled the production of tellurium nanoparticles.
As many cyanobacteria exhibit the natural capability of electrochemical communication with an electrode surface, our group explored the possibility to utilize electrochemical stimulation to enhance ammonia generation from a mutant strain of A. variabilis when immobilized on an indium tin oxide (ITO) electrode.29 The ammonium assimilation step by glutamine synthetase in the A. variabilis mutant strain SA-1 is blocked, hence ammonium will be constantly produced instead of regulated by cellular requirements. A bioelectrocatalytic signal was observed in the presence of light on the A. variabilis SA-1 modified ITO electrode, and nitrogenase was determined as the enzyme that most of the redox moiety is communicating within A. variabilis SA-1. The intriguing part is that nitrogenase is not positioned to be capable of direct electron transfer with the electrode, indicating that an internal mediation system is at play. To clarify the electron transfer process, the electrolyte obtained after the electrochemical characterization of A. variabilis SA-1 in the presence of N2 and light was collected. UV-vis absorption spectra of the concentrated electrolyte closely matched the spectrum of commercial ferredoxin (Fig. 5), which supported the electrochemical evidence of a redox couple attributed to ferredoxin. Accordingly, the electrostimulated ammonia production by A. variabilis SA-1 could be due to endogenously produced ferredoxin exiting cells to mediate electron transfer between the ITO electrode and nitrogenase.
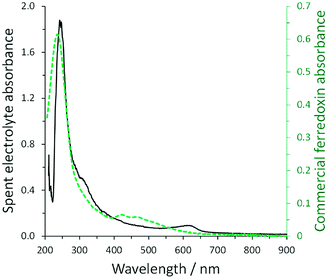 | ||
| Fig. 5 Comparison of UV-vis absorption spectra for concentrated spent electrolyte collected after electrochemical characterization of an ITO electrode modified with A. variabilis SA-1 in N2 purged buffer in the presence of light (solid black line, absorbance plotted on left) and commercial ferredoxin (green dashed line, absorbance plotted on right). Reproduced with permission from K. L. Knoche, E. Aoyama, K. Hasan and S. D. Minteer, Role of Nitrogenase and Ferredoxin in the Mechanism of Bioelectrocatalytic Nitrogen Fixation by the Cyanobacteria A. variabilis SA-1 Mutant Immobilized on Indium Tin Oxide (ITO) Electrodes, Electrochim. Acta, 2017, 232, 396–403. Copyright 2017 Elsevier.29 | ||
DET from cyanobacteria to the electrode through membrane bound cytochromes has also been reported, a process similar to that of metal reducing microorganisms.77,82 A recent study investigated the role of PSII, PSI, and the respiratory metabolism of Synechocystis cells on the obtained photocurrent.83 It was shown that the respiratory metabolism could supply electrons to the PSI which could then photo-reduce the anode, generating the photocurrent. Additionally, bacterial conductive nanowires have also been discovered to facilitate the EET process in the cyanobacterium Synechocystis sp. PCC-6803 using scanning tunnelling microscopy.84
As discussed above for purple bacteria, environmental stress effects need to be elucidated for the successful deployment of photo bioelectrochemical systems, and genome analysis is an excellent platform to study. In a study by Rübsam et al., the mRNA levels of Synechocystis sp. PCC-6803 were analysed in response to saline and iron depletion. It was shown that this strain accumulates the compatible solute glucosylglycerol to overcome saline stress, and an iron-stress activated RNA1 that is directly involved with the photosynthetic electron transport chain.85 Although the resulting electrochemical performance of Synechocystis sp. PCC-6803 has not been evaluated with these environmental stress factors, it is likely that due to the difference in the mRNA expression, both saline and iron could play a role in its photo-bioelectrochemical performance.
4. Artificial approaches for upgraded photo-bioelectrocatalysis
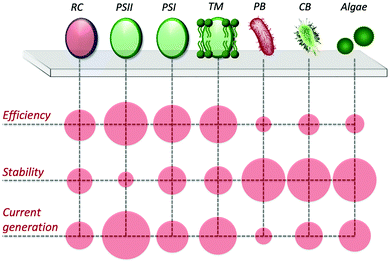
In view of utilizing photo-bioelectrocatalysis for practical applications, it is critical to maximize the efficiency of the process, both in terms of current density and desired product synthesis. Current densities tend to be lower in whole-cell systems than cell-component systems, due to resistances of charge transport and mass transport from additional membranes. Furthermore, sunlight energy conversion efficiencies are also lower, due to respiration competing with the photosynthetic electron transfer pathways and nutrients requirement for growth to continue. However, an advantage of using intact cells is exploiting their regulatory and replicative machinery to increase the overall lifetime and stability of the bioanode (or biocathode). Accordingly, research efforts have been focused on developing artificial approaches to facilitate the PEET process and to drive the electron flux towards desired pathways. A scheme comparing the performance of the various photosynthetic biological entities in biohybrid systems is reported in Fig. 6.4.1 Artificial redox mediating systems
The group of Lo Gorton performed pioneering studies on wiring purple bacteria with an electrode surface utilizing redox polymers. The artificial systems enabling electron-shuttling employed redox moieties constituted by osmium functionalities. Interestingly, it was shown that by modifying the chemical structure of these redox moieties, the photoexcited electron harvesting could be tuned, achieving biophotocurrents in the range of 10 µA cm−2.62,92Our group focused particular efforts on developing artificial mediating systems for diverting the photoexcited electrons to an abiotic electrode. Specifically, inspired by the use of quinone redox moieties in biological processes, we developed a redox polymer containing naphthoquinone redox moieties immobilized on a linear polyethyleneimine backbone.93 The redox polymer was first efficiently employed to enhance the electron transfer between an electrode surface and various enzymes.94 Following, the capability of the polymer to mediate electron transfer from intact bacterial cells was demonstrated as well.95,96 Inspired by these results, we explored the possibility to utilize the redox polymer for harvesting photoexcited electrons.97 Finely controlling the ratio of bacterial cells and redox moieties in a 3-D redox hydrogel geometry allowed establishing PEET with a low overpotential, achieving ∼3 µA cm−2.
We also expanded the applicability of this naphthoquinone-based redox polymer to oxygenic photosynthetic entities, specifically for wiring intact chloroplasts (Fig. 7A).98 Electrical wiring of chloroplasts with the redox polymer allowed a five-fold increase in photocurrent generation compared to the current obtained under DET conditions. Furthermore, a mixed redox polymer-diffusible mediator system was explored, by adding dichlorobenzoquinone to the system. The new artificial redox mediation resulted in an outstanding increase of the photocurrent, enabling an additional five-fold increase for the harvested current, as shown in Fig. 7B.
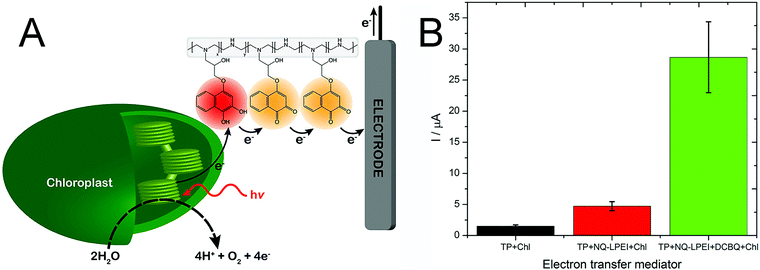 | ||
| Fig. 7 (A) Artificial PEET process through the naphthoquinone-based redox polymer that harvests photoexcited electrons from intact chloroplast and shuttles them to the electrode surface. (B) Comparison of photocurrent generation from chloroplasts in direct electron transfer condition (TP + Chl, black), in the presence of the naphthoquinone redox polymer (TP + NQ-LPEI + Chl, red), and in the presence of both the redox polymer and diffusible dichloro benzoquinone (TP + NQ-LPEI + DCBQ + Chl, green). Adapted with permission from K. Hasan, R. D. Milton, M. Grattieri, T. Wang, M. Stephanz and S. D. Minteer, Photobioelectrocatalysis of Intact Chloroplasts for Solar Energy Conversion, ACS Catal., 2017, 7, 2257–2265. Copyright 2017 American Chemical Society.98 | ||
Expanding to polymers employing different redox moieties, osmium redox polymers have been recently utilized to achieve the electrical wiring of both PSI (used at the biophotocathode) and PSII (used at the biophotoanode) obtaining a fully light-driven semi-artificial electrochemical system that performed water splitting at the anode while obtaining hydrogen evolution at the cathode.99 The combination of an artificial redox mediating system with osmium moieties and an engineered hierarchically-structured indium tin oxide electrode surface was reported by Sokol et al.100 The efficient wiring by the redox polymer and the higher loading enabled by the engineered electrode allowed achieving the outstanding biophotocurrent of 410 µA cm−2.
Regarding cyanobacteria photo-bioelectrocatalysis, major interest has been focused on facilitating cross-membranes electron transportation. Accordingly, various studies on photosynthetic microbial fuel cells employed exogenous electron mediators, using both lipid-insoluble (i.e., ferricyanide), and lipid-soluble (i.e., cytochrome, 2,6-dichloro-1,4-benzoquinone) molecule to harvest photoexcited electrons from cyanobacteria.83,101,102 However, lipid-soluble mediators can disrupt electron transfer processes used for important cellular events within the organism, and eventually cause cell death. Conversely, lipid-insoluble mediators, like ferricyanide ion, accepting electrons from the cell surface offered a more bio-compatible choice over long durations.102 The combination of an osmium-based redox polymer and soluble ferricyanide allowed achieving a biophotocurrent of about 48 µA cm−2 from cyanobacteria Lyptolyngbia sp. (CYN82).103 Finally, for the case of isolated RC–LHC complexes, the combined used of diffusible ubiquinones and cytochrome c as redox mediators allowed achieving photocurrents in the range of 45 µA cm−2.
4.2 Engineering of the biotic/abiotic interface
Crucial to the success of photo-bioelectrochemical systems is improving the performance of the abiotic electrode and its biocompatibility for the photosynthetic biological entity utilized. For each division of biophotocatalysts, pairing of nanoscale materials with a biological entity provides the opportunity for overall current and system enhancement. The use of a hierarchically-structured inverse opal indium tin oxide as support for PSII hybrid electrodes with Os-based redox polymer has been previously introduced.100 Recently, the use of mesoporous NiO surface as a p-type semiconductor in conjunction with PSI has been reported to obtain a biophotocathode capable of tandem operation with PSI-TiO2 biophotoanode, for a biophotovoltaic cell with open-circuit potential of 0.7 V.104To enhance the current density produced by thylakoid photoanodes, multi-walled carbon nanotubes (CNTs) are often mixed in a suspension with thylakoids before depositing on the surface of the electrode. The structure of CNTs increases the electrode surface area, while their hydrophobicity helps to immobilize the thylakoids on the surface, and their combined employment with both diffusible mediators and redox polymer has been reported. Exogenous quinone-based mediators and CNTs-modified thylakoids electrodes were explored by Takeuchi et al., testing a variety of quinone-based and pyridine-based mediators, greatly enhancing the obtained photocurrent.105 The research group of Lo Gorton has employed CNTs in their thylakoid-modified electrodes, where rather than using diffusible quinone-based mediators, an osmium-based redox polymer was used to immobilize thylakoids on graphite electrodes and facilitate mediated electron transfer.106 This quinone-free system allowed biophotocurrents densities similar to those reported with diffusible mediators (on the order of 100 µA cm−2). We also refer the reader to a recent review highlighting the advancements in thylakoid membranes photobioelectrochemical systems.107 The use of carbon nanotubes to enhance photocurrent production was also explored for cyanobacteria photoanodes. Specifically, carbon nanotube modified electrodes served as a tool to bridge the PEET from membranes to the electrode surface.108,109 Additionally, cyanobacterium Leptolyngbia sp. was paired with a Au electrode modified with a conducting polymer and Au nanoparticles to increase current performance in a photo-biofuel cell.110 Recently, Wolfe et al. explored the effect of different three-dimensional electrode morphologies on the photocurrent production from PSI.111 Interestingly, macroporous electrodes (5 µm porous) outperformed mesoporous electrodes (20–100 nm porous) when in the presence of diffusible mediators, achieving biophotocurrents in the range of 40 µA cm−2.
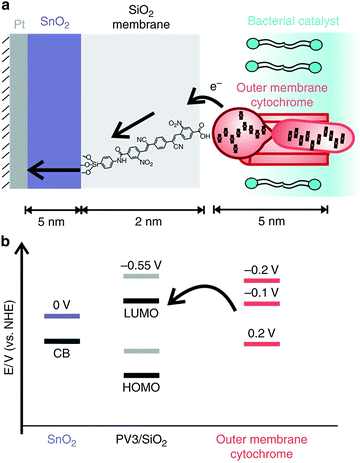
When pairing whole-cell entities to abiotic electrodes, it is of interest to pair the microorganisms as closely to the electrode surface as possible to minimize ohmic losses. Nanoscale engineering of the abiotic electrode surface offers one approach toward this goal. In a recent work by Cornejo et al., the combination was achieved on the nanoscale with the incorporation of a tin oxide inorganic catalyst layer, and a silica oxide layer embedded with molecular wires interfaced with the outer membrane cytochromes of Shewanella oneidensis MR-1, as represented in Fig. 8.112 Although this work was not with photosynthetic microorganisms, the approach was a great advancement in wiring bacterial cells with the electrode surface, which could be extended for applications in biophotovoltaics. Recently, work has also been done to improve the solar-powered CO2 reduction at the cathode of a biophotovoltaic cell for acetate production. Interfacing silicon nanowires with Sporomusa ovata cells allowed for development of a close-packed nanowire-bacteria cathode that, along with optimization of electrolyte buffering capacity and pH, exhibited a 7-day efficiency of 3.6% for solar to acetate conversion.113
4.3 Bioengineering of biological entities for enhancing PEET
Due to the advances in genomic editing tools, the possibility for engineering strains to provide electroactivity by inserting critical genes has attracted particular interest.114 The most well-studied extracellular electron transfer pathways from model electrogenic microorganisms are the mtrCAB pathway from Shewanella oneidensis MR-1,115 and outer membrane cytochrome (omc) genes from Geobacter sulfurreducens, specifically omcS.116 These metabolic pathways can then be engineered into microorganisms, transforming an easily-grown but non-electrogenic strain, such as Escherichia coli, into a strain capable of communicating with and wiring to an electrode. In this section, a brief overview of the recent synthetic biology approaches for purple bacteria and cyanobacteria will be given, and we refer the reader to a recent review describing the bioengineering of photovoltaics in detail.117 For non-photosynthetic microorganisms, electricity generation has been achieved from the pairing of the oxidation of organic substrates to the current generation at an electrode. Specifically, the model genetically modified microorganism, Escherichia coli.118 Similar approaches are being explored to pair the photosynthetic process with an electrode through bioengineering of these electrogenic pathways into photosynthetic microorganisms. As discussed above, the large limitation for purple bacteria and cyanobacteria is the transfer of electrons through the thick cellular membrane to an electrode surface. The mediation approaches discussed face challenges of non-specific electron extraction due to their reactivity with any redox intermediate present with a thermodynamically-favored potential.117 For specific and selective electron harvesting from the photosynthetic electron chain, bioengineering extracellular electron transfer pathways into these microorganisms offer a favorable approach. Genetic bioengineering for enhanced extracellular electron transfer has been achieved in two strains of photosynthetic microorganisms Synechocystis sp. PCC 6803, and Synechococcus elongatus PCC 7942. In a study by Bradley et al.,76 the terminal oxidases, bd-quinol oxidase, cytochrome c oxidase, and an “alternative respiratory terminal oxidase”, were deleted in Synechocystis sp. PCC 6803. Without these terminal oxidases, electrons could not be utilized for O2 reduction and would be available for use in the photosynthetic electron transport chain. The deletion resulted in a 10% increase in power generation in dark conditions. In Synechococcus elongatus PCC 7942, Sekar et al. engineered the strain to contain the omcS gene from Geobacter sulfurreducens in order to express an outer membrane cytochrome protein, OmcS.119 The engineered strain enabled a nine-fold increase in photocurrent (reaching about 15 µA cm−2) compared to the non-engineered strain. In a subsequent work by the same group, OmcS was expressed while also deleting the terminal oxidases in Synechococcus elongatus PCC 7942, resulting in a increased current when compared to both the wildtype strain and the deleted terminal oxidases mutant strain.120Bioengineering of photosynthetic microorganisms has multiple challenges, including the difficulty of synthetic biology techniques, and unknown metabolic effects when compared to model microorganisms such as Escherichia coli.117 Despite these challenges, the advances in understanding these extracellular electron transfer pathways in the model organisms allow methods to be applied to photosynthetic bacteria, offering increased applications in bioelectrochemical systems.
4.4 Tuning product selectivity
The metabolic pathways and enzyme networks that characterize intact microorganisms provide the possibility to perform complex chemistry, channeling substrates along reaction cascades. Accordingly, selectively driving a specific metabolism to obtain a desired product in a biohybrid system with intact microorganisms involves modifying the flux of redox carriers and ions between, and within, the electrode, the electrolyte, and the microorganisms. Both genetic and bioelectrochemical tools can play a critical role in achieving these modifications.121 Genetic tools for metabolic engineering have been extensively developed for model organisms, such as Escherichia coli,122,123 showing the selective bioelectrochemical production of succinate from CO2.124 Conversely, limited toolsets are available for non-model strains, such as photosynthetic microorganisms, and future efforts to develop synthetic biology approaches for these entities will be required to tune photo-bioelectrocatalysis for product synthesis.125The use of bioelectrochemical approaches to finely tune product synthesis has been investigated to a greater degree than genetic approaches. We have previously discussed how electrostimulation allowed the enhancement of ammonia generation from a mutant strain of A. variabilis.29 Recently, the use of electron-regulated flux by means of polarized electrodes has been reported also to increase CH4 formation and CO2 utilization in a bioelectrochemical system for electromethanogenesis.126 Additionally, active modification of electron fluxes through electrochemical polarization was recently utilized to maximize H2 production and CO2 fixation in a photo-microbial electrochemical system with purple bacteria.30
4.5 Artificial light-harvesting complexes and biohybrid systems
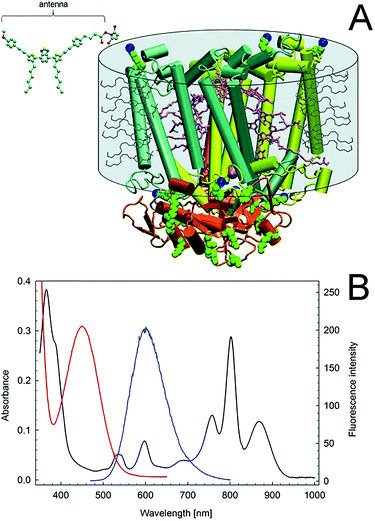
Enhancing the usable portion of light, by expanding the absorption region of photosynthetic entities, could enable higher efficiency for the energy conversion process, further facilitating the implementation of semi-artificial photosynthesis. In biological photosynthetic entities, light is collected and funneled to the photosystems by pigment–protein complexes (i.e., the light-harvesting complexes), which are also referred to as “antennas”. A significant amount of research has been focused on developing artificial antennas to develop hybrid systems with tuned light-harvesting efficiency. In a pioneering study, Nabiev et al. reported the coupling of photoluminescent quantum dots to transfer the energy from an excited electron to the photosynthetic reaction center of the purple bacterium R. spheroides by Förster resonance energy transfer (FRET).127 Following, Milano et al. reported a tailored molecular organic dye, absorbing at 450 nm and emitting at 602 nm, where the RC has an absorption peak (Fig. 9B). A succinimidyl ester group enables selective covalent binding of the synthesized fluorophore to the lysines of the photosynthetic reaction center of R. sphaeroides R26, as shown in Fig. 9A.128Such molecular antennas expanded the tuning possibility for the hybrid system, thanks to the diversity of molecular compounds and their shape and flexibility, reducing the impact of the artificial antennas on the structure and function of the photosynthetic reaction center. The same group further extended the capability to utilize artificial molecular antennas absorbing light in the visible region (400–700 nm) and emitting in correspondence of the near infra-red absorption peak of the reaction center (800 nm) in the following works.129,130 Another study utilized commercially available fluorescent dyes molecules with a genetically modified purple bacteria reaction center.131 The use of organic antennas to increase light absorption was extended to the PSI of the thermophilic cyanobacterium Thermosynechococcus elongates targeting the “green gap” (i.e., the lack of light absorption in the range 450–600 nm),132 as well as for the light-harvesting complex LH2, obtained from a strain of Rhodopseudomonas acidophila, expanding light-harvesting to the 620–750 nm range.133 Artificial antennas have also been coupled to intact organisms. Specifically, the cyanobacterium Synechococcus sp. was combined with quantum dots to enhance the current generation under low light conditions through FRET. A nanocomposite of CdTe quantum dots (broadening the excitation spectrum to 350 nm), graphene nanoplatelets, and silk-fibroin cast on a graphite electrode enabled a 5.7 fold increase in biophotocurrent.134 Recently, Grayson et al. reported that artificial tailor-made light-harvesting antennas can be created in vivo, by genetically modifying the photosystem of R. sphaeroides to incorporate the yellow fluorescent protein.135
In two recent works, Liu et al. utilized a very interesting approach to combine the features of oxygenic photosynthesis in plants with the anoxygenic photosynthesis of purple bacteria, augmenting the near-infrared solar energy conversion of the latter to the visible light spectrum.136,137
5. Conclusions and future outlook for research in photo-bioelectrocatalysis
Employing multidisciplinary approaches has provided unique tools to critically expand our understanding of photo-bioelectrocatalysis. The combination of quantum mechanical calculations and electrochemical experimental evidence enabled us to probe reaction mechanisms taking place in biohybrid systems composed of intact bacterial cells, exogenous redox mediators, and abiotic electrodes, which are otherwise particularly challenging to be studied. While the combination of quantum mechanical methods and microbial electrochemical systems can be considered in its infancy, it could provide critical insights for future developments of the field, both in terms of shedding light on biotic/abiotic materials interactions and reaction mechanisms between biological entities and exogenous molecules. On a similar note, the insights provided by bioinformatics studies allow the understanding of photo-bioelectrocatalysis at a metabolic level. As shown for purple bacteria adaptation to salinity, understanding the metabolic changes resulting from the adaptation process allows artificially tuning the adaptation. Expanding to other environmentally relevant parameters, the knowledge provided by bioinformatic studies can be used to enable rational tuning of detailed metabolisms to drive the biohybrid systems towards desired directions. It is critical to underline that in order to accomplish this goal, bioengineering of the biological entities will play a critical role. The possibility to selectively insert heterologous electron transfer pathways, or to modify the natural metabolic pathways of photosynthetic entities provides unprecedented research possibilities in order to perform multistep reactions in a fashioned way.Accurately combining synthetic biology with engineering of the biotic/abiotic interface and artificial redox mediating systems, as well as implementing artificial light-harvesting complexes, will provide powerful tools to improve the efficiency of the sunlight conversion process. Such results will critically promote the implementation of semi-artificial photosynthesis for practical applications.
In conclusion, a critical aspect to be underlined is that no single approach will enable the successful development and implementation of photo-bioelectrocatalysis, but rather the combination of the multidisciplinary approaches presented will provide the unique features to accomplish this task. A strong collaboration of the community working in the photo-bioelectrochemical field, including (bio)electrochemists, biologists, engineers, organic chemists, computational scientists, material scientists, and biophysicist is vital to provide the required tools to fully explore photo-bioelectrocatalysis of biological entities and enable the development of biohybrid photosynthetic systems with unprecedented capabilities.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the National Science Foundation Grant # 1561427.Notes and references
- D. E. Canfield, Annu. Rev. Earth Planet. Sci., 2005, 33, 1–36 CrossRef CAS.
- S. A. Crowe, L. N. Dossing, N. J. Beukes, M. Bau, S. J. Kruger, R. Frei and D. E. Canfield, Nature, 2013, 501, 535–538 CrossRef CAS PubMed.
- S. Y. Reece, J. A. Hamel, K. Sung, T. D. Jarvi, A. J. Esswein, J. J. H. Pijpers and D. G. Nocera, Science, 2011, 334, 645–648 CrossRef CAS PubMed.
- D. G. Nocera, Acc. Chem. Res., 2012, 45, 767–776 CrossRef CAS PubMed.
- A. Pannwitz and O. S. Wenger, Chem. Commun., 2019, 55, 4004–4014 RSC.
- K. E. Dalle, J. Warnan, J. J. Leung, B. Reuillard, I. S. Karmel and E. Reisner, Chem. Rev., 2019, 119, 2752–2875 CrossRef CAS PubMed.
- A. Sartorel, M. Carraro, F. M. Toma, M. Prato and M. Bonchio, Energy Environ. Sci., 2012, 5, 5592–5603 RSC.
- B. Zhang and L. Sun, Chem. Soc. Rev., 2019, 48, 2216–2264 RSC.
- S. Jarvi, M. Suorsa and E. M. Aro, Biochim. Biophys. Acta, 2015, 1847, 900–909 CrossRef CAS PubMed.
- C. Ota, K. Sugihara, Y. Kinoshita, Y. Kashiyama, Y. Nagasawa and H. Tamiaki, Photochem. Photobiol. Sci., 2019, 18, 64–70 RSC.
- N. Kornienko, J. Z. Zhang, K. K. Sakimoto, P. Yang and E. Reisner, Nat. Nanotechnol., 2018, 13, 890–899 CrossRef CAS PubMed.
- M. Grattieri, Photochem. Photobiol. Sci., 2020, 19, 424–435 RSC.
- K. Hasan, V. Grippo, E. Sperling, M. A. Packer, D. Leech and L. Gorton, ChemElectroChem, 2017, 4, 412–417 CrossRef CAS.
- F. Milano, A. Punzi, R. Ragni, M. Trotta and G. M. Farinola, Adv. Funct. Mater., 2019, 29, 1805521 CrossRef.
- R. E. Blankenship, D. M. Tiede, J. Barber, G. W. Brudvig, G. Fleming, M. Ghirardi, M. R. Gunner, W. Junge, D. M. Kramer, A. Melis, T. A. Moore, C. C. Moser, D. G. Nocera, A. J. Nozik, D. R. Ort, W. W. Parson, R. C. Prince and R. T. Sayre, Science, 2011, 332, 805–809 CrossRef CAS PubMed.
- J. M. Pisciotta, Y. Zou and I. V. Baskakov, PLoS One, 2010, 5, e10821 CrossRef PubMed.
- O. Kruse, J. Rupprecht, J. H. Mussgnug, G. C. Dismukes and B. Hankamer, Photochem. Photobiol. Sci., 2005, 4, 957–970 RSC.
- U.S. Energy Information Administration, U.S. Department of Energy, Washington, DC, 2019. https://www.eia.gov/outlooks/ieo/pdf/ieo2019.pdf.
- N. S. Lewis and D. G. Nocera, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15729–15735 CrossRef CAS PubMed.
- K. Yoshikawa, H. Kawasaki, W. Yoshida, T. Irie, K. Konishi, K. Nakano, T. Uto, D. Adachi, M. Kanematsu, H. Uzu and K. Yamamoto, Nat. Energy, 2017, 2, 17032 CrossRef CAS.
- M. Grattieri and S. D. Minteer, ACS Sens., 2018, 3, 44–53 CrossRef CAS PubMed.
- M. Tucci, M. Grattieri, A. Schievano, P. Cristiani and S. D. Minteer, Electrochim. Acta, 2019, 302, 102–108 CrossRef CAS.
- M. Rasmussen and S. D. Minteer, Anal. Methods, 2013, 5, 1140–1144 RSC.
- M. Chatzipetrou, F. Milano, L. Giotta, D. Chirizzi, M. Trotta, M. Massaouti, M. R. Guascito and I. Zergioti, Electrochem. Commun., 2016, 64, 46–50 CrossRef CAS.
- J. Chouler, M. D. Monti, W. J. Morgan, P. J. Cameron and M. Di Lorenzo, Electrochim. Acta, 2019, 309, 392–401 CrossRef CAS.
- K. L. Saar, P. Bombelli, D. J. Lea-Smith, T. Call, E.-M. Aro, T. Müller, C. J. Howe and T. P. J. Knowles, Nat. Energy, 2018, 3, 75–81 CrossRef CAS.
- M. Grattieri and S. D. Minteer, Nat. Energy, 2018, 3, 8–9 CrossRef.
- J. Leddy and T. M. Pachekewitz, US Pat., US 9506085 B2, University of Iowa Research Fundation, United States, 2016 Search PubMed.
- K. L. Knoche, E. Aoyama, K. Hasan and S. D. Minteer, Electrochim. Acta, 2017, 232, 396–403 CrossRef CAS.
- I. A. Vasiliadou, A. Berná, C. Manchon, J. A. Melero, F. Martinez, A. Esteve-Nuñez and D. Puyol, Front. Energy Res., 2018, 6, 107 CrossRef.
- E. Altamura, F. Milano, R. R. Tangorra, M. Trotta, O. H. Omar, P. Stano and F. Mavelli, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 3837–3842 CrossRef CAS PubMed.
- J. P. Allen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 3790–3791 CrossRef CAS PubMed.
- F. Rappaport, M. Guergova-Kuras, P. J. Nixon, B. A. Diner and J. Lavergne, Biochemistry, 2002, 41, 8518–8527 CrossRef CAS PubMed.
- M. Rasmussen, A. Wingersky and S. D. Minteer, Electrochim. Acta, 2014, 140, 304–308 CrossRef CAS.
- M. Rasmussen and S. D. Minteer, Electrochim. Acta, 2014, 126, 68–73 CrossRef CAS.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- A. Operamolla, R. Ragni, F. Milano, R. Roberto Tangorra, A. Antonucci, A. Agostiano, M. Trotta and G. Farinola, J. Mater. Chem. C, 2015, 3, 6471–6478 RSC.
- D. W. Reed and R. K. Clayton, Biochem. Biophys. Res. Commun., 1968, 30, 471–475 CrossRef CAS PubMed.
- J. P. Allen, G. Feher, T. O. Yeates, H. Komiya and D. C. Rees, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 8487–8491 CrossRef CAS PubMed.
- G. Feher, J. P. Allen, M. Y. Okamura and D. C. Rees, Nature, 1989, 339, 111–116 CrossRef CAS.
- T. O. Yeates, H. Komiya, A. Chirino, D. C. Rees, J. P. Allen and G. Feher, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 7993–7997 CrossRef CAS PubMed.
- R. A. Isaacson, F. Lendzian, E. C. Abresch, W. Lubitz and G. Feher, Biophys. J., 1995, 69, 311–322 CrossRef CAS PubMed.
- J. P. Allen and J. C. Williams, FEBS Lett., 1998, 438, 5–9 CrossRef CAS PubMed.
- F. Milano, A. Agostiano, F. Mavelli and M. Trotta, Eur. J. Biochem., 2003, 270, 4595–4605 CrossRef CAS PubMed.
- F. Ciriaco, R. R. Tangorra, A. Antonucci, L. Giotta, A. Agostiano, M. Trotta and F. Milano, Eur. Biophys. J., 2015, 44, 183–192 CrossRef CAS PubMed.
- L. Kalman, J. C. Williams and J. P. Allen, Photosynth. Res., 2008, 98, 643–655 CrossRef CAS PubMed.
- F. Longobardi, P. Cosma, F. Milano, A. Agostiano, J. Mauzeroll and A. J. Bard, Anal. Chem., 2006, 78, 5046–5051 CrossRef CAS PubMed.
- F. Milano, F. Ciriaco, M. Trotta, D. Chirizzi, V. De Leo, A. Agostiano, L. Valli, L. Giotta and M. R. Guascito, Electrochim. Acta, 2019, 293, 105–115 CrossRef CAS.
- M. Di Lauro, G. Buscemi, M. Bianchi, A. De Salvo, M. Berto, S. Carli, G. M. Farinola, L. Fadiga, F. Biscarini and M. Trotta, MRS Adv., 2020, 5, 985–990 CrossRef CAS.
- M. Kato, J. Z. Zhang, N. Paul and E. Reisner, Chem. Soc. Rev., 2014, 43, 6485–6497 RSC.
- M. Pita, D. M. Mate, D. Gonzalez-Perez, S. Shleev, V. M. Fernandez, M. Alcalde and A. L. De Lacey, J. Am. Chem. Soc., 2014, 136, 5892–5895 CrossRef CAS PubMed.
- J. Z. Zhang, P. Bombelli, K. P. Sokol, A. Fantuzzi, A. W. Rutherford, C. J. Howe and E. Reisner, J. Am. Chem. Soc., 2018, 140, 6–9 CrossRef CAS PubMed.
- F. Zhao, V. Hartmann, A. Ruff, M. M. Nowaczyk, M. Rögner, W. Schuhmann and F. Conzuelo, Electrochim. Acta, 2018, 290, 451–456 CrossRef CAS.
- M. Ciobanu, H. A. Kincaid, V. Lo, A. D. Dukes, G. K. Jennings and D. E. Cliffel, J. Electroanal. Chem., 2007, 599, 72–78 CrossRef CAS.
- G. Chen, G. LeBlanc, G. K. Jennings and D. E. Cliffel, J. Electrochem. Soc., 2013, 160, H315–H320 CrossRef CAS.
- D. Ciornii, M. Riedel, K. R. Stieger, S. C. Feifel, M. Hejazi, H. Lokstein, A. Zouni and F. Lisdat, J. Am. Chem. Soc., 2017, 139, 16478–16481 CrossRef CAS PubMed.
- M. T. Robinson, D. E. Cliffel and G. K. Jennings, J. Phys. Chem. B, 2018, 122, 117–125 CrossRef CAS PubMed.
- M. T. Robinson, E. A. Gizzie, F. Mwambutsa, D. E. Cliffel and G. K. Jennings, Curr. Opin. Electrochem., 2017, 5, 211–217 CrossRef CAS.
- Y. Xiao and F. Zhao, Curr. Opin. Electrochem., 2017, 4, 206–211 CrossRef CAS.
- D. R. Bond and D. R. Lovley, Appl. Environ. Microbiol., 2003, 69, 1548–1555 CrossRef CAS PubMed.
- P. Cristiani, M. L. Carvalho, E. Guerrini, M. Daghio, C. Santoro and B. Li, Bioelectrochemistry, 2013, 92, 6–13 CrossRef CAS PubMed.
- K. Hasan, K. V. R. Reddy, V. Eßmann, K. Górecki, P. Ó. Conghaile, W. Schuhmann, D. Leech, C. Hägerhäll and L. Gorton, Electroanalysis, 2015, 27, 118–127 CrossRef CAS.
- E. M. Gaffney, M. Grattieri, K. Beaver, J. Pham, C. McCartney and S. D. Minteer, Electrochim. Acta, 2020, 337, 135731 CrossRef CAS PubMed.
- C. Cai, B. Liu, M. V. Mirkin, H. A. Frank and J. F. Rusling, Anal. Chem., 2002, 74, 114–119 CrossRef CAS PubMed.
- M. Grattieri, Z. Rhodes, D. P. Hickey, K. Beaver and S. D. Minteer, ACS Catal., 2019, 9, 867–873 CrossRef CAS.
- S. Babanova, I. Matanovic, M. S. Chavez and P. Atanassov, J. Am. Chem. Soc., 2015, 137, 7754–7762 CrossRef CAS PubMed.
- I. Matanovic, S. Babanova, M. S. Chavez and P. Atanassov, J. Phys. Chem. B, 2016, 120, 3634–3641 CrossRef CAS PubMed.
- R. Blasco and F. Castillo, Appl. Environ. Microbiol., 1992, 58, 690–695 CrossRef CAS PubMed.
- B. E. Logan, R. Rossi, A. Ragab and P. E. Saikaly, Nat. Rev. Microbiol., 2019, 17, 307–319 CrossRef CAS PubMed.
- M. F. Siliakus, J. van der Oost and S. W. M. Kengen, Extremophiles, 2017, 21, 651–670 CrossRef CAS PubMed.
- M. Tsuzuki, O. V. Moskvin, M. Kuribayashi, K. Sato, S. Retamal, M. Abo, J. Zeilstra-Ryalls and M. Gomelsky, Appl. Environ. Microbiol., 2011, 77, 7551–7559 CrossRef CAS PubMed.
- M. Grattieri, K. Beaver, E. Gaffney and S. D. Minteer, Faraday Discuss., 2019, 215, 15–25 RSC.
- M. Volpicella, A. Costanza, O. Palumbo, F. Italiano, L. Claudia, A. Placido, E. Picardi, M. Carella, M. Trotta and L. R. Ceci, FEMS Microbiol. Ecol., 2014, 88, 345–357 CrossRef CAS PubMed.
- R. Borghese, M. Malferrari, M. Brucale, L. Ortolani, M. Franchini, S. Rapino, F. Borsetti and D. Zannoni, Bioelectrochemistry, 2020, 133, 107456 CrossRef CAS PubMed.
- M. Paumann, G. Regelsberger, C. Obinger and G. A. Peschek, Biochim. Biophys. Acta, 2005, 1707, 231–253 CrossRef CAS PubMed.
- R. W. Bradley, P. Bombelli, D. J. Lea-Smith and C. J. Howe, Phys. Chem. Chem. Phys., 2013, 15, 13611–13618 RSC.
- J. M. Pisciotta, Y. Zou and I. V. Baskakov, Appl. Microbiol. Biotechnol., 2011, 91, 377–385 CrossRef CAS PubMed.
- A. G. Roberts, M. K. Bowman and D. M. Kramer, Biochemistry, 2004, 43, 7707–7716 CrossRef CAS PubMed.
- J. Yan, G. Kurisu and W. A. Cramer, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 69–74 CrossRef CAS PubMed.
- B. V. Trubitsin, V. V. Ptushenko, O. A. Koksharova, M. D. Mamedov, L. A. Vitukhnovskaya, I. A. Grigor'ev, A. Y. Semenov and A. N. Tikhonov, Biochim. Biophys. Acta, 2005, 1708, 238–249 CrossRef CAS PubMed.
- D. Pils and G. Schmetterer, FEMS Microbiol. Lett., 2001, 203, 217–222 CrossRef CAS PubMed.
- A. J. McCormick, P. Bombelli, A. M. Scott, A. J. Philips, A. G. Smith, A. C. Fisher and C. J. Howe, Energy Environ. Sci., 2011, 4, 4699–4709 RSC.
- G. Saper, D. Kallmann, F. Conzuelo, F. Zhao, T. N. Toth, V. Liveanu, S. Meir, J. Szymanski, A. Aharoni, W. Schuhmann, A. Rothschild, G. Schuster and N. Adir, Nat. Commun., 2018, 9, 2168 CrossRef PubMed.
- Y. A. Gorby, S. Yanina, J. S. McLean, K. M. Rosso, D. Moyles, A. Dohnalkova, T. J. Beveridge, I. S. Chang, B. H. Kim, K. S. Kim, D. E. Culley, S. B. Reed, M. F. Romine, D. A. Saffarini, E. A. Hill, L. Shi, D. A. Elias, D. W. Kennedy, G. Pinchuk, K. Watanabe, S. Ishii, B. Logan, K. H. Nealson and J. K. Fredrickson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11358–11363 CrossRef CAS PubMed.
- H. Rubsam, F. Kirsch, V. Reimann, A. Erban, J. Kopka, M. Hagemann, W. R. Hess and S. Klahn, Environ. Microbiol., 2018, 20, 2757–2768 CrossRef PubMed.
- G. Longatte, H. Y. Fu, O. Buriez, E. Labbe, F. A. Wollman, C. Amatore, F. Rappaport, M. Guille-Collignon and F. Lemaitre, Biophys. Chem., 2015, 205, 1–8 CrossRef CAS PubMed.
- G. Longatte, F. Rappaport, F. A. Wollman, M. Guille-Collignon and F. Lemaitre, Photochem. Photobiol. Sci., 2016, 15, 969–979 RSC.
- G. Longatte, F. Rappaport, F.-A. Wollman, M. Guille-Collignon and F. Lemaître, Electrochim. Acta, 2017, 236, 337–342 CrossRef CAS.
- G. Longatte, A. Sayegh, J. Delacotte, F. Rappaport, F. A. Wollman, M. Guille-Collignon and F. Lemaitre, Chem. Sci., 2018, 9, 8271–8281 RSC.
- L. Beauzamy, J. Delacotte, B. Bailleul, K. Tanaka, F. A. Wollman and F. Lemaitre, Anal. Chem., 2020, 92, 7532–7539 CrossRef CAS PubMed.
- E. Cevik, H. Tombuloglu, I. Anıl, M. Senel, H. Sabit, S. AbdulAzeez, J. F. Borgio and M. Barghouthi, Int. J. Hydrogen Energy, 2020, 45, 11330–11340 CrossRef CAS.
- K. Hasan, S. A. Patil, K. Gorecki, D. Leech, C. Hagerhall and L. Gorton, Bioelectrochemistry, 2013, 93, 30–36 CrossRef CAS PubMed.
- R. D. Milton, D. P. Hickey, S. Abdellaoui, K. Lim, F. Wu, B. Tan and S. D. Minteer, Chem. Sci., 2015, 6, 4867–4875 RSC.
- S. Abdellaoui, R. D. Milton, T. Quah and S. D. Minteer, Chem. Commun., 2016, 52, 1147–1150 RSC.
- K. Hasan, M. Grattieri, T. Wang, R. D. Milton and S. D. Minteer, ACS Energy Lett., 2017, 2, 1947–1951 CrossRef CAS.
- G. Pankratova, D. Pankratov, R. D. Milton, S. D. Minteer and L. Gorton, Adv. Energy Mater., 2019, 9, 1900215 CrossRef.
- M. Grattieri, S. Patterson, J. Copeland, K. Klunder and S. D. Minteer, ChemSusChem, 2020, 13, 230–237 CrossRef CAS PubMed.
- K. Hasan, R. D. Milton, M. Grattieri, T. Wang, M. Stephanz and S. D. Minteer, ACS Catal., 2017, 7, 2257–2265 CrossRef CAS.
- F. Zhao, P. Wang, A. Ruff, V. Hartmann, S. Zacarias, I. A. C. Pereira, M. M. Nowaczyk, M. Rögner, F. Conzuelo and W. Schuhmann, Energy Environ. Sci., 2019, 12, 3133–3143 RSC.
- K. P. Sokol, D. Mersch, V. Hartmann, J. Z. Zhang, M. M. Nowaczyk, M. Rögner, A. Ruff, W. Schuhmann, N. Plumeré and E. Reisner, Energy Environ. Sci., 2016, 9, 3698–3709 RSC.
- J. Tschortner, B. Lai and J. O. Kromer, Front. Microbiol., 2019, 10, 866 CrossRef PubMed.
- A. C. Gonzalez-Aravena, K. Yunus, L. Zhang, B. Norling and A. C. Fisher, RSC Adv., 2018, 8, 20263–20274 RSC.
- K. Hasan, H. Bekir Yildiz, E. Sperling, P. O. Conghaile, M. A. Packer, D. Leech, C. Hagerhall and L. Gorton, Phys. Chem. Chem. Phys., 2014, 16, 24676–24680 RSC.
- Y. Takekuma, N. Ikeda, K. Kawakami, N. Kamiya, M. Nango and M. Nagata, RSC Adv., 2020, 10, 15734–15739 RSC.
- R. Takeuchi, A. Suzuki, K. Sakai, Y. Kitazumi, O. Shirai and K. Kano, Bioelectrochemistry, 2018, 122, 158–163 CrossRef CAS PubMed.
- D. Pankratov, J. Zhao, M. A. Nur, F. Shen, D. Leech, Q. Chi, G. Pankratova and L. Gorton, Electrochim. Acta, 2019, 310, 20–25 CrossRef CAS.
- D. Pankratov, G. Pankratova and L. Gorton, Curr. Opin. Electrochem., 2020, 19, 49–54 CrossRef CAS.
- M. Sawa, A. Fantuzzi, P. Bombelli, C. J. Howe, K. Hellgardt and P. J. Nixon, Nat. Commun., 2017, 8, 1327 CrossRef PubMed.
- N. Sekar, Y. Umasankar and R. P. Ramasamy, Phys. Chem. Chem. Phys., 2014, 16, 7862–7871 RSC.
- E. Cevik, M. Buyukharman and H. B. Yildiz, Biotechnol. Bioeng., 2019, 116, 757–768 CrossRef CAS PubMed.
- K. D. Wolfe, D. Dervishogullari, C. D. Stachurski, J. M. Passantino, G. K. Jennings and D. E. Cliffel, ChemElectroChem, 2019, 7, 596–603 CrossRef.
- J. A. Cornejo, H. Sheng, E. Edri, C. M. Ajo-Franklin and H. Frei, Nat. Commun., 2018, 9, 2263 CrossRef PubMed.
- Y. Su, S. Cestellos-Blanco, J. M. Kim, Y.-X. Shen, Q. Kong, D. Lu, C. Liu, H. Zhang, Y. Cao and P. Yang, Joule, 2020, 4, 800–811 CrossRef CAS.
- F. Li, L. Wang, C. Liu, D. Wu and H. Song, Curr. Opin. Electrochem., 2018, 10, 37–45 CrossRef CAS.
- L. Shi, K. M. Rosso, T. A. Clarke, D. J. Richardson, J. M. Zachara and J. K. Fredrickson, Front. Microbiol., 2012, 3, 50 Search PubMed.
- M. Aklujkar, J. Krushkal, G. DiBartolo, A. Lapidus, M. L. Land and D. R. Lovley, BMC Microbiol., 2009, 9, 109 CrossRef PubMed.
- N. Schuergers, C. Werlang, C. M. Ajo-Franklin and A. A. Boghossian, Energy Environ. Sci., 2017, 10, 1102–1115 RSC.
- H. Jensen, M. Ter Avest, M. Kokish and C. M. Ajo-Franklin, ACS Synth. Biol., 2016, 5, 679–688 CrossRef CAS PubMed.
- N. Sekar, R. Jain, Y. Yan and R. P. Ramasamy, Biotechnol. Bioeng., 2016, 113, 675–679 CrossRef CAS PubMed.
- N. Sekar, J. Wang, Y. Zhou, Y. Fang, Y. Yan and R. P. Ramasamy, Biotechnol. Bioeng., 2018, 115, 1361–1366 CrossRef CAS PubMed.
- L. Su and C. M. Ajo-Franklin, Curr. Opin. Biotechnol., 2019, 57, 66–72 CrossRef CAS PubMed.
- M. A. TerAvest and C. M. Ajo-Franklin, Biotechnol. Bioeng., 2016, 113, 687–697 CrossRef CAS PubMed.
- M. A. TerAvest, T. J. Zajdel and C. M. Ajo-Franklin, ChemElectroChem, 2014, 1, 1874–1879 CrossRef CAS.
- Z. Wu, J. Wang, J. Liu, Y. Wang, C. Bi and X. Zhang, Microb. Cell Fact., 2019, 18, 15 CrossRef PubMed.
- F. Kracke, B. Lai, S. Yu and J. O. Kromer, Metab. Eng., 2018, 45, 109–120 CrossRef CAS PubMed.
- J. S. Sravan, O. Sarkar and S. V. Mohan, Sustainable Energy Fuels, 2020, 4, 700–712 RSC.
- I. Nabiev, A. Rakovich, A. Sukhanova, E. Lukashev, V. Zagidullin, V. Pachenko, Y. P. Rakovich, J. F. Donegan, A. B. Rubin and A. O. Govorov, Angew. Chem., Int. Ed., 2010, 49, 7217–7221 CrossRef PubMed.
- F. Milano, R. R. Tangorra, O. Hassan Omar, R. Ragni, A. Operamolla, A. Agostiano, G. M. Farinola and M. Trotta, Angew. Chem., Int. Ed., 2012, 51, 11019–11023 CrossRef CAS PubMed.
- O. Hassan Omar, S. la Gatta, R. R. Tangorra, F. Milano, R. Ragni, A. Operamolla, R. Argazzi, C. Chiorboli, A. Agostiano, M. Trotta and G. M. Farinola, Bioconjugate Chem., 2016, 27, 1614–1623 CrossRef CAS PubMed.
- R. Ragni, G. Leone, S. la Gatta, G. Rizzo, M. Lo Presti, V. De Leo, F. Milano, M. Trotta and G. M. Farinola, MRS Adv., 2018, 4, 1143–1148 CrossRef.
- P. K. Dutta, S. Lin, A. Loskutov, S. Levenberg, D. Jun, R. Saer, J. T. Beatty, Y. Liu, H. Yan and N. W. Woodbury, J. Am. Chem. Soc., 2014, 136, 4599–4604 CrossRef CAS PubMed.
- P. I. Gordiichuk, D. Rimmerman, A. Paul, D. A. Gautier, A. Gruszka, M. Saller, J. W. de Vries, G. J. Wetzelaer, M. Manca, W. Gomulya, M. Matmor, E. Gloukhikh, M. Loznik, N. Ashkenasy, P. W. Blom, M. Rogner, M. A. Loi, S. Richter and A. Herrmann, Bioconjugate Chem., 2016, 27, 36–41 CrossRef CAS PubMed.
- Y. Yoneda, T. Noji, T. Katayama, N. Mizutani, D. Komori, M. Nango, H. Miyasaka, S. Itoh, Y. Nagasawa and T. Dewa, J. Am. Chem. Soc., 2015, 137, 13121–13129 CrossRef CAS PubMed.
- S. Kaushik, M. K. Sarma and P. Goswami, J. Mater. Chem. A, 2017, 5, 7885–7895 RSC.
- K. J. Grayson, K. M. Faries, X. Huang, P. Qian, P. Dilbeck, E. C. Martin, A. Hitchcock, C. Vasilev, J. M. Yuen, D. M. Niedzwiedzki, G. J. Leggett, D. Holten, C. Kirmaier and C. Neil Hunter, Nat. Commun., 2017, 8, 13972 CrossRef CAS PubMed.
- J. Liu, V. M. Friebe, R. N. Frese and M. R. Jones, Nat. Commun., 2020, 11, 1542 CrossRef CAS PubMed.
- J. Liu, J. Mantell and M. R. Jones, ACS Nano, 2020, 14, 4536–4549 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |