Open Access Article
Open Access ArticleEnzymatic self-assembly of short peptides for cell spheroid formation†‡
Jiaqi
Guo
 ,
Weiyi
Tan
and
Bing
Xu
,
Weiyi
Tan
and
Bing
Xu
 *
*
Department of Chemistry, Brandeis University, 415 South St., Waltham, MA 02453, USA. E-mail: bxu@brandeis.edu
First published on 24th September 2024
Abstract
Cell spheroids, including organoids, serve as a valuable link between in vitro systems and in vivo animal models, offering powerful tools for studying cell biology in a three-dimensional environment. However, existing methods for generating cell spheroids are time consuming or difficult to scale up for large-scale production. Our recent study has revealed that transcytotic peptide assemblies, which transform from nanoparticles to nanofibers by enzymatic reactions, can create an intercellular fibril/gel, accelerating cell spheroid formation from a 2D cell culture or a cell suspension. While this finding presents an alternative approach for generating cell spheroids, the specific structural features required for efficient cell spheroid formation remain unclear. Based on our observation that a phosphotetrapeptide with a biphenyl cap at its N-terminus enables cell spheroid formation, we produced 10 variants of the original peptide. The variants explored modifications to the peptide backbone, length, electronic properties of the biphenyl capping group, and the type of phosphorylated amino acid residue. We then evaluated their ability for inducing cell spheroid formation. Our analysis revealed that, among the tested molecules, peptides with C-terminal phosphotyrosine, low critical micelle concentration, and dephosphorylation-guided nanoparticle to nanofiber morphological transition were the most effective in inducing the formation of cell spheroids. This work represents the first example to correlate the thermodynamic properties (e.g., self-assembling ability) and kinetic behavior (e.g., enzymatic dephosphorylation) of peptides with the efficacy of controlling intercellular interaction, thus offering valuable insights into using enzymatic self-assembly to generate peptide assemblies for biological applications.
Introduction
Cell spheroids,1 which are clusters of cells, can serve as a crucial link between in vitro systems and in vivo animal models, thus offering valuable tools for investigating cell biology within a three-dimensional environment2 and testing drug candidates with the minimal use of animal models.3 Previously, four different approaches have been used to enable cell spheroid formation, such as low-adhesion cell plates to promote cell self-aggregation,4 a hanging drop plate to facilitate cell self-organization,3 a spinner flask or a microgravity bioreactor to drive cells to self-aggregate,5 and a micropatterned pillar for locally enriching cells.6 Although these approaches are capable of enabling cell spheroid formation, they still suffer from several shortcomings. For example, they usually require multiple days for spheroid formation, have difficulty for scaling up, and lack the generality for a wide range of cells. To address these issues, considerable efforts have been made to develop new biomaterials for enabling cell spheroids.7–10In the area of developing new biomaterials, significant progress, over the last two decades, has been made in using peptide assemblies as the scaffolds for hydrogels to mimic the ECM and embed cells for three dimensional cell cultures.11–16 However, most of the hydrogels are made ex situ to embed cells, which differ from an in vivo process where cells generate the ECM in situ to surround themselves. We have unexpectedly found that enzymatic dephosphorylation of peptide assemblies is able to enable two dimensional cell cultures of HS-5 cells to form cell spheroids even when different phosphopeptides were employed.17–20 Our recent study revealed that these phosphopeptide assemblies undergo transcytosis to form fibrils at the intercellular space in situ to promote cell spheroid formation.21 This work suggests that enzyme-instructed peptide assemblies not only mimic ECM biogenesis, but also facilitate ECM fibrillogenesis, such as the fibrogenesis of fibronectin.21 Furthermore, our study indicates that the transcytotic enzyme-triggered nanoparticle-to-fibril shapeshifting of the dynamic continuum is essential for inducing cell spheroids.21 Compared to the current ex situ approach of embedding cells in a gel, this transcytotic creation of in situ intercellular fibrils provides a unique opportunity to allow peptide assemblies to interact with ECM proteins at the beginning of their biogenesis, thus more closely participating the in vivo process of ECM formation. Therefore, it represents an adaptive biomaterial design for better modulating the functionalities of the ECM.16
Although the advancement in the mechanistic understanding of how an enzymatic reaction of phosphopeptide assemblies enables cell spheroid formation, the structural determinant of the peptide assemblies for efficient cell spheroid formation remains largely unexplored. Here, we report the examination of a series of phosphopeptides for enabling the cell spheroids of HS-5 cells. Based on our previous findings, which demonstrated that a phosphotetrapeptide with an N-terminal biphenyl capping motif promotes the formation of cell spheroids,21 we conducted further research to explore the potential of 10 phosphopeptide analogues. These analogues were created by altering various factors, including the peptide backbone, peptide length, electronic properties of the biphenyl capping motif, and substituting phosphorylated amino acid residues. Our objective was to assess their capability to induce the formation of cell spheroids. Our investigation revealed that a low critical micelle concentration (CMC) value, relatively high phosphatase stability, and the nanoparticle-to-nanofiber shapeshifting of the peptide assemblies promote cell spheroid formation. Moreover, the exploration of the structures of the phosphopeptide further supports that the dynamic structural continuum of the peptide assemblies, being controlled by enzymatic reactions, is essential for cell spheroid formation (Scheme 1). This study, together with our work that shows self-assembling ability determining the inhibitory activity of enzyme-instructed self-assembly against cancer cells,22,23 underscores the importance of the enzymatic self-assembling ability of phosphopeptides in controlling the dynamics of peptide assemblies for various biological applications.
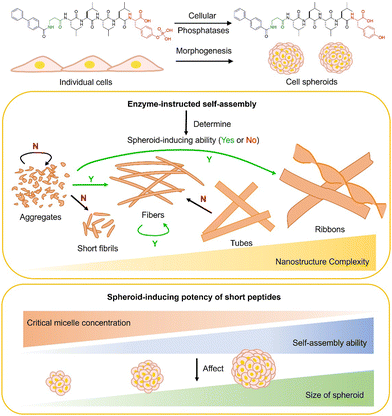 | ||
| Scheme 1 Short peptide assemblies induce cell spheroids and the correlation between enzyme-instructed self-assembly (EISA) and spheroid-inducing effects. | ||
Results and discussion
Molecular design
Fig. 1 shows the chemical structures of the phosphopeptides. We used biphenyl-4-carboxylic acid (BP) as the N-terminal capping motif due to its effectiveness in promoting self-assembly of peptides into filamentous structures.24 Beta-alanine was used as a spacer to provide moderate flexibility between the aromatic motif and the peptide backbone. Unlike the previously reported diphenylalanine motif as the self-assembling backbone,21 we used a tetraleucine motif (L4), which has been demonstrated to be effective in generating ribbon-like nanostructures for inhibiting induced pluripotent stem cells25 and immunosuppressive cancer cells.26,27 For the C-terminus, we connected a phosphotyrosine (pY) to render the peptide molecule phosphatase-responsive. This design strategy leads to the key molecule, BP-L4pY.In addition, we designed 9 analogues that vary in distinct modules and investigated their effects on cell morphogenesis. For example, by removing one or two leucine residues, we synthesized BP-L3pY and BP-L2pY to determine the roles of the peptide length. These two molecules also serve to interrogate the importance of hydrophobic and rigid amino acid side chains in controlling cell spheroid formation. Moreover, we substituted pY with phosphoserine (pS) to investigate the generality of phospho-amino acids in inducing cell spheroids. For the hydrophobic capping motif, we replaced the BP motif with benzoic acid (Bz) to generate Bz-L4pY with weaker self-assembling ability. Furthermore, we introduced various para-substitutions on the BP motif with different sizes and distinct electronic effects. For example, chloride (Cl), nitrogen dioxide (NO2), and trifluoromethyl (CF3) groups have electron-withdrawing effects, with the latter two being much stronger than Cl, while hydroxyl (OH) and methyl (CH3) groups are electron-donating. In terms of sizes, CF3, with fluorine substitutions and its tetrahedral geometry, tends to be larger than the rest. This larger size provides a greater hydrophobic volume, which is significant in self-assembly processes. The aforementioned modifications create a series of phosphopeptides with distinct sizes and electronic effects of the hydrophobic capping motifs, molecule length, and phosphorylated amino acids. By examining their responsiveness to phosphatases and emergent nanostructures through enzyme-instructed self-assembly (EISA),28–34 we can correlate these structural variations with both EISA and the efficacy of cell spheroid induction.
After synthesizing the phosphotyrosine and phosphoserine amino acid building blocks, we connected them via solid-phase peptide synthesis (SPPS) to generate the phosphopeptide molecules (Scheme S1, ESI‡). We used high-performance liquid chromatography (HPLC) for purification and liquid chromatography-mass spectrometry (LC-MS) to verify their identities (Fig. S1–S10, ESI‡).
Concentration-dependent spheroid formation of BP-L4pY
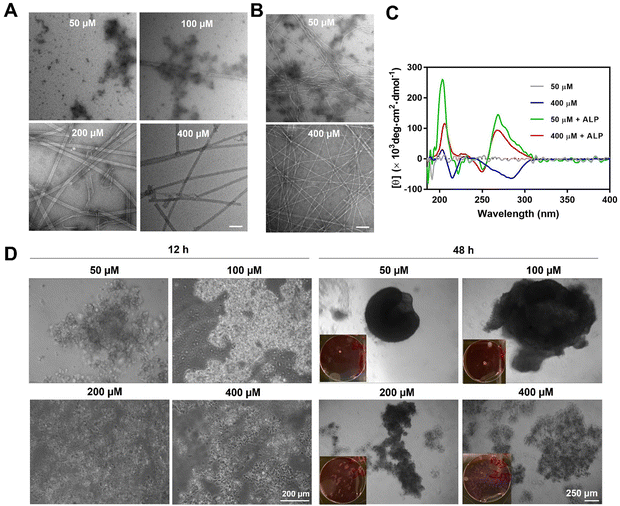
Supramolecular self-assembly of peptides depends on intermolecular noncovalent interactions, like hydrogen bonding and aromatic–aromatic interactions, to maintain nanostructures. This process is highly context-dependent and dynamic, which responds to parameters such as pH, temperature, and ionic strength. Under the influence of these factors, the self-assembly of peptide molecules depends on the concentration of the peptide itself, known as critical micelle concentrations (CMCs). For above CMCs, the peptide molecule spontaneously organizes into specific nanostructures, while, for below CMCs, no significant nanostructure would form. In this work, the rigid aromatic BP motif, together with the hydrophobic tetraleucine side chain, provides BP-L4pY with good self-assembling ability. This self-assembly is concentration-dependent, transitioning from aggregates to tubes as concentration increases (Fig. 2A). ALP-guided dephosphorylation transforms these nanostructures into fibers (Fig. 2B). The self-assembly morphologies correspond to their circular dichroism (CD) spectra, as BP-L4pY at 50 μM, which display aggregates, does not show significant secondary structures (Fig. 2C). In contrast, at 400 μM, BP-L4pY displays negative beta-sheet signals at 210 nm, and negatively induced CD signals from 250 to 300 nm, likely due to the specific arrangement of BP motifs in the nanotubes. Upon dephosphorylation, BP-L4pY at both concentrations display beta-sheets and induced CD signals, but with positive values. These results indicate changes in intermolecular interactions, which alter their arrangement profiles and affect helical arrangements.Since the changes in intermolecular interactions influence physical properties like self-assembly morphologies and helicity, we sought to investigate how these changes affect cellular activities, especially in cell aggregation, in response to concentration changes. After confirming the cell compatibility of BP-L4pY (Fig. S11, ESI‡), we incubated it with HS-5, a human fibroblast cell line proven to be an effective model for examining the morphogenic effects of peptide assemblies.17–19,21 The morphogenic effects of BP-L4pY are also concentration-dependent, as aggregates of BP-L4pY (50 and 100 μM) promote cell aggregation into spheroids, while nanotubes of BP-L4pY (200 and 400 μM) induce loosely packed cell aggregates or sheets (Fig. 2D). The peptide assemblies induced fast spheroid generation in both suspended and adherent cells, with initial aggregation occurring within 12 h and compaction into large spheroids (>500 μm) from 48 to 96 h (Fig. 2D and Fig. S12, ESI‡). We further explored the generality of BP-L4pY's morphogenic effects across different cell lines, including HeLa, SJSA-1, and OVSAHO, confirming its capability in inducing cell aggregation (Fig. S13–S15, ESI‡). These results indicate that the initial nanotube morphology of BP-L4pY impedes spheroid formation, while particular aggregates promote it.
Leucine promotes self-assembly to generate spheroids
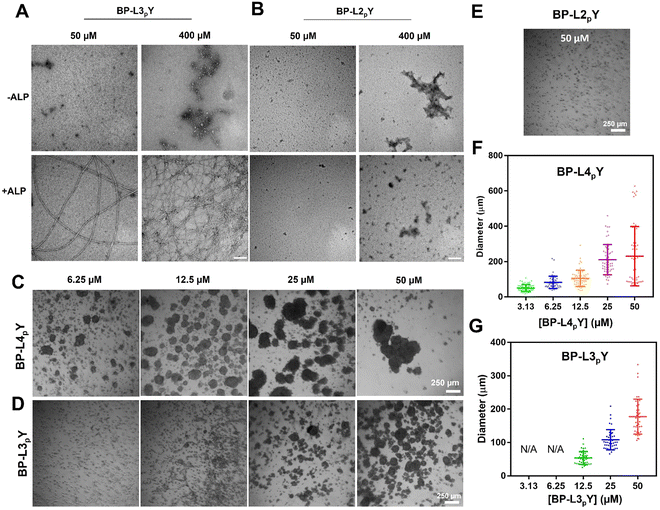
By removing one or two leucine motifs, we created BP-L3pY or BP-L2pY to explore the effects of decreased side chain stiffness, hydrophobicity, and molecule length on spheroid formation. We first examined their ALP-guided self-assembly at the nanoscale. TEM images reveal that BP-L3pY forms aggregates at both low (50 μM) and high (400 μM) concentrations, which transform into nanofibers upon ALP addition (Fig. 3A). The absence of nanotubes at 400 μM indicates reduced self-assembly ability of BP-L3pY. In contrast, BP-L2pY forms aggregates in the presence or absence of ALP, at both concentrations (Fig. 3B), indicating further reduced self-assembly potential. When incubated with HS-5 cells, BP-L4pY efficiently induces spheroids at concentrations ranging from 6.25 to 50 μM (Fig. 3C and Fig. S16, ESI‡). 3D rendering of Z-stack images verifies the spherical morphology and multi-cellular characteristics of the spheroids (Fig. S17, ESI‡). However, due to the inherent limitations of confocal laser scanning microscopy, the signal intensity from deeper sections (typically beyond 80 μm in our case) is too weak to be detected, which makes it difficult to accurately determine cell numbers based on nuclei counting. Since cell numbers can be estimated based on the spheroid volume,35 we estimated that spheroids with diameters ranging from 200 to 300 μm consist of approximately 500–2000 cells, and spheroids of 400–500 μm have around 5000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. The microtubules within the spheroids show a more organized, oriented distribution at the periphery, while exhibiting a random arrangement in the centre (Fig. S18, ESI‡), suggesting spheroid compaction.
000 cells. The microtubules within the spheroids show a more organized, oriented distribution at the periphery, while exhibiting a random arrangement in the centre (Fig. S18, ESI‡), suggesting spheroid compaction.
In contrast, BP-L3pY requires a minimum concentration of 25 μM to generate spheroids (Fig. 3D). BP-L2pY, which lacks morphological transformation upon dephosphorylation, fails to induce any spheroids (Fig. 3E). Quantification of the spheroid diameter shows a positive correlation between the peptide concentration and the spheroid size, with BP-L4pY being more potent than BP-L3pY in terms of lower effective concentrations and larger average and maximum diameters of spheroids under the same concentration (Fig. 3F and G).
Enzymatic self-assembly of BP-L4pY analogues
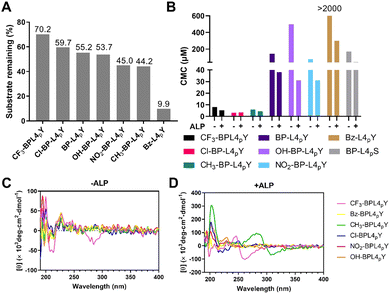
To examine the enzymatic self-assembly of BP-L4pY analogues with N- or C-terminal modifications, we assessed several properties upon enzymatic cleavage, such as enzymatic conversion, CMCs, and CD. Since phosphoserine differs in reactivity to ALP compared to phosphotyrosine, we focused solely on analogues with C-terminal phosphotyrosine to study the phosphatase responsiveness. Modifications in the hydrophobic capping motif change the extent of dephosphorylation in response to ALP. For example, a Bz substitution renders Bz-L4pY more susceptible to ALP (Fig. 4A). In contrast, substitutions on the BP motif have varied effects, as halide substitutions like CF3 and Cl stabilize the phosphopeptides against ALP, while NO2 and CH3 groups make them more labile. OH substitution has negligible effects on enzymatic dephosphorylation. We reasoned that these structural changes may impact the presentation of phosphotyrosine to ALP, potentially through changing the peptide conformation or affecting their self-assembly. In addition to ALP, the phosphopeptides are also susceptible to prostatic acid phosphatase (PAP) (Fig. S19, ESI‡). We then sought to determine the CMCs of analogues in response to dephosphorylation (Fig. S20–S27, ESI‡). Compared with BP-L4pY, analogues with CF3, Cl, and CH3 substitutions lower the CMC values, NO2 substitution and C-terminal pS modification have little impact, and OH and Bz substitutions increase CMC values (Fig. 4B). These results suggest the interplay between the aromatic capping motif and self-assembly ability, where the number of phenyl rings determines the magnitude of CMCs, and modifications to the aromatic motif adjust the value by several tenths of micromoles.To determine the secondary structures of analogues, we conducted CD spectroscopy. The CF3 substitution leads to signals around 210 nm and 260–300 nm, indicating beta-sheets and induced CD, respectively (Fig. 4C), which represent self-assembly before dephosphorylation. After dephosphorylation, ALP converts analogues with CH3, Cl, OH, and NO2 substitutions into beta-sheet-rich structures, with CH3 analogues displaying an induced CD band. However, the CF3 analog shows opposite trends of beta-sheets and induced CD bands, suggesting distinct helical arrangements of its self-assembly (Fig. 4D).
The ECM consists of networks of proteins, with fibrillar ECM proteins being a key topological feature. The fibrous structures of peptide assemblies increase the surface area available for cell–peptide interactions and serve as scaffolds that may mimic the native ECM. These cell–peptide interactions potentially modulate cell adhesion behaviours, thereby promoting cell aggregation and spheroid formation. Moreover, the formation of short peptide assemblies through EISA, due to their small sizes and dynamic properties, allows direct interactions with cells. Therefore, the fibres can engage in specific interactions with cell surface receptors or cell adhesion molecules, like integrins, fibronectin and collagen, which are crucial for regulating cell aggregation and spheroid formation. To evaluate the self-assembly of analogues at the nanoscale, we took their TEM images before and after ALP treatment. As shown in Fig. 5, BP-L4pS, which lacks the phenyl side chain of tyrosine, forms aggregates that self-assemble into sparse and short fibrils after dephosphorylation. Bz-L4pY fails to generate any nanostructures with or without ALP incubation, probably due to the insufficient aromatic interactions provided by the Bz motif. Analogues with electron-withdrawing groups, such as Cl-BP-L4pY and CF3-BP-L4pY, display the mixture of aggregates and fibers and retain nanofibers after ALP addition. In contrast, NO2-BP-L4pY only initiates the formation of an entangling fibrous network after dephosphorylation. Analogues with electron-donating groups show unique morphologies: CH3-BP-L4pY self-assembles into a mixture of nanofibers and aggregates in PBS, which, after enzymatic dephosphorylation, transits into a mixture of nanofibers and twisted ribbons. OH-BP-L4pY undergoes enzymatic morphological transformation from aggregates to ribbons. These distinct morphologies suggest the subtlety of the EISA process, where minor changes in intermolecular interactions result in remarkably different morphologies of nanostructures.
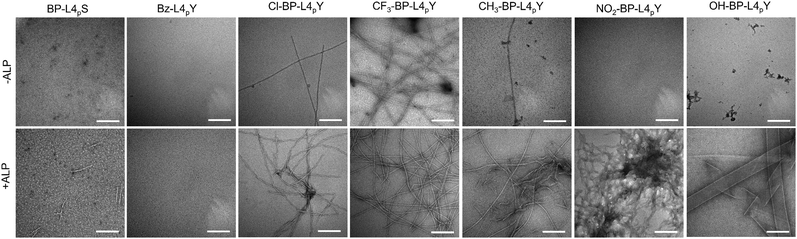 | ||
| Fig. 5 TEM images of analogues of BP-L4pY (50 μM) in PBS in the absence or presence of ALP (0.1 U mL−1) for 24 h. Scale bar: 200 nm. | ||
Morphogenic effects of BP-L4pY analogues
To evaluate the spheroid-inducing ability of analogues, we incubated them with HS-5 cells at a working concentration of 15 μM. It was chosen because BP-L4pY induces moderate sizes of spheroids at this concentration, and therefore changes in spheroid-inducing ability would result in more evident changes in spheroid sizes. Results show that BP-L4pS and Bz-L4pY are ineffective in inducing spheroid formation, with the majority of cells remaining as 2D cell sheets (Fig. 6A). Conversely, OH-BP-L4pY and NO2-BP-L4pY induce small spheroids, while CH3-BP-L4pY induces spheroids comparable in size to those induced by BP-L4pY. CF3-BP-L4pY and Cl-BP-L4pY, on the other hand, lead to the formation of larger spheroids. Quantification of their diameters reveals a distribution profile, where the size variation among large spheroids exhibits more variability, accompanied by larger mean values and standard deviations (Fig. 6B). This suggests that both the mean diameter and the size of the largest spheroid are valuable parameters for evaluating the spheroid inducing potency of these analogues.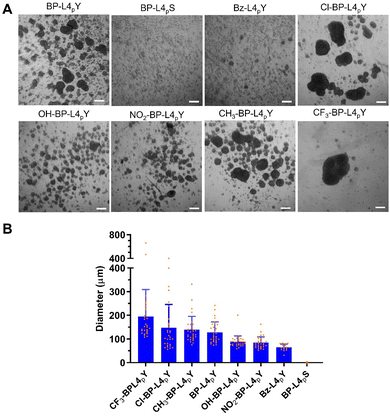 | ||
| Fig. 6 (A) Phase contrast images of suspended HS-5 cells treated with BP-L4pY or analogues (15 μM) for 48 h. Scale bar: 200 μm. (B) Quantification of the diameters of spheroids in (A). | ||
Additionally, we conducted the in vitro reconstitution assay36 of incubating CF3-BPL4pY and ALP with the rhodamine–fibronectin (rFN) protein. The peptide nanofibers induce morphological transformation of fibronectin, a crucial component in modulating cell adhesion, which likely accounts for the observed cell aggregation (Fig. S28, ESI‡).
Correlation between self-assembly and cell morphogenesis
According to the aforementioned results, we summarized the relationships between the cell morphogenic effects and the EISA process of phosphopeptides, primarily focusing on the nanoscale morphology and self-assembly ability. Table 1 summarizes the morphological transformation of phosphopeptides triggered by enzymatic dephosphorylation. Our findings suggest that mature nanotube structures, that is, without dynamic shapeshifting, are unable to induce spheroid formation, and structures like aggregates and short fibrils resulting from dephosphorylation also fail to induce spheroids. In contrast, transitions form aggregates to fibers or ribbons, that is, the dynamic continuum of peptide assemblies facilitates spheroid formation, and the presence of fibers mixed with aggregates or ribbons also supports spheroid formation. Therefore, the morphological shapeshifting from a relatively simple morphology like aggregates to a more extended and ordered morphology like fibers and ribbons is crucial in spheroid formation.| Phosphopeptides | Beofre dephosphorylation | After dephosphorylation | Spheroid formation |
|---|---|---|---|
| BP-L4pY | Aggregate | Fiber | Y |
| Tube | Fiber | N | |
| BP-L3pY | Aggregate | Fiber | Y |
| BP-L2pY | Aggregate | Aggregate | N |
| BP-L4pS | Aggregate | Short fibril | N |
| Bz-L4pY | Aggregate | Aggregate | N |
| Cl-BP-L4pY | Fiber | Fiber | Y |
| CF3-BP-L4pY | Fiber | Fiber | Y |
| CH3-BP-L4pY | Fiber + aggregate | Fiber + ribbon | Y |
| NO2-BP-L4pY | Aggregate | Fiber | Y |
| OH-BP-L4pY | Aggregate | Ribbon | Y |
The morphologies of EISA-generated nanostructures significantly impact cell morphogenic effects. However, the sizes of the resulting spheroids vary among analogues, indicating distinct spheroid-inducing potency. We hypothesized that the self-assembly ability plays a role in this variation. To verify this hypothesis, we calculated the free energy of micellization (−ΔGmicellization) based on the CMC value of peptides, representing self-assembly ability, and correlated these values with their respective mean spheroid diameters. The self-assembly ability of analogues ranks as follows: Cl-BP-L4pY > CH3-BP-L4pY > CF3-BP-L4pY > NO2-BP-L4pY > OH-BP-L4pY > BP-L4pY > BP-L4pS > Bz-L4pY. The first three analogues bear close values around 30 kJ mol−1, while the latter three are around 25 kJ mol−1 (Fig. 7). Although a direct positive correlation with the spheroid diameter is not there, analogues with a −ΔGmicellization value around 30 kJ mol−1 tend to generate larger spheroids than those around 25 kJ mol−1. The discrepancy between −ΔGmicellization values and spheroid diameters suggests that the variation in morphology among analogues via EISA may influence this relationship. BP-L4pS and Bz-L4pY, which fail to induce compact spheroids, are not suitable for correlation with −ΔGmicellization.
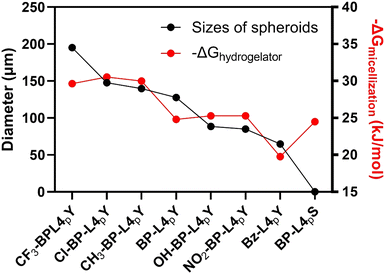 | ||
| Fig. 7 Correlation between the self-assembling ability (−ΔGmicellization) of peptides after incubation with ALP and spheroid-inducing potency (diameter) of BP-L4pY and analogues in HS-5 cells. | ||
Conclusions
In summary, we examined a series of phosphopeptides aimed at enabling the formation of cell spheroids in HS-5 cells. We found that the self-assembly ability and the dynamic shapeshifting of the peptide assemblies are critical to induce cell spheroid formation. Specifically, by using relatively high or low concentrations of BP-L4pY, we created different morphologies of self-assembled phosphopeptides at the nanoscale. Results show that the preformed or mature nanotube structures inhibit the spheroid formation, while aggregates of the phosphopeptides facilitate this process because they undergo dynamic morphological changes. By modifying the N-terminal aromatic motifs, C-terminal phospho-amino acids, or the tetraleucine backbone, we assessed the spheroid-inducing capabilities of different analogues. Results show that the enzymatic shapeshifting from aggregates to fibers or ribbons promotes spheroid formation. Moreover, the spheroid-inducing potency is to some extent positively correlated with the free energy of micellization, particularly when the difference is larger than 5 kJ mol−1. This work, using a biphenyl motif and an L-peptide sequence, exemplifies the use of supramolecular assemblies of L-peptides for spheroid creation with minimum working concentrations below 10 μM, which represents a ten-fold decrease compared to NBD-ffspy. Additionally, the tetraleucine motif, along with the previously reported diphenylalanine, proved effective in generating nanofibers that promote spheroid generation. While this study highlights the potential of enzymatic self-assembly in biomedical applications, several issues remain to be investigated in future research. For example, future studies could determine the structural changes of peptide nanostructures before and after dephosphorylation to elucidate the presentation of C-terminal amino acids to the assembly peripheries among analogues. This is crucial for understanding the interactions between artificial biomaterials and cells. Additionally, the study could benefit from investigating the mechanisms of peptide analogues, especially as the co-assemblies of the phosphopeptides and the peptides, influencing the remodelling of ECM proteins.Author contributions
B. X. and J. G. conceived the study. J. G., under the supervision of B. X., performed molecular design, data acquisition, analysis, and generated images. W. T, under the supervision of B. X., performed the LC-MS analysis. J. G. and B. X. wrote the manuscript.Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the NIH CA142746 and the NSF MRSEC DMR-2011846.Notes and references
- R. M. Sutherland, J. A. McCredie and W. R. Inch, J. Natl. Cancer Inst., 1971, 46, 113–120 CAS.
- S. Guven, P. Chen, F. Inci, S. Tasoglu, B. Erkmen and U. Demirci, Trends Biotechnol., 2015, 33, 269–279 CrossRef CAS PubMed.
- Y.-C. Tung, A. Y. Hsiao, S. G. Allen, Y.-S. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC.
- M. Vinci, S. Gowan, F. Boxall, L. Patterson and M. Zimmermann, BMC Biol., 2012, 10, 29 CrossRef CAS PubMed.
- B. S. Youn, A. Sen, L. A. Behie, A. Girgis-Gabardo and J. A. Hassell, Biotechnol. Prog., 2006, 22, 801–810 CrossRef CAS PubMed.
- Y. Yoshii, A. Waki, K. Yoshida, A. Kakezuka, M. Kobayashi, H. Namiki, Y. Kuroda, Y. Kiyono, H. Yoshii and T. Furukawa, Biomaterials, 2011, 32, 6052–6058 CrossRef CAS PubMed.
- S. Ai, H. Li, H. Zheng, J. Liu, J. Gao, J. Liu, Q. Chen and Z. Yang, Sci. China Mater., 2022, 65, 1655–1661 CrossRef CAS.
- M. Diba, S. Spaans, S. I. S. Hendrikse, M. M. C. Bastings, M. J. G. Schotman, J. F. van Sprang, D. J. Wu, F. J. M. Hoeben, H. M. Janssen and P. Y. W. Dankers, Adv. Mater., 2021, 33, 2008111 CrossRef CAS PubMed.
- L. R. de Mello, V. Carrascosa, E. Rebelato, M. A. Juliano, I. W. Hamley, V. Castelletto, S. V. Vassiliades, W. A. Alves, C. R. Nakaie and E. R. da Silva, Langmuir, 2022, 38, 3434–3445 CrossRef CAS PubMed.
- Y. Zhang, S. Ai, Z. Yu, L. Wang, H. Tao, B. Wang, D. Kong, Z. Yang and Y. Wang, Adv. Funct. Mater., 2024, 2314607 CrossRef CAS.
- M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS PubMed.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. F. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Adv. Mater., 2006, 18, 611–614 CrossRef CAS.
- D. J. Smith, G. A. Brat, S. H. Medina, D. Tong, Y. Huang, J. Grahammer, G. J. Furtmuller, B. C. Oh, K. J. Nagy-Smith, P. Walczak, G. Brandacher and J. P. Schneider, Nat. Nanotechnol., 2016, 11, 95–102 CrossRef CAS PubMed.
- Z. Alvarez, A. N. Kolberg-Edelbrock, I. R. Sasselli, J. A. Ortega, R. Qiu, Z. Syrgiannis, P. A. Mirau, F. Chen, S. M. Chin, S. Weigand, E. Kiskinis and S. I. Stupp, Science, 2021, 374, 848–856 CrossRef CAS PubMed.
- S. M. Winkler, M. R. Harrison and P. B. Messersmith, Biomater. Sci., 2019, 7, 3092–3109 RSC.
- C. Ligorio and A. Mata, Nat. Rev. Bioeng., 2023, 1, 518–536 CrossRef PubMed.
- H. Wang, J. Shi, Z. Feng, R. Zhou, S. Wang, A. A. Rodal and B. Xu, Angew. Chem., Int. Ed., 2017, 56, 16297–16301 CrossRef CAS PubMed.
- H. Wang, Z. Feng and B. Xu, Angew. Chem., Int. Ed., 2019, 58, 5567–5571 CrossRef CAS PubMed.
- H. Wang, Z. Feng and B. Xu, J. Am. Chem. Soc., 2019, 141, 7271–7274 CrossRef CAS PubMed.
- H. Wang, Z. Feng and B. Xu, ChemBioChem, 2019, 20, 2442–2446 CrossRef CAS PubMed.
- J. Guo, F. Wang, Y. Huang, H. He, W. Tan, M. Yi, E. H. Egelman and B. Xu, Nat. Nanotechnol., 2023, 18, 1094–1104 CrossRef CAS PubMed.
- Z. Feng, H. Wang, X. Chen and B. Xu, J. Am. Chem. Soc., 2017, 139, 15377–15384 CrossRef CAS PubMed.
- Z. Feng, H. Wang, R. Zhou, J. Li and B. Xu, J. Am. Chem. Soc., 2017, 139, 3950–3953 CrossRef CAS PubMed.
- M. Yi, F. Wang, W. Tan, J.-T. Hsieh, E. H. Egelman and B. Xu, J. Am. Chem. Soc., 2022, 144, 13055–13059 CrossRef CAS PubMed.
- S. Liu, Q. Zhang, A. N. Shy, M. Yi, H. He, S. Lu and B. Xu, J. Am. Chem. Soc., 2021, 143, 15852–15862 CrossRef CAS PubMed.
- S. Liu, Q. Zhang, H. He, M. Yi, W. Tan, J. Guo and B. Xu, Angew. Chem., Int. Ed., 2022, 61, e202210568 CrossRef CAS PubMed.
- Z. Feng, H. Wang, M. Yi, C. Y. Lo, A. Sallee, J. T. Hsieh and B. Xu, Pept. Sci., 2020, 112, e24123 CrossRef CAS PubMed.
- J. Zhou and B. Xu, Bioconjugate Chem., 2015, 26, 987–999 CrossRef CAS PubMed.
- K. Morita, K. Nishimura, S. Yamamoto, N. Shimizu, T. Yashiro, R. Kawabata, T. Aoi, A. Tamura and T. Maruyama, JACS Au, 2022, 2, 2023–2028 CrossRef CAS PubMed.
- R. A. Pires, Y. M. Abul-Haija, D. S. Costa, R. Novoa-Carballal, R. L. Reis, R. V. Ulijn and I. Pashkuleva, J. Am. Chem. Soc., 2015, 137, 576–579 CrossRef CAS PubMed.
- G. Liang, H. Ren and J. Rao, Nat. Chem., 2010, 2, 54–60 CrossRef CAS PubMed.
- X. Zhang, J. Wang, Y. Zhang, Z. Yang, J. Gao and Z. Gu, Chem. Soc. Rev., 2023, 52, 8126–8164 RSC.
- J. Gao, J. Zhan and Z. Yang, Adv. Mater., 2020, 32, e1805798 CrossRef PubMed.
- Z. Huang, Q. Yao, S. Wei, J. Chen and Y. Gao, Curr. Med. Chem., 2019, 26, 1351–1365 CrossRef CAS PubMed.
- D. P. Ivanov, T. L. Parker, D. A. Walker, C. Alexander, M. B. Ashford, P. R. Gellert and M. C. Garnett, PLoS One, 2014, 9, e103817 CrossRef PubMed.
- J. Guo, A. Zia, Q. Qiu, M. Norton, K. Qiu, J. Usuba, Z. Liu, M. Yi, S. T. Rich-New, M. Hagan, S. Fraden, G. D. Han, J. Diao, F. Wang and B. Xu, J. Am. Chem. Soc., 2024, 146, 26102–26112 CrossRef CAS PubMed.
Footnotes |
| † In dedication to Prof. George Whitesides on the occasion of his 85th birthday. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01154f |
| This journal is © The Royal Society of Chemistry 2024 |