Open Access Article
Open Access ArticleEmerging mycotoxins and preventive strategies related to gut microbiota changes: probiotics, prebiotics, and postbiotics – a systematic review†
Álvaro
Lázaro
,
Pilar
Vila-Donat
 * and
Lara
Manyes
* and
Lara
Manyes

Biotech Agrifood Lab, Faculty of Pharmacy and Food Sciences, University of Valencia, 46100 Burjassot, València, Spain. E-mail: pilar.vila@uv.es
First published on 4th September 2024
Abstract
Recent research has focused on the involvement of the gut microbiota in various diseases, where probiotics, prebiotics, synbiotics, and postbiotics (PPSP) exert beneficial effects through modulation of the microbiome. This systematic review aims to provide insight into the interplay among emerging mycotoxins, gut microbiota, and PPSP. The review was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. In this review, unregulated yet highly recurrent mycotoxins are classified as emerging mycotoxins. The most frequently observed mycotoxins included those from the Fusarium genus—enniatins (n = 11) and beauvericin (n = 11)—and the Alternaria genus—alternariol monomethyl ether, altertoxin, and tentoxin (n = 10). Among probiotics, the most studied genera were Lactobacillus, Bifidobacterium, and the yeast Saccharomyces cerevisiae. Inulin and cellulose were the most found prebiotics. Data on synbiotics and postbiotics are scarce. Studies have shown that both the gut microbiota and PPSP can detoxify and mitigate the harmful effects of emerging mycotoxins. PPSP not only reduced mycotoxin bioaccessibility, but also counteracted their detrimental effects by activating health-promoting pathways such as short-chain fatty acid production, genoprotection, and reduction of oxidative stress. However, both quantitative and qualitative data remain limited, indicating a need for further in vivo and long-term studies. The formulation of PPSP as functional foods, feeds, or nutraceuticals should be considered a preventive strategy against the toxicity of emerging mycotoxins, for which, there is no established regulatory framework.
1. Introduction
Mycotoxins are secondary metabolites produced by various species of fungi, primarily belonging to the genera Aspergillus, Penicillium, and Fusarium. They are estimated to be present in up to 80% of food and feed. Animal feed contamination is particularly concerning, as in developed countries, up to 70% of cereal harvest—a primary source of mycotoxins—is utilized in the daily diet of animals. Mycotoxin regulation for feed is less restrictive than for food. These toxins pose significant public health risks due to their widespread occurrence and their toxic properties in both animals and humans. They can induce immunotoxicity, neurotoxicity, hepatotoxicity, nephrotoxicity, reproductive and developmental toxicity, and carcinogenicity.1Currently, the legislative framework on maximum limits on mycotoxins is established by the recently updated Commission Regulation (EU) 2023/915 of April 25, for aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), aflatoxin G2 (AFG2), aflatoxin M1 (AFM1), ochratoxin A (OTA), patulin (PAT), deoxynivalenol (DON), zearalenone (ZEA), fumonisin B1 (FB1), fumonisin B2 (FB2) and citrinin (CIT), the last one being applied only in food supplements.2 In addition, there are other recent recommendations for emerging or unregulated mycotoxins such as alternariol (AOH), alternariol monomethyl ether (AME), tenuazonic acid (TeA) and the trichothecenes T-2 and HT-2 toxins (T-2 and HT-2).3,4
The term “emerging mycotoxins” was first introduced in 2008 and initially referred primarily to Fusarium metabolites such as fusaproliferin (FP), beauvericin (BEA), enniatins (ENs), and moniliformin (MON). However, more recent scientific publications have defined emerging mycotoxins as those that are neither routinely detected nor regulated by legislation, despite growing evidence of their prevalence. This work is focused on the 22 emerging mycotoxins included in Fig. 1 and 2 that have been recognized for over 20 years, continue to be widespread, and are not yet subject to regulatory oversight, even though their toxic effects are increasingly documented. The mycotoxins covered in this review are: enniatin A (ENA), enniatin (ENB), enniatin A1 (ENA1), enniatin B1 (ENB1), BEA, fusaric acid (FA), MON, fusarenon X (FX), aurofusarin (AUR), FP, diacetoxyscirpenol (DAS), altuene (ALT), AME, TeA, altertoxin (ATX), tentoxin (TEN), CIT, sterigmatocystin (STE), neosolaniol (NEO), citreoviridin, rugulosin and phomopsin.
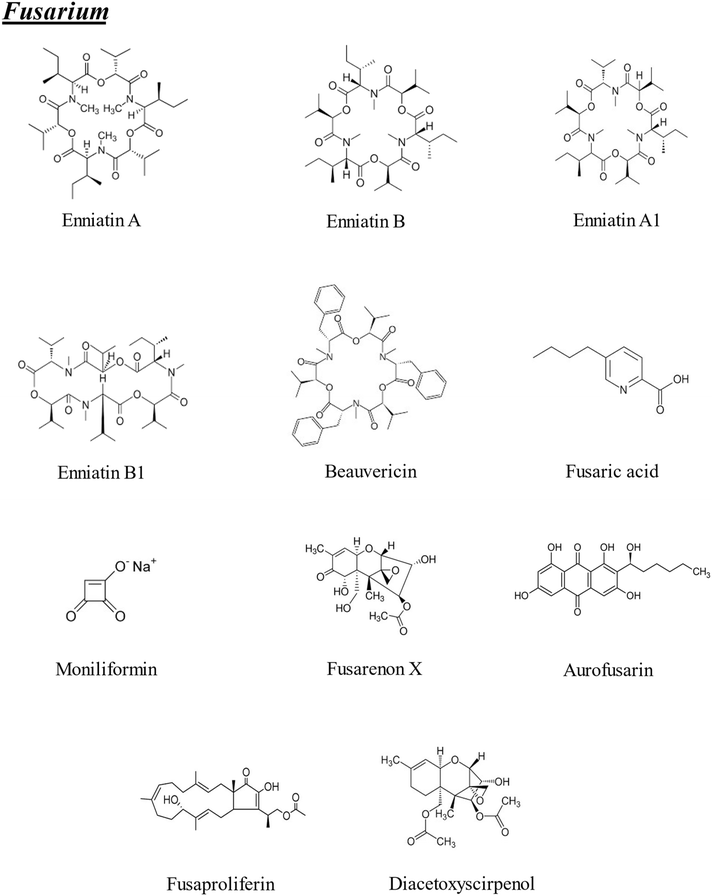 | ||
| Fig. 1 Chemical structures of the emerging mycotoxins of Fusarium included in the bibliographic search (n = 11). | ||
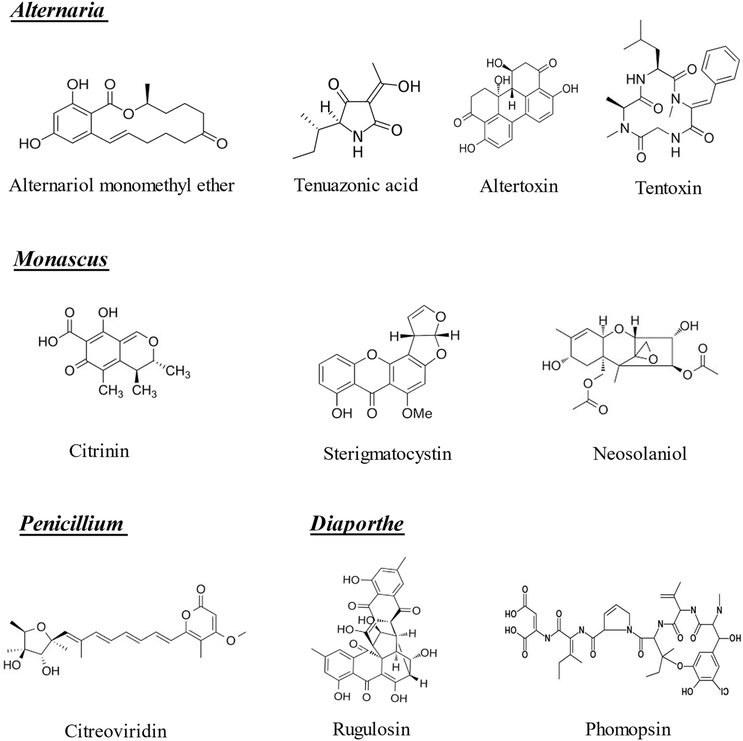 | ||
| Fig. 2 Chemical structures of the emerging mycotoxins of Alternaria, Monascus, Penicillium and Diaporthe included in the bibliographic search (n = 10). | ||
To better understand the relevance of these emerging mycotoxins, the occurrence and co-occurrence of most of them have been studied by Mihalache et al. (2023),5 who evidenced 38 different combinations in foodstuff (maize, rice, wheat, and flours). These combinations changed from binary mixtures (AME + AOH or AME + TeA) to include 12 different emerging mycotoxins (AOH + AME + TeA + TEN + ENA1 + ENB + ENB + BEA + MON + DAS + NIV + STE). Furthermore, the rising of new data regarding their presence in foodstuff has been highly relevant to enhance multiple risk assessment methodologies, developing future evidence-based regulations and public health.5 As shown in Table 1, these emerging mycotoxins are of significant concern due to their involvement in various severe health issues, including cell apoptosis, chromosomal abnormalities, acute cardiac distress, genotoxicity, hemorrhages in legs, skull, and feet, liver injury, cardiovascular collapse, chlorosis, embryotoxicity, teratogenesis, cancer, liver cirrhosis, hepatocellular carcinoma, Shoshin-kakke (acute cardiac beriberi), fatty degeneration, liver cell necrosis and cell death. Although not all emerging mycotoxins were considered in this research, the spectra covered are expected to encompass most of them.
| Mycotoxin | Food sources | Sample | Dose administration | Exposure time | Target organ | Toxicological mechanism | Damage | Ref |
|---|---|---|---|---|---|---|---|---|
| Deoxyribonucleic acid (ADN), alternariol monomethyl ether (AME), adenosine triphosphate (ATP), altertoxin (ATX), aurofusarin (AUR), beauvericin (BEA), body weight (b.w.), citrinin (CIT), diacetoxyscirpenol (DAS), enniatin A (ENA), enniatin A1 (ENA1), enniatin B (ENB), enniatin B (ENB), fusaproliferin (FP), fusarenon X (FX), fusaric acid (FA), moniliformin (MON), neosolaniol (NEO), nicotinamide adenine dinucleotide phosphate oxidase (NADPH), NF-E2-related factor 2/antioxidant responsive element (Nrf2/ARE), not reported (NR), reactive oxygen species (ROS), sterigmatocystin (STE), tenuazonic acid (TeA), and tentoxin (TEN). | ||||||||
| Fusarium | ||||||||
| ENA | Wheat, barley, maize, and rice | Caco-2 and SH-SY5Y cells | 0.66–66.70 μg mL−1 | 24–48 h | Intestine, liver | Generation of ROS and mitochondrial depolarization | Apoptosis | 8 and 9 |
| ENB | Wheat, barley, maize, and rice | Caco-2 and SH-SY5Y cells | 0.63–63.60 μg mL−1 | 24–48 h | Intestine, liver | Generation of ROS and mitochondrial depolarization | Apoptosis | 8 and 9 |
| ENA1 | Wheat, barley, maize, and rice | Caco-2 and SH-SY5Y cells | 0.66–66.70 μg mL−1 | 24–48 h | Intestine, liver | Generation of ROS and NADPH oxidase activation | Apoptosis | 8 and 9 |
| ENB1 | Wheat, barley, maize, and rice | Caco-2 and SH-SY5Y cells | 0.63–63.60 μg mL−1 | 24–48 h | Intestine, liver | Generation of ROS and NADPH oxidase activation | Apoptosis | 9 |
| BEA | Wheat, maize, nuts, and coffee | Caco-2 cells | 1.17–19.6 μg mL−1 | 24–72 h | Intestine, liver | Generation of ROS | Apoptosis | 10 |
| FA | Wheat, barley, corn, and rice | Allium cepa L. bulbs and HepG2 cells | 0.18–1.8 μg mL−1 | 24–168 h | Liver | Inhibition of cytochrome oxidase, decrease in ATP synthesis, change in the electrochemical gradient in the plasma membrane and increase in electrolyte loss | Chromosomal abnormalities | 11 and 12 |
| MON | Maize, wheat | Male Sprague-Dawley rats | 500 μg mL−1 | 28 days | Heart | Pyruvate substitution and carbohydrate metabolism interference | Death and acute cardiac distress | 13 |
| FX | Wheat, barley, and cereal-based products | Male and female mice | 3–15 mg per kg b.w. | 48 h | Liver, kidneys, spleen | Inhibition of protein and DNA synthesis | Apoptosis | 14 and 15 |
| AUR | Maize, barley, oats, and wheat | CHO-K1 cell line | 0.57–5.7 μg mL−1 | 48 h | Intestine | Enhanced levels of p53 protein | Genotoxicity | 16 |
| FP | Maize | Chicken embryo | 0.45–2.22 μg mL−1 | 21 days | Chicken embryos | Reduction in cell viability | Hemorrhages on the surfaces of legs, skull, and feet | 17 and 18 |
| DAS | Wheat, corn, rice, and maize | Newborn chickens and Wistar rats | 1–3 mg per kg bw | 6–24 h | Liver, gallbladder, and small intestine | DNA synthesis and protein biosynthesis inhibitor and heterophil extracellular trap release | Liver injury | 19 and 20 |
| Apicidin | NR | HepG2 cells | 0.06–62.3 μg mL−1 | 24 h | Liver | Cytotoxicity | Cell death | 21 |
| Alternaria | ||||||||
| AME | Wheat and its derivatives, rapeseed oil and peas | Male Sprague-Dawley rats | 7.35 μg per kg bw per day | 28 days | Liver, kidneys, spleen | DNA adducts and production of ROS | Genotoxicity | 21 |
| TeA | Tomatoes, apples, beer, and cereal foods | Mice | 238 μg per kg per day | 56 days | Liver, kidneys | Generation of ROS | Cardiovascular collapse and gastrointestinal hemorrhage | 22 |
| ATX | Cereals, oilseeds, fruits, and vegetables | Male Sprague-Dawley rats | 5.51 μg per kg w per day | 28 days | Liver, kidneys, spleen | Nuclear factor erythroid-derived 2-like 2/antioxidant response element (Nrf2/ARE) | Genotoxicity | 16 and 23 |
| TEN | Rice | Male Sprague Dawley rats | 0.4–41.45 pg mL−1 | 10 min | Liver | ATP synthesis reduction | Chlorosis | 24 and 25 |
| Monascus | ||||||||
| CIT | Cheese, sake, and soy sauce | ProTox-II in humans | 105 mg per kg weight | 24 h | Kidneys | Inhibition of malate dehydrogenase, glutamate dehydrogenase and ATP synthase | Genotoxic, embryotoxic, teratogenic, carcinogenic | 26 |
| Aspergillus | ||||||||
| STE | Maize and peanuts | HepG2 cells | 0.16–2.27 μg mL−1 | 24–48 h | Liver, kidneys | Generation of ROS | Liver cirrhosis hepatocellular carcinoma | 27 |
| NEO | Maize, wheat, and oats | Porcine Langerhans cells | 143.4 mg mL−1 | 24 h | Leydig cells | Decrease in the ATP content, overproduction of ROS, reduction in mitochondrial membrane potential | Cellular apoptosis | 28 |
| Emodin | NR | HepG2 cells | 0.027–27 μg mL−1 | 24 h | Liver | Cytotoxicity | Cell death | 21 |
| Penicillium | ||||||||
| Citreoviridin | Maize, pecan nuts, and wheat products | Mice | 1–10 mg L−1 | 15 days | Heart | Inhibition of mitochondrial ATPase | Shoshin-kakke (acute cardiac beriberi) | 29 |
| Rugulosin | Rice | Mice | 67 mg per kg b.w. | 21 days | Liver | Inhibition in vivo and in vitro of DNA replication, transcription, and reparation | Fatty degeneration and liver cell necrosis | 30 and 31 |
| Cyclopiazonic acid | Corn, peanuts and cheese | Caco-2 cells | 0.014–42 ng mL−1 | 24–48 h | Intestine | Cytotoxicity | Cell death | 33 and 34 |
| Diaporthe | ||||||||
| Phomopsin | Lupins | Sheep | 0.5 mg kg−1 | 4 days | Liver | Cell cycle arrest | Cell death | 32 and 33 |
| Phoma | ||||||||
| Cytochalasin B | Potatoes | EL4 | 2 mg mL−1 | 4 h | Blood | DNA fragmentation | Apoptosis | 36 |
| Cytochalasin D | Potatoes | Mice | 0.3 mg kg−1 | 17 days | Neural tube | Teratogenicity | Fetal death | 37 |
| Cytochalasin E | Potatoes | Brine shrimp | 0.01–5 μg mL−1 | 16–24 h | NR | Cytotoxicity | Cell death | 38 |
Furthermore, climate change is anticipated to become a major environmental concern in the 21st century. Higher temperatures combined with extreme precipitation or prolonged droughts increase the stress experienced by plants, making all plant-based foods more susceptible to fungal infection and mycotoxin contamination. Fusarium mycotoxins are expected to shift towards Northern Europe, while Aspergillus species will primarily affect southern and central Europe. Indeed, it is estimated that in Europe, over the next 50–100 years, mycotoxins will become a significant concern. This includes not only the most common mycotoxins, such as aflatoxins and OTA, but also emerging Fusarium mycotoxins like ENs and BEA.6,7
Recent research has focused on the involvement of the gut microbiota in a considerable number of diseases such as anxiety, depression, inflammatory bowel disease, dementia, Parkinson's disease, diabetes, obesity, and cancer.39–43 To understand how important the gut microbiota is, it should be said that there are around 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 human genes, whereas bacterial genes are more than 4
600 human genes, whereas bacterial genes are more than 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in our body. What is more, the bacterial density in the colon is around 1011–1012 mL−1, being the highest density in any ecosystem on Earth.44 On the other hand, there are some factors, such as diet,45 stress,46 exercise47 and medication,48 having a notable impact on gut microbiota modulation. Among these factors, this systematic review focuses on emerging mycotoxins and probiotics, prebiotics, synbiotics and postbiotics (PPSP); PPSP is a term recently coined by Li et al. (2021).49
000 in our body. What is more, the bacterial density in the colon is around 1011–1012 mL−1, being the highest density in any ecosystem on Earth.44 On the other hand, there are some factors, such as diet,45 stress,46 exercise47 and medication,48 having a notable impact on gut microbiota modulation. Among these factors, this systematic review focuses on emerging mycotoxins and probiotics, prebiotics, synbiotics and postbiotics (PPSP); PPSP is a term recently coined by Li et al. (2021).49
PPSP has shown to regulate the abundances of some intestinal bacteria such as Akkermansia, Bacteroidetes, Blautia, Bifidobacteria, Bifidobacterium (B.) and gut microbial metabolites, such as lactic acid, acetate, butyrate and propionate, while suppressing the bile acid pools and decreasing the production of trimethylamine N-oxide and lipopolysaccharides.49 The significance of PPSP, mainly probiotics and prebiotics, gave rise to the International Scientific Association for Probiotics and Prebiotics (ISAPP) in 2002. ISAPP is a non-profit organization that works to advance the science of probiotics, prebiotics and related substances, such as synbiotics, postbiotics and fermented foods,50 contributing to a better understanding of how biotics are connected to health and disease. Regarding definitions, prebiotics are described as “a substrate that is selectively utilized by host microorganisms, conferring a health benefit”.51 Traditionally, they have been defined as “nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacterial species already resident in the colon, thus attempting to improve host health”.52 Certainly, probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host”.52 Recently, there has been an emerging trend to investigate synbiotics and postbiotics. Synbiotics are considered “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host”, whereas postbiotics are described as “a preparation of inanimate microorganisms and/or their components that confers a health benefit on the host”.53,54
It has been shown that there are three main strategies for alleviating the harm caused by mycotoxins transmitted by microorganisms: (I) directly degrading mycotoxins in food, thanks to microbial pretreatment, and decreasing toxin intake; (II) by boosting probiotic colonization, the creation of bacterial-toxin complexes is suppressed, consequently inhibiting toxin absorption in the intestinal tract; and (III) by modulating the intestinal microecology with probiotics or prebiotics, enhancing the intestinal barrier, remodeling the intestinal microflora, improving intestinal toxicity, and decreasing toxin penetration.55 Previously, the interactions between the gut microbiota and regulated mycotoxins6,56 and the detoxifying capacity of PPSP on some mycotoxins57–61 have been studied. Notwithstanding, the interplay among emerging mycotoxins, gut microbiota and PPSP still remains a framework to systemize.
The aim of this systematic review is to shed light on the potential of PPSP in mitigating the harmful effects of emerging mycotoxins. In addition, it aims to enhance our understanding of the reciprocal relationship between emerging mycotoxins and the gut microbiota, elucidating how emerging mycotoxins can influence the gut microbiota and vice versa. Furthermore, this review aims to contribute to future research endeavors by proposing the development of new functional foods and/or nutraceuticals as a strategy for mitigating toxicity associated with emerging mycotoxins.
2. Materials and methods
2.1 Search strategy
The scientific rigor of this systematic review and the minimization of potential bias were ensured through adherence to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement protocol for screening titles, abstracts, and full texts.62 A comprehensive literature search was conducted in November–December 2023, utilizing three databases (PubMed, Web of Science, and Scopus). The search encompassed the period from January 2012 to December 2023, aiming to include recent data on emerging mycotoxins within the context of the gut microbiota and PPSP. In addition, other relevant terms, such as “bacteria”, were considered in combination with “in vitro” or “in vivo” to capture additional relevant informationThe systematic review was conducted using the following search strings:
(1) Refined research of Pubmed, Web of Science and Scopus. Period: all years.
(“Enniatins” OR “Enniatin A” OR “Enniatin B” OR “Enniatin A1” OR “Beauvericin” OR “Fusaric acid” OR “Moniliformin” OR “Fusarenon X” OR “Aurofusarin” OR “Fusaproliferin” OR “Diacetoxyscirpenol” OR “Alternariol monomethyl ether” OR “Tenuazonic acid” OR “Altertoxin” OR “Tentoxin” OR “Citritin” OR “Sterigmatocystin” OR “Neosolaniol” OR “Citreoviridin” OR “Rugulosin” OR “Phomopsin”) AND (“bacteria”) AND (“in vitro” OR “in vivo”).
(2) Refined research of Pubmed, Web of Science and Scopus. Period: 2012–2023.
(“Enniatins” OR “Enniatin A” OR “Enniatin B” OR “Enniatin A1” OR “Beauvericin” OR “Fusaric acid” OR “Moniliformin” OR “Fusarenon X” OR “Aurofusarin” OR “Fusaproliferin” OR “Diacetoxyscirpenol” OR “Alternariol monomethyl ether” OR “Tenuazonic acid” OR “Altertoxin” OR “Tentoxin” OR “Citritin” OR “Sterigmatocystin” OR “Neosolaniol” OR “Citreoviridin” OR “Rugulosin” OR “Phomopsin”) AND (“microbiota” OR “prebiotics” OR “probiotics” OR “postbiotics”).
The criteria to include non-regulated mycotoxins in the list was based on Mihalache et al. (2023) and the reports issued by the Spanish Agency for Food Safety and Nutrition (AESAN).5,35 It is well known that there are other non-regulated metabolites not been considered when conducting the research. Notwithstanding, it was supposed that the spectra covered would be enough to include all the non-regulated mycotoxins.
2.2 Systematic review process
Among the 811 reports found through the research, 317 were retrieved from PubMed, 295 from Web of Science and 199 from Scopus. After an initial analysis, a total of 401 duplicate records were identified and removed. In addition, during the screening of titles and abstracts, another 381 records were excluded from the systematic review because of different reasons: 124 articles were discarded because they only focused on regulated mycotoxins (AFB1, OTA, fumonisins, etc.); 130 were rejected since they included emerging mycotoxins, but they were not assessed in combination with PPSP and 127 were discarded because they analyzed PPSP, but no emerging mycotoxins. Finally, a total of 29 were considered appropriate for inclusion in the present systematic review, assessed and classified based on: the gut microbiota, probiotics, prebiotics, synbiotics and postbiotics (Fig. 3). The selected articles are key to comprehend more deeply the role of the emerging mycotoxins in their combination with microbiota and PPSP.2.3 Data conversion
All data from the studies discussed in this review were converted to μg kg−1, mg kg−1, ng mL−1, μg mL−1 or mg L−1.3. Results
3.1 Mycotoxins and the gut microbiota
The bidirectionality between regulated mycotoxins and the gut microbiota has been previously described in mice, pigs, rats, broilers, turkey, and ducks. DON has revealed to up-regulate the relative abundance of pathogens highly related to chronic intestinal diseases at different taxonomical levels (phylum, family, and genus). OTA has disrupted the structure and diversity of gut microbial communities and AFB1 to alter gut microbiota-dependent organic acid metabolism. Meanwhile, some gut microbiota strains, such as probiotics Lactobacillus plantarum and Bacillus shackletonii, have also shown mitigation effects by metabolizing, binding, and eliminating these regulated mycotoxins.6,7,56The metabolism of Alternaria mycotoxins such as AOH, AME and ALT by the microbiota present in feces from three human volunteers has also been studied. As a control for the activity of the fecal microbiota, the isoflavone daidzein was incubated with the fecal cultures and was transformed to its expected metabolites. In contrast, no metabolites of AOH, AME and ALT were detected in the fecal cultures from the same volunteers, showing that the gut microbiota did not possess the capacity to metabolize these compounds. Interestingly, AOH was noted to bind non-covalently to the surface of bacteria where the type and composition of this bacterial cell surface played a notable role in the magnitude of this binding.66 An Alternaria alternata extract (5 μg mL−1), containing among others, AOH, AME, altertoxin-II (ATX-II), stemphyltoxin III (STTX-III) and alterperylenol (ALP), all of which are known as genotoxic, was used to assess the impact of short-term fecal incubation on the DNA-damaging effects. It was shown that microorganisms suppressed the DNA-strand-breaking potential promoted by the applied extract. Besides, in fresh fecal samples, even before anaerobic incubation, the concentrations of most of the tested mycotoxins (AOH, AME, ATX-II, STTX-III, and ALP) were lower in those containing microorganisms, especially fecal matter, implying that microbes were engaged in mycotoxin detoxification.67
The ability of the gastrointestinal microflora of pig and chicken to metabolize NIV was studied by Hedman and Pettersson (1997).64 On one hand, before pigs were fed with NIV, this mycotoxin was not de-epoxidated or metabolized in any way. However, when the pigs received either 2.5 or 5 μg g−1 NIV for one week, 97.1% of NIV in feces from five of six pigs was in the de-epoxy-NIV form, while the concentration of unaltered NIV in feces was 0.08 μg g−1. On the other hand, after three weeks on the NIV feed, even the pig that after one week could not metabolize NIV, was now able to form de-epoxides. In the feces from this pig, 90% of NIV was as de-epoxy-NIV, the concentration of it being NIV 0.17 μg g−1. Regarding chicken, it was revealed that de-epoxy-NIV was not detected in any samples of feces collected after three weeks of feeding with either 2.5 or 5 μg g−1 NIV. Interestingly, another unidentified metabolite of NIV was found in all feces samples, except from one bird fed with 5 μg g−1 NIV.64 All the data relating emerging mycotoxins and gut microbiota are shown in Fig. 4 and Table 2. These studies were primarily conducted in vitro using feces from humans, pigs, and chickens, or in cow rumen fluids. The most frequently encountered emerging mycotoxins were ENs, BEA, NIV, and Alternaria mycotoxins (AOH, AME, ALTX-I, and ALT).
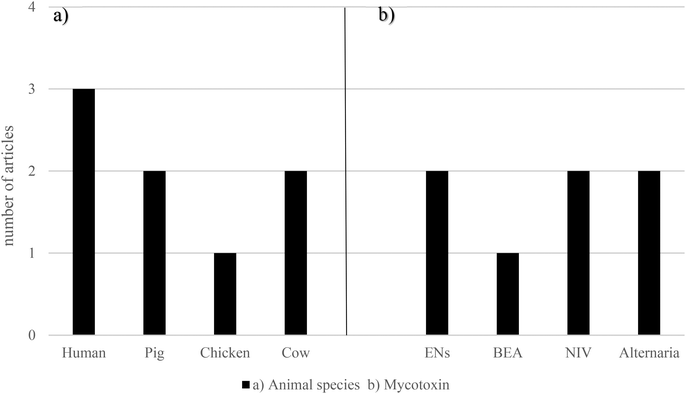 | ||
| Fig. 4 Graphical bars of the gut microbiota publications (n = 6) according to (a) the animal species and (b) emerging mycotoxins studied. Beauvericin – BEA, enniatins – ENs and nivalenol – NIV. | ||
| Mycotoxin | Sample | Dose administration | Exposure time | Experimental assay | Results | Ref. |
|---|---|---|---|---|---|---|
| In vitro | ||||||
| ENB | Cow rumen fluid | 1 mg kg−1 ENB | 0–48 h | In vitro rumen simulation | ENB degradation up to 72% after 48 h and NIV completely detoxified at normal pH (6.8) after 24 h of incubation. In both cases, the degradation was dependent of the microbial activity | 63 |
| NIV | 60 mg kg−1 NIV | |||||
| NIV | Cow rumen fluid | 2 μg mL−1 | 48 h | Anaerobic fecal culture | 78–82% of incubated NIV was transformed into de-epoxy-NIV by the gastrointestinal microbiota in the fluid rumen | 64 |
| Anaerobic incubation of NIV with cow rumen fluid produced de-epoxide at a high proportion | ||||||
| DAS | Human feces | 0.7 ng mL−1 | 2–72 h | Fecal batch culture | DAS was deacetylated into 15-MAS and SCP as an additional metabolite by human fecal microbiota | 65 |
| AOH | Human feces | 12.9 μg mL−1 | 72 h | Anaerobic fecal culture | AOH, AME and ALT were not metabolized by the gut bacteria. AOH bound non-covalently to the bacteria in function according to the type and composition of its surface | 66 |
| AME | ||||||
| ALT | ||||||
| AOH | Human feces | 50 μg mL−1 | 48 h | Fecal anaerobic incubation | Microorganisms suppressed the DNA-strand-breaking potential promoted by the applied extract. AOH, AME, ATX-II, STTX-III, and ALP were lower in the samples containing microorganisms, especially fecal matter, implying that microbes were engaged in detoxification | 67 |
| AME | ||||||
| ATX-II | ||||||
| STTX-III | ||||||
| ALP | ||||||
| Mycotoxin | Sample | Mycotoxin dose administration | Exposure time | Experimental assay | Results | Ref. |
|---|---|---|---|---|---|---|
| Alternariol (AOH), alternariol methyl ether (AME), alterperylenol (ALP), altertoxin I (ATX-I), alterperylenol (ALP), altuene (ALT), beauvericin (BEA), diacetoxyscirpenol (DAS), deoxynivalenol (DON), enniatin B (ENB), Lactobacillus (L), nivalenol (NIV), scirpentriol (SCP), stemphyltoxin III (STTX-III), and 15-monoacetylscirpenol (15-MAS); both SCP and 15-MAS are the products of DAS deacetylation. | ||||||
| In vivo | ||||||
| ENB | Weaning piglets’ feces | 1.345 mg kg−1 (ENB) | 14 days | Metagenomic analysis of the fecal microbiome | L. amylovorus was reduced only in the BEA-fed group, but not in the BEA + DON group | 68 |
| The relative abundance of L. reuteri was increased in the BEA group and, to a lesser extent, in the BEA + DON group compared to the control and the DON group | ||||||
| ENB1 | 1.830 mg kg−1 (ENB1) | |||||
| BEA | 2.570–3.578 mg kg−1 (BEA) | |||||
| NIV | Pig and chicken feces | 2.5–5 mg kg−1 | 1–3 weeks | Chemical analysis of feces | After one week, 97.1% of NIV was in the de-epoxy-NIV form in the feces from five of six pigs. After three weeks, this pig was now able to form de-epoxides (90%) | 64 |
| No de-epoxide of NIV, but another unidentified metabolite was found in the feces from chicken fed with 2.5 or 5 μg g−1 NIV for three weeks | ||||||
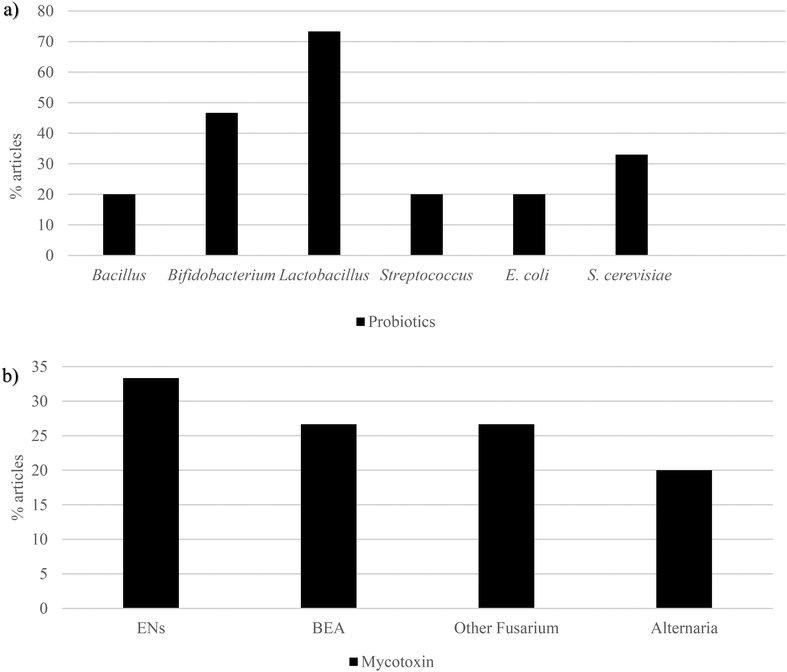
3.2 Emerging mycotoxins and probiotics
Studies in vitro and in vivo have shown the ability of probiotic cultures to bind to and/or even degrade many toxic substances, mitigating their toxicity.64,69 Not only has it been reported that single strain or combination of lactic acid bacteria (LAB) are very useful for removal of heavy metals (copper, lead, cadmium, chromium and arsenic) and cyanotoxins (microcystin-LR, -RR and -LF), but also regarding mycotoxins (AFB1, AFB2, AFM1, aflatoxin M2 (AFM2), AFG1, AFG2, PAT, OTA, DON, FB1 and FB2), 3-acetyldeoxynivalenol, NIV, HT-2 and T-2, ZEA and its derivatives, etc., and their harmful effects.58,61,70Apart from ENs, the interaction between BEA and probiotics is also well documented. Nine yeast strains of S. cerevisiae (LO9, YE-2, YE5, YE-6, YE-4, A34, A17, A42 and A08) have shown a BEA mean degradation of 86.2%. The highest degradation activity (98.8%) was due to S. cerevisiae LO-9, whereas the lowest (39.7%) by S. cerevisiae LS100.75 The protective effect of L. acidophilus against BEA toxicity has been assessed on a Caco-2 cell line. It was revealed that, in the presence of BEA, L. acidophilus significantly increased cell viability at 12 h and 24 h.76L. paracasei CECT 277, L. casei CECT 4180, L. rhamnosus CECT 278T, L. plantarum CECT 220, L. ruminis CECT 4061T, L. casei casei CECT 277, B. breve CCT 4839T, B. adolescentis CECT 5781T, B. bifidum CECT 870T, B. longum CECT 4551, Corynebacterium vitaeruminis CECT 537, Eubacterium crispatus CECT 4840 and S. cerevisiae CECT 1324 have proven to exert a notable impact on BEA bioaccessibility. The highest bioaccessibility decrease was carried out by B. longum showing 45.4%, whereas the lowest data were revealed with the strain L. rhamnosus (27.5%).77 Other 13 bacterial strains belonging to the gastrointestinal tract (B. longum, B. bifidum, B. breve, B. adolescentis, L. rhamnosus, L. casei–casei, L. plantarum, Eubacterium crispatus, Salmonella fecalis, Salmonella thermophilus, L. ruminis, L. casei and L. animalis) were fermented under anaerobic conditions in the liquid medium of De Man–Rogosa–Sharpe agar for 4 h, 12 h, 16 h, 24 h and 48 h at 37 °C. It was revealed that BEA reduction ranged from 66.5% to 83.1%, being the highest decrease caused by the strain L. casei casei.78
Other Fusarium mycotoxins have been studied in combination with probiotics showing similar results. Escherichia coli (ATCC 25922), Salmonella enterica serovar typhimurium 3389–1 (DT12), Staphylococcus aureus (ATCC 29213), L. acidophilus (DSMZ 20079), L. acidophilus (20079), L. salivarius (20555), L. sobrius (16698), B. longum subsp. longum Reuter (20219) and B. breve (20213) received a fast digital time-lapse microscopic method to assess AUR, BEA, ENs and FA antibacterial effects after 6 h of cultivation. The most potent compound was AUR against L. acidophilus, whereas BEA, ENs and FA were characterized by having weak antibiotic effects.79Burkholderia ambifaria T16, a naturally beneficial bacterium with proven biocontrol properties but potential pathogenic risk, has shown to inhibit the growth of several Fusarium species and possess the unique ability to degrade FA as a carbon, nitrogen and energy source, as well as the capacity to detoxify FA in barley seedlings.80 Different S. cerevisiae, Lactobacillus and Bacillus strains were tested for their ability to degrade different mycotoxins. Among the tested mycotoxins, DAS was proven to possess an inhibitory effect on the growth of almost all Lactobacillus and Bacillus strains, whereas no effect on yeast strains could be observed.81 The human microbiome characteristic bacteria Prevotella copri has revealed to deacetylate 51.3% of DAS to 15-MAS after 48 h.82
The same trends have also been reported with Alternaria mycotoxins in Ge et al. (2017).83L. brevis 20023 (LAB-20023) cells have revealed to adsorb TeA from aqueous solution, demonstrating that polysaccharides and protein were important components of the LAB cell wall and were involved in TeA removal. The lowest and highest absorption rates were 12.5% and 90.1%, respectively.83 The effects of a complex extract of Alternaria mycotoxins (AME, ALT, TeA, TEN, ATX-I, ATX-II, ALP, STTX-III, altenusin and altersetin (AST)) were studied in combination with some typical bacterial strains of the gut microbiome such as Bacteroides caccae, Bacteroides eggerthii, Bacteroides thetaiotaomicron, Bacteroides vulgatus, Parabacteroides distasonis, E. coli, Clostridium innocuum, Alistipes finegoldii, Alistipes timonensis, Ruminococcus bicirculans, Akkermansia muciniphila, L. hominis, B. longum and B. sp. The results showed that both Gram types were able to adsorb AOH, AME, and AST, and it was seen that Gram-negative bacteria had higher adsorptive abilities. Notwithstanding, this absorption was not observed for neither ATX-I nor ALP. Interestingly, the mycotoxin tendency to accumulate within bacterial pellets, especially in those of Gram-negative strains, was connected to their lipophilic profile.84
| Mycotoxin | Probiotic | Dose administration | Exposure time | Experimental assay | Results | Ref. | |
|---|---|---|---|---|---|---|---|
| In vitro | |||||||
| ENA | L. johnsonii CECT 289 | B. breve CECT 4839 T | Not reported | 4 h | Dynamic gastrointestinal in vitro digestion | ENA, ENA1, ENB and ENB1 mean bioaccessibility reduced ranging from 21% to 31.2% compared to the control (33.4%–39.6%) | 71 |
| ENA1 | L. rhamnosus CECT | B. adolescentis CECT 5781 T | |||||
| ENB | L. plantarum CECT 220 | B. bifidum CECT 870 T | |||||
| ENB1 | L. reuteri CECT 725 | B. longum CECT 4551 | |||||
| L. casei CECT 47 | |||||||
| ENA | B. longum | L. ruminis | 0.2–20.000 ng per disc | 24 h | Antimicrobial analyses carried out the disc-diffusion method | ENA1 and ENB were the most actives against B. adolescentis 5871, S. thermophilus, 2 strains of Lactobacillus and 2 other strains of Bifidobacterium | 72 |
| ENA1 | B. bifidum | Streptococcus thermophilus | |||||
| ENA2 | B. breve | 22 strains of S. cerevisiae | |||||
| ENB | B. adolescentis | 9 strains of Bacillus subtilis. | |||||
| ENB1 | L. rhamnosus | ||||||
| ENB4 | L. casei–casei | ||||||
| L. plantarum | |||||||
| L. paracasei | |||||||
| EN1 | Bacillus subtilis 168 trpC2 | 2–8 μg mL−1 | 3 weeks | Cocultivation media | ENA1 and ENB1 inhibited the growth of B. subtilis and were also active against Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecalis | 73 | |
| ENB1 | Staphylococcus aureus ATCC 29213 | ||||||
| Streptococcus pneumoniae ATCC 49619 | |||||||
| E. coli ATCC 25922 | |||||||
| Staphylococcus aureus 25697 | |||||||
| Enterococcus faecalis UW 268 | |||||||
| Pseudomonas aeruginosa B 63230 | |||||||
| ENA | B. longum | Streptococcus thermophilus | Not reported | 48 h | In vitro degradation under an anaerobic atmosphere | ENA, ENA1, ENB and ENB1 degradation ranging from 5.2% to 99.5% | 74 |
| ENA1 | B. bifidum | L. ruminis | |||||
| ENB | B. breve | L. casei | |||||
| ENB1 | B. adolescentes | 22 strains of S. cerevisiae | |||||
| L. rhamnosus | |||||||
| L. casei–casei | |||||||
| BEA | S. cerevisiae LO9 | S. cerevisiae YE-6 | 5 mg L−1; 5 mg kg−1 | 48–72 h | Biological degradations under aerobic conditions in the liquid medium of PDB | BEA mean degradation of 86.2%. The highest degradation activity (98.8%) was due to the strain of S. cerevisiae LO-9, whereas the lowest activity was due to LS100 (39.7%) | 75 |
| S. cerevisiae YE-2 | S. cerevisiae A34 | ||||||
| S. cerevisiae YE5 | S. cerevisiae A17 | ||||||
| S. cerevisiae YE-6 | S. cerevisiae A42 | ||||||
| S. cerevisiae YE-4 | S. cerevisiae A08 | ||||||
| S. cerevisiae YE5 | |||||||
| BEA | L. acidophilus | 0.8–15.7 μg mL−1 | 24–72 h | Cell viability assay | L. acidophilus significantly increased cell viability at 12 h and 24 h in the presence of BEA | 76 | |
| BEA | L. paracasei CECT 277 | B. adolescentis CECT 5781T | 5–25 mg kg−1 | 48 h | In vitro dynamic digestion model | BEA bioaccessibility highest reduction (45.4%) was carried out by B. longum, whereas the lowest bioaccessibility data were revealed with the strain of L. rhamnosus, with a 27.5% | 77 |
| L. casei CECT 4180 | B. bifidum CECT 870T | ||||||
| L. rhamnosus CECT 278T | B. Longum CECT 4551 | ||||||
| L. plantarum CECT 220 | Corynebacterium vitaeruminis CECT 537 | ||||||
| L. ruminis CECT 4061T | Eubacterium crispatus CECT 4840 | ||||||
| L. casei casei CECT 277 | S. cerevisinas CECT 1324 | ||||||
| B. breve CECT 4839T | |||||||
| BEA | B. longum | Eubacterium crispatus, Salmonella fecalis and Salmonella thermophilus | 5 mg L−1 | 4–48 h | Fermentations in the liquid medium of MRS | BEA reduction ranged from 66.5% to 83.1%, the highest decrease value being that caused by the strain of L. casei–casei | 78 |
| B. bifidum | L. ruminis | ||||||
| B. breve | L. casei | ||||||
| B. adolescentis | L. animalis | ||||||
| L. rhamnosus | |||||||
| L. casei–casei | |||||||
| L. plantarum | |||||||
| AUR | E. coli (ATCC 25922) | L. acidophilus (DSMZ 20079) | 1.1–146.1 μg mL−1 | 6 h | Fast digital time-lapse microscopic method | The most potent compound was AUR against L. acidophilus. Meanwhile, BEA, ENs and FA were characterized by having weak antibiotic effects | 79 |
| ENs | Salmonella enterica serovar typhimurium 3389-1 (DT12) | L. acidophilus (20079) | 1.4–174.6 μg mL−1 | ||||
| BEA | Staphylococcus aureus (ATCC 29213) | L. salivarius (20555) | 1.6–200.7 μg mL−1 | ||||
| FA | L. sobrius (16698) | 0.2–29.7 μg mL−1 | |||||
| B. longum subsp. longum Reuter (20219) | |||||||
| B. breve (20213) | |||||||
| FA | Burkholderia ambifaria T16 | 0.06 mg mL−1 | 72 h | In vitro detoxification | Burkholderia ambifaria has been shown to inhibit the growth of several Fusarium species and possess the unique ability to use the mycotoxin FA as the sole C, N and energy sources, as well as the capacity to detoxify FA in barley seedlings | 80 | |
| DAS | L. acidophillus 1A | L. bulgaricus 5A | 0.5 μg mL−1 | 48 h | Biodegradation in vitro in agar plates | DAS appeared to possess an inhibitory effect on the growth of almost all Lactobacillus and Bacillus strains, whereas no effect was observed on yeast strains | 81 |
| L. acidophillus 4A | Bacillus licheniformis | ||||||
| L. helveticus 2A | Bacillus subtilis | ||||||
| L. bulgaricus 3A | S. cerevisae | ||||||
| DAS | Akkermansia muciniphila DSM 22959 | Butyrivibrio fibrisolvens 16/4 | 0.7 ng mL−1 | 0–48 h | Anaerobic incubation | The human microbiome characteristic bacteria Prevotella copri revealed to deacetylate 51.3% of DAS to 15-MAS after 48 h | 82 |
| B. adolescentis DSM 20083 | Coprococcus sp. ART55/1 Eubacterium rectale DSM 17629 (A1-86) | ||||||
| Bacteroides thetaiotaomicron DSM 2079 | Roseburia intestinalis L1-82 | ||||||
| Prevotella copri DSM 18205 | Lactiplantibacillus plantarum NCIMB 7220 | ||||||
| Faecalitalea cylindroides T2-87 | Faecalibacterium prausnitzii A2-165 | ||||||
| Anaerobutyricum hallii DSM 3353 | Enterococcus mundtii DSM 4838 | ||||||
| Anaerostipes hadrus SSC/2 | |||||||
| Blautia obeum A2-162 | |||||||
| TeA | L. brevis 20023 (LAB-20023) | 0.5 μg mL−1 | 12 h | Adsorption | L. brevis 20023 (LAB-20023) cells have revealed to adsorb TeA from 12.5% to 90.1% | 83 | |
| AOH | Bacteroides caccae | Alistipes finegoldii | 0.5–50 μg mL−1 | 24 h | Absorption assay | Both Gram types were able to adsorb AOH, AME, and AST. Gram-negative bacteria showed higher adsorptive capacities. This tendency was not observed for neither ATX-I nor ALP | 84 |
| AME | Bacteroides eggerthii | Alistipes timonensis | |||||
| AST | Bacteroides thetaiotaomicron | Ruminococcus bicirculans | |||||
| ATX-I | Bacteroides vulgatus | Akkermansia muciniphila | |||||
| ALP | Parabacteroides distasonis | L. hominis | |||||
| E. coli | B. Longum | ||||||
| Clostridium innocuum (CI) | B. sp. | ||||||
| Mycotoxin | Probiotic | Mycotoxin dose administration | Exposure time | Experimental assay | Results | Ref. |
|---|---|---|---|---|---|---|
| Alternariol methyl ether (AME), alterperylenol (ALP), altertoxin I (ATX-I), alterperylenol (ALP), altuene (ALT), altersetin (AST), aurofusarin (AUR), beauvericin (BEA), Bifidobacterium (B), diacetoxyscirpenol (DAS), enniatin A (ENA), enniatin A1 (ENA1), enniatin A2 (ENA2), enniatin B (ENB), enniatin B1 (ENB), enniatin B4 (ENB4), enniatins (ENs), Escherichia coli (E, coli), fusaric acid (FA), lactic acid bacteria (LAB), Lactobacillus (L), minimal inhibitory concentrations (MICs), De Man–Rogosa–Sharpe agar (MRS), potato dextrose broth (PDB), Saccharomyces (S.), and tenuazonic acid (TeA). | ||||||
| In vivo | ||||||
| AME | L. uvarum LUHS245 | 10.1–17.06 μg kg−1 | 36 days | Fermentation/biotransformation | AME and ALT were found in 61-day-old control piglets’ feces and in fermented feed samples. However, AME was not found in probiotic-treated piglets’ feces | 85 |
| ALT | L. casei LUHS210 | |||||
| Pentosaceus acidilactici LUHS29 | ||||||
| Pediococcus pentosaceus LUHS183 | ||||||
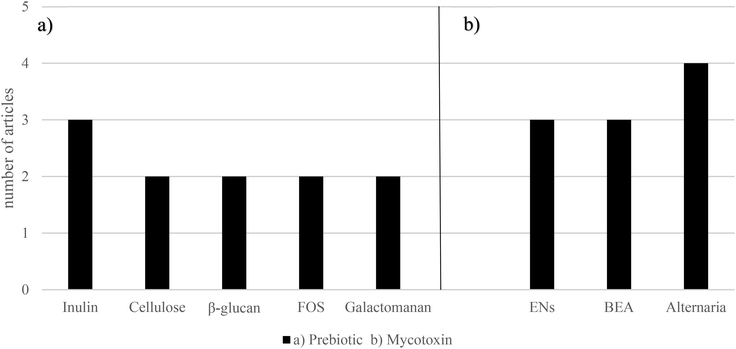
3.3 Mycotoxins and prebiotics
Prebiotics have been described as a potent agent to prevent metabolic disease through microbiome modulation.50 K-carrageenan has revealed to be a promising fiber, thanks to ZEA bioaccessibility reduction, obtaining values lower than 20%. Lactose from fermented whey has shown to protect from AFB1 and OTA toxicity.86,87 Pectin has exerted to decrease DON, HT-2, and T-2 (50–88%) bioaccessibility.59 Garlic, whose prebiotic content is notable, has shown to reduce AFB1 cytotoxicity.88 It has also been reported that flavonoids play an important role in protecting against the toxicity of mycotoxins like OTA, PAT, and DON in different experimental models.89,90BEA bioaccessibility was decreased, ranging from 15.7% to 60.5%, compared to the control (93.2%), with the addition of the following prebiotics: galactomannan 5%, glucomannan high-molecular weight (HMW) 1%, glucomannan HMW 5%, glucomannan fine powder 1%, glucomannan fine powder 5%, citrus fiber 1%, citrus fiber 5%, bamboo fiber 1%, bamboo fiber 5%, carrot fiber 1%, carrot fiber 5%, pie fiber 1%, pie fiber 1%, β-glucan 1%, β-glucan 5%, xilan 1%, xilan 5%, cellulose HMW 1%, cellulose HMW 5%, cellulose medium-molecular weight (MMW) 1% and cellulose MMW 5%.72 BEA has also shown to be decreased by β-1,3 glucan, chitosan LMW, chitosan MMW, FOS, galactomannan, inulin, and pectin at both concentrations (1% and 5%). Each sample was contaminated, reaching a BEA concentration of both 5 and 25 mg L−1. While the mean control bioaccessibility was 92.6% and 90.0% for both BEA concentrations, respectively; when adding prebiotics, it was 13.2% and 50.4%, respectively, galactomannan being the most reducing one (5.2%).93
Similar results have been obtained with Alternaria mycotoxins. AOH, AME, ATX-II, STTX-III, and ALP have been found to be decreased, already before anaerobic incubation, in the samples containing particulate fecal matter. These samples were supposed to be also composed of indigestible fiber fractions.67 The antifungal effects of the volatile compound 2-phenylethyl isothiocyanate (2-PEITC) against A. alternata and its mycotoxins TEN, AOH, AME, and ALT were assessed. 2-PEITC is found in some vegetables and contributes to gastrointestinal health.94 After a 2 h treatment, these four mycotoxins were extracted and detected by high performance liquid chromatography-tandem mass spectrometry (HPLC-TOF-ESI-MS) in the mycelium of A. alternata. The concentrations of TEN, AOH, AME, and ALT at the MIC of 2-PEITC-treated groups were only 27%, 90%, 90%, and 88% of the corresponding control groups, respectively.95 The anthocyanidin delphinidin (30.3 μg mL−1) has exerted to strongly antagonize the genotoxic properties of ATX-II by significantly suppressing the level of DNA strand breaks by up to 75% in HT-29 colon carcinoma cells.84 Conversely, AOH and AME degradation has been assessed after cinnamaldehyde treatment in centrifuge tubes under rotary shaking conditions (150 rpm) at 25 °C for 120 min. It was found that AOH and AME degradation rates were 16.8% and 7.3%, respectively.96 The results are shown in Table 4.
| Mycotoxin | Prebiotic | Dose administration | Exposure time | Experimental assay | Results | Ref. | |
|---|---|---|---|---|---|---|---|
| Alternariol (AOH), alternariol monomethyl ether (AME), alterperylenol (ALP), altertoxin II (ATX-II), altuene (ALT), beauvericin (BEA), enniatin A (ENA), enniatin A1 (ENA1), enniatin B (ENB), enniatin B1 (ENB), fructooligosaccharides (FOS), high-molecular weight (HMW), low-molecular weight (LMW), medium-molecular weight (MMW), minimum inhibitory concentration (MIC), sulforhodamine B/water-soluble tetrazolium salt (SRB/WST-1), tentoxin (TEN), 2-phenylethyl isothiocyanate (2-PEITC), and stemphyltoxin III (STTX-III). | |||||||
| In vitro | |||||||
| ENA | Inulin (1.5%–10%) | 1.02–2.04 mg mL−1 | 8 h | In vitro digestion model | ENA, ENA1, ENB and ENB1 bioaccessibility reduction in a dose-dependent manner at an inulin concentration ranging from 1.5% to 10% | 91 | |
| ENA1 | |||||||
| ENB | |||||||
| ENB1 | |||||||
| ENA | Cellulose (1% and 5%) | Not reported | 4 h | In vitro digestion model | Cellulose (1% and 5%) decreased ENA, ENA1, ENB and ENB1 bioaccessibility a mean of 26.9% and 44.7%, respectively | 71 | |
| ENA1 | BEA bioaccessibility reduction ranged from 60% to 80% | ||||||
| ENB | |||||||
| ENB1 | |||||||
| BEA | |||||||
| ENA | Inulin and FOS (1% and 5%) | Not reported | 4 h | Static gastrointestinal in vitro digestion model | Highest reductions carried out when adding enriched inulin 5% (5.7%) and FOS 5% (3.9%) for ENA and ENA1, respectively | 92 | |
| ENA1 | |||||||
| ENB | |||||||
| ENB1 | |||||||
| BEA | |||||||
| BEA | Galactomanan 1–5% | Pie fiber 1%–5% | 5–25 mg kg−1 | 52 h | In vitro digestion model | BEA bioaccessibility reduction ranging from 15.7% to 60.5% comparing to control (93.2%) | 77 |
| Glucomannan HMW 1%–5% | β-Glucan 1%–5% | ||||||
| Glucomannan FP 1%–5% | Xilan 1–5% | ||||||
| Citrus fiber 1%–5% | Cellulose HMW 1–5% | ||||||
| Bamboo fiber 1%–5% | Cellulose MMW 1–5% | ||||||
| Carrot fiber 1%–5% | |||||||
| BEA | β-1,3 glucan | Galactomannan | 5 mg kg−1 | 52 h | In vitro digestion model | BEA mean bioaccessibility decrease when adding prebiotics (13.2% and 50.4%, respectively for 1% ad 5%), galactomannan being the most reducing one (5.2%) | 94 |
| Chitosan LMW | Inulin | ||||||
| Chitosan MMW | Pectin | ||||||
| FOS | (All 1% and 5%) | ||||||
| AOH | Particulate fecal matter supposed to be composed also by indigestible fiber fractions | 50 μg mL−1 | 48 h | Fecal incubation in vitro | AOH, AME, ATX-II, STTX-III, and ALP reduction by samples supposed to be also composed of indigestible fiber fractions | 67 | |
| AME | |||||||
| ATX-II | |||||||
| STTX-III | |||||||
| ALP | |||||||
| TEN | 2-PEITC | 0.1–1.38 mg kg−1 | 2 h | Qualitative analysis performed using a mass spectrometer after incubation for the MIC | TEN, AOH, AME, and ALT concentrations of 2-PEITC-treated groups were only 27%, 90%, 90%, and 88% of the corresponding control groups, respectively | 95 | |
| AOH | |||||||
| AME | |||||||
| ALT | |||||||
| ATX-II | Delphinidin | 0.35 μg mL−1 | 24 h | Coupled SRB/WST-1 cytotoxicity assays | ATX-II genotoxic properties were antagonized by delphinidin (30.3 μg mL−1) by significantly suppressing the level of DNA strand breaks by up to 75% in HT-29 colon carcinoma cells | 89 | |
| AOH | Cinnamaldehyde | 0.81–0.82 μg mL−1 | 2 h | Degradation assay | AOH and AME degradation rate was 16.8% and 7.3%, respectively, after 120 min of cinnamaldehyde treatment | 96 | |
| AME | |||||||
3.4 Mycotoxins and synbiotics
The use of synbiotics has been previously proved to be effective in the removal of PAT.97,98 The deleterious effects of OTA have also been reversed by using beneficial synbiotics possessing the ability to reduce the number of potential pathogens in the digestive tract and by increasing the activity of α-glucosidase and α-galactosidase, while decreasing the activity of potentially harmful fecal enzymes (β-glucosidase, β-galactosidase and β-glucuronidase).60,993.5 Mycotoxins and postbiotics
Postbiotics are considered to have positive effects on human health.50 Yeast cell wall extract (YCWE) and a post-biotic yeast cell wall-based blend (PYCW) have revealed to reduce the adverse effects related to serum biochemistry, liver function, immune response, altered cecal short chain fatty acids (SCFAs), goblet cell counts and architecture of the intestinal villi. These results were found studying commercial broilers fed with a diet containing DON, T-2 and ZEA at 3.0 mg kg−1, 104 μg kg−1 and 79 μg kg−1, respectively.57 Some characteristic bacterial enzymes, which isolated are considered postbiotics, have exerted to remove AFB1, OTA, DON and ZEA.63 Butyrate, one of the most studied gut microbiota metabolites, has shown to reverse the alterations caused by DON by blocking some genes involved in reactive oxygen species and tumor necrosis factor alpha mediated pathways.100 Likewise, AFB1 and ZEA have been reported to reduce the content of SCFAs (butyrate, propionate and acetate).101,1024 Discussion
The interactions along the gut are very complex due to the quantity of factors, which play a notable role in its physiology, so that there are times where it is so likely not to know completely which specific pathways or molecules are the consequence of a particular fact. However, there are some stablished correlations which help us to better understand how biological systems work. One of these strong correlations is the preventive role of PPSP and the gut microbiota in disease and its health-promotion effects.49 As has already been said,62 PPSP and the gut microbiota possess the capacity to bind and metabolize a vast quantity of substances entering the gastrointestinal tract. Therefore, despite theoretically reducing beneficial compounds, PPSP can also decrease the exposure to detrimental ones, such as toxins, including emerging mycotoxins.As presented, European regulations only encompass a small set of mycotoxins (AFB1, AFB2, AFG1, AFG2, AFM1, OTA, PAT, DON, ZEA, FB1, FB2, CIT, AOH, AME, TeA, T-2 and HT-2 and ergoalkaloids).2 Due to their regulation, these mycotoxins tend to be more studied – also including their interactions with the gut microbiota and PSPP. In fact, studies show that studied mycotoxins disrupt the gut microbiota, while concomitant intake of PSPP mitigates the effects of the mycotoxins and contributes to maintain gut microbiota homeostasis.56,58,90,99,101 Although similar trends are expected with unregulated mycotoxins, it has to be carried out as a one-on-one mycotoxin analysis, since every mycotoxin possesses a specific chemical structure and a different response in the environment where it is located.62 Actually, there are some 400 known mycotoxins, so the vast majority of them are not still regulated and need a more profound comprehension.1 Furthermore, it should not be forgotten that mycotoxins tend to be found in combinations, so the most common mixtures found in food and feed are the ones interesting to be tested.
4.1 Emerging mycotoxins and the gut microbiota
Regarding the interaction between the gut microbiota and emerging mycotoxins, studies have revealed a clear tendency: the gut microbiota contributes to emerging mycotoxin detoxification through binding and/or metabolization63–65,67 and emerging mycotoxins alter the gut microbiota composition.68 Despite being the trend confirmed by these studies, the difficulty stems from the heterogeneity of the analyzed samples. The microbiota exhibits significant variability, with the microbiome differing by species. In humans, microbiota changes are influenced by factors such as the level of urbanization of the location and even the time of day within the same individual.105–107 For example, rumen microbiome is characterized by possessing many enzymes involved in the de novo synthesis of vitamin B12.108According to experimental feasibility, it has been shown that gut microbiota samples are obtained from feces in most of the experiments, since the fecal microbiota is more accessible than the colon one. Interestingly, it has been revealed that colon microbiota has a different profile than the fecal one, the microbiome in the large intestine being much more diverse than those of the small intestine and feces. For instance, while in the large intestine there were 4080 operational taxonomic units, in the feces only 443 were found.109 Indeed, bacterial biomass also changes along the gastrointestinal tract and the feces: duodenum (103 bacteria per g), jejunum (104 g−1), ileum (107 g−1), colon (1012 g−1) and feces (510 g−1).110,111 In addition, short-term trials are likely to hide that long-term mycotoxin exposure may lead to the development of the ability to degrade these mycotoxins despite being unable to do it in the first moment.64
There are other limitations that should be highlighted. Although in Debevere et al. (2020)63 the incubation time (48 h) was in line with other experiments studying mycotoxin degradation in vitro, it is considerably longer than the in vivo retention time of the digesta in the rumen, implying that under normal physiological conditions the results would be different. Despite Crudo et al. (2020)67 having shown that AOH, AME, ATX-II, STTX-III and ALP were lower in the samples containing microorganisms than in those without them, it should be noted that the sample size was very small (n = 4). In this study, all donors shared some characteristics: they were healthy omnivorous volunteers (two males and two females) ranging from 26 to 34 years old and with a normal body mass index (18.5–24.9 kg m−2), none of them had been treated with antibiotics, probiotics, and prebiotics for the previous three months and they did not present previous intestinal diseases. However, the entire diet, playing a notable role in gut metabolism and in the interactions with mycotoxins, was not considered. Moreover, as mentioned in the same article, there are other limitations such as the short-term trial (3 h) and the fact that the growth media used might alter the initial microbial composition of the feces, contributing to the growth of some microorganisms at the expense of others. Conversely, in Novak et al. (2021),68 only the mycotoxins ENB, ENB1, BEA and DON were assessed in weaning piglets’ feces in contrast to the control. It was shown that L. amylovorus and L. reuteri, both beneficial microbes, were up-regulated and down-regulated, respectively, when adding BEA in comparison with DON and the control. Furthermore, the combination of ENs and BEA increased the total protein synthesis, but the addition of DON had an inhibitory effect.68 Although it is important to know the interactions among these four mycotoxins, the real conditions are more complex, since a meal may contain a variety of contaminants, so the results under normal conditions are so likely to differ from these ones. While microbes could metabolize emerging mycotoxins, these toxins can also influence the composition of the microbiome, creating a potential double-edged sword scenario. In addition, it is important to consider the varying doses of emerging mycotoxins used in experiments, ranging from nanograms to milligrams.
Viewing mycotoxins as toxic compounds or as potential drugs is indeed a question that arises. On the one hand, they can reduce beneficial microorganisms, such as Lactobacillus, S. ceverisiae and Bacillus,72,73,79,81 whose depletion is generally connected to disease promotion.112 On the other hand, they can play a role as antibiotics by eliminating microorganisms involved in diseases.112–116 For instance, AOH and AME have exerted to be promising inhibitors of glycogen-synthase-kinase 3, being an important target in drug discovery. In fact, this enzyme has revealed to participate in signaling pathways of type 2 diabetes, cancer, neurological disorders, and other diseases.117 In addition, ergotamine with its α-adrenoreceptor-blocking mode of action has also been used for acute migraine treatments for over 50 years.118 Further investigation is needed in the field to clarify the results and assess the real implications of these emerging mycotoxins towards health and diseases, also taking into consideration that the dose and the type of mycotoxin may be decisive in the health or disease-promoting outcomes.
4.2 Emerging mycotoxins and probiotics
The bidirectional fluxes between probiotics and emerging mycotoxins have been widely confirmed. A vast quantity of probiotics (Lactobacillus, Bifidobacterium and S. cerevisiae) has shown to reduce emerging mycotoxins’ bioaccessibility (ENs, BEA and Alternaria).74–78,92,93 At the same time, emerging mycotoxins have exerted to alter probiotic compositions (B. subtilis and Staphylococcus aureus).72,73,79,81 This bioaccessibility decrease may be due to the bacterial mycotoxin binding capacity83,84 and/or their ability to contribute to detoxification by originating derivatives characterized by possessing less toxicity.82,83 However, in most of these studies, probiotics are not assessed in combination, but one by one and using in vitro conditions. Only Vadopalas et al. (2020)85 assessed probiotics’ effects on emerging mycotoxins under in vivo conditions. In this study, not only did the addition of some microbial starters modified some blood parameters, but also contributed to modulate the gut microbiota and detoxify AME. Despite these beneficial effects, it should be noted that the experiment only lasted two months and the piglets’ organs were not fully mature. Combining probiotics and working in vivo would be a more realistic approach to the human environment, since there are a lot of microorganisms that form a part of the gut microbiota.119 What is more, it is very well documented that the interplay along the gut concerns not only different bacteria, but also fungi, archaea, viruses, and helminths.1204.3 Emerging mycotoxins and prebiotics
It has been clearly shown that prebiotics reduce emerging mycotoxin bioaccessibility. Although the term prebiotic is very vast, the collected data cover a high quantity of these types of compounds at a different concentration.67,71,77,91–93,95,96 As in the probiotic case, there are two mechanisms explaining this reduction. The first one is the capacity of prebiotics to bind emerging mycotoxins, and the second one is the ability to metabolize them, and it is very likely to have both processes working at the same time. However, most studies do not consider an analysis of secondary metabolites of emerging mycotoxins, so this metabolization cannot be fully confirmed. Regarding mycotoxin reduction, if the metabolites have not been degraded, it is assumed that they have bound to the structures of the prebiotics, although they are also subject to metabolization anyway. Conversely, apart from reducing bioaccessibility, prebiotics can also directly suppress the detrimental effects of mycotoxins. As it is shown in Aichinger et al. (2018), the anthocyanidin delphinidin blocks significantly ATX-II genotoxic properties by suppressing the level of DNA strand breaks.89 The action of these prebiotics may derive from blocking the mycotoxin, preventing its action or from having a beneficial agonist impact on some physiological pathways. Furthermore, it should also be noted that in all these experiments, prebiotics are evaluated only one by one. The reality of the human diet includes a wide combination of them, so more research needs to be carried out taking this complexity into account.4.4 Emerging mycotoxins, synbiotics and postbiotics
The role of synbiotics in the modulation of emerging mycotoxins has been assessed only in two studies.92,93 In both, the results showed that the addition of synbiotics increased mycotoxin bioaccessibility when compared to prebiotics. It has been hypothesized that probiotics degraded prebiotics, so that the binding ability of the last ones was lost. Although it may be possible, it cannot be extended as a generality, since only small fractions of prebiotics and probiotics were used in these two trials, and they do not represent either all probiotics or complete prebiotics. Conversely, those synbiotics applied to non-emerging mycotoxins reveal opposite results. For instance, Zoghi et al. (2017)97 showed that the removal of PAT by probiotic bacteria from apple juice depended significantly on the FOS content (as a prebiotic) and the addition of ascorbic acid. Under the best conditions, 91.3% of the initial patulin concentration was removed from juice for 6 weeks.97 Zoghi et al. (2019) also reported that by adding FOS, ascorbic and citric acid to apple juice inoculated with L. plantarum, the efficiency of PAT removal was improved to 95.9% for 6 weeks cold storage.98 On the other hand, despite being supposed to enhance clinical outcomes compared to probiotics or prebiotics alone due to their synergistic effect, synbiotics are very difficult to formulate and few have shown to possess clinical efficacy. It is also worth mentioning that most clinical trials are not characterized by including relevant probiotic or prebiotic controls, and many have not performed relevant microbiota analyses.121 Research needs to be expanded to more (emerging) mycotoxins in combination with synbiotics.Finally, postbiotics have shown to bind, metabolize and alleviate the harmful effects of some emerging and non-emerging mycotoxins57,63,100,103 as well as these mycotoxins have revealed to alter the composition of some postbiotics.101,102 Notwithstanding, it is needed to extend these experiments to more (emerging) mycotoxins and to more postbiotics, since only the combination of PYCW with BEA and CIT has been assessed. In fact, it will be very interesting to focus on the interplay between (emerging) mycotoxins and SCFAs.
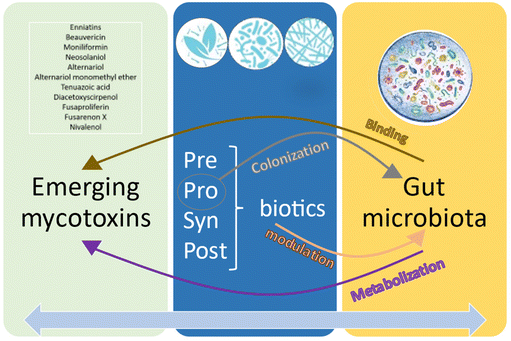
4.5 Interplay between emerging mycotoxins, the gut microbiota and PSPP
Although the results have revealed a clear trend, data on this interaction have only been found in 12 of the 20 emerging mycotoxins investigated. However, apart from these 20 ones, other emerging mycotoxins were found. The collected data encompassed a lot of experimental differences (mycotoxin/biotic type and concentration, species, time exposure, assay type, etc.), making the comparisons difficult among them. As Fig. 7 illustrates, the interaction between the gut microbiota and emerging mycotoxins involves metabolization and binding processes. The role of PPSP in relation to emerging mycotoxins primarily revolves around colonization (probiotics) and modulation (all). Furthermore, there exists a two-way communication between microbiota and PPSP, where the gut microbiota can degrade and metabolize PPSP, activating secondary bioactive compounds that may contribute to the detoxification and/or alleviation of emerging mycotoxins. Similarly, PPSP can enhance certain gut microbiota strains involved in the removal of emerging mycotoxins. Indeed, this reciprocal relationship can also be applied to the entire spectrum of PPSP, as the addition of probiotics, prebiotics, and synbiotics may enhance the production of certain postbiotics (such as short-chain fatty acids), and vice versa.5 Conclusions
Emerging mycotoxins have been implicated in numerous harmful biological processes. Despite their diverse nature, studies have demonstrated that the gut microbiota and PPSP can detoxify and/or alleviate the adverse effects resulting from exposure to these emerging mycotoxins. ENs, BEA, NIV, and Alternaria mycotoxins were among the most frequently encountered emerging mycotoxins. The genera Lactobacillus, Bifidobacterium, and Saccharomyces were the most extensively studied probiotics, while inulin, cellulose, β-glucan, FOS, and galactomannan were among the most evaluated prebiotics. Data on synbiotics and postbiotics were limited. Evidence clearly demonstrates the existence of interplay between the gut microbiota and emerging mycotoxins, as well as between PPSP and emerging mycotoxins, involving processes such as metabolization, colonization, modulation, and binding. In addition, connections between the gut microbiota and PPSP, as well as within the same PPSP, were identified, all contributing to the alleviation of the effects of the emerging mycotoxins.However, quantitative and qualitative data remain limited. Further in vivo and long-term trials are necessary to better comprehend the complexity of these interactions in both animals and humans. The formulation of PPSP as functional food and nutraceuticals should be considered a preventive strategy against the toxicity of emerging mycotoxins in human diets and animal feed.
Author contributions
Á. Lázaro: writing – original draft, methodology, investigation and validation. P. Vila-Donat: methodology, investigation, and review and editing. L. Manyes: investigation, review and editing, project administration, and conceptualization.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This publication is part of the project PID2022-140722OB-I00 funded by MCIU/AEI/AEI/10.13039/501100011033/FEDER, EU.References
- L. Manyes and G. Font, Mycotoxins: Toxicity, occurrence, risk assessment and prevention, in Encyclopedia of Human Nutrition, Elsevier, New York, NY, USA, 4th edn, 2023, pp. 492–500. DOI:10.1016/B978-0-12-821848-8.00147-5.
- European Commission, Commission Regulation (EU) 2023/915 of 25 April 2023 on maximum levels for certain contaminants in food and repealing Regulation (EC) No 1881/2006, Off. J. Eur. Union, 2023, 119, 103–157 Search PubMed.
- European Commission, Commission Recommendation (EU) 2013/165 of 27 March 2013 on the presence of T–2 and HT–2 toxin in cereals and cereal products, Off. J. Eur. Union, 2013, 91, 12–15 Search PubMed.
- European Commission, Commission recommendation (EU) 2022/553 of 5 April 2022 on monitoring the presence of Alternaria toxins in food, Off. J. Eur. Communities, 2022, 107, 90 Search PubMed.
- O. A. Mihalache, M. De Boevre, L. Dellafiora, S. De Saeger, A. Moretti, L. Pinson-Gadais, N. Ponts, F. Richard-Forget, A. Susca and C. Dall'Asta, The occurrence of non-regulated mycotoxins in foods: A systematic review, Toxins, 2023, 15(9), 583, DOI:10.3390/toxins15090583.
- W. P. P. Liew and S. Mohd-Redzwan, Mycotoxin: Its impact on gut health and Microbiota, Front. Cell. Infect. Microbiol., 2018, 8, 60, DOI:10.3389/fcimb.2018.00060.
- J. Kos, M. Anić, B. Radić, M. Zadravec, E. Janić, E. Hajnaland and J. Pleadin, Climate change—A global threat resulting in increasing mycotoxin occurrence, Foods, 2023, 12(14), 2704, DOI:10.3390/foods12142704.
- H. Olleik, C. Nicoletti, M. Lafond, E. Courvoisier-Dezord, P. Xue, A. Hijazi, E. Baydoun, J. Perrier and M. Maresca, Comparative structure–activity analysis of the antimicrobial activity, cytotoxicity, and mechanism of action of the fungal cyclohexadepsipeptides enniatins and beauvericin, Toxins, 2019, 11(9), 514, DOI:10.3390/toxins11090514.
- N. Pérez-Fuentes, R. Alvariño, A. Alfonso, J. González-Jartín, S. Gegunde, M. R. Vieytes and L. M. Botana, Enniatins A1 and B1 alter calcium homeostasis of neuronal cells leading to apoptotic death, Food Chem. Toxicol., 2022, 168, 113361, DOI:10.1016/j.fct.2022.113361.
- F. Caloni, P. Fossati, A. Anadón and A. Bertero, Beauvericin: The beauty and the beast, Environ. Toxicol. Pharmacol., 2020, 75, 103349, DOI:10.1016/j.etap.2020.103349.
- D. Çavuşoğlu, K. Çavuşoğlu, E. Yalçin and K. Çavuşoğlu, Potential toxicity assessment of mycotoxin fusaric acid with the spectral shift profile on DNA, Environ. Sci. Pollut. Res., 2023, 30(29), 73506–73517, DOI:10.1007/s11356-023-27436-w.
- N. Sheik Abdul, S. Nagiah and A. A. Chuturgoon, Fusaric acid induces mitochondrial stress in human hepatocellular carcinoma (HepG2) cells, Toxicon, 2016, 119, 336–344, DOI:10.1016/j.toxicon.2016.07.002.
- M. Jonsson, J. Atosuo, M. Jestoi, A. V. Nathanail, U. M. Kokkonen, M. Anttila, P. Koivisto, E. M. Lilius and K. Peltonen, Repeated dose 28-day oral toxicity study of moniliformin in rats, Toxicol. Lett., 2015, 233(1), 38–44, DOI:10.1016/j.toxlet.2014.11.006.
- S. Aupanun, S. Poapolathep, M. Giorgi, K. Imsilp and A. Poapolathep, An overview of the toxicology and toxicokinetics of fusarenon-X, a type B trichothecene mycotoxin, J. Vet. Med. Sci., 2017, 79(1), 6–13, DOI:10.1292/jvms.16-0008.
- S. Sutjarit and A. Poapolathep, Fusarenon-X-induced apoptosis in the liver, kidney, and spleen of mice, J. Toxicol. Pathol., 2016, 29(3), 207–211, DOI:10.1293/tox.2015-0063.
- K. Jarolim, G. Del Favero, G. Pahlke, V. Dostal, K. Zimmermann, E. Heiss, D. Ellmer, T. D. Stark, T. Hofmann and D. Marko, Activation of the Nrf2-ARE pathway by the Alternaria alternata mycotoxins altertoxin I and II, Arch. Toxicol., 2017, 91(1), 203–216, DOI:10.1007/s00204-016-1726-7.
- C. Gruber-Dorninger, B. Novak, V. Nagl and F. Berthiller, Emerging mycotoxins: Beyond traditionally determined food contaminants, J. Agric. Food Chem., 2017, 65(33), 7052–7070, DOI:10.1021/acs.jafc.6b03413.
- A. Ritieni, S. M. Monti, G. Randazzo, A. Logrieco, A. Moretti, G. Peluso, R. Ferracane and V. Fogliano, Teratogenic effects of fusaproliferin on chicken embryos, J. Agric. Food Chem., 1997, 45(8), 3039–3043, DOI:10.1021/jf960890v.
- D. Wu, S. Li, P. Li, A. Jiang, Z. Liu, Y. Zhang, J. Wang, Z. Yang and Z. Wei, Diacetoxyscirpenol-induced heterophil extracellular traps contribute to the immune toxicity of liver injury in chickens, Food Chem. Toxicol., 2021, 148, 111926, DOI:10.1016/j.fct.2020.111926.
- S. Yang, M. De Boevre, H. Zhang, K. De Ruyck, F. Sun, Z. Wang, X. Cao, J. Shen, S. De Saeger and S. Zhang, Unraveling the in vitro and in vivo metabolism of diacetoxyscirpenol in various animal species and human using ultrahigh-performance liquid chromatography-quadrupole/time-of-flight hybrid mass spectrometry, Anal. Bioanal. Chem., 2015, 407(28), 8571–8583, DOI:10.1007/s00216-015-9016-4.
- A. L. Hasuda, E. Person, A. Khoshal, S. Bruel, S. Puel, I. P. Oswald, A. P. F. R. L. Bracarense and P. Pinton, Emerging mycotoxins induce hepatotoxicity in pigs’ precision-cut liver slices and HepG2 cells, Toxicon, 2023, 231, 107195, DOI:10.1016/j.toxicon.2023.107195.
- X. Tang, Y. Chen, X. Zhu, Y. Miao, D. Wang, J. Zhang, R. Li, L. Zhang and J. Chen, Alternariol monomethyl ether toxicity and genotoxicity in male Sprague-Dawley rats: 28-Day in vivo multi-endpoint assessment, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2022, 873, 503435, DOI:10.1016/j.mrgentox.2021.503435.
- A. Kumari and K. Singh, Evaluation of prophylactic efficacy of cinnamaldehyde in murine model against Paradendryphiella arenariae mycotoxin tenuazonic acid-induced oxidative stress and organ toxicity, Sci. Rep., 2021, 11(1), 19420, DOI:10.1038/s41598-021-98319-8.
- X. Zhu, Y. Chen, X. Tang, D. Wang, Y. Miao, J. Zhang, R. Li, L. Zhang and J. Chen, General toxicity and genotoxicity of altertoxin I: A novel 28-day multiendpoint assessment in male Sprague–Dawley rats, J. Appl. Toxicol., 2022, 42(8), 1310–1322, DOI:10.1002/jat.4297.
- L. De Bruyne, C. Van Poucke, D. J. Di Mavungu, N. A. I. M. Zainudin, L. Vanhaecke, D. De Vleesschauwer, B. G. Turgeon, S. De Saeger and M. Höfte, Comparative chemical screening and genetic analysis reveal tentoxin as a new virulence factor in Cochliobolus miyabeanus, the causal agent of brown spot disease on rice, Mol. Plant Pathol., 2016, 17(6), 805–817, DOI:10.1111/mpp.12329.
- M. Delaforge, F. Andre and M. Jaouen, Metabolism of tentoxin by hepatic cytochrome P–450 3A isozymes, Eur. J. Biochem., 1997, 250(1), 150–157, DOI:10.1111/j.1432-1033.1997.00150.x.
- S. Zargar and T. A. Wani, Food toxicity of mycotoxin citrinin and molecular mechanisms of its potential toxicity effects through the implicated targets predicted by computer-aided multidimensional data analysis, Life, 2023, 13(4), 880, DOI:10.3390/life13040880.
- V. Zingales, M. Fernández-Franzón and J. M. Ruiz, Sterigmatocystin: Occurrence, toxicity and molecular mechanisms of action – A review, Food Chem. Toxicol., 2020, 146, 111802, DOI:10.1016/j.fct.2020.111802.
- J. Xu, Z. Zhao, W. Guo, A. Ling, J. Wang, X. Wang and J. Yang, Potential role of individual and combined effects of T-2 toxin, HT-2 toxin and neosolaniol on the apoptosis of porcine Leydig cells, Toxins, 2022, 14(2), 145, DOI:10.3390/toxins14020145.
- C. M. Maragos, Y. Uchiyama, N. Kobayashi, F. Kominato and Y. Sugita-Konishi, Development and characterization of monoclonal antibodies for the mycotoxin citreoviridin, Toxins, 2019, 11(11), 630, DOI:10.3390/toxins11110630.
- M. W. Sumarah, G. W. Adams, J. Berghout, G. J. Slack, A. M. Wilson and J. D. Miller, Spread and persistence of a rugulosin-producing endophyte in Picea glauca seedlings, Mycol. Res., 2008, 112(6), 731–736, DOI:10.1016/j.mycres.2008.01.007.
- J. C. Bouhet and P. Pham van Chuong, Specific and non-specific interactions of two carcinogenic mycotoxins, luteoskyrin and rugulosin with nucleic acids, Ann. Nutr. Aliment., 1977, 31(4–6), 811–830 CAS.
- B. M. Kunz, L. Pförtner, S. Weigel, S. Rohn, A. Lehmacher and R. Maul, Growth and toxin production of phomopsin A and ochratoxin A forming fungi under different storage conditions in a pea (Pisum sativum) model system, Mycol. Res., 2022, 38(1), 37–50, DOI:10.1007/s12550-021-00446-8.
- N. Hymery, F. Masson, G. Barbier and E. Coton, Cytotoxicity and immunotoxicity of cyclopiazonic acid on human cells, Toxicol. in Vitro, 2014, 28(5), 940–947, DOI:10.1016/j.tiv.2014.04.003.
- Agencia Española de Seguridad Alimentaria y Nutrición. (n.d.). Gob.es. Retrieved January 21, 2024, from https://www.aesan.gob.es/AECOSAN/web/seguridad_alimentaria/subdetalle/micotoxinas.ht.
- A. Ancona, C. Petito, I. Iavarone, V. Petito, L. Galasso, A. Leonetti, L. Turchini, D. Belella, D. Ferrarrese, G. Addolorato, A. Armuzzi, A. Gasbarrini and F. Scaldaferri, The gut-brain axis in irritable bowel syndrome and inflammatory bowel disease, Dig. Liver Dis., 2021, 53(3), 298–305, DOI:10.1016/j.dld.2020.11.026.
- M. A. Kolber, K. O. Broschat and B. Landa-Gonzalez, Cytochalasin B induces cellular DNA fragmentation, FASEB J., 1990, 4(12), 3021–3027, DOI:10.1096/fasebj.4.12.2394319.
- W. L. Austin, M. Wind and K. S. Brown, Differences in the toxicity and teratogenicity of cytochalasins D and E in various mouse strains, Teratology, 1982, 25(1), 11–18, DOI:10.1002/tera.1420250103.
- T. M. Lopez-Diaz and B. Flannigan, Mycotoxins of Aspergillus clavatus: Toxicity of cytochalasin E, patulin, and extracts of contaminated barley malt, J. Food Prot., 1997, 60(11), 1381–1385, DOI:10.4315/0362-028x-60.11.1381.
- J. W. Daily, S. Kang and S. Park, Protection against Alzheimer's disease by luteolin: Role of brain glucose regulation, anti–inflammatory activity, and the gut microbiota–liver–brain axis, BioFactors, 2021, 47(2), 218–231, DOI:10.1002/biof.1703.
- E. Menozzi, J. Macnaughtan and A. H. V. Schapira, The gut-brain axis and Parkinson disease: clinical and pathogenetic relevance, Ann. Med., 2021, 53(1), 611–625, DOI:10.1080/07853890.2021.1890330.
- D. Micic, Obesity and gut-brain axis, Acta Endocrinol., 2023, 19(2), 234–240, DOI:10.4183/aeb.2023.234.
- M. M. Moughnyeh, K. M. Brawner, B. A. Kennedy, V. A. Yeramilli, N. Udayakumar, J. A. Graham and C. A. Martin, Stress and the gut-brain axis: Implications for cancer, inflammation and sepsis, J. Surg. Res., 2021, 266, 336–344, DOI:10.1016/j.jss.2021.02.055.
- X. Hu, T. Wang and F. Jin, Alzheimer's disease and gut microbiota, Sci. China: Life Sci., 2016, 59(10), 1006–1023, DOI:10.1007/s11427-016-5083-9.
- M. S. Desai, A. M. Seekatz, N. M. Koropatkin, N. Kamada, C. A. Hickey, M. Wolter, N. A. Pudlo, S. Kitamoto, N. Terrapon, A. Muller, V. B. Young, B. Henrissat, P. Wilmes, T. S. Stappenbeck, G. Núñez and E. C. Martens, A dietary fiber-deprived gut Microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility, Cell, 2016, 167(5), 1339–1353, DOI:10.1016/j.cell.2016.10.043.
- J. P. Karl, L. M. Margolis, E. H. Madslien, N. E. Murphy, J. W. Castellani, Y. Gundersen, A. V. Hoke, M. W. Levangie, R. Kumar, N. Chakraborty, A. Gautam, R. Hammamieh, S. Martini, S. J. Montain and S. M. Pasiakos, Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress, Am. J. Physiol.: Gastrointest. Liver Physiol., 2017, 312(6), G559–G571, DOI:10.1152/ajpgi.00066.2017.
- L. C. M. Antunes, J. Han, R. B. R. Ferreira, P. Lolić, C. H. Borchers and B. B. Finlay, Effect of antibiotic treatment on the intestinal metabolome, Antimicrob. Agents Chemother., 2011, 55(4), 1494–1503, DOI:10.1128/AAC.01664-10.
- A. Madison and J. K. Kiecolt-Glaser, Stress, depression, diet, and the gut microbiota: human–bacteria interactions at the core of psychoneuroimmunology and nutrition, Curr. Opin. Behav. Sci., 2019, 28, 105–110, DOI:10.1016/j.cobeha.2019.01.011.
- H. Y. Li, D. D. Zhou, R. Y. Gan, S. Y. Huang, C. N. Zhao, A. Shang, X. Y. Xu and H. B. Li, Effects and mechanisms of probiotics, prebiotics, synbiotics, and postbiotics on metabolic diseases targeting gut Microbiota: A narrative review, Nutrients, 2021, 13(9), 3211, DOI:10.3390/nu13093211.
- ISAPP – International Scientific Association for Probiotics and Prebiotics. (2018, October 31). Retrieved January 21, 2024, from https://isappscience.org/.
- G. R. Gibson and M. B. Roberfroid, Dietary modulation of the human colonic Microbiota: Introducing the concept of prebiotics, J. Nutr., 1995, 125(6), 1401–1412, DOI:10.1093/jn/125.6.1401.
- C. Hill, F. Guarner, G. Reid, R. Eliakim, A. Gangl, A. Thomson, J. Krabshuis, T. Lemair, P. Kaufmann, J. A. de Paula, R. Fedorak, F. Shanahan, M. E. Sanders, H. Szajewska, B. S. Ramakrishna, T. Karakan, N. Kim and World Gastroenterology Organization, The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic, Nat. Rev. Gastroenterol. Hepatol., 2014, 11(8), 506–514, DOI:10.1038/nrgastro.2014.66.
- K. S. Swanson, G. R. Gibson, R. Hutkins, R. A. Reimer, G. Reid, K. Verbeke, K. Scott, H. D. Holscher, M. B. Azad, N. M. Delzenne and M. E. Sanders, The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics, Nat. Rev. Gastroenterol. Hepatol., 2020, 17(11), 687–701, DOI:10.1038/s41575-020-0344-2.
- F. Guarner, A. G. Khan, J. Garisch, R. Eliakim, A. Gangl, A. Thomson, J. Krabshuis, T. Lemair, P. Kaufmann, J. A. de Paula, R. Fedorak, F. Shanahan, M. E. Sanders, H. Szajewska, B. S. Ramakrishna, T. Karakan, N. Kim and World Gastroenterology Organization, Global Guidelines: probiotics and prebiotics October 2011, J. Clin. Gastroenterol., 2012, 46(6), 468–481, DOI:10.1097/MCG.0b013e3182549092.
- D. Xia, Q. Mo, L. Yang and W. Wang, Crosstalk between mycotoxins and intestinal Microbiota and the alleviation approach via microorganisms, Toxins, 2022, 14(12), 859, DOI:10.3390/toxins14120859.
- M. B. Kudupoje, V. Malathi and A. Yiannikouris, Impact of a natural fusarial multi-mycotoxin challenge on broiler chickens and mitigation properties provided by a yeast cell wall extract and a postbiotic yeast cell wall-based blend, Toxins, 2022, 14(5), 315, DOI:10.3390/toxins14050315.
- P. Petrova, A. Arsov, F. Tsvetanova, T. Parvanova-Mancheva, E. Vasileva, L. Tsigoriyna and K. Petrov, The complex role of lactic acid bacteria in food detoxification, Nutrients, 2022, 14(10), 2038, DOI:10.3390/nu14102038.
- R. López-Ruiz, J. Marin-Saez, S. C. Cunha, A. Fernandes, V. de Freitas, O. Viegas and I. M. P. L. V. O. Ferreira, Investigating the impact of dietary fibers on mycotoxin bioaccessibility during in vitro biscuit digestion and metabolites identification, Foods, 2023, 12(17), 3175, DOI:10.3390/foods12173175.
- K. Śliżewska, P. Markowiak-Kopeć, A. Sip, K. Lipiński and M. Mazur-Kuśnirek, The effect of using new synbiotics on the turkey performance, the intestinal Microbiota and the fecal enzymes activity in turkeys fed ochratoxin A contaminated feed, Toxins, 2020, 12(9), 578, DOI:10.3390/toxins12090578.
- A. Zoghi, K. Khosravi-Darani and S. Sohrabvandi, Surface binding of toxins and heavy metals by probiotics, Mini-Rev. Med. Chem., 2014, 14(1), 84–98, DOI:10.2174/1389557513666131211105554.
- M. J. Page, J. E. McKenzie, P. M. Bossuyt, I. Boutron, T. C. Hoffmann, C. D. Mulrow, L. Shamseer, J. M. Tetzlaff, E. A. Akl, S. E. Brennan, R. Chou, J. Glanville, J. M. Grimshaw, A. Hróbjartsson, M. M. Lalu, T. Li, E. W. Loder, E. Mayo-Wilson, S. McDonald and D. Moher, The PRISMA 2020 statement: an updated guideline for reporting systematic reviews, Br. Med. J., 2021, 372, n71, DOI:10.1136/bmj.n71.
- P. Guerre, Mycotoxin and gut Microbiota interactions, Toxins, 2020, 12(12), 769, DOI:10.3390/toxins12120769.
- S. Debevere, A. Cools, S. D. Baere, G. Aesaert, M. Rychlik, S. Croubels and V. Fievez, In vitro rumen simulations show a reduced disappearance of deoxynivalenol, nivalenol and enniatin B at conditions of Rumen acidosis and lower microbial activity, Toxins, 2020, 12(2), 101, DOI:10.3390/toxins12020101.
- R. Hedman and H. Pettersson, Transformation of nivalenol by gastrointestinal microbes, Arch. Tierernahr., 1997, 50(4), 321–329, DOI:10.1080/17450399709386142.
- N. Daud, V. Currie, G. Duncan, M. Busman and S. W. Gratz, Intestinal hydrolysis and microbial biotransformation of diacetoxyscirpenol-α-glucoside, HT-2-β-glucoside and N-(1-deoxy-d-fructos-1-yl) fumonisin B1 by human gut microbiota in vitro, Int. J. Food Sci. Nutr., 2020, 71(5), 540–548, DOI:10.1080/09637486.2019.1698015.
- A. Lemke, B. Burkhardt, D. Bunzel, E. Pfeiffer, M. Metzler, M. Huch, S. E. Kulling and C. M. A. P. Franz, Alternaria toxins of the alternariol type are not metabolised by human faecal microbiota, World Mycotoxin J., 2016, 9(1), 41–50, DOI:10.3920/wmj2014.1875.
- F. Crudo, G. Aichinger, J. Mihajlovic, L. Dellafiora, E. Varga, H. Puntscher, B. Warth, C. Dall'Asta, D. Berry and D. Marko, Gut microbiota and undigested food constituents modify toxin composition and suppress the genotoxicity of a naturally occurring mixture of Alternaria toxins in vitro, Arch. Toxicol., 2020, 94(10), 3541–3552, DOI:10.1007/s00204-020-02831-1.
- B. Novak, A. Lopes Hasuda, M. Ghanbari, V. Mayumi Maruo, A. P. F. R. L. Bracarense, M. Neves, C. Emsenhuber, S. Wein, I. P. Oswald, P. Pinton and D. Schatzmayr, Effects of Fusarium metabolites beauvericin and enniatins alone or in mixture with deoxynivalenol on weaning piglets, Food Chem. Toxicol., 2021, 158, 112719, DOI:10.1016/j.fct.2021.112719.
- K. Baralić, K. Živančević, D. Bozic and D. Đukić-Ćosić, Probiotic cultures as a potential protective strategy against the toxicity of environmentally relevant chemicals: State-of-the-art knowledge, Food Chem. Toxicol., 2023, 172, 113582, DOI:10.1016/j.fct.2022.113582.
- A. Zoghi, S. D. Todorov and K. Khosravi-Darani, Potential application of probiotics in mycotoxicosis reduction in mammals and poultry, Crit. Rev. Toxicol., 2020, 52(9), 731–741, DOI:10.1080/10408444.2023.2168176.
- M. Ferrer, L. Manyes, J. Mañes and G. Meca, Influence of prebiotics, probiotics and protein ingredients on mycotoxin bioaccessibility, Food Funct., 2015, 6(3), 987–994, 10.1039/c4fo01140f.
- M. Roig, G. Meca, R. Marín, E. Ferrer, E. Ferrer and J. Mañes, Antibacterial activity of the emerging Fusarium mycotoxins enniatins A, A1, A2, B, B1, and B4 on probiotic microorganisms, Toxicon, 2014, 85, 1–4, DOI:10.1016/j.toxicon.2014.04.007.
- A. R. B. Ola, D. Thomy, D. Lai and H. Brötz-Oesterhelt, et al., Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis, J. Nat. Prod., 2013, 76(11), 2094–2099, DOI:10.1021/np400589h.
- M. Roig, G. Meca, E. Ferrer and J. Mañes, Reduction of the enniatins A, A1, B, B1 by an in vitro degradation employing different strains of probiotic bacteria: identification of degradation products by LC-MS-LIT, Toxicon, 2013, 70, 44–53, DOI:10.1016/j.toxicon.2013.04.001.
- G. Meca, T. Zhou, X. Z. Li, A. Ritieni and J. Mañes, Ciclohexadespipeptide beauvericin degradation by different strains of Saccharomyces cerevisiae, Food Chem. Toxicol., 2013, 59, 334–338, DOI:10.1016/j.fct.2013.06.010.
- A. Salim, S. Nadri, M. J. Hosseini, H. Rokni-Zadeh and M. Mohseni, Protective effect of probiotic Lactobacillus acidophilus against the toxicity of beauvericin mycotoxin on the Caco-2 cell line, Toxicon, 2020, 185, 184–187, DOI:10.1016/j.toxicon.2020.07.003.
- B. Mallebrera, G. Meca, L. Manyes, J. Mañes and G. Font, Influence of pro- and prebiotics on gastric, duodenal and colonic bioaccessibility of the mycotoxin beauvericin, J. Food Compos. Anal., 2013, 32(2), 141–149, DOI:10.1016/j.jfca.2013.09.003.
- G. Meca, A. Ritieni and J. Mañes, Reduction in vitro of the minor Fusarium mycotoxin beauvericin employing different strains of probiotic bacteria, Food Control, 2012, 28(2), 435–440, DOI:10.1016/j.foodcont.2012.04.002.
- T. Sondergaard, M. Fredborg and A. M. Oppenhagen Christensen, et al., Fast screening of antibacterial compounds from fusaria, Toxins, 2016, 8(12), 355, DOI:10.3390/toxins8120355.
- E. Simonetti, I. N. Roberts, M. S. Montecchia, F. H. Gutierrez-Boem, F. M. Gomez and J. A. Ruiz, A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth, Microbiol. Res., 2018, 206, 50–59, DOI:10.1016/j.micres.2017.09.008.
- J. Böhm, J. Grajewski, H. Asperger, B. Cecon, B. Rabus and E. Razzazi, Study on biodegradation of some A- and B-trichothecenes and ochratoxin A by use of probiotic microorganisms, Mycotoxin Res., 2000, 16(Suppl 1(S1)), 70–74, DOI:10.1007/BF02942985.
- N. Daud, V. Currie, G. Duncan, F. Farquharson, T. Yoshinari, P. Louis and S. Gratz, Prevalent human gut bacteria hydrolyse and metabolise important food-derived mycotoxins and masked mycotoxins, Toxins, 2020, 12(10), 654, DOI:10.3390/toxins12100654.
- N. Ge, J. Xu, B. Peng and S. Pan, Adsorption mechanism of tenuazonic acid using inactivated lactic acid bacteria, Food Control, 2017, 82, 274–282, DOI:10.1016/j.foodcont.2017.07.009.
- F. Crudo, G. Aichinger, J. Mihajlovic, E. Varga, L. Dellafiora, B. Warth, C. Dall'Asta, D. Berry and D. Marko, In vitro interactions of Alternaria, mycotoxins, an emerging class of food contaminants, with the gut microbiota: a bidirectional relationship, Arch. Toxicol., 2021, 95(7), 2533–2549, DOI:10.1007/s00204-021-03043-x.
- L. Vadopalas, M. Ruzauskas, V. Lele, V. Starkute, P. Zavistanaviciute, E. Zokaityte, V. Bartkevics, I. Pugajeva, I. Reinolds, S. Badaras, D. Klupsaite, E. Mozuriene, A. Dauksiene, R. Gruzauskas and E. Bartkiene, Combination of antimicrobial starters for feed fermentation: Influence on piglet feces Microbiota and health and growth performance, including mycotoxin biotransformation in vivo, Front. Vet. Sci., 2020, 7, 528990, DOI:10.3389/fvets.2020.528990.
- A. Cimbalo, M. Alonso-Garrido, G. Font and L. Manyes, Toxicity of mycotoxins in vivo on vertebrate organisms: A review, Food Chem. Toxicol., 2020, 137, 111161, DOI:10.1016/j.fct.2020.111161.
- M. Frangiamone, M. Lozano, A. Cimbalo, A. Lazaro, G. Font and L. Manyes, The protective effect of pumpkin and fermented whey mixture against AFB1 and OTA immune toxicity in vitro. A transcriptomic approach, Mol. Nutr. Food Res., 2023, 67(19), e2200902, DOI:10.1002/mnfr.202200902.
- A. Lázaro, M. Frangiamone, A. Maietti, A. Cimbalo, P. Vila-Donat and L. Manyes, Allium sativum L. var. Voghiera Reduces Aflatoxin B1 Bioaccessibility and Cytotoxicity In Vitro, Foods, 2024, 13(3), 487, DOI:10.3390/foods13030487.
- G. Aichinger, H. Puntscher, J. Beisl, M. L. Kütt, B. Warth and D. Marko, Delphinidin protects colon carcinoma cells against the genotoxic effects of the mycotoxin altertoxin II, Toxicol. Lett., 2018, 284, 136–142, DOI:10.1016/j.toxlet.2017.12.002.
- M. Frangiamone, A. Lázaro, A. Cimbalo, G. Font and L. Manyes, In vitro and in vivo assessment of AFB1 and OTA toxic effects and the beneficial role of bioactive compounds. A systematic review, Food Chem., 2024, 447, 138909, DOI:10.1016/j.foodchem.2024.138909.
- G. Meca, A. Giri, G. Sagratini, S. Vittori, G. Font and J. Mañes, The soluble dietary fiber inulin can influence the bioaccessibility of enniatins, Food Funct., 2012, 3(8), 853, 10.1039/c2fo00004k.
- M. Manzini, M. T. Rodriguez-Estrada, G. Meca and J. Mañes, Reduction of beauvericin and enniatins bioaccessibility by prebiotic compounds, evaluated in static and dynamic simulated gastrointestinal digestion, Food Control, 2015, 47, 203–211, DOI:10.1016/j.foodcont.2014.07.016.
- G. Meca, G. Meneghelli, A. Ritieni, J. Mañes and G. Font, Influence of different soluble dietary fibers on the bioaccessibility of the minor Fusarium mycotoxin beauvericin, Food Chem. Toxicol., 2012, 50(5), 1362–1368, DOI:10.1016/j.fct.2012.02.038.
- E. R. Coscueta, A. S. Sousa, C. A. Reis and M. M. Pintado, Phenylethyl isothiocyanate: A bioactive agent for gastrointestinal health, Molecules, 2022, 27(3), 794, DOI:10.3390/molecules27030794.
- M. Zhang, Y. Li, Y. Bi, T. Wang, Y. Dong, Q. Yang and T. Zhang, 2-phenylethyl isothiocyanate exerts antifungal activity against Alternaria alternata by affecting membrane integrity and mycotoxin production, Toxins, 2020, 12(2), 124, DOI:10.3390/toxins12020124.
- L. Xu, N. Tao, W. Yang and G. Jing, Cinnamaldehyde damaged the cell membrane of Alternaria alternata and induced the degradation of mycotoxins in vivo, Ind. Crops Prod., 2018, 112, 427–433, DOI:10.1016/j.indcrop.2017.12.038.
- A. Zoghi, K. Khosravi-Darani, S. Sohrabvandi, H. Attar and S. A. Alavi, Effect of probiotics on patulin removal from synbiotic apple juice, J. Sci. Food Agric., 2017, 97(8), 2601–2609, DOI:10.1002/jsfa.8082.
- A. Zoghi, K. Khosravi-Darani, S. Sohrabvandi and H. Attar, Patulin removal from synbiotic apple juice using Lactobacillus plantarum ATCC, 8014, J. Appl. Microbiol., 2019, 126(4), 1149–1160, DOI:10.1111/jam.14172.
- P. Markowiak, K. Śliżewska, A. Nowak, A. Chlebicz, A. Żbikowski, K. Pawłowski and P. Szeleszczuk, Probiotic microorganisms detoxify ochratoxin A in both a chicken liver cell line and chickens, J. Sci. Food Agric., 2019, 99(9), 4309–4318, DOI:10.1002/jsfa.9664.
- Q. Zong, K. Li, H. Qu, P. Hu, C. Xu, H. Wang, S. Wu, S. Wang, H. Y. Liu, D. Cai and W. Bao, Sodium butyrate ameliorates deoxynivalenol-induced oxidative stress and inflammation in the porcine liver via NR4A2-mediated histone acetylation, J. Agric. Food Chem., 2023, 71(27), 10427–10437, DOI:10.1021/acs.jafc.3c02499.
- J. Cai, N. Wang, J. Chen, A. Wu, E. Nepovimova, M. Valis, M. Long, W. Wu and K. Kuca, Bacillus velezensis A2 inhibited the cecal inflammation induced by zearalenone by regulating intestinal flora and short-chain fatty acids, Front. Nutr., 2022, 9, 806115, DOI:10.3389/fnut.2022.806115.
- J. Zhou, L. Tang, J. Wang and J. S. Wang, Aflatoxin B1 disrupts gut-microbial metabolisms of short-chain fatty acids, long-chain fatty acids, and bile acids in male F344 rats, Toxicol. Sci., 2018, 164(2), 453–464, DOI:10.1093/toxsci/kfy102.
- R. Xu, A. Yiannikouris, U. K. Shandilya and N. A. Karrow, Comparative assessment of different yeast cell wall-based mycotoxin adsorbents using a model- and bioassay-based in vitro approach, Toxins, 2023, 15(2), 104, DOI:10.3390/toxins15020104kudu.
- O. Kolawole, J. Meneely, B. O. Greer, O. Chevallier, D. S. Jones, L. Connolly and C. Elliott, Comparative in vitro assessment of a range of commercial feed additives with multiple mycotoxin binding claims, Toxins, 2019, 11(11), 659, DOI:10.3390/toxins11110659.
- P. Trinh, J. R. Zaneveld, S. Safranek and P. M. Rabinowitz, One health relationships between human, animal, and environmental microbiomes: A mini-review, Public Health Front., 2018, 6, 235, DOI:10.3389/fpubh.2018.00235.
- C. A. Thaiss, D. Zeevi, M. Levy, E. Segal and E. Elinav, A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host, Gut Microbes, 2015, 6(2), 137–142, DOI:10.1080/19490976.2015.1016690.
- S. Subramanian, H. Thiruvengadamani and M. Sathiavelu, Comparison of Human gut Microbiota with other Animals, Res. J. Pharm. Technol., 2022, 5541–5547, DOI:10.52711/0974-360x.2022.00935.
- R. Seshadri, Hungate1000 project collaborators, S. C. Leahy, G. T. Attwood, K. H. Teh, S. C. Lambie, A. L. Cookson, E. A. Eloe-Fadrosh, G. A. Pavlopoulos, M. Hadjithomas, N. J. Varghese, D. Paez-Espino, R. Perry, G. Henderson, C. J. Creevey, N. Terrapon, P. Lapebie, E. Drula, V. Lombard and W. J. Kelly, Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection, Nat. Biotechnol., 2018, 36(4), 359–367, DOI:10.1038/nbt.4110.
- J. S. Ahn, E. Lkhagva, S. Jung, H. J. Kim, H. J. Chung and S. T. Hong, Fecal microbiome does not represent whole gut microbiome, Cell. Microbiol., 2023, 1–14, DOI:10.1155/2023/686841.
- F. F. Campos, P. A. Sales Junior, A. J. Romanha, M. S. S. Araújo, E. P. Siqueira, J. M. Resende, T. M. A. Alves, O. A. Martins-Filho, V. L. Santos, C. A. dos Rosa, C. L. Zani and B. B. Cota, Bioactive endophytic fungi isolated from Caesalpinia echinata Lam. (Brazilwood) and identification of beauvericin as a trypanocidal metabolite from Fusarium sp., Mem. Inst. Oswaldo Cruz, 2015, 110(1), 65–74, DOI:10.1590/0074-02760140243.
- D. D. Heeney, M. G. Gareau and M. L. Marco, Intestinal Lactobacillus in health and disease, a driver or just along for the ride?, Curr. Opin. Biotechnol, 2018, 49, 140–147, DOI:10.1016/j.copbio.2017.08.004.
- T. T. Thanh, T. N. Quoc and H. L. Xuan, Fusaric acid and derivatives as novel antimicrobial agents, Med. Chem. Res., 2020, 29(9), 1689–1696, DOI:10.1007/s00044-020-02596-3.
- G. Wang, W. Dong, H. Lu, W. Lu, J. Feng, X. Wang, H. Chen, M. Liu and C. Tan, Enniatin A1, A natural compound with bactericidal activity against Mycobacterium tuberculosis in vitro, Molecules, 2019, 25(1), 38, DOI:10.3390/molecules25010038.
- Q. Wu, J. Patocka, E. Nepovimova and K. Kuca, A review on the synthesis and bioactivity aspects of beauvericin, a Fusarium mycotoxin, Front. Pharmacol., 2018, 9, 1338, DOI:10.3389/fphar.2018.01338.
- J. Wiese, J. F. Imhoff, T. A. M. Gulder, A. Labes and R. Schmaljohann, Marine fungi as producers of benzocoumarins, a new class of inhibitors of glycogen-synthase-kinase 3β, Mar. Drugs, 2016, 14(11), 200, DOI:10.3390/md14110200.
- M. Frangiamone, A. Yemelin, A. Cimbalo, G. Font, E. Thines and L. Manyes, Fermented whey modulated AFB1 and OTA-induced hepatotoxicity and nephrotoxicity in vivo. A relative and absolute quantification about sex differences, Toxicol. Mech. Methods, 2023, 33(6), 529–540, DOI:10.1080/15376516.2023.2195488.
- R. Vemuri, E. M. Shankar, M. Chieppa, R. Eri and K. Kavanagh, Beyond just bacteria: Functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths, Microorganisms, 2020, 8(4), 483, DOI:10.3390/microorganisms8040483.
- D. F. Gómez Quintero, C. R. Kok and R. Hutkins, The Future of Synbiotics: Rational Formulation and Design, Front. Microbiol., 2022, 13, 919725, DOI:10.3389/fmicb.2022.919725.
- D. Mulac and H. U. Humpf, Cytotoxicity and accumulation of ergot alkaloids in human primary cells, Toxicology, 2011, 282(3), 112–121, DOI:10.1016/j.tox.2011.01.019.
- M. Frangiamone, A. Yemelin, A. Cimbalo, G. Font, E. Thines and L. Manyes, Fermented whey modulated AFB1 and OTA-induced hepatotoxicity and nephrotoxicity in vivo. A relative and absolute quantification about sex differences, Toxicol. Mech. Methods, 2023, 33(6), 529–540, DOI:10.1080/15376516.2023.2195488.
- R. Vemuri, E. M. Shankar, M. Chieppa, R. Eri and K. Kavanagh, Beyond just bacteria: Functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths, Microorganisms, 2020, 8(4), 483, DOI:10.3390/microorganisms8040483.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo01705f |
| This journal is © The Royal Society of Chemistry 2024 |