Open Access Article
Open Access ArticleThe lipidomic profile of the nanoparticle-biomolecule corona reflects the diversity of plasma lipids†
Lana
Papafilippou
 a,
Anna
Nicolaou
a,
Anna
Nicolaou
 *bc,
Alexandra C.
Kendall
*bc,
Alexandra C.
Kendall
 b,
Dolores
Camacho-Muñoz
b,
Dolores
Camacho-Muñoz
 b and
Marilena
Hadjidemetriou
b and
Marilena
Hadjidemetriou
 *a
*a
aNanomedicine Lab, Faculty of Biology, Medicine & Health, AV Hill Building, University of Manchester, Manchester Academic Healthy Science Centre, Manchester, M13 9PT, UK. E-mail: marilena.hadjidemetriou@manchester.ac.uk
bLaboratory for Lipidomics and Lipid Biology, Division of Pharmacy and Optometry, School of Health Sciences, Faculty of Biology, Medicine & Health, University of Manchester, Manchester Academic Healthy Science Centre, Manchester, M13 9PT, UK. E-mail: anna.nicolaou@manchester.ac.uk
cLydia Becker Institute of Immunology and Inflammation, School of Biological Sciences, University of Manchester, Manchester, M13 9PT, UK
First published on 20th June 2023
Abstract
The spontaneous self-assembly of biomolecules around the surface of nanoparticles (NPs) once exposed to plasma and other biofluids, has been termed the ‘biomolecule corona’. While the protein composition of the biomolecule corona has been widely characterised, the interaction of NPs with the plasma lipidome has not been fully investigated. Here, we use targeted and untargeted lipidomics to analyse a wide spectrum of bioactive lipids adsorbed onto the surface of liposome NPs post-incubation with human plasma. Our data indicate that the biomolecule corona contains a diverse mixture of simple and complex lipid species, including sphingolipids such as ceramides and sphingomyelins, glycerolipids, glycerophospholipids, cholesteryl esters, as well as oxylipin and N-acyl ethanolamine derivatives of fatty acids. Although the corona lipidomic profiles reflected the overall composition of the plasma lipidome, monohydroxy- and oxo-fatty acid oxylipins, mono-, di- and tri- acylglycerols, sphingomyelins and ceramides showed a preferential binding for liposome NP surface. Interestingly, the biomolecule corona lipid profiles appeared to mirror those of the lipoprotein lipid cargo, suggesting that lipid species may be carried within the lipoprotein complexes attached to the corona. Proteomic analysis of corona-associated proteins showed the presence of several apolipoproteins (A-I, A-II, A-IV, B, C-I, C-III, C-IV, C2-C4, D, E, L, M and lipoprotein Lp(A)), supporting this notion. Our findings reveal the wide lipid diversity of the biomolecule corona and indicate a potential lipoprotein-mediated adsorption mechanism of lipids onto liposome NPs, highlighting the importance of bridging proteomics with lipidomics to fully comprehend the interactions at the bio-nano interface.
Introduction
Once nanoparticles (NPs) are exposed to biological fluids, they are immediately covered by a shell of biomolecules. This spontaneous self-assembly of biomolecules onto the surface of NPs is termed the ‘biomolecule corona’.1–3 The formation of the biomolecule corona alters the pristine physicochemical properties of NPs, confers them with a new identity and becomes their interface with biological systems.4,5 As the clinical development and use of NP-based formulations is rising,6 it has become imperative to determine the molecular composition of the biomolecule corona and understand its impact on the bio-nano interactions. To date, the majority of studies focus on the protein composition of the NP-biomolecule corona, with only a few investigating its lipid7–17 and/or other components.18–28Lipids are a diverse group of biomolecules with distinct physicochemical properties allowing them to exhibit a range of bioactivities. They are key components of the cellular membrane, having both structural and signaling roles, and are known to be involved in physiological and pathophysiological processes, including inflammation, neuropathy, metabolic disorders, cardiovascular diseases, tumorigenesis and cancer.29–34 Human plasma contains an array of lipid species produced by the endothelium, platelets and other blood cells, adipose tissue, liver and various organs. These plasma lipids are transported via lipoproteins, albumin and other protein complexes.35,36 Recent plasma lipidomic studies have identified an array of glycerophospholipids, glycerolipids, sphingolipids including ceramides, sterol lipids, prenol lipids, as well as fatty acids (FA) and, their oxylipin and N-acyl ethanolamine (NAE) derivatives.34,37–43 As plasma lipid profiles can be affected by disease, they have been targeted as potential biomarkers.30,33,40–43 Particularly attractive for biomarker discovery are bioactive lipid mediators such as ceramides and oxylipins. Ceramides are potent sphingolipids involved in apoptotic pathways, and currently considered markers of cardiovascular disease with potential heritable characteristics, whilst oxylipins are oxygenated derivatives of polyunsaturated fatty acids (PUFA) with diverse properties and critical biological roles in a wide array of homeostatic and pathophysiological events, including systemic inflammation.31,40,44–46
The large number and structural diversity of lipids found in biofluids necessitates the lipidomic analysis of the NP-biomolecule corona, in order to decipher the role and potential impact of corona-associated lipids on critical biological events occurring at the bio-nano interface, including NP cellular internalisation, active targeting, clearance and cytotoxicity. Hellstrand et al. were the first to report that copolymer NPs interact with cholesterol, triacylglycerols and phospholipids, as well as with high-density lipoprotein (HDL) complexes from human plasma.7 This work triggered more investigations into the lipid composition of the biomolecule corona formed in various biofluids (e.g. plasma, serum, pulmonary surfactant), using several experimental methods (e.g. cholesterol assays, glycerol assays, metabolomic- and lipidomic-based liquid chromatography mass spectrometry) and different NP types (e.g. titanium dioxide, iron oxide, 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) liposomes, magnetic, gold, single-walled carbon nanotubes).8–23 These studies demonstrated the presence of glycerophospholipids (e.g. phosphatidylcholines (PC), phosphatidylinositols (PI), lysophosphatidylcholines (LPC), phosphatidylethanolamines (PE), lysophosphatidylethanolamines (LPE)), glycerolipids (e.g. mono (MAG)-, di (DAG)-, tri (TAG)-acylglycerols), sphingolipids (e.g. sphingomyelins (SM)), cholesterol and free fatty acids (FFA), as well as lipoprotein complexes. The lipid cargo of lipoproteins comprises simple lipids, such as ceramides, oxylipins and cholesterol, as well as complex glycerophospholipids, sphingolipids and acylglycerols,47,48 that may contribute to the lipid diversity of the NP-biomolecule corona.10,19 However, more work to fully characterise the lipidome of the biomolecule corona is needed to support this notion. Studies combining proteomics and lipidomics have also provided a better understanding of the interaction mechanism between the plasma components and NPs, as well as its impact at the bio-nano interface,9,11,12 suggesting that the complete characterisation of the biomolecule corona may be key to improving our knowledge on its formation and evolution.
To date, most studies examining the lipidome of the NP-biomolecule corona have reported the presence of glycerophospholipids, glycerolipids, sphingomyelins, cholesterol and some FFA species, while there is a notable lack of information on the presence of bioactive lipid mediators including ceramides and oxylipins.7–17 Liposome NPs are considered the most clinically established nanoplatform for drug delivery, yet their interaction with plasma lipids has been scarcely explored, with only one published study to date.10 La Barbera et al. demonstrated the presence of glycerophospholipids (PC, PE and PI), sphingolipids and a few FFA on positively-charged DOTAP liposomes using a metabolomics approach. Nevertheless, more work is required to assess the interactions of clinically used liposomal formulations with plasma lipids.
In this work, we present an in-depth lipidomic analysis of the plasma-derived liposome-biomolecule corona using both targeted and untargeted lipidomics (Table 1 and ESI Fig. S1A†). Our findings show that the liposome NP-corona is rich in simple and complex bioactive lipids, including an array of previously unexplored oxylipins, NAEs and ceramides, reflecting the diversity of the human plasma lipidome. Interestingly, liposome NPs show a preference towards monohydroxy- and oxo-FA oxylipins, 2-MAG, DAG, TAG, SM and ceramides. In addition, proteomic analysis of the biomolecule corona was carried out to examine potential links between the corona identified lipids and proteins. Our data reveal the presence of lipid-binding proteins in the corona, including lipoproteins and albumin, suggesting a potential protein-mediated interaction of liposome NPs with plasma lipids. Our results demonstrate that the biomolecule corona composition mimics the lipidomic fingerprints of the biofluid of origin and highlight the importance of integrating lipidomics with proteomics in future investigations of the bio-nano interface.
| Lipid category | Lipid class | Lipidomic analysis approach | Data shown in |
|---|---|---|---|
| Fatty acyls and derivatives | Oxylipins | Targeted (UPLC/ESI-MS/MS) | Fig. 1 |
| N-Acyl-ethanolamines (NAE) | Targeted (UPLC/ESI-MS/MS) | Fig. 1 | |
| Free fatty acids (FFA) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 1 | |
| Sphingolipids | Ceramides | Targeted (UPLC/ESI-MS/MS) | Fig. 2 |
| Sphingomyelins (SM) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Glycerolipids | Mono-acylglycerols (2-MAG) | Targeted (UPLC/ESI-MS/MS) | Fig. 1 |
| Di-acylglycerols (DAG) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Tri-acylglycerols (TAG) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Glycerophospholipids | Phosphatidylcholines (PC) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 |
| LysoPC (LPC) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Phosphatidylethanolamines (PE) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Phosphatidylinositol (PI) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 | |
| Sterol lipids | Cholesteryl esters (CE) | Untargeted (UHPSFC/ESI-QTOF-MSE) | Fig. 3 |
Results & discussion
Although there has been a strong interest and intense effort to comprehensively characterise the protein content of the NP-biomolecule corona, to date only a few studies have interrogated the spontaneous interaction of NPs with plasma lipids.7–12,28 Blood contains a wide range of lipid species,34,37–43 yet our current knowledge of the lipidomic content of the NP-biomolecule corona is limited to the analysis of glycerophospholipids and glycerolipids.7–12,18 In this study we report an extensive lipidomic profile of the human plasma liposome NP-biomolecule corona that includes a range of glycerophospholipids (PC, LPC, PE, PI), sphingolipids (ceramides, SM), glycerolipids (DAG, TAG), FFA and fatty acyl derivatives of the oxylipin, NAE and 2-MAG classes (Table 1). We also perform a lipidomic analysis of whole plasma, plasma processed by the two-step purification protocol used to isolate the corona-coated NP (referred to as ‘purified plasma’) and of bare liposomes, as controls (see experimental section for details). Our results provide the first experimental evidence that the liposome NP-biomolecule corona contains bioactive lipid mediators such as ceramides and oxylipins, known to be involved in systemic inflammatory disease and also explored for biomarker discovery.30,32,40,43Overall, only very small amounts of some lipid species were detected in the purified plasma and/or bare liposomes controls (marked with # in Fig. 1–3), indicating that the corona lipid profiles mainly resulted from the adsorption of plasma lipids onto liposome NPs and were not artefacts of incomplete purification or components from the employed liposomal formulation. The liposome NP-corona purification protocol employed ensures retention of the tightly adsorbed molecules onto the NP surface, also referred to as the ‘hard corona’.
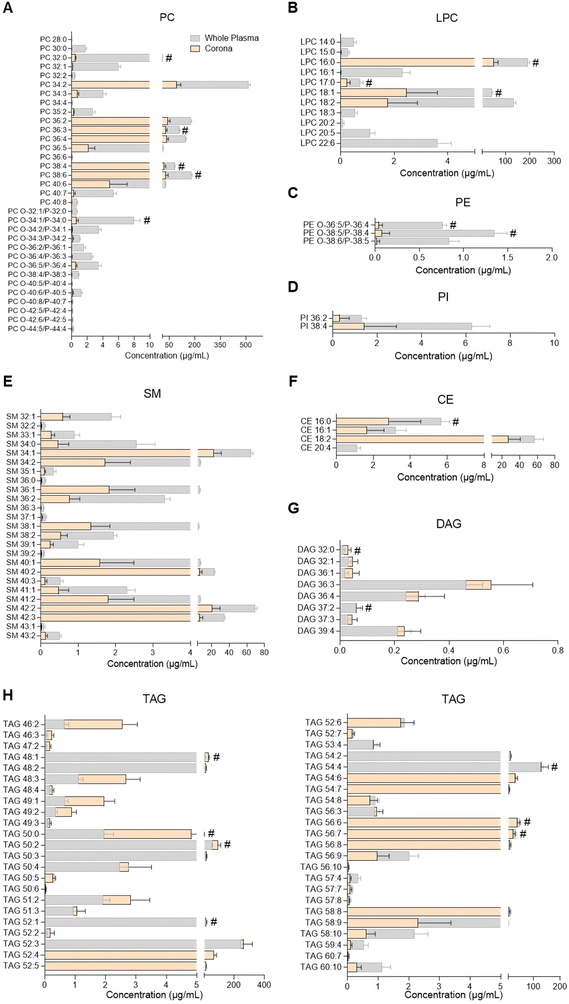 | ||
| Fig. 3 Profiles of glycerophospholipids, sphingomyelins, glycerolipids and cholesteryl esters in human plasma and the liposome nanoparticle (NP)-biomolecule corona. (A) Phosphatidylcholines (PC); (B) lysophosphatidylcholines (LPC); (C) phosphatidylethanolamines (PE); (D) phosphatidylinositols (PI); (E) sphingomyelins (SM); (F) cholesteryl esters (CE); (G) diacylglycerols (DAG); (H) triacylglycerols (TAG). Lipids were analysed by UHPSFC/ESI-QTOF-MSE. Data shown as the mean ± SD of n = 5 analyses; # denotes lipid species also found in bare liposomes. Constituent lipids of the liposomes have been excluded from the figures but can be seen in ESI Table S2.† | ||
It should be mentioned that any lipids found in the NP-corona that were also identified at equal concentrations in bare liposomes were excluded from Fig. 1–3. These were either components identified by the supplier or impurities of the HSPC lipid used (ESI Table S2†) and include the NAE stearoyl ethanolamine (STEA), the CER[NP] species N(26)P(17) and N(26)P(18), PC 34:0, PC 36:0, LPC 18:0, LPC 20:0, LPC 20:4, the FFA stearic acid (C18:0) and palmitic acid (C16:0), as well as the linoleic and alpha-linolenic acid oxylipin derivatives 9-HOTrE, 9(10)-EpOME, 9,10-DiHOME, 12,13-DiHOME (ESI Tables S3–S7†).
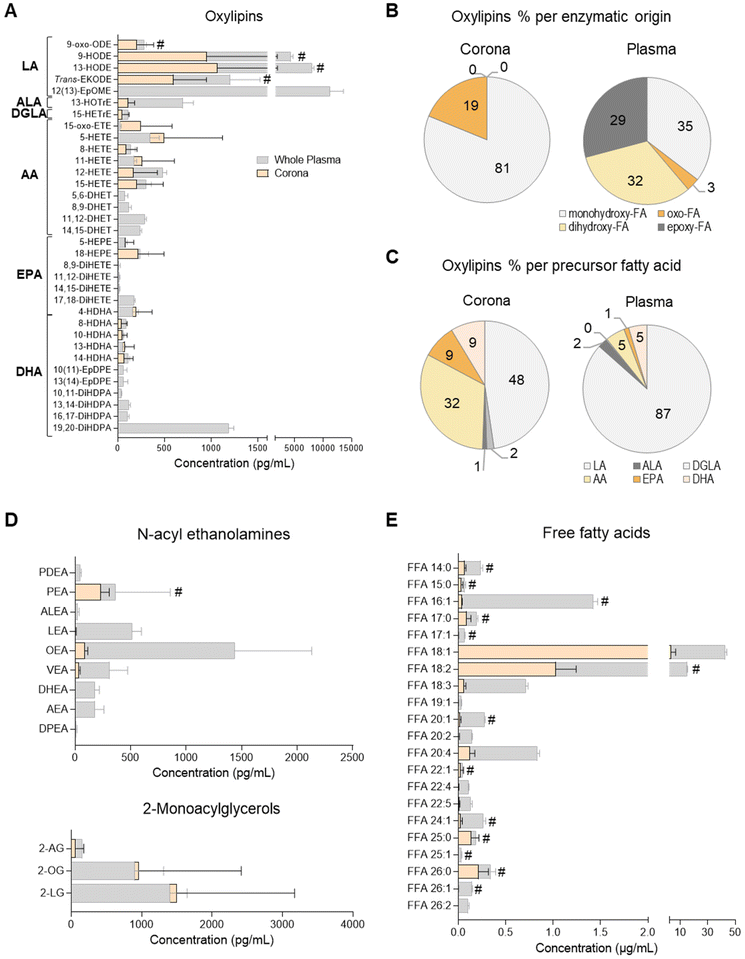
Oxylipin composition of the plasma liposome NP-biomolecule corona
A total of 38 oxylipins, derivatives of the PUFAs linoleic acid (LA; C18:2n-6), alpha-linolenic acid (ALA; C18:3n-3), dihomo-gamma-linolenic acid (DGLA; C20:3n-6), arachidonic acid (AA; C20:4n-6), eicosapentaenoic acid (EPA; C20:5n-3) and docosahexaenoic acid (DHA; C22:6n-3) were quantitated in human plasma samples using targeted lipidomics (UPLC/ESI-MS/MS)49 (Fig. 1A and ESI Table S3†). These species were mainly derivatives of lipoxygenase (LOX) and cytochrome P450 monooxygenase/soluble epoxide hydrolase (CYP/sEH) enzymatic metabolism of PUFA. Although a number of cyclooxygenase (COX)-derived mediators were also detected, they were not consistently found to be above the limit of quantitation of the assay (data not shown).Of these 38 plasma oxylipins, 19 species were found in the liposome NP-corona (Fig. 1A). A number of CYP/sEH epoxy- and dihydroxy-fatty acids (i.e. EpOME, EpDPE, DiHOME, DHET, DiHETE, DiHDPA) were found mainly in whole plasma, while monohydroxy- and oxo-fatty acids (i.e. oxo-ODE, HODE, Trans-EKODE, HOTrE, oxo-ETE, HETE, HEPE and HDHA) were also identified in the liposome NP-corona. These differences are reflected in the relative abundance of oxylipins, when grouped according to their enzymatic origin. Although 35% of the whole plasma oxylipins were LOX-derived monohydroxy-FA, their proportion in the liposome NP-corona increased to 81%, with no CYP/sEH-derived dihydro-FA species being identified in the corona (Fig. 1B). When grouped according to their precursor PUFA, 87% of the oxylipins found in whole plasma were LA-derivatives with only 5% derived from AA. However, the liposome NP-corona profiles differed, with 48% LA-derivatives and 32% AA-derivatives (Fig. 1C). Furthermore, the liposome NP-corona had no PUFA dihydroxy- and epoxy-FA compared with whole plasma (32% and 29% of these found in whole plasma, respectively), but had a higher abundance of oxo-FA derivatives (19% compared with 3% in whole plasma).
Oxylipins are a group of low abundance oxygenated derivatives of PUFA, better known for their involvement in inflammation.44–46 The presence of various oxylipins in blood reflects endothelial, platelet and immune cell biosynthesis, as well as lipoprotein cargo, and can be influenced by nutritional status and systemic disease.37,47,49 This group of lipid mediators includes the COX-derived prostanoids, LOX-derived mono- and poly-hydroxy-FA, and CYP/sEH epoxy- and hydroxy-FA. Our data reveal that the liposome NP-corona carries a range of oxylipins, with some showing a preferential binding to liposome NPs surfaces (Fig. 1). Interestingly, the relative abundance of monohydroxy-FA found in the NP-corona was higher than whole plasma (81% vs. 35%, respectively), suggesting their preferential binding to liposomes, while dihydroxy- and epoxy-FA were detected exclusively in plasma (Fig. 1A). Furthermore, even though LA-derived mediators dominated the plasma profile, the corona oxylipins included a large component of both LA- and AA-derivatives (Fig. 1C). These differential profiles may be reflecting the lipoprotein cargo typically consisting of mono-hydroxy-FA species including the LA-derived HODE (VLDL, LDL and HDL).47,50
Endocannabinoid N-acyl ethanolamine and 2-monoacyl glycerol species of the plasma liposome NP-biomolecule corona
N-Acyl ethanolamines (NAE) and 2-monoacyl glycerols (2-MAG) are FA derivatives found in plasma, cells, tissues and other biofluids, many of which have endocannabinoid-like activities. We quantitated 10 species of NAE and 3 species of 2-MAG in whole plasma using a targeted UPLC/ESI-MS/MS lipidomic assay (Fig. 1D and ESI Table S4†). All 2-MAG species found in plasma were also present in the liposome NP-corona. However, the NP-corona contained only a few NAEs and these were palmitoyl ethanolamine (PEA), linoleoyl-EA (LEA), oleoyl ethanolamine (OEA) and vaccenoyl ethanolamine (VEA). As previously discussed, STEA was identified in the liposome NP-corona, however as it was also found at an equal concentration in bare liposomes (ESI Table S4†). PEA was also found in bare liposomes (ESI Table S4†), but in lower abundance than in the liposome NP-corona (Fig. 1D; marked with #). Both these NAE species are known to contain the precursor FA palmitic acid and stearic acid, respectively, which are indeed identified as components of the HSPC by the supplier (ESI Table S2†).Free fatty acids (FFA) of the plasma liposome NP-biomolecule corona
The presence of FFA was also assessed by UHPSFC/ESI-QTOF-MSE using an untargeted approach with relative quantitation (Fig. 1E and ESI Table S5†). This analysis revealed that both whole plasma and corona carry with a wide array of FFA, with different degree of unsaturation and acyl chain lengths. The most abundant corona FFA species were C18:1, C18:3, C20:2, C20:4 and C22:5. Whole plasma had higher concentrations of FFA compared to the liposome NP-corona, whilst 15 of the 23 corona FFA were also identified in bare liposomes, with stearic acid (C18:0) and palmitic acid (C16:0) being the most abundant species (Fig. 1E). As discussed, this was attributed to the composition of the HSPC phospholipid used for the preparation of liposomes (ESI Table S2†).The presence of FFA has been reported in the serum biomolecule corona of positively charged liposomes (DOTAP) and mineralo-organic NPs.10,18 In our study, we report a wider range of FFA including saturated and unsaturated species with medium, long and very long acyl chains (Fig. 1E). These differences in the profile of FFA may be attributed to the use of plasma instead of serum, the composition of NPs and/or the use of a different lipidomic platform, as our approach is based on neutral phospholipid liposomes and our method on supercritical fluid chromatography. Exploring the affinity of FFA for NPs prepared from different liposome formulations may support the development of lipid-selective NPs, as the properties of the liposomes may influence the composition of the lipid corona, similar to what has already been reported for NP-protein interactions.1
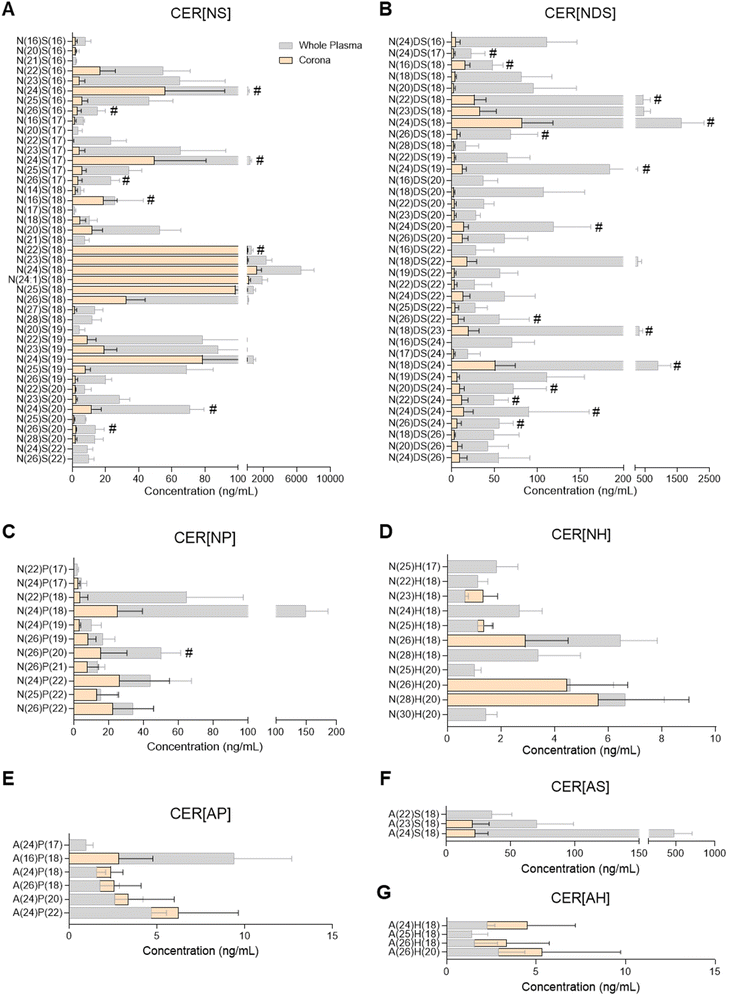
Ceramide composition of the plasma liposome NP-biomolecule corona
Ceramide species were analysed by UPLC/ESI-MS/MS using a targeted approach with relative quantitation (Fig. 2 and ESI Table S6†). The majority of ceramides found in the liposome NP-corona were derivatives of the sphingoid bases sphingosine and dihydro-sphingosine with non-hydroxy FA (ceramide classes CER[NS] and CER[NDS]; Fig. 2A and B). A number of minor species with phyto-sphingosine, 6-hydroxy sphingosine and sphingosine bases with non-hydroxy FA (CER[NP], CER[NH]) or alpha-hydroxy FA (CER[AP], CER[AH], CER[AS]) were also detected (Fig. 2C–F). The profile of liposome NP-corona ceramides reflected the prevalence of whole plasma ceramides and their concentration levels were mostly lower than plasma. Overall, ceramide species of the CER[NS] and CER[NDS] classes were the most abundant in both whole plasma and corona samples, while ceramides of the CER[NH], CER[AP] and CER[AH] classes were minor species (ESI Fig. S2A†).Glycerophospholipid, sphingomyelin, glycerolipid and sterol lipid composition of the plasma liposome NP-biomolecule corona
PC, LPC, PE, PI, SM, cholesteryl ester (CE), DAG and TAG species were analysed by UHPSFC/ESI-QTOF-MSE using an untargeted approach with relative quantitation (individual lipid species shown in Fig. 3 and ESI Table S7†). The concentration of each lipid class in whole plasma, liposome NP-corona, purified plasma and bare liposome samples is shown in ESI Fig. S2B.† The range of lipids found on the liposome NP-corona was similar to that of whole plasma lipids, however the levels of certain lipid classes differed. A total of 33 PC species were found in whole plasma and 31 in liposome NP-corona, all detected at higher concentrations in plasma than liposome NP-corona (Fig. 3A). Interestingly, most of the alkyl ether and alkenyl ether PC species (PC-O/PC-P) species were only found at very low concentrations in the liposome NP-corona compared to whole plasma. 46 TAG species were measured in all samples and interestingly, the liposome NP-corona contained species with shorter acyl chains (TAG 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2–52
2–52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) at higher levels compared with whole plasma (Fig. 3H).
3) at higher levels compared with whole plasma (Fig. 3H).
Considering the importance of ceramides and other sphingolipids in the central nervous system, signal transduction, cell proliferation, differentiation and death, as well as their implication in inflammatory diseases including neuroinflammation, cancer and cardiovascular disease, their potential interaction with NPs could support biomedical applications.32,43,51 For example, ceramide biosynthesis is activated upon cancer chemotherapy, and thus ceramide-rich NPs could potentially be used to selectively deliver ceramides to malignant cells, aiming to supress cell growth or inducing apoptosis.51 Another potential application stems from the dysregulation of sphingolipid metabolism in the presence of tumours.51 Despite the biological significance of ceramides and their presence in blood and other biofluids, there has been only one previous study investigating the presence of sphingolipids in the NP-biomolecule corona, reporting 23 different SMs and only one ceramide species interacting with the surface of DOTAP liposomes.10 In the present study, 25 SM species and more than 90 ceramides were found in the liposome NP-corona (Fig. 2, 3E and ESI Tables S6, S7†). The ceramide profile of the liposome NP-corona ceramides reflected that of whole plasma, albeit at lower concentrations.
Most previous studies have focused on the identification of glycerophospholipids (PC, LPC, PE, LPE, PI), glycerolipids (TAG, DAG, MAG) and cholesterol in the corona upon exposure of NP to various biofluids.7–12,18 These revealed the presence of a number of these lipid species in the corona, with PC, TAG and DAG being the most abundant. In agreement with the literature, we identified several glycerophospholipids (PC, LPC, PE, PI), sterol lipids (CE) and glycerolipids (TAG, DAG, 2-MAG) in the corona (Fig. 1D, 3 and ESI Tables S4, S7†). Among these, liposomes showed a preference towards glycerolipids, with the concentrations of their individual species in the corona being comparable to whole plasma (Fig. 1D, 3G and H). Further studies could explore this selectivity and assess whether it is related to specific lipoproteins preferentially interacting with the liposomes used. Although to a lower extent than glycerolipids, CE and glycerophospholipids also interacted with liposomes, with corona concentrations lower than whole plasma (Fig. 3A–D and F). All these suggest a preferential binding of liposomes to neutral lipids (TAG, DAG, 2-MAG). Interestingly, the lipid corona profile found here differs from that previously seen in DOTAP liposomes, with only a few PC species (PC 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, PC 36
2, PC 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) found to be common among glycerolipids and glycerophospholipids in each case.10 This potentially stems from the positive charge and/or the different chemical composition of the DOTAP liposomes, suggesting that the physicochemical properties of the NP could play a significant role in the corona lipid configuration and could be tuned to target specific lipid corona patterns.
4) found to be common among glycerolipids and glycerophospholipids in each case.10 This potentially stems from the positive charge and/or the different chemical composition of the DOTAP liposomes, suggesting that the physicochemical properties of the NP could play a significant role in the corona lipid configuration and could be tuned to target specific lipid corona patterns.
Proteomic analysis of the liposome NP-biomolecule corona
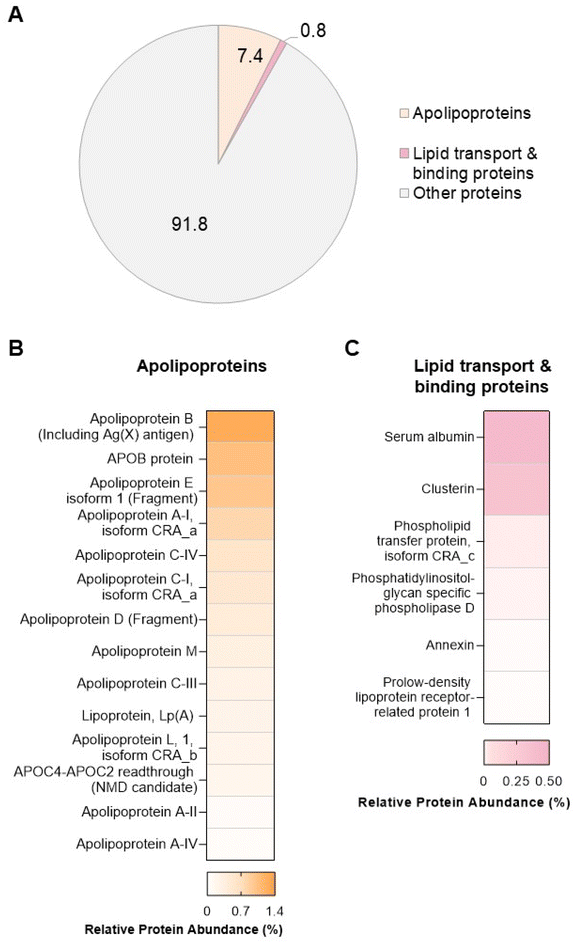
In order to explore potential links between the proteins and lipids found in the biomolecule corona, we performed proteomic analysis by LC-MS/MS. A total of 285 proteins (ESI Table S8†) were found in the liposome NP-corona. Of these proteins, we identified 14 apolipoproteins (apolipoproteins A-I, A-II, A-IV, B, C-I, C-III, C-IV, C4-C2, D, E, L, M), including lipoprotein Lp(A) – a key constituent of low-density lipoprotein (LDL) particles that carry free cholesterol (Fig. 4). The identified apolipoproteins are known to associate with HDL, LDL and very-LDL (VLDL) particles, as well as chylomicrons – large triacylglycerol-rich lipoproteins, with the most abundant species (apolipoprotein B, E and A-I) being major structural components of LDL (APOB), chylomicrons-VLDL-LDL (APOE) and HDL (APOA-I). Apart from apolipoproteins, 6 lipid-associated proteins were also identified in the liposome NP-corona: 2 lipid-transport proteins (albumin, phospholipid transfer protein) and 4 lipid-binding proteins (clusterin, phosphatidylinositol-glycan-specific phospholipase d, annexin, prolow density lipoprotein receptor-related protein 1). To elucidate the contribution of these proteins to the total protein content of the biomolecule corona, we calculated the total mean relative protein abundance (RPA%) of these two categories (Fig. 4A). Apolipoproteins accounted for 7% of the total liposome NP-corona proteins, while lipid transport/binding proteins accounted for approximately 1% of the total liposome NP-corona protein content. Our data suggest a protein-mediated interaction of lipids with the surface of liposomes NPs. This observation does not exclude direct binding of the lipid molecules onto the NP surface and more work is needed to fully comprehend the self-assembly mechanism of the biomolecule corona formation.During the past decade, we have learnt that a complex biomolecule corona forms rapidly on the surfaces of nanoscale materials to varying degrees, depending on their physicochemical properties. A diverse array of biomolecules has now been found in the multi-layered structure of the corona including proteins,3 lipids,7–17 metabolites18–21,23,25–28 and cell free DNA (cfDNA).24,52 In this study, we developed a workflow for the comprehensive investigation of the lipidomic and proteomic content of the biomolecule corona. Such multi-omics approaches are essential to fully comprehend the effect of the biomolecule corona on the overall clinical performance of NPs. For example, a recent proof-of-principle study by Rabel et al.11 combined proteomics with lipidomics to provide a mechanistic understanding of the effect of the biomolecule corona formation on the colloidal stability, degradation and toxicity of iron oxide NPs.11 More work is also needed to comprehensively characterise the biomolecule corona under dynamic in vivo conditions, upon intravenous administration of NPs.53,54 Understanding the multi-layered composition of the NP biomolecule corona opens up possibilities to engineer NP formulations that bind certain protein and lipid species, in order to target specific organs.55
It is now well established that the composition of the biomolecule corona varies in the presence of a disease,24,56–58 and this has led to the exploitation of the biomolecule protein corona as a novel protein biomarker discovery tool to meet pressing clinical needs.56–62 The emerging Nano-omics paradigm seeks to apply the knowledge garnered at the bio-nano interface in order to unveil novel disease specific fingerprints in blood and other biological fluids.52 Proteogenomic analysis of the liposome NP biomolecule corona has recently revealed a protein-mediated interaction of liposomes with cfDNA and highlighted the potential exploitation of the biomolecule corona as a novel blood-analysis nanoscale tool for cancer diagnostics.24 Considering the diversity of the NP-biomolecule corona lipids identified in this study, we anticipate that this work will provide impetus for further studies aiming to explore the use of NPs as a scavenging tool for the enrichment and analysis of lipid biomarkers. Overall, the complex multi-omics fingerprint of the biomolecule corona offers an exciting opportunity to develop nanoscale multi-omics platforms for biofluid based biomarker discovery.
Conclusion
In this study, we describe the lipidomic profile of the biomolecule corona formed around human plasma-incubated liposome NPs, using targeted and untargeted approaches. Our findings reveal a complex lipidomic corona fingerprint that reflects to a great extent the composition of the plasma lipidome. We found that the corona lipidome comprises a range of free fatty acids, glycerolipids, glycerophospholipids and lysoglycerolipids, sphingomyelins, cholesteryl esters, ceramides, oxylipins and N-acyl ethanolamines. Compared to plasma, the NP-biomolecule corona demonstrated preferential binding for monohydroxy-fatty acids, mono-, di- and tri- acylglycerols, sphingomyelins and an array of ceramides. Proteomic analysis of the NP-biomolecule corona revealed the precence of lipoproteins and other lipid-transport proteins suggesting a protein-mediated mechanism of lipid adsorption onto the NP surface. Our work highlights the importance of integrative multi-omics to fully comprehend the multi-molecular composition of the biomolecule corona and opens up new possibilities for the exploitation of the bio-nano interactions as a novel multi-omic blood analysis nanoscale tool.Experimental
Human plasma samples
K2 EDTA plasma samples from healthy female donors were supplied by Cambridge Bioscience, UK (Batches: 385365, 385429, 385446, 386259 and 386260; ESI Table S1†). Frozen plasma samples were received on dry ice and stored at −80 °C. Samples were thawed, centrifuged for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, pooled together, aliquoted in 1 mL aliquots and stored at −80 °C. These pooled samples were used for all experiments (n = 5 aliquots per assay).
000 rpm, 4 °C, pooled together, aliquoted in 1 mL aliquots and stored at −80 °C. These pooled samples were used for all experiments (n = 5 aliquots per assay).
Preparation and physicochemical characterisation of liposome NPs (HSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) Cholesterol 59.6
Cholesterol 59.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 40.4 w/w%)
40.4 w/w%)
Liposome NPs (12.5 mM) were prepared using phosphatidylcholine from hydrogenated soy bean (HSPC; Lipoid, Germany; ESI Table S2†) and cholesterol (Sigma Aldrich, UK) by the thin lipid film method followed by hydration and extrusion, as previously described.53 This liposome composition was chosen as it forms the basis of the clinically-used liposomal doxorubicin formulation (Doxil®). The size and surface charge of liposome NPs were measured by Dynamic Light Scattering and Electrophoretic Light Scattering, respectively, using a Zetasizer Nano ZS (Malvern Panalytical, Instruments, UK) (ESI Fig. S3A†).
Ex vivo liposome NP-biomolecule corona formation
The biomolecule corona was allowed to form by adding liposome NPs (180 μL, 12.5 mM) to human plasma samples (820 μL) and incubating for 10 min at 37 °C in an orbital shaker (ThermoFisher, MaxQ™ 4450 Benchtop Orbital Shaker) at 250 rpm. Samples were then subjected to a two-step purification protocol, comprising size exclusion chromatography and membrane ultrafiltration, as previously described, to isolate the corona-coated liposome NPs from unbound plasma biomolecules.24,53,54,56–59,63 Corona lipids were then extracted using organic solvents and analysed by targeted (UPLC/ESI-MS/MS) and untargeted lipidomics (UHPSFC/ESI-QTOF-MSE) (Table 1). Plasma samples from the same healthy donors were also analysed (whole plasma) and compared with the resulting liposome-corona profiles. To confirm that lipids identified as part of the corona lipidome were indeed liposome-NP adsorbed lipids and not unbound, co-eluting species, or artefacts of the purification process, plasma samples without liposome NPs added, were subjected to the same two-step purification protocol (purified plasma) and analysed as a control. As the NPs used in this study were lipid-based, lipidomic analysis of liposome NPs without any interaction with plasma (bare liposomes) was performed, as another control. It should be noted that for all sample preparations described here (liposome NP-corona, whole and purified plasma), we started with the same volume (820 μL) of plasma.Quantitation of liposome NPs
The concentration of phospholipids (mM) in the liposome preparations was determined by the Stewart assay (ESI Fig. S3B†), as previously described.24,53,54,56–59,63 In brief, liposome NPs were treated with ammonium ferrothiocyanate [(NH4)4Fe(SCN)6] and the absorbance of the resulting solution was measured at 485 nm using a Cary 50 Bio Spectrophotometer (Agilent Technologies, USA).Quantitation of liposome NP-corona proteins
The total amount of proteins adsorbed onto the surface of liposome NPs was quantitated by the BCA Protein assay kit (ThermoFisher Scientific), as previously desrcibed.24,53,54,56–59,63 Protein binding (Pb) values, defined as amount of protein per mole of liposome NP, were then calculated per sample. Data shown as the mean ± SD (μg of protein/μmole of liposome NP) n = 25 technical repeats per treatment group (ESI Fig. S3B†).Analysis of oxylipins by ultra-performance liquid chromatography tandem mass spectrometry with an electrospray ionisation source (UPLC/ESI-MS/MS)
Extraction and analysis of oxylipins was performed, as previously described.39,49 Briefly, plasma samples (1 mL), corona-coated liposome NPs (100 μL) and controls (purified plasma 100 μL; bare liposome NPs 100 μL) (n = 5 technical repeats per group) were spiked with deuterated internal standards: 20 ng each of prostaglandin PGB2-d4, 12-HETE-d8, 15-HETE-d8, 8,9-EET-d11, 8,9-DHET-d11 and 12,13-DiHOME-d4; Cayman Chemicals, Ann Arbor, MI, (USA). Oxylipins were extracted using ice-cold methanol (15% v/v) followed by solid phase extraction (SPE). The extracted oxylipins were separated on a C18 column (Acquity UPLC ® BEH C18, 2.1 × 50 mm, 1.7 μm; Waters, UK) and analysed by UPLC/ESI-MS/MS (UPLC Acquity and Xevo TQ-S, Waters, USA). Detection was performed by multiple reaction monitoring (MRM) in the negative ionisation mode (ESI−). Quantitation was performed using calibration lines constructed with commercially available standards. Instrument and data analysis were carried out by MassLynx and TargetLynx software (Waters, UK).Analysis of NAE and 2-MAG by UPLC/ESI-MS/MS
Extraction and analysis of NAE and 2-MAG were performed, as previously described.39,49 Briefly, samples (500 μL each) of plasma, corona-coated liposome NPs and controls (purified plasma and bare liposome NPs) (n = 5 technical repeats per group) were spiked with 20 ng of AEA-d8 and 40 ng of 2-AG-d8; Cayman Chemicals, Ann Arbour, MI, (USA) and extracted using ice-cold chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (2
methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). NAE and 2-MAG were recovered in the organic layer and analysed by UPLC/ESI-MS/MS (UPLC Acquity and Xevo TQ-S, Waters, USA) using a C18 column (Acquity UPLC ® BEH C18, 2.1 × 50 mm, 1.7 μm; Waters, UK) and gradient elution. Detection was performed in the positive ionisation mode (ESI+) using MRM. Lipid species were quantitated using calibration lines constructed using commercially available standards. MassLynx and TargetLynx software (Waters, UK) were used for instrument control and data analysis.
1 v/v). NAE and 2-MAG were recovered in the organic layer and analysed by UPLC/ESI-MS/MS (UPLC Acquity and Xevo TQ-S, Waters, USA) using a C18 column (Acquity UPLC ® BEH C18, 2.1 × 50 mm, 1.7 μm; Waters, UK) and gradient elution. Detection was performed in the positive ionisation mode (ESI+) using MRM. Lipid species were quantitated using calibration lines constructed using commercially available standards. MassLynx and TargetLynx software (Waters, UK) were used for instrument control and data analysis.
Analysis of ceramides by UPLC/ESI-MS/MS
Extraction and analysis of ceramides was performed, as previously described.39,49 Briefly, 50 μL of plasma samples, corona-coated liposome NPs and controls (purified plasma and bare liposome NPs) (n = 5 technical repeats per group) were spiked with 4 ng each of CER[N(16)S(18)]-d9, CER[N(16)DS(18)]-d9, CER[N(16)H(18)]-d9, CER[N(16)P(18)]-d9, CER[A(16)S(18)]-d9, CER[A(16)DS(18)]-d9, CER[A(16)H(18)]-d9 and CER[A(16)P(18)]-d9, Avanti Polar Lipids, Alabaster, AL, (USA) and extracted using ice-cold chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (2
methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) before clean-up using silica SPE cartridges (100 mg Strata SI-1 Silica, Phenomenex, UK). The analysis of ceramides was performed by UPLC/ESI-MS/MS. Ceramides were separated on a C8 column (UPLC BEH C8, 2.1 × 100 mm, 1.7 μm; Waters, UK). Detection was performed by MRM using ESI+. Ceramides were identified by retention time and characteristic fragmentation patterns. Relative quantitation of ceramide species was carried out using class-specific deuterated internal standards. Instrument and data analysis were carried out by MassLynx and TargetLynx softwate (Waters, UK).
1 v/v) before clean-up using silica SPE cartridges (100 mg Strata SI-1 Silica, Phenomenex, UK). The analysis of ceramides was performed by UPLC/ESI-MS/MS. Ceramides were separated on a C8 column (UPLC BEH C8, 2.1 × 100 mm, 1.7 μm; Waters, UK). Detection was performed by MRM using ESI+. Ceramides were identified by retention time and characteristic fragmentation patterns. Relative quantitation of ceramide species was carried out using class-specific deuterated internal standards. Instrument and data analysis were carried out by MassLynx and TargetLynx softwate (Waters, UK).
Analysis of complex lipids (glycerophospholipids, sphingomyelins, glycerolipids, cholesteryl esters) and FFA by ultra-high performance supercritical fluid chromatography coupled to quadrupole time of flight mass spectrometry with electrospray ionisation (UHPSFC/ESI-QTOF-MSE)
Extraction and analysis of complex lipids was performed, as previously described.46,64 Briefly, 50 μL of plasma samples, corona-coated liposome NPs and controls (purified plasma and bare liposome NPs) (n = 5 technical repeats per group) were spiked with 4 μg each of CE 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d7, SM 16
0-d7, SM 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d31, PC 16
0-d31, PC 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d31-18
0-d31-18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, LPC 26
1, LPC 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d4, PE 16
0-d4, PE 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d31-18
0-d31-18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, TAG 17
1, TAG 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-17
0-17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 117
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d5, DAG 18
0-d5, DAG 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1-d5, lyso-PE (LPE) 18
1-d5, lyso-PE (LPE) 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1-d7, 12 μg of phosphatidylglycerol (PG 16
1-d7, 12 μg of phosphatidylglycerol (PG 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0-d31-18
0-d31-18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Avanti Polar Lipids, AL, USA); and 4 μg of palmitic acid-d31 (Sigma Aldrich, UK) and extracted using ice-cold chloroform
1) (Avanti Polar Lipids, AL, USA); and 4 μg of palmitic acid-d31 (Sigma Aldrich, UK) and extracted using ice-cold chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (2
methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). After the addition of ice-cold water, the organic layer was retrieved by centrifugation, evaporated to dryness and reconstituted in chloroform/2-propanol (1
1 v/v). After the addition of ice-cold water, the organic layer was retrieved by centrifugation, evaporated to dryness and reconstituted in chloroform/2-propanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The complex lipids and FFA were analysed by untargeted UHPSFC/ESI-QTOF-MSE (UHPSFC, Acquity UPC2, Waters; Synapt G2 High Definition QTOF, Waters, Milford, MA, USA) in both ESI+ and ESI−. Chromatographic separation was carried out using an Acquity UPC2 Torus 2-PIC column (100 mm × 3.0 mm × 1.7 μm; Waters, UK) and a gradient elution with CO2 and methanol with 1% ultrapure water and 10 mM ammonium acetate. Data acquisition was done by MSE in full scan mode over the range m/z 150–1200 Da. Mass data were corrected during acquisition using leucine encephalin (m/z 556.2771 for ESI+ and 554.2615 for ESI-). MassLynx software (Waters, UK) and Progenesis QI (Nonlinear Dynamics, Newcastle, UK) were used for data acquisition and processing, respectively. Relative quantitation was based on the concentration of class-specific deuterated internal standards.
1, v/v). The complex lipids and FFA were analysed by untargeted UHPSFC/ESI-QTOF-MSE (UHPSFC, Acquity UPC2, Waters; Synapt G2 High Definition QTOF, Waters, Milford, MA, USA) in both ESI+ and ESI−. Chromatographic separation was carried out using an Acquity UPC2 Torus 2-PIC column (100 mm × 3.0 mm × 1.7 μm; Waters, UK) and a gradient elution with CO2 and methanol with 1% ultrapure water and 10 mM ammonium acetate. Data acquisition was done by MSE in full scan mode over the range m/z 150–1200 Da. Mass data were corrected during acquisition using leucine encephalin (m/z 556.2771 for ESI+ and 554.2615 for ESI-). MassLynx software (Waters, UK) and Progenesis QI (Nonlinear Dynamics, Newcastle, UK) were used for data acquisition and processing, respectively. Relative quantitation was based on the concentration of class-specific deuterated internal standards.
Analysis of proteins by LC-MS/MS
Digestion of corona proteins was performed prior to LC-MS/MS analysis by suspension trapping (S-trap). Briefly, proteins (10 μg) were mixed overnight with lysis buffer (5% SDS, 50 mM triethylammonium bicarbonate TEAB pH 7.5), reduced with 5 mM dithiothreitol (DTT, Fisher) and alkylated with 15 mM iodoacetamide (IAM, Sigma Aldrich). Samples were then acidified by aqueous phosphoric acid (12% w/w) and S-trap binding buffer (90% aqueous methanol containing 100 mM TEAB pH 7.1) was added to the protein lysates. Subsequently, samples were loaded onto S-trap columns (ProtiFi, LLC, USA) with trypsin (0.1 μg μL−1) at 47 °C for 1 hour and digested peptides were collected. Peptide samples were finally desalted using oligo R3 beads in 50% acetonitrile and dried in a vacuum centrifuge (Heto Speedvac). Following digestion, samples were analysed by LC-MS/MS using an UltiMate® 3000 Rapid Separation LC (RSLC, Dionex Corporation, Sunnyvale, CA) coupled to a Q Exactive™ Hybrid Quadrupole-Orbitrap™ (ThermoFisher Scientific, Waltham, MA) mass spectrometer. LC-MS/MS data processing was carried out using Scaffold software (version 4.8.5, Proteome Software Inc., Portland, OR), as previously described.53,63Author contributions
All authors have given approval to the final version of the manuscript. L. P. designed and performed the experiments, analysed all data and took responsibility for planning and writing the manuscript. A. K. performed lipidomics experiments and contributed to data analysis. D. C.-M. performed lipidomics experiments and data analysis. A. N. and M. H. initiated, designed, directed, supervised and provided intellectual input throughout the study and contributed to the writing of the manuscript. L. P., A. N. and M. H. wrote the manuscript.Abbreviations
| 2-AG | 2-Arachidonoyl-glycerol |
| 2-LG | 2-Linoleoyl-glycerol |
| 2-MAG | 2-Monoacylglycerol |
| 2-OG | 2-Oleoyl-glycerol |
| AA | Arachidonic acid |
| AEA | Arachidonoyl ethanolamine /anandamide |
| ALA | α-Linolenic acid |
| ALEA | Alpha-linolenoyl ethanolamine |
| CE | Cholesteryl ester |
| CER[AH] | Alpha-hydroxy fatty acid/6-hydroxy-sphingosine base ceramide |
| CER[AP] | Alpha-hydroxy fatty acid/phyto-sphingosine base ceramide |
| CER[AS] | Alpha-hydroxy fatty acid/sphingosine base ceramide |
| CER[NDS] | Non-hydroxy fatty acid/dihydro-sphingosine base ceramide |
| CER[NH] | Non-hydroxy fatty acid/6-hydroxy-sphingosine base ceramide |
| CER[NP] | Non-hydroxy fatty acid/phyto-sphingosine base ceramide |
| CER[NS] | Non-hydroxy fatty acid/sphingosine base ceramide |
| COX | Cyclooxygenase |
| CYP/sEH | Cytochrome P450 monooxygenase/soluble epoxide hydrolase |
| DAG | Diacylglycerol |
| DGLA | Dihomo-gamma-linolenic acid |
| DHA | Docosahexaenoic acid |
| DHET | Dihydroxyeicosatrienoic acid |
| DiHDPA | Dihydroxydocosapentaenoic acid |
| DiHETE | Dihydroxyeicosatetraenoic acid |
| DiHOME | Dihydroxyoctadecenoic acid |
| DOTAP | 1,2-Dioleoyl-3-trimethylammonium-propane |
| DHEA | Docosahexaneoyl ethanolamine |
| DPEA | Docosapentaenoyl ethanolamine |
| EPA | Eicosapentaenoic acid |
| EpDPE | Epoxydocosapentaenoic acid |
| EpOME | Epoxyoctadecenoic acid |
| FFA | Free fatty acids |
| HDHA | Hydroxydocosaheptaenoic acid |
| HDL | High-density lipoprotein |
| HEPE | Hydroxyeicosapentaenoic acid |
| HETE | Hydroxyeicosatetraenoic acid |
| HETrE | Hydroxyeicosatrienoic acid |
| HODE | Hydroxyoctadecadienoic acid |
| HOTrE | Hydroxyoctadecatrienoic acid |
| HSPC | Hydrogenated soy bean |
| LA | Linoleic acid |
| LDL | Low-density lipoprotein |
| LEA | Linoleoyl ethanolamine |
| LOX | Lipoxygenase |
| LPC | Lysophosphatidylcholine |
| LPE | Lysophosphatidylethanolamine |
| MAG | Monoacylglycerol |
| NAE | N-Acyl-ethanolamine |
| NPs | Nanoparticles |
| OEA | Oleoyl ethanolamine |
| oxo-ETE | Oxo-eicosatetraenoic acid |
| oxo-ODE | Oxo-octadecadienoic acid |
| PC | Phosphatidylcholine |
| PC-O | Ether-phosphatidyl glycerol |
| PDEA | Pentadecenoyl ethanolamine |
| PE | Phosphatidylethanolamine |
| PEA | Palmitoyl ethanolamine |
| PI | Phosphatidylinositol |
| PUFA | Polyunsaturated fatty acids |
| RPA | Relative protein abundance |
| SEA | Stearoyl ethanolamine |
| S-trap | Suspension-trapping |
| SM | Sphingomyelin |
| TAG | Triacylglycerol |
| TEAB | Triethylammonium bicarbonate |
| trans-EKODE | trans-Epoxyketooctadecenoic acid |
| UHPSFC/ESI-QTOF-MSE | Ultra-high performance supercritical fluid chromatography coupled to quadrupole time of flight mass spectrometry with electrospray ionisation |
| UPLC/ESI-MS/MS | Ultra-performance liquid chromatography tandem mass spectrometry with electrospray ionisation |
| VEA | Vaccenoyl ethanolamine |
| VLDL | Very low-density lipoprotein. |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors wish to thank the Faculty of Life Sciences Mass Spectrometry Facility staff at the University of Manchester for their assistance and technical support in proteomic LC-MS/MS. We also thank Neil O'Hara and Marta Koszyczarek (Laboratory for Lipidomics and Lipid Biology, University of Manchester) for excellent technical support in lipidomic analyses. We thank Non-linear Dynamics/Waters for their support of this work. Finally, we would like to thank Prof Kostas Kostarelos for his intellectual input at the initial stages of this work. This work has been funded by the Faculty of Biology, Medicine and Health, University of Manchester and the Cancer Research UK International Alliance for Cancer Early Detection (ACED; EICEDAAP\100013).References
- M. Hadjidemetriou and K. Kostarelos, Nat. Nanotechnol., 2017, 12, 288–290 CrossRef CAS PubMed
.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS
.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin and H. Nilsson,
et al.
, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed
.
- D. Nierenberg, A. R. Khaled and O. Flores, Rep. Pract. Oncol. Radiother., 2018, 23, 300–308 CrossRef PubMed
.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed
.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–15574 CrossRef CAS PubMed
.
- E. Hellstrand, I. Lynch, A. Andersson, T. Drakenberg, B. Dahlback and K. A. Dawson,
et al.
, FEBS J., 2009, 276, 3372–3381 CrossRef CAS PubMed
.
- J. Y. Lee, H. Wang, G. Pyrgiotakis, G. M. DeLoid, Z. Zhang and J. Beltran-Huarac,
et al.
, Anal. Bioanal. Chem., 2018, 410, 6155–6164 CrossRef CAS PubMed
.
- T. Lima, K. Bernfur, M. Vilanova and T. Cedervall, Sci. Rep., 2020, 10, 1129 CrossRef CAS PubMed
.
- G. La Barbera, A. L. Capriotti, G. Caracciolo, C. Cavaliere, A. Cerrato and C. M. Montone,
et al.
, Talanta, 2020, 209, 120487 CrossRef CAS PubMed
.
- M. Rabel, P. Warncke, M. Thurmer, C. Gruttner, C. Bergemann and H. D. Kurland,
et al.
, Nanoscale, 2021, 13, 9415–9435 RSC
.
- L. M. Kobos, S. Alqatani, C. R. Ferreira, U. K. Aryal, V. Hedrick and T. J. P. Sobreira,
et al.
, Appl. In Vitro Toxicol., 2019, 5, 150–166 CrossRef CAS PubMed
.
- M. Gasser, B. Rothen-Rutishauser, H. F. Krug, P. Gehr, M. Nelle and B. Yan,
et al.
, J. Nanobiotechnol., 2010, 8, 31 CrossRef CAS PubMed
.
- M. Gasser, P. Wick, M. J. Clift, F. Blank, L. Diener and B. Yan,
et al.
, Part. Fibre Toxicol., 2012, 9, 17 CrossRef CAS PubMed
.
- A. A. Kapralov, W. H. Feng, A. A. Amoscato, N. Yanamala, K. Balasubramanian and D. E. Winnica,
et al.
, ACS Nano, 2012, 6, 4147–4156 CrossRef CAS PubMed
.
- S. S. Raesch, S. Tenzer, W. Storck, A. Rurainski, D. Selzer and C. A. Ruge,
et al.
, ACS Nano, 2015, 9, 11872–11885 CrossRef CAS PubMed
.
- E. Maretti, C. Rustichelli, M. Lassinantti Gualtieri, L. Costantino, C. Siligardi and P. Miselli,
et al.
, Pharmaceutics, 2019, 11, 508 CrossRef CAS PubMed
.
- J. Martel, C. Y. Wu, C. Y. Hung, T. Y. Wong, A. J. Cheng and M. L. Cheng,
et al.
, Nanoscale, 2016, 8, 5537–5545 RSC
.
- J. Muller, D. Prozeller, A. Ghazaryan, M. Kokkinopoulou, V. Mailander and S. Morsbach,
et al.
, Acta Biomater., 2018, 71, 420–431 CrossRef PubMed
.
- N. N. Nam, T. L. Bui, S. J. Son and S. W. Joo, Bull. Korean Chem. Soc., 2015, 37, 3–4 CrossRef
.
- M. Pink, N. Verma, C. Kersch and S. Schmitz-Spanke, Environ. Sci.: Nano, 2018, 1420–1427 RSC
.
- S. Lara, F. Alnasser, E. Polo, D. Garry, M. C. Lo Giudice and D. R. Hristov,
et al.
, ACS Nano, 2017, 11, 1884–1893 CrossRef CAS PubMed
.
- S. B. Gunnarsson, K. Bernfur, A. Mikkelsen and T. Cedervall, Nanoscale, 2018, 10, 4246–4257 RSC
.
- L. Gardner, J. Warrington, J. Rogan, D. G. Rothwell, G. Brady and C. Dive,
et al.
, Nanoscale Horiz., 2020, 5, 1476–1486 RSC
.
- A. J. Chetwynd, W. Zhang, K. Faserl, J. A. Thorn, I. Lynch and R. Ramautar,
et al.
, J. Visualized Exp., 2020, 164, e61760 Search PubMed
.
- A. J. Chetwynd, E. J. Guggenheim, S. M. Briffa, J. A. Thorn, I. Lynch and E. Valsami-Jones, Nanomaterials, 2018, 16(21), 2000295 Search PubMed
.
- A. J. Chetwynd, W. Zhang, J. A. Thorn, I. Lynch and R. Ramautar, Small, 2020, 16, e2000295 CrossRef PubMed
.
- A. J. Chetwynd and I. Lynch, Environ. Sci.: Nano, 2020, 1041–1060, 10.1039/c9en00938h
.
- M. T. Snaebjornsson, S. Janaki-Raman and A. Schulze, Cell Metab., 2020, 31, 62–76 CrossRef CAS PubMed
.
- P. Proitsi, M. Kim, L. Whiley, A. Simmons, M. Sattlecker and L. Velayudhan,
et al.
, Alzheimers Dement, 2017, 13, 140–151 CrossRef PubMed
.
- M. Masoodi, A. Gastaldelli, T. Hyötyläinen, E. Arretxe, C. Alonso and M. Gaggini,
et al.
, Nat. Rev. Gastroenterol. Hepatol., 2021, 18, 835–856 CrossRef PubMed
.
- K. A. McGurk, B. D. Keavney and A. Nicolaou, Atherosclerosis, 2021, 327, 18–30 CrossRef CAS PubMed
.
- D. Wolrab, R. Jirásko, E. Cífková, M. Höring, D. Mei and M. Chocholoušková,
et al.
, Nat. Commun., 2022, 13, 124 CrossRef CAS PubMed
.
- P. J. Meikle and S. A. Summers, Nat. Rev. Endocrinol., 2017, 13, 79–91 CrossRef CAS PubMed
.
- L. H. Wong, A. T. Gatta and T. P. Levine, Nat. Rev. Mol. Cell Biol., 2019, 20, 85–101 CrossRef CAS PubMed
.
- G. J. van der Vusse, Drug Metab. Pharmacokinet., 2009, 24, 300–307 CrossRef CAS PubMed
.
- O. Quehenberger and E. A. Dennis, N. Engl. J. Med., 2011, 365, 1812–1823 CrossRef CAS PubMed
.
- O. Quehenberger, A. M. Armando, A. H. Brown, S. B. Milne, D. S. Myers and A. H. Merrill,
et al.
, J. Lipid Res., 2010, 51, 3299–3305 CrossRef CAS PubMed
.
- A. C. Kendall, S. M. Pilkington, S. A. Murphy, F. Del Carratore, A. L. Sunarwidhi and M. Kiezel-Tsugunova,
et al.
, FASEB J., 2019, 33, 13014–13027 CrossRef CAS PubMed
.
- A. Yasumoto, S. M. Tokuoka, Y. Kita, T. Shimizu and Y. Yatomi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1068–1069, 98–104 CrossRef CAS PubMed
.
- K. A. McGurk, S. G. Williams, H. Guo, H. Watkins, M. Farrall and H. J. Cordell,
et al.
, Hum. Mol. Genet., 2021, 30, 500–513 CrossRef CAS PubMed
.
- T. Ayakannu, A. H. Taylor, T. H. Marczylo, M. Maccarrone and J. C. Konje, Front. Oncol., 2019, 9, 430 CrossRef PubMed
.
- M. M. Mielke, V. V. Bandaru, N. J. Haughey, P. V. Rabins, C. G. Lyketsos and M. C. Carlson, Neurobiol. Aging, 2010, 31, 17–24 CrossRef CAS PubMed
.
- K. A. Massey and A. Nicolaou, Free Radicals Biol. Med., 2013, 59, 45–55 CrossRef CAS PubMed
.
- G. Astarita, A. C. Kendall, E. A. Dennis and A. Nicolaou, Biochim. Biophys. Acta, 2015, 1851, 456–468 CrossRef CAS PubMed
.
- A. C. Kendall, M. M. Koszyczarek, E. A. Jones, P. J. Hart, M. Towers and C. E. M. Griffiths,
et al.
, Exp. Dermatol., 2018, 27, 721–728 CrossRef PubMed
.
- A. Surendran, H. Zhang, T. Winter, A. Edel, H. Aukema and A. Ravandi, Atherosclerosis, 2019, 288, 101–111 CrossRef CAS PubMed
.
- J. W. Heinecke, J. Clin. Lipidol., 2010, 4, 371–375 CrossRef PubMed
.
- D. Camacho-Munoz, M. Kiezel-Tsugunova, O. Kiss, M. Uddin, M. Sunden and M. Ryaboshapkina,
et al.
, FASEB J., 2021, 35, e21976 CrossRef CAS PubMed
.
- J. W. Newman, T. L. Pedersen, V. R. Brandenburg, W. S. Harris and G. C. Shearer, PLoS One, 2014, 9, e111471 CrossRef PubMed
.
- A. Kroll, H. E. Cho and M. H. Kang, Front. Oncol., 2020, 10, 833 CrossRef PubMed
.
- L. Gardner, K. Kostarelos, P. Mallick, C. Dive and M. Hadjidemetriou, Nat. Rev. Clin Oncol., 2022, 19, 551–561 CrossRef PubMed
.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Mazza, R. F. Collins, K. Dawson and K. Kostarelos, ACS Nano, 2015, 9, 8142–8156 CrossRef CAS PubMed
.
- M. Hadjidemetriou, S. McAdam, G. Garner, C. Thackeray, D. Knight and D. Smith,
et al.
, Adv. Mater., 2018, 31(4), e1803335 CrossRef PubMed
.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(52), 2109256118 CrossRef PubMed
.
- M. Hadjidemetriou, J. Rivers-Auty, L. Papafilippou, J. Eales, K. A. B. Kellett and N. M. Hooper,
et al.
, ACS Nano, 2021, 15, 7357–7369 CrossRef CAS PubMed
.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Buggio, J. Swift and K. Kostarelos, Biomaterials, 2018, 188, 118–129 CrossRef PubMed
.
- L. Papafilippou, A. Claxton, P. Dark, K. Kostarelos and M. Hadjidemetriou, Nanoscale, 2020, 12, 10240–10253 RSC
.
- M. Hadjidemetriou, L. Papafilippou, R. D. Unwin, J. Rogan, A. Clamp and K. Kostarelos, Nano Today, 2020, 34, 100901 CrossRef CAS
.
- D. Caputo and G. Caracciolo, Cancer Lett., 2020, 470, 191–196 CrossRef CAS PubMed
.
- M. Del Pilar Chantada-Vazquez, A. C. Lopez, M. G. Vence, S. Vazquez-Estevez, B. Acea-Nebril and D. G. Calatayud,
et al.
, J. Proteomics, 2020, 212, 103581 CrossRef CAS PubMed
.
- D. Caputo, L. Digiacomo, C. Cascone, D. Pozzi, S. Palchetti and R. Di Santo,
et al.
, Cancers, 2020, 13(1), 93 CrossRef PubMed
.
- M. Hadjidemetriou, Z. Al-Ahmady and K. Kostarelos, Nanoscale, 2016, 8, 6948–6957 RSC
.
- D. Cucchi, D. Camacho-Munoz, M. Certo, J. Niven, J. Smith and A. Nicolaou,
et al.
, Cardiovasc. Res., 2020, 116, 1006–1020 CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr05982g |
| This journal is © The Royal Society of Chemistry 2023 |