Open Access Article
Open Access ArticleMonitoring SARS-CoV-2 in wastewater during New York City's second wave of COVID-19: sewershed-level trends and relationships to publicly available clinical testing data†
Catherine
Hoar
 a,
Francoise
Chauvin
b,
Alexander
Clare
b,
Hope
McGibbon
b,
Esmeraldo
Castro
b,
Samantha
Patinella
b,
Dimitrios
Katehis
b,
John J.
Dennehy
a,
Francoise
Chauvin
b,
Alexander
Clare
b,
Hope
McGibbon
b,
Esmeraldo
Castro
b,
Samantha
Patinella
b,
Dimitrios
Katehis
b,
John J.
Dennehy
 cd,
Monica
Trujillo
e,
Davida S.
Smyth‡
f and
Andrea I.
Silverman
cd,
Monica
Trujillo
e,
Davida S.
Smyth‡
f and
Andrea I.
Silverman
 *a
*a
aDepartment of Civil and Urban Engineering, New York University Tandon School of Engineering, Brooklyn, NY, USA. E-mail: andrea.silverman@nyu.edu
bNew York City Department of Environmental Protection, New York, NY, USA
cBiology Department, Queens College, The City University of New York, Queens, NY, USA
dBiology Doctoral Program, The Graduate Center, The City University of New York, New York, NY, USA
eDepartment of Biology, Queensborough Community College, The City University of New York, Bayside, NY, USA
fDepartment of Natural Sciences and Mathematics, Eugene Lang College of Liberal Arts at The New School, New York, NY, USA
First published on 16th March 2022
Abstract
New York City's wastewater monitoring program tracked trends in sewershed-level SARS-CoV-2 loads starting in the fall of 2020, just before the start of the city's second wave of the COVID-19 outbreak. During a five-month study period, from November 8, 2020 to April 11, 2021, viral loads in influent wastewater from each of New York City's 14 wastewater treatment plants were measured and compared to new laboratory-confirmed COVID-19 cases for the populations in each corresponding sewershed, estimated from publicly available clinical testing data. We found significant positive correlations between viral loads in wastewater and new COVID-19 cases. The strength of the correlations varied depending on the sewershed, with Spearman's rank correlation coefficients ranging between 0.38 and 0.81 (mean = 0.55). Based on a linear regression analysis of a combined data set for New York City, we found that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the SARS-CoV-2 viral load in wastewater corresponded to a 0.6
log10 change in the SARS-CoV-2 viral load in wastewater corresponded to a 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed. An estimated minimum detectable case rate between 2–8 cases per day/100
log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed. An estimated minimum detectable case rate between 2–8 cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people was associated with the method limit of detection in wastewater. This work offers a preliminary assessment of the relationship between wastewater monitoring data and clinical testing data in New York City. While routine monitoring and method optimization continue, information on the development of New York City's wastewater monitoring program may provide insights for similar wastewater-based epidemiology efforts in the future.
000 people was associated with the method limit of detection in wastewater. This work offers a preliminary assessment of the relationship between wastewater monitoring data and clinical testing data in New York City. While routine monitoring and method optimization continue, information on the development of New York City's wastewater monitoring program may provide insights for similar wastewater-based epidemiology efforts in the future.
Water impactExpanding the use of wastewater-based epidemiology (WBE) to inform public health responses requires an understanding of its performance across various communities. Results from New York City's SARS-CoV-2 wastewater monitoring program indicate associations between wastewater data and clinical data in a large urban setting and provide insights for the development of long-term WBE monitoring efforts. |
Introduction
In March 2020, New York City became an epicenter of the coronavirus disease 2019 (COVID-19) pandemic. In response to this first wave of COVID-19 cases, the New York City Department of Environmental Protection (NYC DEP) – the city agency responsible for wastewater collection and treatment – launched a wastewater monitoring program with the goal of tracking sewershed-level trends in the concentration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19. The program was developed in partnership with researchers at New York University, Queens College, Queensborough Community College, and The New School, with all routine analysis conducted in the NYC DEP's existing microbiology laboratory under the management of the NYC DEP.Wastewater-based epidemiology (WBE) programs for COVID-19, including the one in New York City (NYC), were established on the premise that SARS-CoV-2 virions are excreted in the human waste of individuals infected with SARS-CoV-2 and that the resulting concentrations of viral RNA measured in wastewater are indicative of disease incidence or prevalence in the contributing sewershed. Significant associations between SARS-CoV-2 RNA concentrations measured in wastewater and metrics of COVID-19 disease incidence–including case rates–have been shown at scales ranging from single buildings to entire sewersheds.1–3 Early reports from WBE programs suggested promising applications that could help inform COVID-19 response measures,2,4 sparking widespread interest in SARS-CoV-2 monitoring programs around the world.5,6 While the extent to which wastewater data is a leading indicator of trends in COVID-19 incidence ahead of clinical data may vary depending on clinical testing rates,7,8 WBE data do offer the advantage of providing information representative of entire populations, free from clinical testing-related biases. In NYC, where communities of color and high-poverty areas were disproportionately impacted by the first wave of the COVID-19 pandemic,9 testing rates varied spatially, with significant demographic-based disparities.10 In situations where clinical testing does not adequately sample vulnerable populations, WBE may help inform modifications to testing strategies and provide supplemental information regarding COVID-19 trends. Wastewater monitoring is therefore a potential tool to identify new outbreaks of COVID-19 after high clinical testing rates associated with major “waves” of disease incidence have subsided or when resources and technical capacity for extensive clinical testing of individuals are limited.
These opportunities make WBE an attractive option for many municipalities, including NYC, to confirm findings from clinical testing about population-level COVID-19 dynamics and to monitor for new outbreaks in instances when testing is inadequate. In August 2020, the NYC DEP's SARS-CoV-2 wastewater monitoring program began routine analysis of influent wastewater collected from NYC's 14 wastewater treatment plants (referred to as wastewater resource recovery facilities (WRRFs) by the NYC DEP) (ESI† Table S1), capturing data during the region's second wave of COVID-19 cases, which started in the fall of 2020. The sewershed catchment areas contributing to each of the 14 WRRFs vary markedly in size, serving populations ranging from approximately 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1.2 million residents. To assess the relationship between NYC sewershed-level SARS-CoV-2 RNA concentrations and confirmed cases of COVID-19 within each sewershed, wastewater data were compared to publicly available case data provided by the NYC Department of Health and Mental Hygiene (DOHMH). In presenting findings from the NYC DEP, we also aim to provide insights into the development of a wastewater monitoring program designed for long-term, routine tracking of trends in virus loads for multiple sewersheds serving a large urban population.
000 to 1.2 million residents. To assess the relationship between NYC sewershed-level SARS-CoV-2 RNA concentrations and confirmed cases of COVID-19 within each sewershed, wastewater data were compared to publicly available case data provided by the NYC Department of Health and Mental Hygiene (DOHMH). In presenting findings from the NYC DEP, we also aim to provide insights into the development of a wastewater monitoring program designed for long-term, routine tracking of trends in virus loads for multiple sewersheds serving a large urban population.
Methods
Sample collection and processing
24 h flow-weighted composite influent wastewater samples were collected from each of NYC's 14 WRRFs twice weekly beginning August 31, 2020. From January 31, 2021 to April 18, 2021 sampling was reduced to once weekly. Each composite sample consisted of eight grab samples collected every three hours beginning at 7:00 AM on the sampling date. The volume of each grab sample added to the composite was determined based on the flowrate during the associated 3 h collection period. Samples were transported on ice and stored at 4 °C until processing, which started within twelve hours after the final grab sample was collected. For each sampling date, one of the 14 samples was analyzed in duplicate and the remainder were analyzed as single samples; facilities were selected for duplicate analysis on a rotating basis. A method blank containing type I deionized water was included with each set of samples to confirm the absence of contamination during sample processing. Detailed descriptions of materials, methods, and data analysis are provided in the ESI.† In brief, 40 mL aliquots of the 24 h composite samples were first pasteurized (60 °C, 90 min), and then centrifuged (5000 × g, 4 °C, 10 min) to remove solids. The supernatant was filtered (0.22 μm, cellulose acetate) and then subjected to virus concentration using polyethylene glycol (PEG) precipitation (addition of 4.0 g PEG and 0.9 g NaCl followed by overnight incubation at 4 °C, and centrifugation at 12000 × g at 4 °C for 120 min to pellet viruses).11 The supernatant was discarded and RNA (along with any DNA present) was extracted from the concentrated PEG pellet using the Qiagen QiaAmp Viral RNA Mini Kit with modifications (described in the ESI†).SARS-CoV-2 quantification by RT-qPCR
A one-step RT-qPCR assay was used to quantify copies of the SARS-CoV-2 nucleocapsid (N) gene, targeting the N1 region (CDC RUO Primers and Probes, Integrated DNA Technologies)12 in triplicate reactions on a StepOnePlus real-time PCR system (Thermo Fisher Scientific). Synthetic SARS-CoV-2 RNA covering >99.9% of the viral genome (Twist Bioscience Control 1, GENBANK ID MT007544.1), quantified using reverse transcription droplet digital PCR (RT-ddPCR) as described by Al-Duroobi et al.,13 with a minor modification (details are provided in the ESI†), served as both a positive control and standard used in a decimal serial dilution for quantification of N1 gene copies.The limit of detection (LOD) and limit of quantification (LOQ) for the assay were estimated from replicate standard curves as described by Forootan et al. 2017 (ref. 14) and found to be 180 copies per L of wastewater sample and 590 copies per L of wastewater sample, respectively. We elected to use a pooled standard curve to quantify samples on all plates to ameliorate variability in standard preparation by different analysts from plate to plate. A description of the analysis used to motivate this decision is presented in the ESI† (Fig. S1). The absence of contamination during RT-qPCR preparation was confirmed through no template controls included on all RT-qPCR plates. Only samples quantified above the LOQ were included in subsequent analysis. From September 8, 2020 to June 8, 2021, samples were collected from each facility on 72 sampling dates, with samples from only two dates associated with method blanks having N1 concentrations above the LOD; samples collected on these two dates were flagged as contaminated and were not included in subsequent analysis.
An attenuated bovine coronavirus (BCoV) (Calf-Guard® Bovine Rota-Coronavirus Vaccine, Zoetis) was used as a process control.15,16 BCoV was inoculated into samples after the pasteurization step (details provided in the ESI†). A one-step RT-qPCR assay, adapted from previously published assays,15–17 targeting the transmembrane-protein gene of BCoV was used to qualitatively assess BCoV recovery for each sample using an aliquot of the extracted RNA (primers and probes purchased from Integrated DNA Technologies). Detection of BCoV was used to confirm whether viruses were recovered in samples for which the N1 target was not detected. Additional details regarding the RT-qPCR assays, standard curves, and QA/QC procedures are provided in the ESI.†
Data analysis
The concentration of the N1 RNA target in wastewater (CWW) was determined for each sample in units of N1 gene copies (GC) per L according to eqn (1), where Nr is the number of N1 GC measured by RT-qPCR, VRNA,s is the volume of RNA extracted from each sample (60 μL), VRNA,r is the volume of template RNA added to the RT-qPCR reaction (5 μL), and Vs is the volume of wastewater sample analyzed (0.04 L).| CWW = (Nr × VRNA,s)/(VRNA,r × Vs) | (1) |
| LWW = CWW × Q × CF | (2) |
Statistical analyses of relationships between SARS-CoV-2 loads in wastewater and laboratory-confirmed COVID-19 cases
Relationships between SARS-CoV-2 wastewater data in each sewershed and laboratory-confirmed COVID-19 cases for the associated sewershed population were evaluated through correlation and linear regression analyses. Clinical data were obtained from publicly available data provided by the NYC DOHMH.18 In particular, the data set “last7days-by-modzcta.csv”, which was posted online daily, was used to obtain daily reports of the cumulative clinical molecular testing results over the previous seven days for each modified ZIP code tabulation area (MODZCTA) in NYC.18 Specifically, data on the total clinical COVID-19 tests administered and the total number of positive tests (not including individuals who previously tested positive), reported based on date of specimen collection, were obtained. Note that molecular tests included diagnostic PCR tests and did not include antigen or antibody tests. This data set was used to calculate 7-day averages of new COVID-19 cases (i.e., positive molecular tests) per day, organized by the last date in the 7-day range. For example, the 7-day average reported on February 14 represents the daily average of new cases calculated based on the total number of positive molecular tests collected from February 8 to February 14. Data were available starting on November 7, 2020, with data from March 15, 2021 to March 21, 2021 omitted due to technical issues related to data transmission during this period (Fig. S2†). While alternative data sets were available with cumulative new COVID-19 case counts prior to November 2020, these data were organized by the date that test results were reported, as opposed to date of specimen collection, and were therefore not recommended by NYC DOHMH for use in calculating the number of daily new COVID-19 cases.18Each of the 177 MODZCTAs were assigned to one of NYC's 14 sewersheds. Of the 177 MODZCTAs, 44 straddled multiple sewershed areas and were assigned to only the sewershed in which it had the greatest overlapping land area, determined based on visual inspection of sewershed boundary maps provided by the NYC DEP Bureau of Water and Sewer Operations. Total new cases in each sewershed for each 7-day period were calculated by summing the cumulative 7-day positive test counts in each MODZCTA assigned to that sewershed. The same data set was used to calculate 7-day averages of COVID-19 testing rates (i.e., the number of tests administered divided by the total population) and the percentages of COVID-19 tests that were positive for each sewershed (Fig. S2†).
Spearman correlations between SARS-CoV-2 viral loading rates in wastewater (N1 GC per day) and 7-day averages of new daily COVID-19 cases were determined for each individual sewershed for a five-month study period (November 8, 2020 to April 11, 2021). Correlations were also determined for a combined data set that included each data pair (i.e., SARS-CoV-2 viral loading rates and 7-day average of new COVID-19 cases on each date) for all facilities, excluding the Port Richmond and Oakwood Beach WRRFs (see the Results and discussion section). For the combined data, correlations were also evaluated after removing data pairs associated with potentially inadequate clinical testing rates: data for dates with percentages of positive molecular tests (7-day average) that exceeded 10% in the sewershed were excluded. A general benchmark suggested by the World Health Organization in the Spring of 2020 indicated that clinical testing is less likely to represent all infections in a population when the percentage of positive tests exceeds approximately 10%;19,20 we therefore excluded these data in an effort to best approximate the incidence of SARS-CoV-2 infections.
To assess whether trends in SARS-CoV-2 viral loading rates in wastewater preceded trends in clinical testing data, correlations between the two data sets were also evaluated for each sewershed with the clinical data shifted back in time with lags ranging from −7 to 21 days. Additional details for this analysis are provided in the ESI.†
Simple linear regressions were performed using log10-transformed SARS-CoV-2 viral loading rates (N1 GC per day) and log10-transformed 7-day averages of new COVID-19 cases (new COVID-19 cases per day) for each individual sewershed. For each sewershed, Spearman correlations were determined between the slope of the resulting linear regression line and the (1) average testing rate for the study period, (2) average wastewater flow rate, (3) population, and (4) average per capita wastewater flow rate. Linear regressions were also performed using log10-transformed SARS-CoV-2 viral loading rates (N1 GC per day) and log10-transformed 7-day averages of new COVID-19 cases (new COVID-19 cases per day) for the combined data set, both with and without the testing rate filter described above. Linear regressions were used to estimate the equivalent number of cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people associated with the method LOD (CLOD), equal to 180 N1 GC L−1. This estimate was calculated for each facility using individual, sewershed-specific linear regressions and using the linear regression for the combined data set. First, the LOD was converted to a SARS-CoV-2 viral loading rate in wastewater (LWW,LOD) for each sewershed in units of N1 GC per day using eqn (3), where Qavg is the average of daily flow rates at the facility over the study period (Table S1†), in MGD.
000 people associated with the method LOD (CLOD), equal to 180 N1 GC L−1. This estimate was calculated for each facility using individual, sewershed-specific linear regressions and using the linear regression for the combined data set. First, the LOD was converted to a SARS-CoV-2 viral loading rate in wastewater (LWW,LOD) for each sewershed in units of N1 GC per day using eqn (3), where Qavg is the average of daily flow rates at the facility over the study period (Table S1†), in MGD.
| LWW,LOD = CLOD × Qavg × CF | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people using MODZCTA-level population estimates from the NYC DOHMH NYC Coronavirus Disease 2019 (COVID-19) data.18
000 people using MODZCTA-level population estimates from the NYC DOHMH NYC Coronavirus Disease 2019 (COVID-19) data.18| log10(CaseLOD) = m × log10(LWW,LOD) + b | (4) |
Results and discussion
Methodological considerations for SARS-CoV-2 quantification in wastewater
The public health emergency caused by the emergence of COVID-19 required the expedited development of NYC DEP's SARS-CoV-2 wastewater monitoring program. As such, several methodological choices for virus quantification were considered, and the ultimate standard operating procedure (SOP) described herein was developed reflecting NYC DEP's program goals of monitoring trends in SARS-CoV-2 viral loads in wastewater, accounting for equipment availability, existing expertise of personnel, and considerations of material procurement. Selections were also made to minimize analyst-based variability. For example, commercially-available kits for RNA extraction were considered over alternatives that may be more sensitive to analyst skill and consistency. Data analysis and internally-developed QA/QC guidelines were established in line with programmatic goals. Additional methodological considerations, such as the inclusion of a filtration step in sample preparation, are discussed in the ESI.†Long-term routine monitoring to assess virus trends through quantification with RT-qPCR requires reliable comparison of data originating from different RT-qPCR plates prepared by different analysts, which presents several challenges. First, this program relied on the use of a synthetic RNA control as a standard for the N1 RNA target. Because the concentration of this RNA control was found to differ between lots purchased at different times, one lot of the RNA control was quantified using RT-ddPCR13 (details are provided in the ESI†). We then determined the concentration of all other RNA control lots relative to this quantified lot. In addition, standard curves for routine RT-qPCR assays were prepared by different analysts on different days, with separate serial dilutions of standards performed for each individual RT-qPCR plate. To account for any resulting variability caused by these aspects of the RT-qPCR quantification method and allow comparison of measured concentrations over the course of many months, we applied a pooled standard curve for quantification of all samples (Fig. S1†). Challenges associated with RT-qPCR-based quantification using a standard curve highlight the benefits of alternative methods, such as digital PCR for absolute RNA quantification, which eliminates the need for a standard curve and may offer more sensitive detection for environmental samples.23 Nonetheless, the methodology employed in this work allowed us to compare relative viral loads and confidently assess of trends of SARS-CoV-2 in wastewater over time.
SARS-CoV-2 viral loads in influent wastewater
SARS-CoV-2 viral loads in NYC's 14 sewersheds between September 8, 2020 and June 8, 2021 were determined from quantifiable N1 gene copy (GC) concentrations in influent samples and are presented normalized by sewershed population (Table S1† (ref. 24)) in Fig. 1. Maximum population-normalized SARS-CoV-2 viral loads for each facility during this period ranged from 6.2 × 106 to 2.7 × 107 N1 GC per day/population, with many of these values occurring around the time when a peak in COVID-19 cases was observed (January 2021). Note that in September of 2020, prior to the increase in COVID-19 cases associated with NYC's second wave of the outbreak, N1 concentrations in wastewater remained below the LOQ in several sewersheds.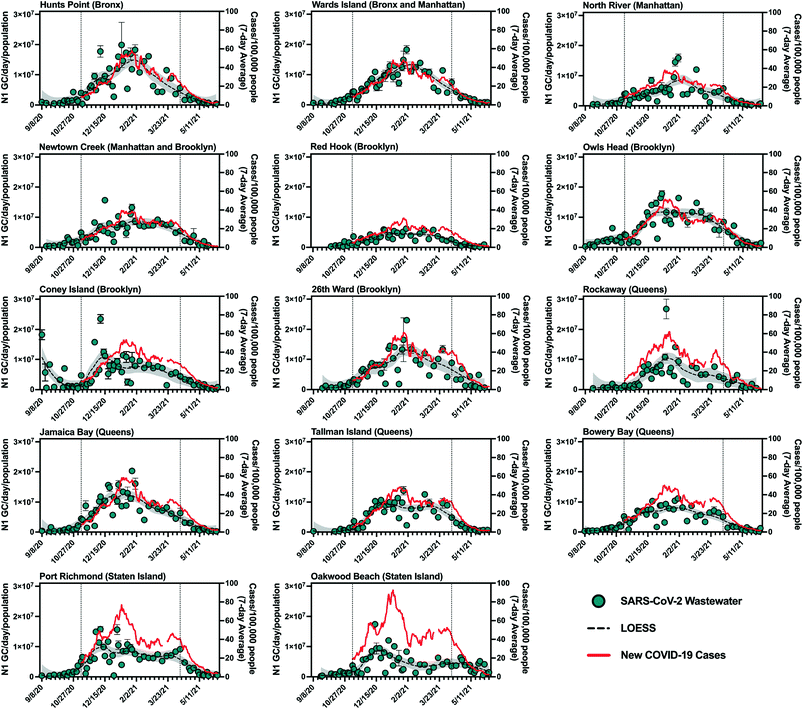 | ||
Fig. 1 Summary of SARS-CoV-2 wastewater data for New York City's 14 sewersheds. Data from September 8, 2020 to June 8, 2021 is shown, with the period for which statistical analysis was conducted (November 8, 2020 to April 11, 2021) bounded by vertical dotted lines. Primary (left) y-axis, blue circles: influent SARS-CoV-2 viral loads normalized by sewershed populations. Error bars indicate standard deviations from triplicate RT-qPCR reactions as well as standard deviations of duplicate samples, where applicable. Dashed black lines represent LOESS curve fits (span = 0.4), with the 95% confidence intervals shaded in grey. Secondary (right) y-axis, red line: 7-day average of new COVID-19 cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people in the previous 7 days normalized using MODZCTA-level population estimates from the NYC DOHMH's NYC Coronavirus Disease 2019 (COVID-19) data.18 Normalization by population was used for visual comparison across different sewersheds only and was not used for statistical analysis. Note that Newtown Creek WRRF also serves a small section of Queens. 000 people in the previous 7 days normalized using MODZCTA-level population estimates from the NYC DOHMH's NYC Coronavirus Disease 2019 (COVID-19) data.18 Normalization by population was used for visual comparison across different sewersheds only and was not used for statistical analysis. Note that Newtown Creek WRRF also serves a small section of Queens. | ||
Visual inspection of trends in SARS-CoV-2 quantities in wastewater and new laboratory-confirmed COVID-19 cases indicates an association between the wastewater and clinical data. The strength of this association varied across sewersheds, as reflected in results from statistical analysis presented in the next section. Additionally, most sewersheds exhibited peaks for both data sets in January 2021 (Fig. 1), with two notable exceptions being Oakwood Beach and Port Richmond, discussed below. Sewersheds with lower incidence rates (new cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people) of COVID-19 (e.g., Red Hook WRRF) generally had lower per capita SARS-CoV-2 viral loads in wastewater than those with higher incidence rates of COVID-19 (e.g., Hunts Point WRRF).
000 people) of COVID-19 (e.g., Red Hook WRRF) generally had lower per capita SARS-CoV-2 viral loads in wastewater than those with higher incidence rates of COVID-19 (e.g., Hunts Point WRRF).
SARS-CoV-2 viral loads in the Coney Island WRRF influent in September 2020 and October 2020 displayed a high degree of variability, with some measured virus loads that were greater than those in all other sewersheds during that period, despite a consistent processing method applied for all samples and confirmed COVID-19 case rates that were consistently low across NYC (Fig. 1). While there were relatively low rates of clinical testing in New York City in September 2020 and COVID-19 clusters emerged in some neighborhoods served by the Coney Island WRRF at that time,25 it is unclear if these factors contributed to the high viral loads measured in some Coney Island WRRF samples. For example, COVID-19 clusters were also identified in other sewersheds at this time, yet did not result in high SARS-CoV-2 loads in influent samples collected from other WRRFs, and it is difficult to determine whether clinical testing was adequate. It should also be noted that given its large geographic resolution, sewershed-level monitoring may not fully capture the effect of disease clusters (such as those identified at high spatiotemporal resolution using clinical data26) that may be relatively small compared to the sewershed or may straddle multiple sewersheds.
A smaller extent of variability in measured SARS-CoV-2 viral loads was observed to varying degrees across all facilities and can stem from several sources. Evaluation of duplicate samples analyzed during the study period allowed for an assessment of potential variability due to sample processing and RNA quantification. Relative standard deviations for N1 concentrations of duplicate samples (i.e., the standard deviation of concentrations from duplicate samples, each with triplicate RT-qPCR reactions, as a percent of the average concentration) ranged from 3% to 44% (mean = 18%, median = 14%); these values are comparable to those reported elsewhere for measurement of N1 concentrations in influent wastewater.16,27 Aside from methodological sources of variability, additional sources of variability or uncertainty could include (1) dilution of wastewater from non-domestic water inputs and variations in domestic water use habits, (2) wastewater chemical composition, which may interfere with sample processing or RNA quantification methods, (3) variability in SARS-CoV-2 shedding intensity and duration for infected individuals28–30 and (4) the extent and consistency of viral RNA degradation in sewers.27,31
To account for variability in wastewater flow rates and minimize the effect of (1), viral loads calculated using measured wastewater flow rates (eqn (2)) were used for analysis instead of N1 concentrations. The impact of factor (2), namely, RT-qPCR inhibition, was assessed by evaluating ten-fold dilutions of a selection of samples from each WRRF; inhibition was considered minimal in the evaluated samples based on comparison of Cq values between diluted samples and associated undiluted samples (details provided in the ESI†). Regular assessment of inhibition with control assays was not feasible during routine monitoring due to resource constraints. In addition, dilution of RNA, a strategy used to reduce PCR inhibition, was avoided in order to maintain consistency in sample processing, given the risk of diluting samples to viral concentrations below the limits of quantification or detection during periods with low COVID-19 case rates. While beyond the scope of this work, assessment of viral recovery and wastewater matrix effects should be considered for future research aiming to characterize uncertainty in WBE data. For example, identifying and characterizing external factors related to (3) and (4) is the focus of ongoing SARS-CoV-2 WBE research efforts. Considering these uncertainties and variabilities in wastewater data, which likely increase with scale,32 we did not attempt to quantify the number of SARS-CoV-2 infections in each sewershed based on wastewater data, but instead explored the relationship between viral quantities in wastewater and publicly available clinical data to assess trends and associations, and examined differences between sewersheds. Nonetheless, poorly characterized variability in WBE data can hamper the critical goal of relating viral loads in wastewater to disease dynamics. Clear characterization of uncertainties related to analytical methodologies would therefore facilitate interpretation of wastewater data by public health agencies.33
As mentioned above, SARS-CoV-2 viral loads in wastewater from the Port Richmond and Oakwood Beach WRRFs (both located in the borough of Staten Island) did not capture the peak in COVID-19 cases that was observed in January 2021 across all sewersheds. In the Port Richmond and Oakwood Beach sewersheds there was a marked increase in COVID-19 cases in December 2020 that was accompanied by an associated peak in the SARS-CoV-2 viral load in wastewater during this time. However, as new COVID-19 cases in Staten Island increased by 60% in January 2021, the virus loads in wastewater stayed constant or decreased. Compared to sewersheds in the other boroughs, those in Staten Island had relatively high clinical test positivity in December and January (7–14%), despite having an average testing rate (i.e., number of clinical tests administered per capita) for the study period that was greater than that of over half of the other sewersheds (Fig. S2†). This observation suggests that testing may not have adequately captured all infections in Staten Island during this period. While inadequate clinical testing rates could potentially reduce the accuracy of the observed relationships between clinical and wastewater data for these sewersheds, it does not explain the lower-than-expected SARS-CoV-2 viral loads measured in Staten Island wastewater in January 2020. A more likely explanation could stem from the composition or operation of the wastewater system in the borough. For example, a portion of the Staten Island population is not served by the sewer system and instead uses septic systems. As such, a segment of this population does not contribute to the sewer system, and viruses excreted by these residents would not have been present in the influent wastewater at the Oakwood Beach and Port Richmond WRRFs. Nonetheless, given that the population served by septic systems on Staten Island is thought to be smaller than those served by the sewer system, it is unlikely that this hypothesis can entirely explain the discrepancy between measured SARS-CoV-2 viral loads and new COVID-19 cases. In addition, much of Staten Island uses separated rather than combined stormwater–sewer systems, which could potentially impact the wastewater matrix and influence viral recovery during concentration and quantification steps in sample analysis. Because of these discrepancies, the Staten Island sewersheds were excluded from analysis of the combined data set and the estimation of minimum COVID-19 case rates associated with the LOD.
Relationships between SARS-CoV-2 viral loads in wastewater and new laboratory-confirmed COVID-19 cases
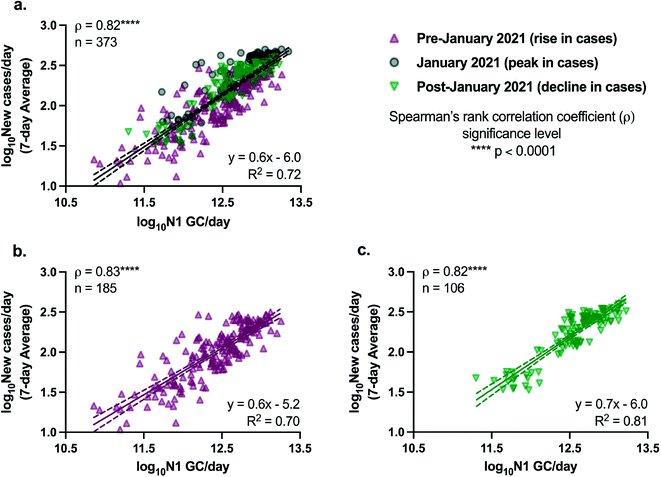
The relationships between SARS-CoV-2 viral loads in wastewater and new laboratory-confirmed COVID-19 cases in the corresponding sewershed populations were assessed between November 8, 2020 and April 11, 2021. By early June 2021, city-wide weekly averages of the percentage of positive COVID-19 clinical tests declined below l%, and over 50% of NYC residents had received at least one dose of a COVID-19 vaccine.18,34 To minimize the potential impact of mass vaccination on the evaluation of relationships between case rates and SARS-CoV-2 concentrations in wastewater presented in this work, we chose to conduct the statistical analyses described below for a period ending in early April, shortly after New York State extended vaccination availability to individuals of 16 years and older.Significant positive correlations between SARS-CoV-2 viral loads in wastewater and new laboratory-confirmed COVID-19 cases in the corresponding populations were found for all individual sewersheds and for a combined data set that included all sewersheds other than Port Richmond and Oakwood Beach (Spearman, p < 0.05), indicating, as expected, that an increase in COVID-19 cases was associated with an increase in SARS-CoV-2 concentrations in wastewater (Fig. 2). Correlation coefficients (ρ) for the individual sewersheds ranged from 0.38 (Coney Island WRRF) to 0.81 (Wards Island WRRF), with an average of 0.55 and a median of 0.55; correlation coefficients for four sewersheds were greater than or equal to 0.60. Similar correlation coefficients between SARS-CoV-2 wastewater concentrations and clinical case data have been reported elsewhere.16,35 Note that analysis of correlations between virus concentrations (N1 GC L−1, as opposed to virus loads) and new COVID-19 case rates (cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, as opposed to cases per day) yielded similar results (Table S3†). The correlation coefficient for the combined data set (ρ = 0.82) was higher than for any of the individual sewersheds (Fig. 3a).
000, as opposed to cases per day) yielded similar results (Table S3†). The correlation coefficient for the combined data set (ρ = 0.82) was higher than for any of the individual sewersheds (Fig. 3a).
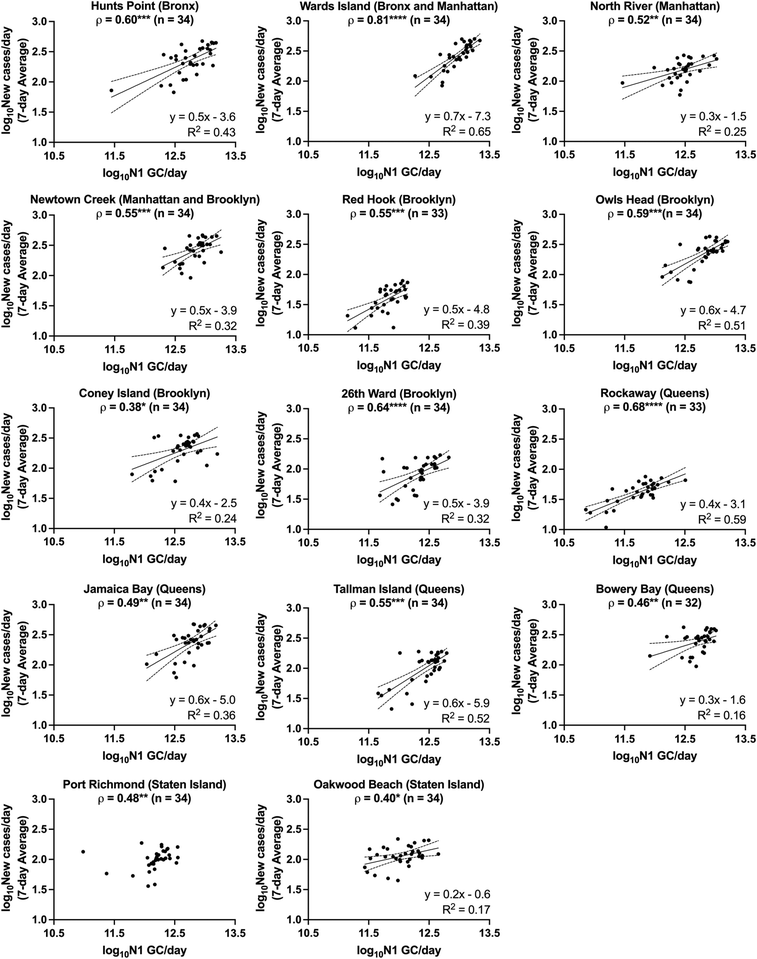 | ||
| Fig. 2 Linear regressions of log10-transformed SARS-CoV-2 viral loads in wastewater (N1 GC per day) and log10-transformed 7-day averages of new COVID-19 cases per day for each sewershed in New York City. Linear regressions (solid lines) and associated 95% confidence intervals (dashed lines) are shown along with goodness of fit R2 values for those data sets with significantly non-zero slopes. Note that linear regression for Port Richmond has been excluded as the slope was not significantly non-zero (see ESI†). The Spearman's rank correlation coefficient (ρ) between N1 GC per day and new COVID-19 cases per day is shown at the top of each sewershed plot, with significance levels indicated (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). | ||
For each sewershed, minimal differences were observed between the magnitudes of the Spearman's rank correlation coefficients determined using data sets with and without lag times applied (Fig. S4†). Some previous studies, applying a variety of assessment methods, have suggested that there is a time lag for the identification of clinical cases after the measurement of SARS-CoV-2 in wastewater that ranges on the order of days to weeks, while others have indicated that the SARS-CoV-2 concentration in wastewater is not a leading indicator of COVID-19 diagnosis.8 Inconsistent findings for lag times may be attributed to whether clinical data are presented (1) by the date of specimen collection, (2) by the date that results are reported, or (3) as a rolling average of either over multiple days, as well as the adequacy of COVID-19 testing rates, which vary in different regions and shift across time. Clinical data collected during periods with low testing rates are less likely to capture all infections in a region, and individuals may be more likely to be tested after symptom onset, at a time when viral shedding in feces may have already begun. These conditions can result in a lag behind wastewater monitoring data, which provides viral load information independent from clinical testing rates. Data for this work was collected during a time when testing rates were significantly higher than those during the first wave of the pandemic in NYC, and weekly median turnaround times for test results were 1 to 2 days.18 In addition, we could not confidently rule out that the small improvements in correlations observed when applying a lag time for some sewersheds was an artifact of variability in the measured wastewater data. Furthermore, our preliminary assessment of lag time did not include analysis of autocorrelation, which could be considered in future work. A rigorous assessment of lag time would need to account for contributions of previous as well as newly infected individuals to viral loads in wastewater, which was beyond the scope of this work. For these reasons, we compared data sets without a time lag for subsequent comparisons and linear regression analysis. It should also be noted that reported observations regarding lag times may not apply after mass vaccination or the spread of new viral variants, as the potential effects of these factors on fecal shedding of SARS-CoV-2 are currently unknown.
Because the nonparametric Spearman's rank correlation was used for this analysis, results suggest that there is, at minimum, a monotonic, direct relationship between SARS-CoV-2 quantified in wastewater and clinically confirmed COVID-19 cases. Linear relationships between the two log10-transformed datasets were assessed through analysis of linear regressions, with the best fit found for the Wards Island WRRF (R2 = 0.65) and some of the poorest fits found for the sewersheds in Staten Island (Fig. 2). Inconsistent relationships between sewershed-level SARS-CoV-2 viral loads in wastewater and COVID-19 cases observed across sewersheds may be due to differences in the sewer systems for each sewershed, including sewershed areas, residence times of wastewater in the sewer system, the presence of non-domestic wastewater inputs, proportions of the population made up by transient individuals or commuters, and per capita water use. Differences could also be related to clinical testing rates for each sewershed, though no significant correlation was found between the slopes of the linear regression lines and the average testing rates for the study period for each sewershed (Spearman, p > 0.05). Similarly, no significant correlations were found between the slopes of the linear regression lines for each sewershed and the (1) average wastewater flow rate, (2) sewershed population, or (3) average per capita wastewater flow rate (Spearman, p > 0.05), which was expected given that N1 concentrations were normalized by flow rate. Nonetheless, the linear regression found between log10-transformed SARS-CoV-2 viral loads in wastewater and log10-transformed 7-day averages of new COVID-19 cases per day using the combined data set had a strong fit (R2 = 0.72) relative to the fits of regressions for the individual sewersheds.
Evaluations of the utility of SARS-CoV-2 wastewater monitoring data has largely involved comparison of viral RNA in wastewater to COVID-19 case counts based on clinical testing.36 Given that the accuracy of confirmed case rates as a measure of the number of infected individuals is dependent on COVID-19 testing rates, this comparison must be made with a consideration of clinical testing biases. Moreover, if multiple clinical data types are available, one must determine which is most appropriate for comparison to wastewater data. The analysis applied herein utilized a data set containing 7-day averages of new COVID-19 cases based on testing in each approximated sewershed area. Uncertainties surrounding such clinical testing data include (1) whether there were regional biases in testing results (Fig. S2†), potentially due to testing disparities;10 (2) whether testing rates were adequate and what constitutes adequate testing; and (3) how long before specimen collection infected individuals contracted COVID-19 and started shedding the virus. Others have reported correlations of wastewater data with COVID-19 surveillance data sets other than clinical case rates, such as clinical test positivity or hospitalization rates.2 Hospital admissions data, although not without its own biases,37 may be an alternative epidemiological metric to compare to or to validate wastewater monitoring data if significant inadequacies in clinical testing are suspected, though the relationship between hospitalization data and wastewater data may differ for vaccinated and unvaccinated populations. While hospitalization data at the MODZCTA level were not publicly available for NYC, visual comparison at the borough level indicates that trends in daily hospitalizations generally reflected trends in case rates for sewersheds within each borough (Fig. S3†). The limitations of clinical testing are in fact a major driver for the application of WBE, which aims to provide community-level information free from clinical testing bias.38–40 Continued population-level monitoring from wastewater data could become increasingly useful in areas where clinical testing rates decline or resources for clinical testing are limited.
Linear regressions for the combined data set are presented in Fig. 3, with data collected on dates with over 10% positive COVID-19 testing rates removed. Removing data associated with potentially inadequate testing from the combined data set did not significantly change the regression (analysis of covariance, p > 0.05) compared to the full data set without filtering (Fig. S5†). After the peak in COVID-19 cases in NYC in January 2021, there was a decline in cases across all sewersheds. The relationship between SARS-CoV-2 loads in wastewater and new clinical COVID-19 cases during the period of declining cases (after January 2021) was not found to be significantly different from the relationship during the period when cases were increasing (prior to January 2021), based on a comparison of separate linear regressions for the data associated with the rise in case rates and the decline in case rates (analysis of covariance comparing slopes, p > 0.05; Fig. 3b and c).
The slope of the linear regression line for the full combined data set was found to be 0.6 (Fig. 3a), indicating that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the number of N1 GC per day corresponded to a 0.6
log10 change in the number of N1 GC per day corresponded to a 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed. Metrics such as these are derived from relative changes in viral load, and therefore do not require absolute quantification of viral concentrations in wastewater, allowing for comparison to other studies and alleviating challenges related to absolute quantification of standard curves. However, this metric comparing SARS-CoV-2 loads and daily new COVID-19 cases has not been consistently reported in studies monitoring SARS-CoV-2 in influent wastewater. Harmonizing data analysis strategies to include such a metric would improve efforts to compare results across different locations. The slope of 0.6 observed herein is greater than that reported previously by Wolfe et al. (slope = 0.24), who compared SARS-CoV-2 concentrations measured in primary wastewater settled solids and COVID-19 incidence in seven publicly owned treatment works located across the United States, including one of the NYC facilities described in this work.36 However, it is important to note that Wolfe et al. examined daily incident cases/population and primary settled solids samples for SARS-CoV-2 concentrations, which differ from the influent wastewater evaluated herein. In addition, Wolfe et al. normalized measured SARS-CoV-2 concentrations in wastewater solids by concentrations of pepper mild mottle virus (PMMoV),36 a normalization biomarker that has also been used by others,16,41 rather than using influent flow rates to calculate viral loads. The differences in the slopes may be due to either of these factors, to variations in the relationship between SARS-CoV-2 wastewater loads and COVID-19 cases in different regions, or to a difference in the overall sensitivity of the methodology applied by Wolfe et al.
log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed. Metrics such as these are derived from relative changes in viral load, and therefore do not require absolute quantification of viral concentrations in wastewater, allowing for comparison to other studies and alleviating challenges related to absolute quantification of standard curves. However, this metric comparing SARS-CoV-2 loads and daily new COVID-19 cases has not been consistently reported in studies monitoring SARS-CoV-2 in influent wastewater. Harmonizing data analysis strategies to include such a metric would improve efforts to compare results across different locations. The slope of 0.6 observed herein is greater than that reported previously by Wolfe et al. (slope = 0.24), who compared SARS-CoV-2 concentrations measured in primary wastewater settled solids and COVID-19 incidence in seven publicly owned treatment works located across the United States, including one of the NYC facilities described in this work.36 However, it is important to note that Wolfe et al. examined daily incident cases/population and primary settled solids samples for SARS-CoV-2 concentrations, which differ from the influent wastewater evaluated herein. In addition, Wolfe et al. normalized measured SARS-CoV-2 concentrations in wastewater solids by concentrations of pepper mild mottle virus (PMMoV),36 a normalization biomarker that has also been used by others,16,41 rather than using influent flow rates to calculate viral loads. The differences in the slopes may be due to either of these factors, to variations in the relationship between SARS-CoV-2 wastewater loads and COVID-19 cases in different regions, or to a difference in the overall sensitivity of the methodology applied by Wolfe et al.
At present, limitations regarding the accuracy of COVID-19 clinical testing data and uncertainties related to SARS-CoV-2 measurements in wastewater–including SARS-CoV-2 shedding rates and RNA stability in different sewersheds–preclude development and validation of a universal, quantitative model to predict disease incidence based on viral RNA concentrations in wastewater. Ongoing research continues to expand our understanding of critical model parameters and factors contributing to uncertainty, owing particularly to SARS-CoV-2 monitoring work completed at smaller scales (e.g., building-level),42 from which information about the contributing population can be obtained more easily than from larger sewersheds. An attempt to quantify COVID-19 case rates in NYC's sewersheds based on wastewater data at this time would be inaccurate, and is not currently recommended for application in the realm of public health.43 However, based on our analysis and others, there is utility in using wastewater data to monitor trends in COVID-19 incidence.
Estimated case rates associated with method LOD
The utility of SARS-CoV-2 wastewater data depends on whether virions are present in wastewater at detectable concentrations (i.e., above the LOD and LOQ). It is therefore useful to approximate the minimum number of contributing COVID-19 cases per day required for detection of the SARS-CoV-2 N1 gene target in wastewater using the methodology described here. When estimated using individual, sewershed-specific linear regressions (Fig. 2), the minimum new COVID-19 case rate that corresponds to the method LOD varied for each sewershed, ranging between 2 and 8 cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people (Table S4†). Minimum detectable case rates were also estimated for each sewershed using the linear regression from the combined data set and the average daily influent flow rates for each WRRF during the study period. These estimates fell within the same range as those derived from sewershed-specific linear regressions (Table S4†).
000 people (Table S4†). Minimum detectable case rates were also estimated for each sewershed using the linear regression from the combined data set and the average daily influent flow rates for each WRRF during the study period. These estimates fell within the same range as those derived from sewershed-specific linear regressions (Table S4†).
The minimum detectable case rate estimates presented here should be taken as order-of-magnitude approximations rather than absolute quantities, especially considering the varying strength of the linear relationships between data for certain sewersheds (e.g., data sets for Coney Island, Bowery Bay, Oakwood Beach, and Port Richmond WRRFs had Pearson correlation coefficients below 0.5). Furthermore, these findings hold only for the specific SARS-CoV-2 concentration, RNA extraction, and RT-qPCR methodology applied herein (as different methodologies will likely have different sensitivities and resulting LODs), and may not be transferable to locations with different per capita wastewater flow rates, even if testing rates and case rates are similar to those described here. The estimates may also be limited by the assumption that the dominant source of the SARS-CoV-2 viral load in the wastewater is from recent cases as opposed to prolonged fecal shedding, which is consistent with assumptions made in previous studies.36,44 The relationships found are also limited by the accuracy of clinical testing data, as discussed above. Furthermore, variability in virus shedding rates were not considered for the simple linear models in our study. By April 2021, Iota (B.1.526) and Alpha (B.1.1.7) had become the dominant SARS-CoV-2 variants observed in NYC, based on data summarizing the percent of sequenced clinical samples associated with each variant identified in NYC starting in January 2021.18 Delta (B.1.617.2) and Omicron (B.1.1.529) variants were not present in notable numbers during the study period, based on the same data set.18 It is still unknown how different SARS-CoV-2 variants affect fecal shedding rates of the virus; therefore, it is possible that the relationships described herein may not be applicable during periods when variants other than those present during the study period are dominant.
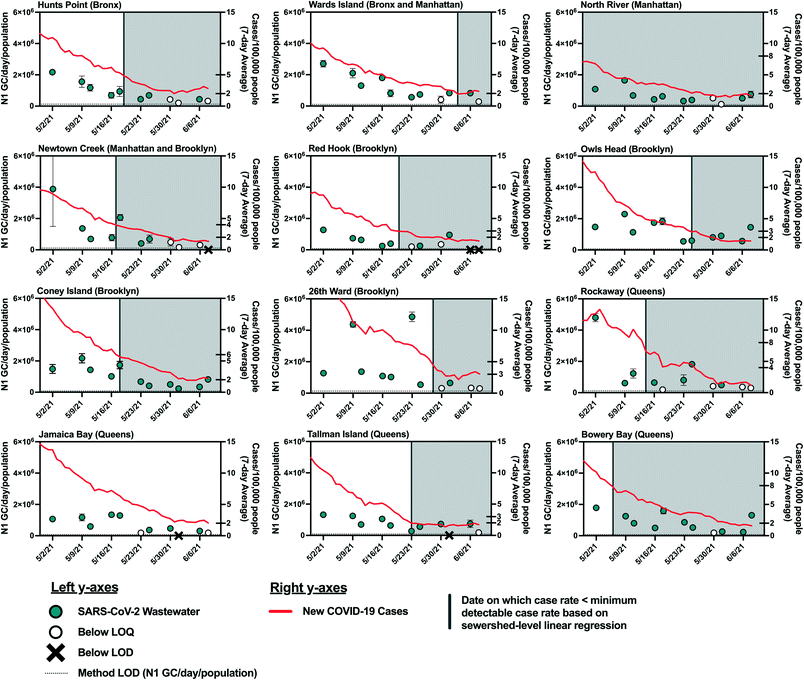
As COVID-19 cases declined in NYC in the spring and early summer of 2021, the estimated minimum detectable COVID-19 case rates were reached in most sewersheds by May and June 2021. As such, we expected that SARS-CoV-2 viral loads in wastewater would have decreased to below the LOQ and LOD at this time. However, viral RNA was still detectable in influent wastewater collected from all sewersheds in mid June 2021 (Fig. 4). While this discrepancy may be explained by the limitations described above, it may also be due to decreasing COVID-19 testing rates, which could result in reduced diagnosis of individuals with asymptomatic infections, who are less likely to seek out COVID-19 tests. For example, the average COVID-19 testing rate in NYC during the period from May 2, 2021 to June 8, 2021 (390 test per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people) decreased 30% from the average in January 2021 (560 tests per day/100
000 people) decreased 30% from the average in January 2021 (560 tests per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people). Additionally, widespread vaccination of adults in New York may have resulted in asymptomatic and mild infections that were not diagnosed. While individuals with asymptomatic SARS-CoV-2 infections may not be captured by clinical testing, viral shedding by asymptomatic individuals would still contribute to the viral load in wastewater, given that SARS-CoV-2 has been detected in fecal samples associated with asymptomatic or mild cases of COVID-19.45–47 Viral loads may have also been elevated in wastewater because of prolonged fecal shedding of the virus. Finally, it is possible that the linear relationship found in this work does not hold at low SARS-CoV-2 infection levels as the study period used for statistical analysis included only case rates above the minimum detectable case rates estimated for each sewershed.
000 people). Additionally, widespread vaccination of adults in New York may have resulted in asymptomatic and mild infections that were not diagnosed. While individuals with asymptomatic SARS-CoV-2 infections may not be captured by clinical testing, viral shedding by asymptomatic individuals would still contribute to the viral load in wastewater, given that SARS-CoV-2 has been detected in fecal samples associated with asymptomatic or mild cases of COVID-19.45–47 Viral loads may have also been elevated in wastewater because of prolonged fecal shedding of the virus. Finally, it is possible that the linear relationship found in this work does not hold at low SARS-CoV-2 infection levels as the study period used for statistical analysis included only case rates above the minimum detectable case rates estimated for each sewershed.
The estimated minimum numbers of COVID-19 cases required before SARS-CoV-2 can be detected in wastewater from NYC sewersheds are associated with considerable disease incidence that may be captured if some degree of clinical testing continues. Nonetheless, these estimates could aid public health agencies in understanding what COVID-19 incidence to expect if SARS-CoV-2 loads measured in wastewater influent cross the threshold from being below the detection limit to being detected. Improvements to analytical methods that lower the LOD48–50 would expand the utility of WBE in indicating low levels of disease incidence.
Conclusion
Results presented herein demonstrate that relative trends in SARS-CoV-2 loads in NYC wastewater can be evaluated and associated with trends in clinical COVID-19 testing data, and have potential to contribute to situational awareness of disease incidence in large urban sewersheds. SARS-CoV-2 loads were strongly correlated with reported rates of new COVID-19 cases (Spearman's ρ = 0.82) based on a combined data set including 12 of NYC's 14 sewersheds during a study period that included both the rise and decline of the City's second major peak in COVID-19 cases. Ours is the first study to confirm a direct, linear relationship between SARS-CoV-2 wastewater data and clinical testing data for NYC, a finding consistent with reports for other large urban areas. Our results indicate that the relationship we found between SARS-CoV-2 loads and COVID-19 cases held during both the rise and the decline in cases, under the conditions of the study period (e.g., rates of vaccination, proportions of dominant SARS-CoV-2 variants). Specifically, we found that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the number of N1 GC per day corresponded to a 0.6
log10 change in the number of N1 GC per day corresponded to a 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed, a metric that provides qualitative insights into COVID-19 trends, even in the absence of absolute quantification of SARS-CoV-2 viral loads.
log10 change in the number of new laboratory-confirmed COVID-19 cases per day in a sewershed, a metric that provides qualitative insights into COVID-19 trends, even in the absence of absolute quantification of SARS-CoV-2 viral loads.
The strengths of the correlations observed for each of NYC's sewersheds differed (Spearman's ρ between 0.38 and 0.81), underscoring that the utility of wastewater monitoring for estimating disease incidence may be localized and that further research is critical to understanding and addressing this variability and uncertainty. Moreover, Iota and Alpha became the dominant SARS-CoV-2 variants observed in NYC by the end of the study period; if different variants cause different fecal shedding rates of the virus, these findings may not hold when variants such as Omicron, which was not dominant during the study period, are more prevalent in a contributing population.
Additionally, establishing the range of disease incidence associated with the limit of detection for SARS-CoV-2—or any WBE target—as we have done herein (e.g., 2–8 COVID-19 cases per day/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people), may be useful for understanding the public health implications of initial detection of a target in wastewater. These values serve as order-of-magnitude estimates and are dependent on the methodology applied by the WBE program. Nonetheless, establishing this association may be particularly helpful during early outbreaks of new diseases, before clinical testing is widely available, or during periods of low disease incidence, when rates of clinical testing may wane.
000 people), may be useful for understanding the public health implications of initial detection of a target in wastewater. These values serve as order-of-magnitude estimates and are dependent on the methodology applied by the WBE program. Nonetheless, establishing this association may be particularly helpful during early outbreaks of new diseases, before clinical testing is widely available, or during periods of low disease incidence, when rates of clinical testing may wane.
Finally, some lessons from the development of NYC's SARS-CoV-2 wastewater monitoring program may be useful for agencies interested in implementing wastewater monitoring programs for emerging pathogens. First, collaborating parties–including academic partners and NYC DEP personnel–worked in partnership to develop a monitoring program centered around NYC DEP's priorities. Second, sample analysis was conducted in the NYC DEP microbiology laboratory, which allowed the program to take advantage of existing equipment, expertise, wastewater sampling and transport protocols and infrastructure, and resources related to wastewater analysis, while maximizing use of the NYC DEP's extensive knowledge base and data. Doing so expedited the initiation of the wastewater monitoring program, allowed protocol adjustments to respond to practical challenges as well as technical ones, and supported a rich training experience, in which analysts shared insights from hands-on experience, contributed to workflow decisions, and were exposed to the empirical reasoning behind methodological choices.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Funding for this work was provided by the New York City Department of Environmental Protection and the Alfred P. Sloan Foundation. An extensive team at the NYC DEP made this monitoring program possible, including Samantha MacBride, Peter Williamsen, Gina Behnke, Jasmin Torres, and Jorge Villacis; members of the NYC DEP Microbiology Lab, including William Kelly, Naudet Joasil, Patrick Hoyes, Donnovan Johnson, Manzura Kopusov, Oren Sachs, and Samantha Cruickshank; the NYC DEP transportation team, including Lateef Franklin, Samuel Young, and John Congemi; and Abeba Negatu, Patrick Jagessar, Max Verastegui and their process control laboratory teams at NYC DEP. Several researchers at CUNY provided support and assistance for protocol development, optimization, and training, including Sherin Kannoly, Kaung Myat “Zach” San, Kristen Cheung, Anna Gao, Michelle Markman, Nanami Kubota, and Irene Hoxie. We thank Vikram Kapoor, Associate Professor in the Department of Civil and Environmental Engineering at the University of Texas San Antonio, and the Kapoor Lab for generously quantifying the synthetic RNA control using RT-ddPCR. We thank Professors Alexandria Boehm (Stanford University) and Sandra McLellan (University of Wisconsin-Milwaukee) for their support and guidance during program development. We also acknowledge the many insights gained from the interactions through the NSF Research Coordination Network (RCN) on Wastewater Surveillance for SARS-CoV-2. A script automating the download of New York City's publicly available COVID-19 clinical testing data was generously provided by Charlie Mydlarz (NYU Center for Urban Science and Progress). The GIS data used for the NYC sewershed map in the table of contents entry art was retrieved from Open Sewer Atlas NYC.51References
- G. Medema, F. Been, L. Heijnen and S. Petterson, Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: Opportunities and challenges, Curr. Opin. Environ. Sci. Health, 2020, 17, 49–71, DOI:10.1016/j.coesh.2020.09.006.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan, A. Casanovas-Massana, A. I. Ko, A. A. Malik, D. Wang, M. Wang, J. L. Warren, D. M. Weinberger, W. Arnold and S. B. Omer, Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics, Nat. Biotechnol., 2020, 38, 1164–1167, DOI:10.1038/s41587-020-0684-z.
- L. C. Scott, A. Aubee, L. Babahaji, K. Vigil, S. Tims and T. G. Aw, Targeted wastewater surveillance of SARS-CoV-2 on a University Campus for COVID-19 outbreak detection and mitigation, Environ. Res., 2021, 200, 111374, DOI:10.1016/j.envres.2021.111374.
- F. Wu, J. Zhang, A. Xiao, X. Gu, W. L. Lee, F. Armas, K. Kauffman, W. Hanage, M. Matus, N. Ghaeli, N. Endo, C. Duvallet, M. Poyet, K. Moniz, A. D. Washburne, T. B. Erickson, P. R. Chai, J. Thompson and E. J. Alm, SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases, mSystems, 2020, 5, e00614–e00620, DOI:10.1128/mSystems.00614-20.
- F. Kreier, The myriad ways sewage surveillance is helping fight COVID around the world, Nature, 2021 DOI:10.1038/d41586-021-01234-1.
- World Health Organization, Status of environmental surveillance for SARS-CoV-2 virus: Scientific Brief, https://www.who.int/news-room/commentaries/detail/status-of-environmental-surveillance-for-sars-cov-2-virus, (accessed 31 May 2021) Search PubMed.
- X. Fernandez-Cassi, A. Scheidegger, C. Bänziger, F. Cariti, A. Tuñas Corzon, P. Ganesanandamoorthy, J. C. Lemaitre, C. Ort, T. R. Julian and T. Kohn, Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high, Water Res., 2021, 200, 117252, DOI:10.1016/j.watres.2021.117252.
- S. W. Olesen, M. Imakaev and C. Duvallet, Making waves: Defining the lead time of wastewater-based epidemiology for COVID-19, Water Res., 2021, 202, 117433, DOI:10.1016/j.watres.2021.117433.
- C. N. Thompson, J. Baumgartner, C. Pichardo, B. Toro, L. Li, R. Arciuolo, P. Y. Chan, J. Chen, G. Culp, A. Davidson, K. Devinney, A. Dorsinville, M. Eddy, M. English, A. M. Fireteanu, L. Graf, A. Geevarughese, S. K. Greene, K. Guerra, M. Huynh, C. Hwang, M. Iqbal, J. Jessup, J. Knorr, R. Lall, J. Latash, E. Lee, K. Lee, W. Li, R. Mathes, E. McGibbon, N. McIntosh, M. Montesano, M. S. Moore, K. Murray, S. Ngai, M. Paladini, R. Paneth-Pollak, H. Parton, E. Peterson, R. Pouchet, J. Ramachandran, K. Reilly, J. Sanderson Slutsker, G. Van Wye, A. Wahnich, A. Winters, M. Layton, L. Jones, V. Reddy and A. Fine, COVID-19 Outbreak — New York City, February 29–June 1, 2020, US Department of Health and Human Services/Centers for Disease Control and Prevention, Morb. Mortal. Wkly. Rep., 2020, 69, 1725–1729, DOI:10.15585/mmwr.mm6946a2.
- W. Lieberman-Cribbin, S. Tuminello, R. M. Flores and E. Taioli, Disparities in COVID-19 Testing and Positivity in New York City, Am. J. Prev. Med., 2020, 59, 326–332, DOI:10.1016/j.amepre.2020.06.005.
- M. Trujillo, K. Cheung, A. Gao, I. Hoxie, S. Kannoly, N. Kubota, K. M. San, D. S. Smyth and J. J. Dennehy, Protocol for safe, affordable, and reproducible isolation and quantitation of SARS-CoV-2 RNA from wastewater, PLoS One, 2021, 16, e0257454, DOI:10.1371/journal.pone.0257454.
- X. Lu, L. Wang, S. K. Sakthivel, B. Whitaker, J. Murray, S. Kamili, B. Lynch, L. Malapati, S. A. Burke, J. Harcourt, A. Tamin, N. J. Thornburg, J. M. Villanueva and S. Lindstrom, US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2, Emerging Infect. Dis., 2020, 26, 1654–1665, DOI:10.3201/eid2608.201246.
- H. Al-Duroobi, S. V. Moghadam, D. C. Phan, A. Jafarzadeh, A. Matta and V. Kapoor, Wastewater surveillance of SARS-CoV-2 corroborates heightened community infection during the initial peak of COVID-19 in Bexar County, Texas, FEMS Microbes, 2021, 2, xtab015, DOI:10.1093/femsmc/xtab015.
- A. Forootan, R. Sjöback, J. Björkman, B. Sjögreen, L. Linz and M. Kubista, Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR), Biomol. Detect. Quantif., 2017, 12, 1–6, DOI:10.1016/j.bdq.2017.04.001.
- S. Loeb, K. Graham, D. Catoe, M. Wolfe, A. B. Boehm and K. Wigginton, One-Step RT-ddPCR for Detection of SARS-CoV-2, Bovine Coronavirus, and PMMoV RNA in RNA Derived from Wastewater or Primary Settled Solids, protocols.io, 2020, DOI:10.17504/protocols.io.bi6vkhe6.
- S. Feng, A. Roguet, J. S. McClary-Gutierrez, R. J. Newton, N. Kloczko, J. G. Meiman and S. L. McLellan, Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities, ACS ES&T Water, 2021, 1, 1955–1965, DOI:10.1021/acsestwater.1c00160.
- N. Decaro, G. Elia, M. Campolo, C. Desario, V. Mari, A. Radogna, M. L. Colaianni, F. Cirone, M. Tempesta and C. Buonavoglia, Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay, J. Virol. Methods, 2008, 151, 167–171, DOI:10.1016/j.jviromet.2008.05.016.
- NYC Department of Health and Mental Hygiene, NYC Coronavirus Disease 2019 (COVID-19) Data, https://github.com/nychealth/coronavirus-data, (accessed 20 May 2021) Search PubMed.
- World Health Organization, COVID-19 - virtual press conference - March 30, 2020, COVID-19 - virtual press conference - March 30, 2020, https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-press-conference-full-30mar2020.pdf?sfvrsn=6b68bc4a_2, (accessed 25 May 2021) Search PubMed.
- A. Aubrey, Which States Are Doing Enough Testing? This Benchmark Helps Settle The Debate, NPR, https://www.npr.org/sections/health-shots/2020/04/22/840526338/is-the-u-s-testing-enough-for-covid-19-as-debate-rages-on-heres-how-to-know, (accessed 25 May 2021) Search PubMed.
- R. Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2019 Search PubMed.
- GraphPad Prism version 9.1.1 for macOS, GraphPad Software, La Jolla California USA, www.graphpad.com Search PubMed.
- M. Ciesielski, D. Blackwood, T. Clerkin, R. Gonzalez, H. Thompson, A. Larson and R. Noble, Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater, J. Virol. Methods, 2021, 297, 114230, DOI:10.1016/j.jviromet.2021.114230.
- New York Metropolitan Transportation Council, 2050 SED Forecasts, https://www.nymtc.org/DATA-AND-MODELING/SED-Forecasts/2050-Forecasts, (accessed 12 April 2021) Search PubMed.
- New York City Department of Health and Mental Hygiene, Press Notice About COVID-19 Areas of Concern: Tuesday, September 22, 2020, https://www1.nyc.gov/assets/doh/downloads/pdf/covid/dear-reporter-letter-09222020.pdf Search PubMed.
- S. K. Greene, E. R. Peterson, D. Balan, L. Jones, G. M. Culp, A. D. Fine and M. Kulldorff, Detecting COVID-19 Clusters at High Spatiotemporal Resolution, New York City, New York, USA, June–July 2020, Emerging Infect. Dis., 2021, 27, 1500–1504, DOI:10.3201/eid2705.203583.
- X. Li, S. Zhang, J. Shi, S. P. Luby and G. Jiang, Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology, Chem. Eng. J., 2021, 415, 129039, DOI:10.1016/j.cej.2021.129039.
- D. L. Jones, M. Q. Baluja, D. W. Graham, A. Corbishley, J. E. McDonald, S. K. Malham, L. S. Hillary, T. R. Connor, W. H. Gaze, I. B. Moura, M. H. Wilcox and K. Farkas, Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19, Sci. Total Environ., 2020, 749, 141364, DOI:10.1016/j.scitotenv.2020.141364.
- S. Mallett, A. J. Allen, S. Graziadio, S. A. Taylor, N. S. Sakai, K. Green, J. Suklan, C. Hyde, B. Shinkins, Z. Zhelev, J. Peters, P. J. Turner, N. W. Roberts, L. F. di Ruffano, R. Wolff, P. Whiting, A. Winter, G. Bhatnagar, B. D. Nicholson and S. Halligan, At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data, BMC Med., 2020, 18, 346, DOI:10.1186/s12916-020-01810–8.
- M. Cevik, M. Tate, O. Lloyd, A. E. Maraolo, J. Schafers and A. Ho, SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis, Lancet Microbe, 2021, 2, e13–e22, DOI:10.1016/S2666-5247(20)30172–5.
- A. Bivins, J. Greaves, R. Fischer, K. C. Yinda, W. Ahmed, M. Kitajima, V. J. Munster and K. Bibby, Persistence of SARS-CoV-2 in Water and Wastewater, Environ. Sci. Technol. Lett., 2020, 7, 937–942, DOI:10.1021/acs.estlett.0c00730.
- D. A. Larsen and K. R. Wigginton, Tracking COVID-19 with wastewater, Nat. Biotechnol., 2020, 38, 1151–1153, DOI:10.1038/s41587-020-0690–1.
- J. S. McClary-Gutierrez, M. C. Mattioli, P. Marcenac, A. I. Silverman, A. B. Boehm, K. Bibby, M. Balliet, F. L. de los Reyes, D. Gerrity, J. F. Griffith, P. A. Holden, D. Katehis, G. Kester, N. LaCross, E. K. Lipp, J. Meiman, R. T. Noble, D. Brossard and S. L. McLellan, SARS-CoV-2 Wastewater Surveillance for Public Health Action, Emerging Infect. Dis., 2021, 27, e210753, DOI:10.3201/eid2709.210753.
- NYC Department of Health and Mental Hygiene, COVID-19 Vaccination Reporting, https://github.com/nychealth/covid-vaccine-data, (accessed 11 August 2021) Search PubMed.
- J. Weidhaas, Z. T. Aanderud, D. K. Roper, J. VanDerslice, E. B. Gaddis, J. Ostermiller, K. Hoffman, R. Jamal, P. Heck, Y. Zhang, K. Torgersen, J. V. Laan and N. LaCross, Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds, Sci. Total Environ., 2021, 775, 145790, DOI:10.1016/j.scitotenv.2021.145790.
- M. K. Wolfe, A. Archana, D. Catoe, M. M. Coffman, S. Dorevich, K. E. Graham, S. Kim, L. M. Grijalva, L. Roldan-Hernandez, A. I. Silverman, N. Sinnott-Armstrong, D. J. Vugia, A. T. Yu, W. Zambrana, K. R. Wigginton and A. B. Boehm, Scaling of SARS-CoV-2 RNA in Settled Solids from Multiple Wastewater Treatment Plants to Compare Incidence Rates of Laboratory-Confirmed COVID-19 in Their Sewersheds, Environ. Sci. Technol. Lett., 2021, 8, 398–404, DOI:10.1021/acs.estlett.1c00184.
- K. Sherratt, S. Abbott, S. R. Meakin, J. Hellewell, J. D. Munday, N. Bosse, M. Jit and S. Funk, Exploring surveillance data biases when estimating the reproduction number: with insights into subpopulation transmission of COVID-19 in England, Philos. Trans. R. Soc., B, 2021, 376, 20200283, DOI:10.1098/rstb.2020.0283.
- M. Murakami, A. Hata, R. Honda and T. Watanabe, Letter to the Editor: Wastewater-Based Epidemiology Can Overcome Representativeness and Stigma Issues Related to COVID-19, Environ. Sci. Technol., 2020, 54, 5311–5311, DOI:10.1021/acs.est.0c02172.
- A. Zahedi, P. Monis, D. Deere and U. Ryan, Wastewater-based epidemiology—surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia, Parasitol. Res., 2021, 1–22, DOI:10.1007/s00436-020-07023–5.
- N. Sims and B. Kasprzyk-Hordern, Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level, Environ. Int., 2020, 139, 105689, DOI:10.1016/j.envint.2020.105689.
- P. M. D'Aoust, E. Mercier, D. Montpetit, J.-J. Jia, I. Alexandrov, N. Neault, A. T. Baig, J. Mayne, X. Zhang, T. Alain, M.-A. Langlois, M. R. Servos, M. MacKenzie, D. Figeys, A. E. MacKenzie, T. E. Graber and R. Delatolla, Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence, Water Res., 2021, 188, 116560, DOI:10.1016/j.watres.2020.116560.
- B. W. Schmitz, G. K. Innes, S. M. Prasek, W. Q. Betancourt, E. R. Stark, A. R. Foster, A. G. Abraham, C. P. Gerba and I. L. Pepper, Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology, Sci. Total Environ., 2021, 801, 149794, DOI:10.1016/j.scitotenv.2021.149794.
- Centers for Disease Control and Prevention (CDC), National Wastewater Surveillance System, https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.html, (accessed 1 June 2021) Search PubMed.
- D. Gerrity, K. Papp, M. Stoker, A. Sims and W. Frehner, Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: Methodology, occurrence, and incidence/prevalence considerations, Water Res.: X, 2021, 10, 100086, DOI:10.1016/j.wroa.2020.100086.
- S. Park, C.-W. Lee, D.-I. Park, H.-Y. Woo, H. S. Cheong, H. C. Shin, K. Ahn, M.-J. Kwon and E.-J. Joo, Detection of SARS-CoV-2 in Fecal Samples From Patients With Asymptomatic and Mild COVID-19 in Korea, Clin. Gastroenterol. Hepatol., 2021, 19, 1387–1394.e2, DOI:10.1016/j.cgh.2020.06.005.
- A. Mesoraca, K. Margiotti, A. Viola, A. Cima, D. Sparacino and C. Giorlandino, Evaluation of SARS-CoV-2 viral RNA in fecal samples, Virol. J., 2020, 17, 86, DOI:10.1186/s12985-020-01359–1.
- X. Jiang, M. Luo, Z. Zou, X. Wang, C. Chen and J. Qiu, Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days, J. Med. Virol., 2020, 92, 1807–1809, DOI:10.1002/jmv.25941.
- W. Ahmed, P. M. Bertsch, A. Bivins, K. Bibby, K. Farkas, A. Gathercole, E. Haramoto, P. Gyawali, A. Korajkic, B. R. McMinn, J. F. Mueller, S. L. Simpson, W. J. M. Smith, E. M. Symonds, K. V. Thomas, R. Verhagen and M. Kitajima, Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater, Sci. Total Environ., 2020, 739, 139960, DOI:10.1016/j.scitotenv.2020.139960.
- S. E. Philo, E. K. Keim, R. Swanstrom, A. Q. W. Ong, E. A. Burnor, A. L. Kossik, J. C. Harrison, B. A. Demeke, N. A. Zhou, N. K. Beck, J. H. Shirai and J. S. Meschke, A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance, Sci. Total Environ., 2021, 760, 144215, DOI:10.1016/j.scitotenv.2020.144215.
- A. Pérez-Cataluña, E. Cuevas-Ferrando, W. Randazzo, I. Falcó, A. Allende and G. Sánchez, Comparing analytical methods to detect SARS-CoV-2 in wastewater, Sci. Total Environ., 2021, 758, 143870, DOI:10.1016/j.scitotenv.2020.143870.
- Open Sewer Atlas NYC, Open Sewer Atlas NYC: Data ‘Sewersheds’, https://openseweratlas.tumblr.com/data, (accessed 7 September 2021) Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ew00747e |
| ‡ Present affiliation: Department of Life Sciences, Texas A&M University San Antonio, San Antonio, Texas, USA. |
| This journal is © The Royal Society of Chemistry 2022 |